Abstract
Rvb1 and Rvb2 are highly conserved and essential eukaryotic AAA+ proteins linked to a wide range of cellular processes. AAA+ proteins are ATPases associated with diverse cellular activities and are characterized by the presence of one or more AAA+ domains. These domains have the canonical Walker A and Walker B nucleotide binding and hydrolysis motifs. Rvb1 and Rvb2 have been found to be part of critical cellular complexes: the histone acetyltransferase Tip60 complex, chromatin remodelling complexes Ino80 and SWR-C, and the telomerase complex. In addition, Rvb1 and Rvb2 are components of the R2TP complex that was identified by our group and was determined to be involved in the maturation of box C/D small nucleolar ribonucleoprotein (snoRNP) complexes. Furthermore, the Rvbs have been associated with mitotic spindle assembly, as well as phosphatidylinositol 3-kinase-related protein kinase (PIKK) signalling. This review sheds light on the potential role of the Rvbs as chaperones in the assembly and remodelling of these critical complexes.
Keywords: Rvb1, Rvb2, R2TP, Ino80, PIKK, RNA polymerase II
1. What are Rvb1 and Rvb2?
Rvb1 and its paralogue Rvb2, with 43 per cent sequence identity and 65 per cent sequence similarity to each other (for the human proteins), belong to the AAA+ (adenosine triphosphases associated with diverse cellular activities) superfamily of ATPases. This class of ATPases is present in all kingdoms of life and is divided into numerous groups, clades and families based on structural and sequence analyses [1–3]. AAA+ proteins usually form hexameric ring structures and are characterized by the presence of the AAA+ module, which contains the highly conserved Walker A and Walker B motifs responsible for nucleotide binding and hydrolysis, respectively [4].
Rvb1 and Rvb2 are known under diverse names such as Pontin/Reptin, TIP49/TIP48, RuvBL1/RuvBL2 and ECP54/ECP51, respectively, reflecting their appearance in many cellular protein complexes and their discovery by unrelated approaches in multiple organisms [5–9]. In this review, we refer to these two proteins as Rvb1 and Rvb2.
2. Discovery and roles of Rvb1 and Rvb2
Rvb1 was originally discovered in 1997 as part of a complex with the TATA-binding protein (TBP) in rat [10]. Rvb1 and Rvb2 were found in complex with the large RNA polymerase II holoenzyme oligomer in 1998 [11], and, subsequently, Rvb2 was identified as an interacting partner of Rvb1 in human cells in 1999 [12]. Rvb1 and Rvb2 share limited sequence similarity (approx. 30%) to the bacterial RuvB helicase [13,14]. RuvB drives the branch migration and resolution of the Holliday junction in complex with RuvA and RuvC during homologous recombination and DNA repair [15]. This sequence similarity suggested that the Rvbs might have helicase activity using ATP binding and hydrolysis, since the deletion of RVB1 and RVB2 genes in Saccharomyces cerevisiae was complemented by the overexpression of the bacterial RuvAB complex [16] and since Rvb1 was found to be associated with the human replication protein (RP)A3 [11]. Indeed, the purified proteins exhibit weak helicase activity [12,17].
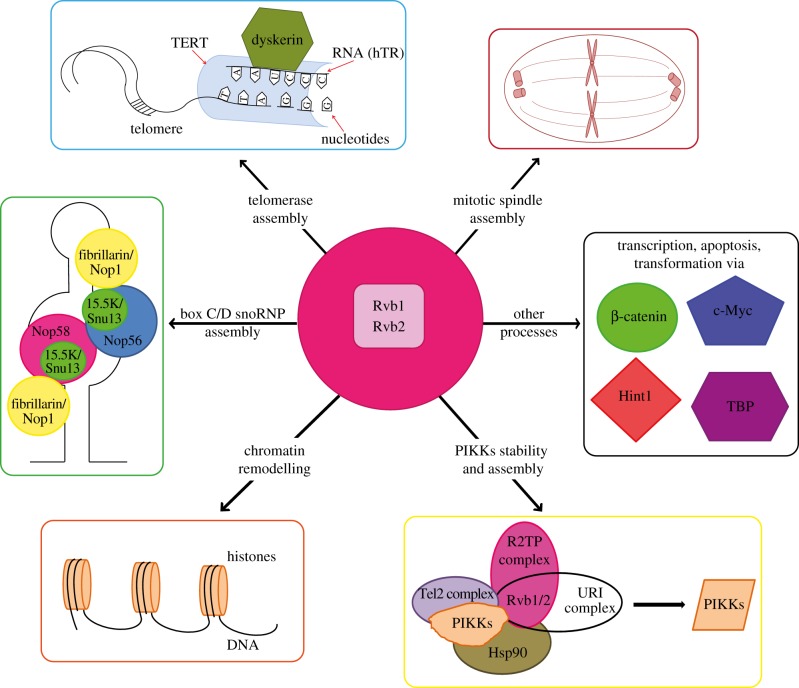
The RVB1 and RVB2 genes were found to be essential for viability in all model organisms examined so far, including S. cerevisiase [11], Drosophila melanogaster [8] and Caenorhabditis elegans [18], and are speculated to be also essential in mammalian cells. Since their discovery, the Rvbs have been found to be associated with many cellular pathways [19], including chromatin remodelling [9,20–23], transcription regulation [9,24], ribonuleoprotein complex biogenesis [25–29], mitotic assembly [30–32], telomerase complex assembly [33], RNA polymerase II assembly [26,34] and phosphatidylinositol 3-kinase-related protein kinase (PIKK) signalling [29] (figure 1).
Figure 1.
Overview of Rvb1/2 function. Rvb1 and Rvb2 function in the assembly of multiple cellular complexes/processes. They are involved in the assembly of mitotic spindles, telomerase complex, box C/D snoRNPs, chromatin remodelling complexes, and PIKKs. They also exhibit other roles/functions in processes such as transcription, transformation and apoptosis by interacting with factors including β-catenin, c-Myc, Hint1 and TBP. TERT, telomerase reverse transcriptase.
3. The structure of Rvb1 and Rvb2
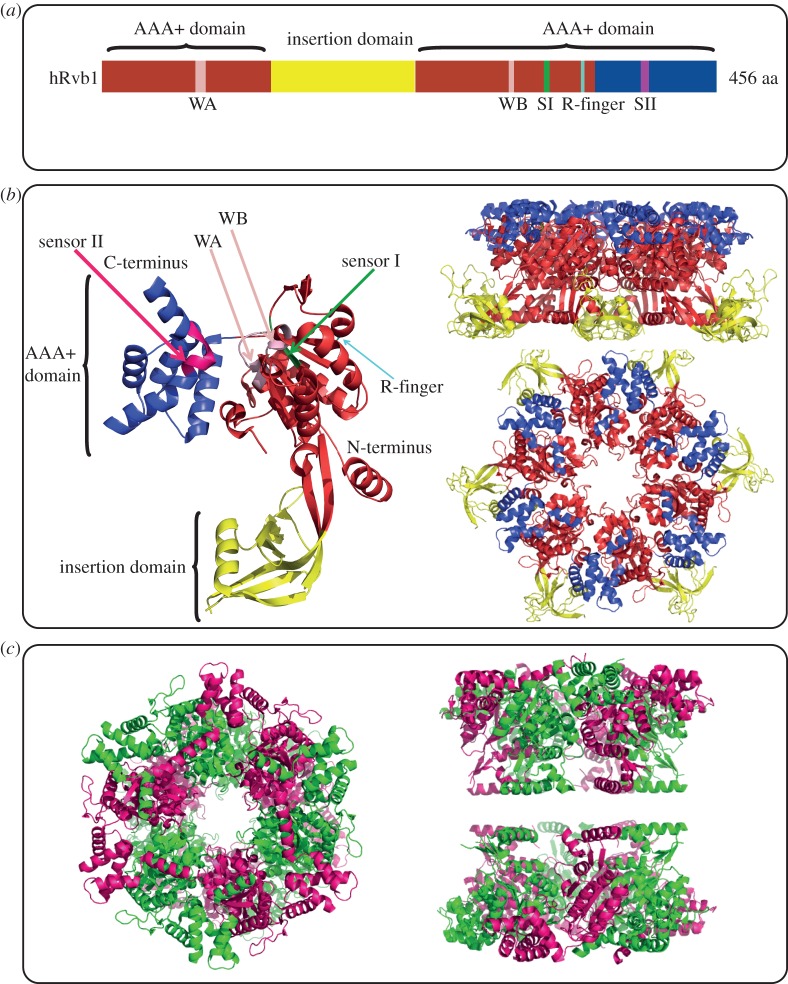
Based on the X-ray structure of human Rvb1 [18], the Rvb sequence can be divided into three domains (figure 2a,b): (i) an N-terminal αβα subdomain of the AAA+ domain, (ii) a 170 amino acid-insertion domain unique to the Rvbs among the AAA+ proteins which mediates DNA/RNA binding and shows similarity to the ssDNA binding domain of the replication factor replication protein A (RPA), and (iii) an all α subdomain of the AAA+ domain. In the AAA+ domain (figure 2a,b), the Walker A and Walker B motifs are responsible for ATP binding and hydrolysis, respectively, while sensor I and sensor II motifs sense whether the protein is bound to di- or tri-phosphates. The arginine finger (Arg-finger) of one subunit extends into the ATPase site of the neighbouring subunit and allows coordination of ATP hydrolysis between the subunits in the hexamer [3].
Figure 2.
Overview of the Rvb1/2 structure. (a) Bar graph of the domain organization of human Rvb1. In red is the N-terminal αβα subdomain of the AAA+ domain, in blue is the C-terminal all α subdomain of the AAA+ domain, and in yellow is the insertion domain. Conserved motifs with the AAA+ domain are also highlighted: WA, Walker A; WB, Walker B; SI, Sensor I; SII, sensor II; R-finger, arginine finger. (b) Crystal structure of human Rvb1 monomer on the left-hand side. Side and top view of human Rvb1 hexamer are shown on the right-hand side. The colour scheme used is the same as the one in (a). (c) Top and side views of the crystal structure of dodecameric human Rvb1/2 complex with truncation in part of Domain II. Human Rvb1 monomers are shown in green and human Rvb2 monomers are shown in pink.
The crystal structure of human Rvb1 has been solved as a hexamer [18] (figure 2b), however, the homohexamer was found to be inactive as a helicase and ATPase, suggesting that this might not be the physiologically relevant complex. There is no crystal structure of Rvb2 alone; however, more recently, the crystal structure of the human Rvb1–Rvb2 complex, with truncation of the insertion domain in both proteins, was solved [35] (figure 2c). The complex was found to be a dodecamer composed of two hetero-hexameric rings with alternating Rvb1 and Rvb2 monomers [35]. The study showed that the truncated version of the complex exhibits an enhanced ATPase and helicase activity compared with the wild-type (WT) complex, thus, suggesting that the insertion domain functions as a regulator of the activity of the complex.
Using multiple biophysical techniques including analytical ultracentrifugation, size exclusion chromatography, mass spectrometry and electron microscopy, it has been found that human Rvb1 and Rvb2 can form various oligomeric states that are modulated by the insertion domain [36,37]. The oligomeric state of yeast Rvb1 and Rvb2 was also found to be modulated by the presence of a tag at the N-terminus [17,38,39]. These observations seem to indicate that the Rvbs are capable of forming different oligomeric states depending on the complex or cellular process in which they are involved and that other proteins and cofactors might modulate the oligomeric state and, consequently, the activity of the Rvbs.
4. Chaperone-like activity of the Rvbs
Several studies have demonstrated a role of the Rvbs in the assembly of various complexes in different organisms suggesting that they might have a chaperone-like activity. The low abundance of Rvb1 and Rvb2 in eukaryotic cells relative to other components of several complexes which they are part of suggests that the Rvbs are not permanently associated with each complex, therefore providing further support for a general chaperone-like activity of the Rvbs rather than a defined catalytic activity within each complex [40]. In order to understand the exact role of the Rvbs in each process/complex, many studies mutated different domains/motifs in the Rvbs and assessed their effects on the activity of the complexes or on the processes being studied. Table 1 summarizes the main reported mutations and their effects as relevant to this review. The chaperone-like activity of the Rvbs in different complexes is further discussed below.
Table 1.
Summary of main reported mutations in Rvb1 and Rvb2 and their effect on the respective processes/complexes.
| mutated protein: Rvb1 or Rvb2 | complex studied | cellular process involved | mutation | effect | organism | reference |
|---|---|---|---|---|---|---|
| Rvb1 | interaction with c-Myc | c-Myc-mediated cellular transformation | Walker B mutant D302N | inhibition of c-Myc-mediated cellular transformation but did not affect general growth of cells | rat | Wood et al. [7] |
| Rvb1 | interaction with c-Myc | c-Myc-mediated cellular transformation | deletion of Walker A (Δ63–135) and Walker B (Δ290–366) motifs | no effect on the binding to c-Myc | rat | Wood et al. [7] |
| both | — | growth | Walker A and Walker B | incapable of supporting growth | yeast | Jonsson et al. [9] |
| Rvb1 | R2TP? | snoRNP biogenesis | Walker A (K81A) | inhibiting snoRNA production | yeast | King et al. [41] |
| Rvb2 | interaction with ATF-2 | DNA damage repair, apoptosis | Walker B | does not affect interaction with ATF-2 and still represses function of ATF-2 | human cell lines | Cho et al. [42] |
| Rvb2 | Interaction with β-catenin | expression of endogenous β-catenin/TCF target genes | Liebeskummer (lik) : 9 bp insertion (FCR a.a.) | enhanced ATPase activity leading to enhanced repression of β-catenin/TCF signalling leading to hyperplastic heart growth | zebrafish | Rottbauer et al. [43] |
| Rvb1 | interaction with β-catenin | β-catenin-mediated neoplastic transformation | Walker B mutant D302N | blocked expression of endogenous β-catenin/TCF target genes (e.g. ITF-2 and Axil) | rat | Feng et al. [44] |
| Rvb2 | Ino80 | chromatin remodelling | Walker B | no effect on recruitment of Arp5 to the Ino80 complex. No effect on chromatin remodelling function of the complex | yeast | Jonsson et al. [45] |
| both | interaction with c-Myc | cell proliferation | deletion of Walker A (Δ70–77 in Rvb1, Δ76–83 in Rvb2) and Walker B (Δ302–306 in Rvb1 and Δ299–303 in Rvb2) motifs | deletion mutants lacking WA or WB motifs did not affect cell division. Therefore not crucial in stimulating cell proliferation | Xenopus | Etard et al. [46] |
| both | PcG and TrxG | Hox gene transcription | ATPase domain | abolish normal control of Hox gene expression | Drosophila | Diop et al. [47] |
| Rvb1 | — | telomerase biogenesis | Walker B D302N | telomerase synthesis could not be rescued by expression of ATPase-deficient Rvb1 | human cell lines | Venteicher et al. [33] |
| Rvb2 | interaction with Influenza virus A polymerase | viral RNA synthesis | Walker B | does not affect interaction with the polymerase and still inhibits its activity | human cell lines | Kakugawa et al. [48] |
| both | — | PIKKs signalling | Walker B | PIKKs expression could not be rescued by the ATPase-deficient Rvbs | human cell lines | Izumi et al. [29] |
| Rvb1 | — | PIKKs signalling | Walker B | SMG-1–mediated Upf1 phosphorylation could not be rescued by the expression of Walker B mutant of Rvb1 | human cell lines | Izumi et al. [29] |
| both | — | — | truncation of one third of insertion domain | enhanced ATPase and helicase activity of the complex | in vitro | Gorynia et al. [35] |
5. Role of Rvb1 and Rvb2 in the assembly of chromatin remodelling complexes
Organisms use DNA as their genetic substance, therefore, DNA-related processes such as transcription, recombination, replication and repair are very critical. The eukaryotic DNA is packaged into chromatin in the nucleus. Nucleosomes form the fundamental repeating units of eukaryotic chromatin. The canonical nucleosome includes about 147 base pairs of DNA wrapped in approximately two superhelical turns around a histone octamer composed of two histone H2A–H2B heterodimers and a histone (H3–H4)2 heterotetramer [49]. Non-canonical nucleosomes have one or more histone variants (e.g. H2A.Z) replacing the canonical histones [49]. The compaction of DNA into a smaller volume is critical for the regulation of the above-mentioned DNA-related processes; however, it also impedes DNA transcription, replication and repair. Several chromatin remodelling complexes modulate these processes by using one of the following mechanisms to facilitate the access of proteins/cofactors to the underlying DNA: (i) using the energy of ATP hydrolysis to slide nucleosomes along the DNA, (ii) adding or removing covalent modifications on the tails of the histones in the nucleosome core or (iii) exchanging canonical histones with histone variants [50,51]. Over the last few years, several studies revealed that Rvb1 and Rvb2 are associated with various chromatin remodelling complexes such as the Ino80 complex in S. cerevisiae, Homo sapiens and D. melanogaster [9,20,21], the SWR-C complex in S. cerevisiae [52], and its homologous SRCAP in H. sapiens [53–56], and the Tip60 complex in H. sapiens and D. melanogaster [53,57–59] (figure 1).
(a). The Ino80 complex
The multisubunit Ino80 complex is very well studied and was first purified from yeast by immunoprecipitation [20]. This complex is involved in transcription regulation, replication and repair of DNA double strand breaks by catalysing ATP-dependent mobilization of nucleosomes along the DNA [20,21]. The core subunits of the Ino80 complex are common between yeast and human: the SNF2 family helicase Ino80, which is the catalytic subunit of the complex, Rvb1, Rvb2, Act1, Ino80 subunit (Ies)2 and Ies6 [52], and the actin-related proteins Arp4, Arp5 and Arp8. In addition, the yeast and human Ino80 complexes have their own distinct set of additional subunits. Both yeast and human Ino80 complexes exhibit ATP-dependent nucleosome remodelling activity and DNA and nucleosome-activated ATPase activity [21].
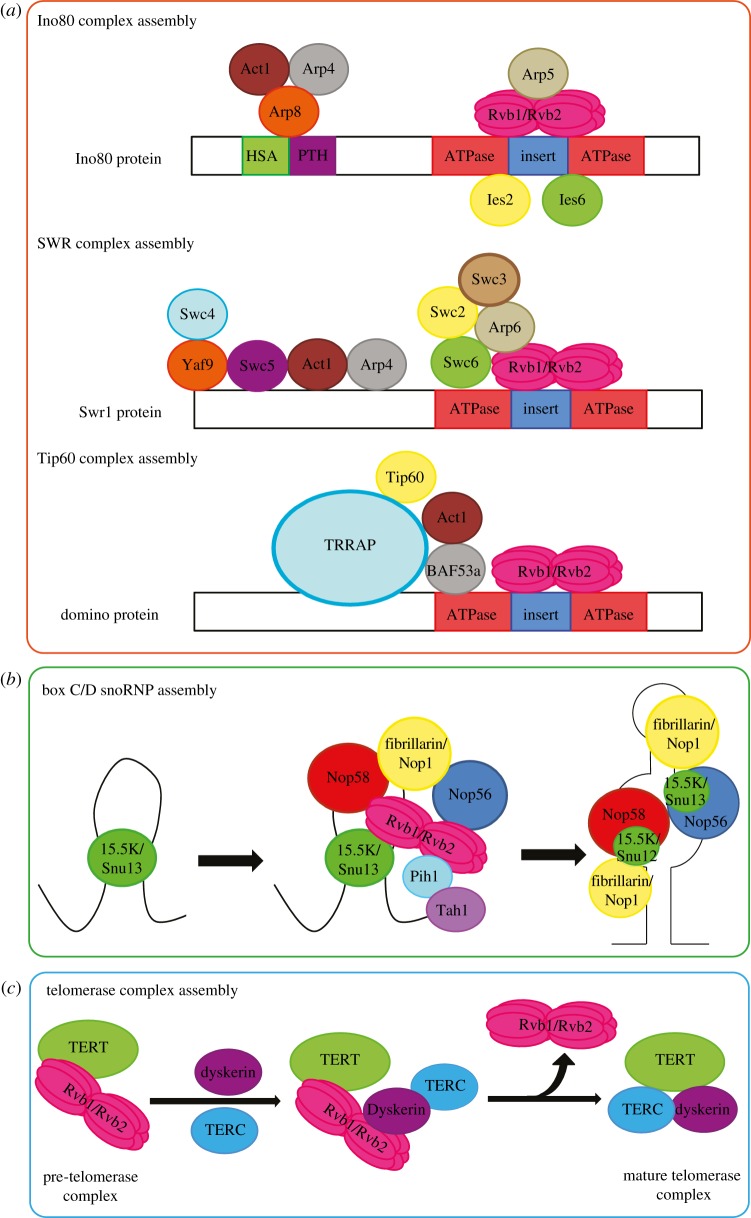
In yeast, considerable overlap was found between genes regulated by Ino80 protein and those regulated by Rvb1 and Rvb2 [9]. The promoters of those genes were found to be associated with the Ino80 protein but not with Rvb1 or Rvb2 [9]. The Ino80 complex has ATPase activity ascribed largely to the Ino80 protein rather than the Rvbs since mutating the ATP-binding site of the Ino80 protein results in significant reduction in the ATPase activity of the complex without affecting the subunit composition of the complex [9]. However, loss of the Rvbs leads to the loss of Arp5 protein from the complex, and, consequently, the loss of the chromatin remodelling activity of the Ino80 complex [45]. The association between Arp5 and the Rvbs requires ATP but not the ATPase activity of the Rvbs [45]. Recently, Chen et al. [60] showed that in human Ino80 complex, Rvb1 and Rvb2 together with Arp5, Ies2 and Ies6 associate with an insertion region within the ATPase domain of the Ino80 protein (figure 3a).
Figure 3.
Chaperone-like activities of the Rvbs. (a) Rvb1 and Rvb2 assemble the Ino80 complex by recruiting Arp5 to the Ino80 protein. Rvb1 and Rvb2 have a possible role in the assembly of the SWR complex by binding and recruiting subunits integral for the activity of the complex. Finally, Rvb1 and Rvb2 interact with the ATPase domain of domino/p400, and help in the assembly of the Tip60 complex (TRRAP, transformation/transcription domain-associated protein). (b) Rvb1 and Rvb2 function in the assembly of box C/D snoRNP by bridging interactions between 15.5K/Snu13 and the other core proteins. (c) Rvb1 and Rvb2 bring together TERT, dyskerin and TERC (telomerase RNA component) and remodel the pre-telomerase complex into a mature TERT–TERC–dyskerin complex.
The Ino80 complex in yeast causes the proximal eviction of nucleosomes surrounding double strand breaks [52]. The Rvb proteins were found to be recruited to the homothallic switching (HO) endonuclease-induced DNA double-strand break along with Arp8, Arp5 and Ino80 protein [52]. This recruitment of the Ino80 complex was dependent upon the phosphorylation of the histone variant H2AX. Deletion of Arp4 and Nhp10 (two subunits of the Ino80 complex) caused a reduction in the recruitment of the complex, including the Rvbs, to the double strand breaks, therefore suggesting that these two proteins are necessary for the recognition of the phosphorylated histones and for the interaction of the Ino80 complex with the double strand break [52]. The exact role of the Rvbs in this recruitment process is yet to be determined. It can be speculated that the Rvbs are required to recruit the rest of the Ino80 subunits to the double strand breaks to form a functional complex.
(b). SWR/SRCAP complex
The Swi/Snf2-related (SWR) complex in yeast, also known as the Snf2-related CREBBP activator protein (SRCAP) complex in mammalian cells, is yet another chromatin remodelling complex that contains both Rvb1 and Rvb2 as integral subunits. Both the SWR and SRCAP complexes were found to remodel chromatin by catalysing the ATP-dependent replacement of H2A–H2B histone dimers in nucleosomes by dimers containing the histone variant Htz1 in yeast or H2AZ in mammalian cells [54,55,61]. This mechanism is essential in a range of cellular processes, such as transcriptional regulation, chromosome segregation, cell cycle progression and DNA damage response. The catalytic subunit of the complex is the Swr1/SRCAP protein, which is an SNF2 helicase. Rvb1, Rvb2, Act1, Arp4, Arp6 and Yaf9/GAS41 are shared subunits between the SWR complex in yeast and the SRCAP complex in mammalian cells [20,52,56]. The SWR/SRCAP complex shares several subunits with the Ino80 complex, namely Act1, Arp4, Rvb1 and Rvb2. In yeast, it was shown that the ATPase domain of the Swr1 protein binds Rvb1, Rvb2, Arp6, Swc2, Swc3 and Swc6 [62] (figure 3a), reflecting yet another similarity with the Ino80 complex. The exact function of the Rvbs in the SWR/SRCAP complex remains unexplored. However, given the significant similarity between the SWR complex and the Ino80 complex, it can be speculated that the Rvb proteins perform a role in the assembly of the SWR complex by binding and recruiting a subunit integral for the activity of the complex, just as they recruit Arp5 to the Ino80 complex [45].
(c). Tip60 complex
This complex, which is a histone acetyltransferase (HAT) found in both human and fly cells, remodels chromatin by acetylating histones converting chromatin to euchromatin, which is a relaxed, transcriptionally active DNA [59,63]. It has been shown that this complex also acetylates proteins such as the ataxia telangiectasia mutated (ATM) protein kinase after DNA damage, therefore activating ATM [64]. The Tip60 complex is involved in transcription, DNA repair and apoptosis [65]. The catalytic subunit in the complex is Tip60 (Tat interactive protein 60). The complex has Rvb1 and Rvb2 as its integral subunits. The Tip60 complex shares several subunits with the SWR complex and several other subunits with the NuA4 (Nucleosomal Acetyltransferase of H4) complex, which is an acetyltransferase complex found in yeast but that does not contain Rvb1 or Rvb2, suggesting that the Tip60 complex is a fusion of those two complexes [53]. Esa1 in yeast, which is the orthologue of the Tip60 protein in mammals, is the catalytic subunit of the NuA4 complex. Eaf1, a subunit found in the NuA4 complex, is the orthologue of the mammalian p400/domino protein found in the Tip60 complex; however, Eaf1 lacks the ATPase domain found in p400/domino protein [53,57]. The absence of Rvb1 and Rvb2 in the NuA4 complex may be because of the absence of the ATPase domain in Eaf1 since Rvb1 and Rvb2 were shown to interact with the ATPase domain of p400/domino [53] (figure 3a), similarly to the way they interact with the ATPase domain of the Ino80 protein.
As mentioned above, Tip60 is involved in DNA damage repair. DNA damage causes histone variant H2AX to be phosphorylated by ATM and ATR protein kinases. The phospho-H2AX acts as a marker that recruits other proteins to the sites of DNA damage to amplify the damage signal and repair the damage [52,63]. HAT activity of Tip60 is required to acetylate H4 before the phospho-H2AX can be remodelled and dephosphorylated in DNA damage response [63]. It has been shown that depletion of either Rvb1 or Tip60 causes an increase in the phosphorylated H2AX and that the Rvbs are required for the HAT activity of the Tip60 complex, suggesting that Rvb1 is required for the assembly of the Tip60 complex [63].
The role of Rvb1 is also linked to apoptosis through Tip60. Tip60 is required for the acetylation of p53, and the acetylation of p53 is required for its binding to promoters of proapoptotic genes [66]. In another example, Feng et al [44] showed that the stable expression of the Walker B mutant of Rvb1 blocked the expression of endogenous β-catenin/T-cell factor (TCF) target genes, which is because of inhibition of histone acetylation of β-catenin/TCF target gene sequences, thus suggesting that Rvb1 exerts its effect through Tip60 [44]. Also, Rvb1, along with Tip60, binds to and acetylates histones at the promoter of KAI1, which is a metastasis suppressor gene, resulting in the induction of the expression of KAI1 [67].
6. Role of Rvb1 and Rvb2 in box C/D snoRNP biogenesis
In an attempt to identify the interactors of yeast Hsp90, which is a ubiquitous molecular chaperone that is essential in many signalling pathways, our group conducted systematic genome-wide screens and found Rvb1 and Rvb2 to be components of a complex interacting with Hsp90 that we termed the R2TP complex [68]. In yeast, this complex consists of two Hsp90 interactors, which we identified and termed Tah1 (tetratricopeptide repeat (TPR)-containing protein associated with Hsp90) and Pih1 (protein interacting with Hsp90), and the two AAA+ helicases Rvb1 and Rvb2 [68], hence the name R2TP. Tah1, which was uncharacterized at the time and whose structure we solved recently [69], consists of two TPR motifs and a C-terminal helix. Tah1 was found to bind to the MEEVD peptide corresponding to the C-terminus of Hsp90, while the C-terminus of Tah1 binds to the C-terminus of Pih1 [69]. Pih1, also uncharacterized at the time, is a 40 kDa protein which was found to be unstable on its own, and stable upon binding to the C-terminus of Tah1 [69].
The R2TP complex is highly conserved in eukaryotes. In humans, R2TP contains Rvb1, Rvb2, RPAP3 (protein equivalent to Tah1 although not similar) and PIH1D1 (Pih1 orthologue) [26]. The R2TP complex has been implicated in small nucleolar ribonucleoprotein (snoRNP) assembly and pre-ribosomal RNA processing in human and yeast cells [26,27]. The complex also plays essential roles in apoptosis, PIKK signalling [29] and RNA polymerase II assembly [70].
snoRNP complexes are made up of either box C/D or box H/ACA small nucleolar RNAs (snoRNAs) complexed with proteins. snoRNPs are involved in cleavage and modification of small nuclear RNA (snRNA), ribosomal RNA (rRNA) and tRNAs [71]. Box C/D snoRNPs catalyse ribose 2′-O methylation of pre-ribosomal RNA (pre-rRNA), while box H/ACA snoRNPs mediate pseudo-uridylation of pre-rRNA [28]. Mature box C/D snoRNAs in eukaryotes are associated with four common core proteins: 15.5K (Snu13 in yeast), NOP56, NOP58 and the methyltransferase fibrillarin (Nop1 in yeast) [28]. The core box C/D proteins bind a conserved sequence termed the box C/D motif that folds into a stem-internal loop-stem structure known as a k-turn (figure 3b). 15.5K, an RNA binding protein, binds directly to the k-turn to recruit the other core proteins [72–74]. The assembly of the complete complex is essential for the nucleolar localization of the complex [74]. Several proteins are required for this assembly, including the R2TP complex, as well as, NUFIP, TAF9 and BCD1 [28,41,75,76] (figure 3b). It has been shown that Rvb1 and Rvb2 weakly interact with NOP56, NOP58 and fibrillarin, and that the presence of ATP stimulates the interaction of Rvb1 and Rvb2 with 15.5K [28]. Rvb1 and Rvb2 appear to bridge the interaction between 15.5K and both NOP56 and NOP58 proteins [28]. In both yeast [27,41] and mammalian cells [28], depletion of the Rvbs results in the mislocalization of the snoRNP core proteins. The data to date indicate that the Rvb proteins play an important role in the assembly and remodelling of the snoRNP complex during biogenesis (figure 3b) mainly as components of the R2TP complex.
7. Role of Rvb1 and Rvb2 in PIKK signalling
Recent studies revealed that Rvb1 and Rvb2 are common regulators of all phosphatidylinositol 3-kinase-related protein kinase (PIKK) members [77]. PIKKs are serine–threonine protein kinases with catalytic domains homologous to those of phosphatidylinositol 3-kinases. PIKKs regulate DNA damage responses, nutrient-dependent signalling, and nonsense-mediated mRNA decay (NMD) [77]. The PIKK family includes DNA-PKcs (DNA-dependent protein kinase catalytic subunit), ATM and ATR (ATM- and Rad3-related), which are collectively responsible for signalling the presence of DNA damage [77]. They phosphorylate proteins that have roles in regulation of cell cycle progression, DNA repair, apoptosis and cellular senescence [77]. The PIKK family also includes SMG-1 (suppressor with morphological effect on genitalia 1), mTOR (mammalian target of rapamycin) and TRRAP (transformation/transcription domain-associated protein) in mammals [77]. SMG-1 is an essential factor of NMD and TRRAP regulates transcription as a subunit of HAT complexes [78]. SMG-1 and TRRAP are also involved in DNA damage signalling and repair [78]. A multiprotein complex called SMG1C, which is composed of SMG-1, SMG-8 and SMG-9, is essential for NMD. SMG1C detects and degrades mRNAs to prevent the production of potentially harmful premature proteins [29]. mTOR regulates nutrient-dependent signalling.
Knockdown of human Rvb1 or Rvb2 has been shown to lead to decreased phosphorylation of direct downstream effectors of ATM, ATR, mTOR and SMG-1, and also to decreased abundance of mRNA and proteins for ATM, ATR, DNA-PKcs, TRRAP and mTOR but not the abundance of other kinases [29]. WT Rvb1 or Rvb2 were able to rescue the reduced PIKK abundance, however, ATPase-deficient mutants failed to rescue the reduced abundance, indicating that the ATPase activities of both Rvb1 and Rvb2 are required to control the abundance of PIKKs [29]. It was also revealed that human Rvb1 and Rvb2 are required for SMG-1-mediated Upf1 phosphorylation, which occurs on a spliced mRNP in the cytoplasm, and that the phosphorylation was dependent on the ATPase activity of Rvb1. This phosphorylation is induced by remodelling of the mRNA surveillance complex that involves first the formation of the SURF complex, which is composed of SMG1, UPF1, eRF1 and eRF3, on a ribosome recognizing premature termination codon(s) and then the formation of the decay-inducing (DECID) complex on an mRNP. Immunoprecipitation experiments suggested that the Rvb1/2 complex associates with SURF playing a role in the remodelling of the surveillance complex and, thus, in forming a DECID complex [29].
In addition, human Rvb1/2, as part of the R2TP complex, plays a role in the assembly and stabilization of the PIKKs. This stability and assembly is achieved when the R2TP-Hsp90/Prefoldin-like complex interacts with PIKKs via the Tel2 complex (also known as the TTT complex), which is composed of Tel2, Tti1 and Tti2 [78]. A recent study in yeast linked the Tel2 complex and Asa1p to PIKKs [79].
8. Role of Rvb1 and Rvb2 in telomerase complex assembly
Telomeres are repetitive nucleotide sequences located at the ends of chromosomes, capping and protecting them from degradation and recombinogenic activities. They are un-replicated and lost during cell division owing to the ‘end replication problem’ exhibited during DNA replication, and are replenished by the telomerase [80]. The end replication problem is a problem the DNA polymerase runs into because the leading strand in the double-stranded DNA can be replicated to the very end, but the lagging strand cannot. The polymerase needs RNA primers to replicate the lagging strand DNA; however, use of the RNA primer is not possible at the end of the DNA because there is nothing for the primer to bind to, therefore, the last section of the lagging strand cannot be synthesized. Thus, after several cycles of replication, the DNA would continue to get smaller.
Telomerase is a multisubunit RNP complex that adds DNA repeats to telomeres. The complex is composed of the catalytic subunit TERT (telomerase reverse transcriptase), TERC (telomerase RNA component) and the TERC-binding protein dyskerin [33]. In humans, Rvb1 and Rvb2 were identified as subunits of the telomerase complex, and they were found to be required for telomerase assembly/biogenesis through maintaining the telomerase RNA stability [33]. It was demonstrated that Rvb1 directly interacts with TERT, recruiting Rvb2 and bridging its interaction to the TERT complex. It was also shown that Rvb1 and Rvb2 interact with dyskerin [33]. Depletion of Rvb1 and Rvb2 caused a loss of TERC and dyskerin from the complex suggesting that dyskerin bridges the interaction between the Rvb proteins and TERC [33]. The Walker B mutant of Rvb1 could not rescue TERC and dyskerin loss from the complex, thus indicating that Rvb1 and Rvb2 are essential for telomerase activity and for TERC and dyskerin accumulation through a mechanism that requires ATPase activity [33]. Rvb proteins seem to help bring together TERT, dyskerin and TERC and remodel the TERT–Rvb1–Rvb2 complex into a mature TERT–TERC–dyskerin complex [33] (figure 3c).
In addition to their role in the assembly of the telomerase complex, Rvb1 and Rvb2 seem to be also involved in the transcription of TERT [81]. Knocking down Rvb1 or its partner Rvb2 using siRNA in gastric and cervical cancer cells led to significant decreases in TERT mRNA. In addition, human Rvb2 depletion resulted in a significant decrease in the activity of TERT promoter that is dependent on c-MYC [81]. Therefore, TERT transcription requires the constitutive expression of Rvb2 and its cooperation with c-MYC.
In yeast, the Rvbs are also subunits of what is called the ASTRA complex [79,82]. ASTRA (ASsembly of Tel, Rvb, and Atm-like kinase) complex is composed of Tra1 (TRRAP homolog), Rvb1, Rvb2, Tel2 (telomere binding protein), Tti1p, Tti2p and Asa1p (a WD-repeat-containing protein). The ASTRA complex is poorly studied, but it is proposed to play a role in telomeric maintenance and its components (Tti1p, Tti2p, Tel2 and Asa1p) have been shown to be linked to PIKKs as mentioned previously. The role of the Rvbs within this complex is not yet characterized.
9. Role of Rvb1 and Rvb2 in mitotic spindle assembly
Several studies reported the involvement of Rvb1 and Rvb2 in mitosis. Human Rvb1 was found to copurify with tubulin isolated from U937 cells [30]. Furthermore, human Rvb1 was found to colocalize with tubulin at the centrosome and at the mitotic spindle in addition to being present in the nucleus. Using an in vitro tubulin assembly assay, it was demonstrated that Rvb1 is involved in the formation of microtubules [30]. Subsequently, another study showed that Rvb2 associates with the centrosome and the mitotic spindle [31]. However, it was demonstrated that, unlike Rvb1, Rvb2 localizes to the midzone during telophase and to the midbody during cytokinesis [31]. In 2008, Ducat et al. [32] demonstrated that depletion of Rvb1 using siRNA causes a defect in spindle assembly in Drosophila and mammalian cell lines. The same result was observed when depleting Rvb1 in Xenopus egg extracts. Moreover, Rvb1 and Rvb2 were found to interact with the γ-tubulin ring complex in Xenopus, which is involved in nucleating spindle formation, suggesting that both Rvb proteins are involved in mitotic spindle assembly.
10. Role of Rvb1 and Rvb2 in cancer
In mammalian cells, Rvb1 and Rvb2, separately and together, were found to have a crucial role in pathways linked closely to cancer. Several studies have shown that both Rvb1 and Rvb2 are overexpressed in 80 per cent of colon cancer specimens. Rvb2 is found to be overexpressed in human hepatocellular carcinoma cells, while Rvb1 transcript levels are found to be increased in non-small cell lung cancer [83]. The transcription of both genes is deregulated in several cancers such as liver, bladder and melanoma. In addition, it has been demonstrated that decreasing the expression of Rvb1 or Rvb2 results in reduced tumor cell growth and increased apoptosis in vitro and that decreasing Rvb2 expression results in growth arrest of established tumours in xenograft experiments in mice [83].
The roles of the Rvbs associated with modulating cellular transformation, signalling, apoptosis and response to DNA damage is mediated through their interaction with a multitude of proteins such as the tumor suppressor protein Hint1 and the transcription factors β-catenin, c-Myc, E2F (only Rvb1) and ATF2 (only Rvb2) [6–9].
11. Role of Rvb1 and Rvb2 in transcription regulation
Rvb1 and Rvb2 can function together but in several cases have also been shown to function independently and to exhibit antagonistic effects on the regulation of transcription of several target genes. Rvb1 and Rvb2 interact with β-catenin, which is a major player in Wnt signalling that affects TCF-mediated transcription [8]. In the nucleus, stable unphosphorylated β-catenin binds to the TCF family of transcription factors and increases the expression of downstream genes (e.g. c-Myc, ITF-2 and Cox-2) [44]. Rvb1 and Rvb2 have opposing effects on β-catenin-TCF transcriptional activity. Rvb1 increases the transcriptional activation of target genes, while Rvb2 represses the β-catenin/TCF transactivation complex and thus decreases the transcription of downstream genes [8]. The Walker B mutant of Rvb1 was found to block β-catenin-mediated transcription of TCF-dependent genes owing to inhibition of acetylation of histones near β-catenin target gene sequences suggesting that Rvb1/Tip60 mediates the regulation of this transcription [44]. On the other hand, Rvb2 represses gene activation mediated by β-catenin and TCF through its interaction with histone deacetylase HDAC1 and 2, and corepressor TLE (transducin-like enhancer) [8]. In another example, Rvb1/Tip60 are recruited on the promoter of the KAI1 (a metastasis suppressor which inhibits metastasis by promoting cell adhesion) gene as a co-activator complex, while Rvb2/β-catenin act as a co-repressor of the transcription which recruits HDAC1 as well [84]. In addition, Hint1 (histidine triad nucleotide-binding protein 1), which acts as a co-regulator of β-catenin-TCF-mediated transcription, was shown to bind to the insertion domain in Rvb1 and Rvb2 [85]. It was demonstrated that Hint1 prevents formation of hetero and homo complexes of the Rvbs, but not the interaction between the Rvb proteins with β-catenin. Hint1 was found to be a regulator of the Rvbs/Wnt-catenin signalling pathway since its overexpression was found to modulate Rvbs/β-catenin regulated genes.
Rvb1 and Rvb2 were found to bind to and regulate the function of the transcription factor c-Myc [7] (table 1). c-Myc, which is involved in oncogenic transformation, apoptosis and stimulation of cell proliferation, contains two conserved regions: Myc homology box I (MBI) and MBII, with the latter being the region where both Rvb1 and Rvb2 bind. The Walker B mutant form of Rvb1 was found to inhibit c-Myc oncogenic activity but did not inhibit cellular growth indicating that Rvb1 is essential for c-Myc-mediated oncogenic transformation [7].
12. Concluding remarks
Rvb1 and Rvb2 are involved in various cellular complexes and processes in different organisms. They exhibit different roles and functions specific to the processes in which they are involved. Besides being ATPases that provide energy for several processes and helicases with potential DNA/RNA unwinding activity, many studies have shown that the Rvbs seem to act as chaperones. They have been found to recruit proteins/DNA/RNA to their respective complexes and to remodel these complexes by bridging the interactions between the different components within the complex. Hence, we propose that the Rvbs are potential chaperones for the assembly and maturation of protein–protein and protein–DNA/RNA complexes. However, further studies need to be conducted to determine the exact role of the Rvbs in the assembly of these complexes.
Note added in proof
While this review was in preparation for publication, the crystal structure of human Rvb2 with truncation in part of Domain II was published by Petukhov et al. [86] and the cryo-electron microscopy structures of human double-ring Rvb1-Rvb2 complexes were published by López-Perrote et al. [87]. In addition, the role of human Rvbs (through the R2TP complex) in H/ACA RNP biogenesis was established by Machado-Pinilla et al. [88]
Acknowledgements
We thank Dr Yoshito Kakihara, Jennifer Huen and Liang Zhao for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research (MOP-93778) to W.A.H.
References
- 1.Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 2.Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 10.1016/j.jsb.2003.10.010 (doi:10.1016/j.jsb.2003.10.010) [DOI] [PubMed] [Google Scholar]
- 3.Snider J, Thibault G, Houry WA. 2008. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9, 216. 10.1186/gb-2008-9-4-216 (doi:10.1186/gb-2008-9-4-216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makino Y, Mimori T, Koike C, Kanemaki M, Kurokawa Y, Inoue S, Kishimoto T, Tamura T. 1998. TIP49, homologous to the bacterial DNA helicase RuvB, acts as an autoantigen in human. Biochem. Biophys. Res. Commun. 245, 819–823 10.1006/bbrc.1998.8504 (doi:10.1006/bbrc.1998.8504) [DOI] [PubMed] [Google Scholar]
- 6.Bauer A, Huber O, Kemler R. 1998. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc. Natl Acad. Sci. USA 95, 14 787–14 792 10.1073/pnas.95.25.14787 (doi:10.1073/pnas.95.25.14787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood MA, McMahon SB, Cole MD. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5, 321–330 10.1016/S1097-2765(00)80427-X (doi:10.1016/S1097-2765(00)80427-X) [DOI] [PubMed] [Google Scholar]
- 8.Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. 2000. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 19, 6121–6130 10.1093/emboj/19.22.6121 (doi:10.1093/emboj/19.22.6121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. 2001. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 276, 16 279–16 288 10.1074/jbc.M011523200 (doi:10.1074/jbc.M011523200) [DOI] [PubMed] [Google Scholar]
- 10.Kanemaki M, et al. 1997. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 235, 64–68 10.1006/bbrc.1997.6729 (doi:10.1006/bbrc.1997.6729) [DOI] [PubMed] [Google Scholar]
- 11.Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. 1998. A eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 273, 27 786–27 793 10.1074/jbc.273.43.27786 (doi:10.1074/jbc.273.43.27786) [DOI] [PubMed] [Google Scholar]
- 12.Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. 1999. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 274, 22 437–22 444 10.1074/jbc.274.32.22437 (doi:10.1074/jbc.274.32.22437) [DOI] [PubMed] [Google Scholar]
- 13.Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. 2001. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 311, 297–310 10.1006/jmbi.2001.4852 (doi:10.1006/jmbi.2001.4852) [DOI] [PubMed] [Google Scholar]
- 14.Yamada K, Kunishima N, Mayanagi K, Ohnishi T, Nishino T, Iwasaki H, Shinagawa H, Morikawa K. 2001. Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc. Natl Acad. Sci. USA 98, 1442–1447 10.1073/pnas.98.4.1442 (doi:10.1073/pnas.98.4.1442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsaneva IR, Muller B, West SC. 1993. RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc. Natl Acad. Sci. USA 90, 1315–1319 10.1073/pnas.90.4.1315 (doi:10.1073/pnas.90.4.1315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radovic S, Rapisarda VA, Tosato V, Bruschi CV. 2007. Functional and comparative characterization of Saccharomyces cerevisiae RVB1 and RVB2 genes with bacterial Ruv homologues. FEMS Yeast Res. 7, 527–539 10.1111/j.1567-1364.2006.00205.x (doi:10.1111/j.1567-1364.2006.00205.x) [DOI] [PubMed] [Google Scholar]
- 17.Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. 2008. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. 376, 1320–1333 10.1016/j.jmb.2007.12.049 (doi:10.1016/j.jmb.2007.12.049) [DOI] [PubMed] [Google Scholar]
- 18.Matias PM, Gorynia S, Donner P, Carrondo MA. 2006. Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 281, 38 918–38 929 10.1074/jbc.M605625200 (doi:10.1074/jbc.M605625200) [DOI] [PubMed] [Google Scholar]
- 19.Jha S, Dutta A. 2009. RVB1/RVB2: running rings around molecular biology. Mol. Cell 34, 521–533 10.1016/j.molcel.2009.05.016 (doi:10.1016/j.molcel.2009.05.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X, Mizuguchi G, Hamiche A, Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 10.1038/35020123 (doi:10.1038/35020123) [DOI] [PubMed] [Google Scholar]
- 21.Jin J, et al. 2005. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280, 41 207–41 212 10.1074/jbc.M509128200 (doi:10.1074/jbc.M509128200) [DOI] [PubMed] [Google Scholar]
- 22.Bakshi R, Mehta AK, Sharma R, Maiti S, Pasha S, Brahmachari V. 2006. Characterization of a human SWI2/SNF2 like protein hINO80: demonstration of catalytic and DNA binding activity. Biochem. Biophys. Res. Commun. 339, 313–320 10.1016/j.bbrc.2005.10.206 (doi:10.1016/j.bbrc.2005.10.206) [DOI] [PubMed] [Google Scholar]
- 23.Choi J, Heo K, An W. 2009. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 37, 5993–6007 10.1093/nar/gkp660 (doi:10.1093/nar/gkp660) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohdate H, Lim CR, Kokubo T, Matsubara K, Kimata Y, Kohno K. 2003. Impairment of the DNA binding activity of the TATA-binding protein renders the transcriptional function of Rvb2p/Tih2p, the yeast RuvB-like protein, essential for cell growth. J. Biol. Chem. 278, 14 647–14 656 10.1074/jbc.M213220200 (doi:10.1074/jbc.M213220200) [DOI] [PubMed] [Google Scholar]
- 25.McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. 2007. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell Biol. 27, 6782–6793 10.1128/MCB.01097-07 (doi:10.1128/MCB.01097-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulon S, et al. 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180, 579–595 10.1083/jcb.200708110 (doi:10.1083/jcb.200708110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao R, et al. 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578 10.1083/jcb.200709061 (doi:10.1083/jcb.200709061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKeegan KS, Debieux CM, Watkins NJ. 2009. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell Biol. 29, 4971–4981 10.1128/MCB.00752-09 (doi:10.1128/MCB.00752-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. 2010. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci. Signal. 3, ra27. 10.1126/scisignal.2000468 (doi:10.1126/scisignal.2000468) [DOI] [PubMed] [Google Scholar]
- 30.Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, Waldhausl W, Pasternack MS, Wagner L. 2003. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil. Cytoskeleton 56, 79–93 10.1002/cm.10136 (doi:10.1002/cm.10136) [DOI] [PubMed] [Google Scholar]
- 31.Sigala B, Edwards M, Puri T, Tsaneva IR. 2005. Relocalization of human chromatin remodeling cofactor TIP48 in mitosis. Exp. Cell Res. 310, 357–369 10.1016/j.yexcr.2005.07.030 (doi:10.1016/j.yexcr.2005.07.030) [DOI] [PubMed] [Google Scholar]
- 32.Ducat D, Kawaguchi S, Liu H, Yates JR, III, Zheng Y. 2008. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol. Biol. Cell 19, 3097–3110 10.1091/mbc.E07-11-1202 (doi:10.1091/mbc.E07-11-1202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–957 10.1016/j.cell.2008.01.019 (doi:10.1016/j.cell.2008.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forget D, et al. 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteom. 9, 2827–2839 10.1074/mcp.M110.003616 (doi:10.1074/mcp.M110.003616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorynia S, et al. 2011. Structural and functional insights into a dodecameric molecular machine—the RuvBL1/RuvBL2 complex. J. Struct. Biol. 176, 279–291 10.1016/j.jsb.2011.09.001 (doi:10.1016/j.jsb.2011.09.001) [DOI] [PubMed] [Google Scholar]
- 36.Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. 2007. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J. Mol. Biol. 366, 179–192 10.1016/j.jmb.2006.11.030 (doi:10.1016/j.jmb.2006.11.030) [DOI] [PubMed] [Google Scholar]
- 37.Niewiarowski A, Bradley AS, Gor J, McKay AR, Perkins SJ, Tsaneva IR. 2010. Oligomeric assembly and interactions within the human RuvB-like RuvBL1 and RuvBL2 complexes. Biochem. J. 429, 113–125 10.1042/BJ20100489 (doi:10.1042/BJ20100489) [DOI] [PubMed] [Google Scholar]
- 38.Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O. 2008. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure 16, 1511–1520 10.1016/j.str.2008.08.009 (doi:10.1016/j.str.2008.08.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung KL, Huen J, Kakihara Y, Houry WA, Ortega J. 2010. Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J. Mol. Biol. 404, 478–492 10.1016/j.jmb.2010.10.003 (doi:10.1016/j.jmb.2010.10.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallant P. 2007. Control of transcription by Pontin and Reptin. Trends Cell Biol. 17, 187–192 10.1016/j.tcb.2007.02.005 (doi:10.1016/j.tcb.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 41.King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. 2001. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell Biol. 21, 7731–7746 10.1128/MCB.21.22.7731-7746.2001 (doi:10.1128/MCB.21.22.7731-7746.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho SG, Bhoumik A, Broday L, Ivanov V, Rosenstein B, Ronai Z. 2001. TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell Biol. 21, 8398–8413 10.1128/MCB.21.24.8398-8413.2001 (doi:10.1128/MCB.21.24.8398-8413.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rottbauer W, et al. 2002. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111, 661–672 10.1016/S0092-8674(02)01112-1 (doi:10.1016/S0092-8674(02)01112-1) [DOI] [PubMed] [Google Scholar]
- 44.Feng Y, Lee N, Fearon ER. 2003. TIP49 regulates β-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res. 63, 8726–8734 [PubMed] [Google Scholar]
- 45.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16, 465–477 10.1016/j.molcel.2004.09.033 (doi:10.1016/j.molcel.2004.09.033) [DOI] [PubMed] [Google Scholar]
- 46.Etard C, Gradl D, Kunz M, Eilers M, Wedlich D. 2005. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech. Dev. 122, 545–556 10.1016/j.mod.2004.11.010 (doi:10.1016/j.mod.2004.11.010) [DOI] [PubMed] [Google Scholar]
- 47.Diop SB, et al. 2008. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 9, 260–266 10.1038/embor.2008.8 (doi:10.1038/embor.2008.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakugawa S, Shimojima M, Neumann G, Goto H, Kawaoka Y. 2009. RuvB-like protein 2 is a suppressor of influenza A virus polymerases. J. Virol. 83, 6429–6434 10.1128/JVI.00293-09 (doi:10.1128/JVI.00293-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conaway RC, Conaway JW. 2009. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci. 34, 71–77 10.1016/j.tibs.2008.10.010 (doi:10.1016/j.tibs.2008.10.010) [DOI] [PubMed] [Google Scholar]
- 50.Flaus A, Owen-Hughes T. 2001. Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev. 11, 148–154 10.1016/S0959-437X(00)00172-6 (doi:10.1016/S0959-437X(00)00172-6) [DOI] [PubMed] [Google Scholar]
- 51.Flaus A, Owen-Hughes T. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14, 165–173 10.1016/j.gde.2004.01.007 (doi:10.1016/j.gde.2004.01.007) [DOI] [PubMed] [Google Scholar]
- 52.Morrison AJ, Shen X. 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 10, 373–384 10.1038/nrm2693 (doi:10.1038/nrm2693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell Biol. 24, 1884–1896 10.1128/MCB.24.5.1884-1896.2004 (doi:10.1128/MCB.24.5.1884-1896.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131. 10.1371/journal.pbio.0020131 (doi:10.1371/journal.pbio.0020131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 10.1126/science.1090701 (doi:10.1126/science.1090701) [DOI] [PubMed] [Google Scholar]
- 56.Cai Y, et al. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280, 13 665–13 670 10.1074/jbc.M500001200 (doi:10.1074/jbc.M500001200) [DOI] [PubMed] [Google Scholar]
- 57.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278, 42 733–42 736 10.1074/jbc.C300389200 (doi:10.1074/jbc.C300389200) [DOI] [PubMed] [Google Scholar]
- 58.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, III, Abmayr SM, Washburn MP, Workman JL. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 10.1126/science.1103455 (doi:10.1126/science.1103455) [DOI] [PubMed] [Google Scholar]
- 59.Qi D, Jin H, Lilja T, Mannervik M. 2006. Drosophila Reptin and other TIP60 complex components promote generation of silent chromatin. Genetics 174, 241–251 10.1534/genetics.106.059980 (doi:10.1534/genetics.106.059980) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Cai Y, Jin J, Florens L, Swanson SK, Washburn MP, Conaway JW, Conaway RC. 2011. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 286, 11 283–11 289 10.1074/jbc.M111.222505 (doi:10.1074/jbc.M111.222505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krogan NJ, et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576 10.1016/S1097-2765(03)00497-0 (doi:10.1016/S1097-2765(03)00497-0) [DOI] [PubMed] [Google Scholar]
- 62.Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, Wu C. 2005. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12, 1064–1071 10.1038/nsmb1023 (doi:10.1038/nsmb1023) [DOI] [PubMed] [Google Scholar]
- 63.Jha S, Shibata E, Dutta A. 2008. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell Biol. 28, 2690–2700 10.1128/MCB.01983-07 (doi:10.1128/MCB.01983-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl Acad. Sci. USA 102, 13 182–13 187 10.1073/pnas.0504211102 (doi:10.1073/pnas.0504211102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 10.1016/S0092-8674(00)00051-9 (doi:10.1016/S0092-8674(00)00051-9) [DOI] [PubMed] [Google Scholar]
- 66.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 10.1016/j.molcel.2006.11.026 (doi:10.1016/j.molcel.2006.11.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell 110, 55–67 10.1016/S0092-8674(02)00809-7 (doi:10.1016/S0092-8674(02)00809-7) [DOI] [PubMed] [Google Scholar]
- 68.Zhao R, et al. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727 10.1016/j.cell.2004.12.024 (doi:10.1016/j.cell.2004.12.024) [DOI] [PubMed] [Google Scholar]
- 69.Jimenez B, Ugwu F, Zhao R, Orti L, Makhnevych T, Pineda-Lucena A, Houry WA. 2012. Structure of minimal tetratricopeptide repeat domain protein tah1 reveals mechanism of its interaction with pih1 and hsp90. J. Biol. Chem. 287, 5698–5709 10.1074/jbc.M111.287458 (doi:10.1074/jbc.M111.287458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boulon S, et al. 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924 10.1016/j.molcel.2010.08.023 (doi:10.1016/j.molcel.2010.08.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matera AG, Terns RM, Terns MP. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220 10.1038/nrm2124 (doi:10.1038/nrm2124) [DOI] [PubMed] [Google Scholar]
- 72.Watkins NJ, et al. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103, 457–466 10.1016/S0092-8674(00)00137-9 (doi:10.1016/S0092-8674(00)00137-9) [DOI] [PubMed] [Google Scholar]
- 73.Szewczak LB, DeGregorio SJ, Strobel SA, Steitz JA. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9, 1095–1107 10.1016/S1074-5521(02)00239-9 (doi:10.1016/S1074-5521(02)00239-9) [DOI] [PubMed] [Google Scholar]
- 74.Watkins NJ, Dickmanns A, Luhrmann R. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell Biol. 22, 8342–8352 10.1128/MCB.22.23.8342-8352.2002 (doi:10.1128/MCB.22.23.8342-8352.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16, 789–798 10.1016/j.molcel.2004.11.012 (doi:10.1016/j.molcel.2004.11.012) [DOI] [PubMed] [Google Scholar]
- 76.Kakihara Y, Houry WA. 2012. The R2TP complex: discovery and functions. Biochim. Biophys. Acta 1823, 101–107 10.1016/j.bbamcr.2011.08.016 (doi:10.1016/j.bbamcr.2011.08.016) [DOI] [PubMed] [Google Scholar]
- 77.Lovejoy CA, Cortez D. 2009. Common mechanisms of PIKK regulation. DNA Repair (Amst.) 8, 1004–1008 10.1016/j.dnarep.2009.04.006 (doi:10.1016/j.dnarep.2009.04.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Izumi N, Yamashita A, Hirano H, Ohno S. 2012. Heat shock protein 90 regulates phosphatidylinositol 3-kinase-related protein kinase family proteins together with the RUVBL1/2 and Tel2-containing co-factor complex. Cancer Sci. 103, 50–57 10.1111/j.1349-7006.2011.02112.x (doi:10.1111/j.1349-7006.2011.02112.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stirling PC, et al. 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7, e1002057. 10.1371/journal.pgen.1002057 (doi:10.1371/journal.pgen.1002057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olovnikov AM. 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41, 181–190 10.1016/0022-5193(73)90198-7 (doi:10.1016/0022-5193(73)90198-7) [DOI] [PubMed] [Google Scholar]
- 81.Li W, Zeng J, Li Q, Zhao L, Liu T, Bjorkholm M, Jia J, Xu D. 2010. Reptin is required for the transcription of telomerase reverse transcriptase and over-expressed in gastric cancer. Mol. Cancer 9, 132. 10.1186/1476-4598-9-132 (doi:10.1186/1476-4598-9-132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shevchenko A, et al. 2008. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 9, R167. 10.1186/gb-2008-9-11-r167 (doi:10.1186/gb-2008-9-11-r167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huber O, Menard L, Haurie V, Nicou A, Taras D, Rosenbaum J. 2008. Pontin and reptin, two related ATPases with multiple roles in cancer. Cancer Res. 68, 6873–6876 10.1158/0008-5472.CAN-08-0547 (doi:10.1158/0008-5472.CAN-08-0547) [DOI] [PubMed] [Google Scholar]
- 84.Kim JH, et al. 2005. Transcriptional regulation of a metastasis suppressor gene by Tip60 and β-catenin complexes. Nature 434, 921–926 10.1038/nature03452 (doi:10.1038/nature03452) [DOI] [PubMed] [Google Scholar]
- 85.Weiske J, Huber O. 2005. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-β-catenin-mediated transcription. J. Cell Sci. 118, 3117–3129 10.1242/jcs.02437 (doi:10.1242/jcs.02437) [DOI] [PubMed] [Google Scholar]
- 86.Petukhov M, et al. 2012. Large-scale conformational flexibility determines the properties of AAA+ TIP49 ATPases. Structure 20, 1321–1331 10.1016/j.str.2012.05.012 (doi:10.1016/j.str.2012.05.012) [DOI] [PubMed] [Google Scholar]
- 87.López-Perrote A, Muñoz-Hernández H, Gil D, Llorca O. 2012. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 40, 11 086–11 099 10.1093/nar/gks871 (doi:10.1093/nar/gks871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Machado-Pinilla R, Liger D, Leulliot N, Meier UT. 2012. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA 18, 1833–1845 10.1261/rna.034942.112 (doi:10.1261/rna.034942.112) [DOI] [PMC free article] [PubMed] [Google Scholar]