Abstract
We introduce an approach based on particle lithography to prepare spatially selective surface platforms of organosilanes that are suitable for nanoscale studies of protein binding. Particle lithography was applied for patterning fibrinogen, a plasma protein that has a major role in the clotting cascade for blood coagulation and wound healing. Surface nanopatterns of mercaptosilanes were designed as sites for the attachment of fibrinogen within a protein-resistant matrix of 2-[methoxy(polyethyleneoxy)propyl] trichlorosilane (PEG-silane). Preparing site-selective surfaces was problematic in our studies, because of the self-reactive properties of PEG-organosilanes. Certain organosilanes presenting hydroxyl head groups will cross react to form mixed surface multi-layers. We developed a clever strategy with particle lithography using masks of silica mesospheres to protect small, discrete regions of the surface from cross reactions. Images acquired with atomic force microscopy (AFM) disclose that fibrinogen attached primarily to the surface areas presenting thiol head groups, which were surrounded by PEG-silane. The activity for binding anti-fibrinogen was further evaluated using ex situ AFM studies, confirming that after immobilization the fibrinogen nanopatterns retained capacity for binding immunoglobulin G. Studies with AFM provide advantages of achieving nanoscale resolution for detecting surface changes during steps of biochemical surface reactions, without requiring chemical modification of proteins or fluorescent labels.
Keywords: atomic force microscopy, fibrinogen, particle lithography, organosilane, self-assembled monolayers, biosensors
1. Introduction
Surface patterning is essential for the integration of biomolecules into miniature bioelectronic and sensing devices because the sensing element consists of a layer of biomolecules for the capture of target molecules and analytes. Micro- and nanoscale sensing devices require the immobilization of active proteins onto flat substrates [1–3]. Patterning of proteins has been accomplished at the micrometre level using microcontact printing [4,5], photolithography [6–9], electron-beam lithography [10–12] and microfluidic channels [13,14]. Advancements for patterning proteins at the nanoscale on surfaces of self-assembled monolayers (SAMs) have been achieved using dip-pen nanolithography [15–19], bias-induced oxidation lithography [20,21], imprint lithography [22,23] and nanografting [24–27]. Innovative approaches for patterning proteins using particle lithography have also been reported, achieving high throughput across areas spanning millimetre-to-centimetre dimensions [28–34].
In this study, we investigate the immobilization of fibrinogen on designed surfaces of nanopatterned organosilanes. Fibrinogen has a central role in both blood coagulation and blood-based infections and has intensively been studied because of its fundamental role in blood clotting, thrombosis, angiogenesis, wound healing, platelet adhesion and biocompatibility [35–37]. Surface studies using fluorescently labelled fibrinogen were reported for micro-line patterns of 3-mercaptopropyltriethoxylsilane prepared by photopatterning [38]. Microcontact printing was used to directly stamp patterns of fibrinogen for studies of platelet adhesion and activation [39]. Micropatterned domains of fibrinogen within a micropatterned surface film of serum albumin were detected using tapping-mode atomic force microscopy (AFM) by Soman et al. [40]. Height and phase changes were used to map the regions containing fibrinogen after the patterns were infused with nanogold labels conjugated to antifibrinogen polycolonal antibody.
To further advance to molecular-level investigations of protein binding, technologies to produce even smaller patterns must be developed that enable control over protein density and orientation. Robust immobilization is required for AFM studies to prevent displacement of proteins by the motion of the scanning probe [41]. The chemistry selected for immobilizing biomolecules to surfaces should not use denaturing conditions that destroy the active protein structure. Miniaturization to the nanometre scale offers opportunities for studying surface-mediated biological reactions with AFM, particularly for investigating the parameters that influence mechanisms of protein activity, function and molecular recognition. The resolution of AFM is comparable to that of the electron microscope, however, with additional benefits for studying proteins in liquid or ambient environments with minimal sample preparation [42–46]. Studies of the conformational changes in fibrinogen attributed to effects of surface hydrophobicity were reported using AFM in a liquid environment [47,48]. High-resolution visualization of fibrinogen was achieved with AFM in liquid environments as well as measurements of the adhesion forces of fibrinogen towards different surfaces [49]. Yermolenko et al. [50] achieved molecular-level resolution with visualization of the αC region of fibrinogen.
In this investigation, a compositionally patterned Si(111) surface of mercaptosilanes was designed to provide selective sites for the attachment of fibrinogen within a protein-resistant matrix of 2-[methoxy(polyethyleneoxy)propyl] trichlorosilane (PEG-silane). Previously, we introduced several particle lithography-based protocols for patterning organosilanes using steps of vapour deposition, immersion or contact printing [51,52]. The surface selectivity of organosilane nanopatterns prepared with particle lithography approaches can be further applied to define the placement of other molecules and nanoparticles [53,54]. Particle lithography offers advantages for high-throughput patterning and enables control of the surface coverage, geometry and lateral dimensions of nanopatterns with relatively inexpensive reagents. However, when backfilling the patterns with a second organosilane, the self-reactive properties of hydroxyl terminated PEG-organosilanes were problematic. When using successive immersion steps in solutions of organosilanes, the hydroxyl groups of PEG-silanes were found to cross react and form mixed surface multi-layers.
In this study, details will be disclosed for an innovative strategy of masking areas of a mercaptosilane film to protect small, discrete regions of the surface from cross reaction with PEG-silane. Images acquired with AFM disclose surface changes throughout the steps of preparing organosilane patterns, binding fibrinogen and for studying the activity of the immobilized protein for binding anti-fibrinogen. The size of the fibrinogen nanopatterns achieved with particle lithography is of the order of 100–200 nm, enabling resolution of surface changes at the nanoscale. The simple steps of bench chemistry used for particle lithography protocols are accessible for most laboratories and are well suited for studies of proteins using small quantities (microlitres) of highly dilute protein solutions.
2. Material and methods
2.1. Materials and reagents
Pieces of single-side polished Si(111) doped with boron (Ted Pella, Inc., Redding, CA) were used as substrate. The silicon substrates were cleaned with sulfuric acid (ACS reagent 95%) and hydrogen peroxide (30%) purchased from Sigma-Aldrich. Silane reagents of PEG-silane and 3-mercaptopropyl-trimethoxysilane (MPTMS) were purchased from Gelest (Morrisville, PA) and used without further purification. Anhydrous toluene was obtained from Sigma-Aldrich. A suspension of 500 nm silica mesoparticles was obtained from Fisher Scientific. Monodisperse silica powder (250 and 100 nm) was acquired from Fiber Optic Center, Inc. (New Bedford, MA) and prepared in ethanol (ACS grade; Pharmco-Aaper, TX). Deionized water used in experiments and to prepare phosphate-buffered saline (PBS, pH 7.4) was obtained from a Direct-Q3 system (18 MΩ; Millipore, Bedford, MA). Bovine fibrinogen was used for surface studies (Sigma-Aldrich). For antigen–antibody studies, human fibrinogen and polyclonal rabbit anti-human fibrinogen were acquired and used as received from Calbiochem (La Jolla, CA). Sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC) was used as a cross-linking agent for attaching fibrinogen to MPTMS (Thermo Fisher Scientific, Inc., Rockford, IL).
2.2. Particle lithography procedure
An overview of the steps for particle lithography with organosilanes is shown in figure 1. Silicon substrates (5 × 5 mm2) were rinsed with deionized water, dried with argon and placed in a 3 : 1 (v/v) solution of sulfuric acid and hydrogen peroxide (piranha solution) for cleaning. The piranha solution should be handled with care, it is highly corrosive. After 90 min, the substrates were removed from the cleaning solution, rinsed with deionized water and dried under a stream of argon. The substrates were placed on a platform within a sealed reaction vessel containing 400 µl of neat MPTMS. The vessel was heated in an oven at 70°C to generate a vapour of MPTMS (figure 1a). Over time, the vapour reacted with the silicon substrate to produce a thin film of MPTMS throughout the surface areas of Si(111). After 4 h of exposure to vapour, the samples were removed from the vessel, washed with ethanol, further rinsed by sonication in ethanol for 30 min and then dried under a stream of argon. A suspension of silica mesoparticles was prepared by adding 0.1 g of dry, powdered mesospheres to 10 ml of ethanol followed by 30 min sonication. Solutions of silica mesoparticles were cleaned by washing four times using centrifugation with resuspension in deionized water. To prepare a surface mask, 10 µl of silica mesoparticles was deposited on the MPTMS/Si sample and dried at 4°C for 16 h. The samples were then treated with UV–ozone for 20 min to oxidize exposed areas between the particle masks (figure 1b). Molecules of MPTMS within the areas exposed to UV–ozone treatment were decomposed, forming silanol functional groups [55]. Immediately after UV–ozone treatment, the samples were immersed in a 1% (v/v) solution of PEG-silane in anhydrous toluene for 5 h. During the immersion step, the areas exposed to UV–ozone refilled with PEG-silane, whereas the areas underneath the silica spheres with MPTMS remained protected (figure 1c). Water was used to initially rinse the sample and quench the silanization reaction. Next, the samples were rinsed with ethanol with 30 min sonication to remove the silica mesospheres. A further rinsing step with sonication in water for 30 min ensured complete removal of the mesospheres and then the samples were dried under argon.
Figure 1.
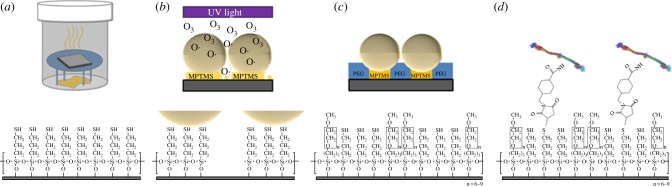
Steps for preparing protein nanostructures with particle lithography. (a) A surface film of MPTMS was prepared by vapour deposition on Si(111) substrates; (b) after coating the MPTMS surface with a mask of silica mesospheres, the samples were treated with UV–ozone. (c) Samples were immersed in a PEG-silane solution to refill the exposed surface sites with a protein resistive matrix. The mask of silica mesospheres was removed by solvent rinsing. (d) Proteins were coupled to MPTMS sites after immersing samples in sulfo-SMCC.
2.3. Attachment of fibrinogen to nanopatterned surfaces
The sulfhydryl groups located within the nanopatterned pores of MPTMS were activated through sulfo-SMCC coupling [56]. The samples were submerged in a 1 mM solution of sulfo-SMCC prepared in PBS (pH 7.4) for 1 h. Next, the samples were rinsed with PBS to remove excess sulfo-SMCC. The samples were then immersed in a 0.5 mg ml−1 solution of fibrinogen in PBS for 50 min. Lysine residues of the protein structure attach covalently to the reactive N-hydroxysuccinimide ester (NHS-ester) located on the exposed region of sulfo-SMCC (figure 1d). The samples were rinsed successively with PBS followed by detergent (0.1% solution Tween 20) to remove non-specifically adsorbed protein.
2.4. Antigen–antibody binding studies
PBS was used to rinse the surface of fibrinogen nanopatterns. Next, the samples were immersed in a 0.25 mg ml−1 solution of anti-fibrinogen in PBS for 30 min. After removal from the antibody solution, samples were rinsed successively with PBS, 0.1 per cent Tween 20 and deionized water. The samples were dried under a stream of argon and then characterized using tapping-mode AFM.
2.5. Atomic force microscopy
Scanning probe studies were done with either a model 5420 or 5500 instrument equipped with PicoView v. 1.8 software (Agilent Technologies, Tempe, AZ). Probes from Applied Nanostructures (ACTA, resonant frequency 300 kHz, k = 37 N m−1) were used for imaging in tapping mode. Images were processed and analysed using Gwyddion (v. 2.15), which is freely available on the Internet and supported by the Czech Metrology Institute [57]. The surface coverage of nanopatterned areas was evaluated using UTHSCA Image Tool software [58]. To estimate the surface coverage, the AFM frames were converted to black and white images by manually selecting a threshold value and counting pixels.
3. Results and discussion
3.1. Nanopatterns of 3-mercaptopropyl-trimethoxysilane within a resistive 2-[methoxy(polyethyleneoxy)propyl] trichlorosilane matrix
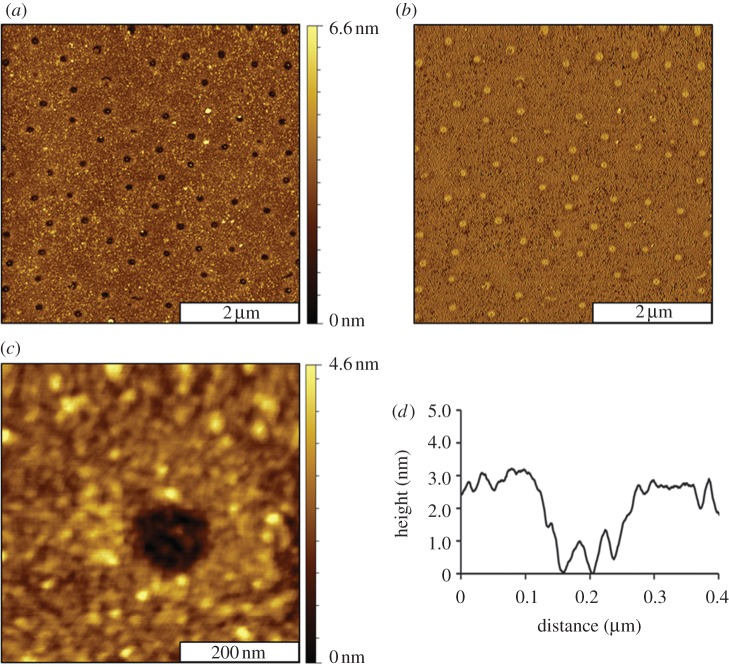
Images of the surface changes were captured with ambient AFM studies after key reaction steps of organosilane patterning, protein immobilization and antibody binding. Representative images of the MPTMS nanodots within a PEG-silane matrix are shown in figure 2. Images of the clean substrate, MPTMS film and silica mesosphere samples are presented in the electronic supplementary material, figure S1. The silica spheres were completely removed by rinsing steps, to disclose designed nanopatterns of organosilanes. The nanopores are shallower than the surrounding areas of PEG-silane and appear as dark spots (figure 2a). There are 70 nanopores visible within the 5 × 5 µm2 topograph, which scales to a surface density of approximately 108 nanopatterns cm−2, corresponding to 5.1 per cent surface coverage of MPTMS. The MPTMS nanopatterns are produced at the regions of contact that were directly underneath the silica mesospheres, which were protected from UV–ozone treatment. The nanopores are spaced regularly at 500 nm intervals, as determined by the periodicity of the mesosphere masks. The diameters of the nanopatterns measure 160 ± 20 nm and reflect the geometry of the areas of close physical contact between the spheres and the Si(111) substrates. The corresponding phase image (figure 2b) further reveals the differences in surface chemistry. The colour contrast is reversed for the phase frame, with brighter spots shown for the nanopores terminated with sulfhydryl groups; whereas the surrounding matrix areas of PEG-silane are darker. The phase images result from mapping slight incremental changes in the oscillation of the AFM tip caused by damping of the motion when the tip interacts with the surface. Thus, phase images are sensitive maps of changes in film thickness and tip–surface adhesion. A magnified view of a single nanopore is shown in the AFM topography frame of figure 2c. Within the 500 × 500 nm2 frame, the clustered morphology of the surrounding PEG-silane matrix is apparent. The uneven morphology located at the bottom of the nanopore is attributable to the roughness for the underlying MPTMS film. The height of the nanopores measures 3.0 ± 0.3 nm above the matrix areas of PEG-silane (figure 2d). Assuming that the MPTMS film is a monolayer with a thickness of 0.7 nm [59], then the multi-layer thickness of the PEG-silane film is approximately 3.7 nm. This suggests that the surrounding film of PEG-silane probably forms a cross-linked bilayer.
Figure 2.
Views of MPTMS nanopores obtained with ambient tapping-mode AFM. (a) Topograph of the nanopores and (b) corresponding phase image. (c) Close-up topography view of a single nanopore and (d) height profile for the white line in (c).
The hydroxyl functional groups of PEG-silane are reported to be suitable for designing protein-resistant films [60,61]. There are few surfaces that resist protein adsorption, and considerable research has addressed studies of the mechanisms of protein resistance or adhesion to surfaces [62–65]. Systematic studies have been carried out to evaluate the molecular characteristics of functionalized SAMs for resisting protein adsorption [66,67].
Immersion of PEG-silane films into solutions of organosilanes generated mixed layers and multi-layer surface structures, which were not spatially selective for patterning proteins (data not shown). The rationale for using UV–ozone treatment to remove the selected areas of an organosilane surface layer follows from our studies of the reactivity of PEG-silane with other organosilanes. When using multi-step protocols to generate organosilane nanopatterns, experiments with particle lithography showed that hydroxyl head groups of PEG-silane surfaces react with other silanes during immersion steps. To prepare MPTMS nanopores, UV–ozone treatment through surface masks was found to provide selectivity in combination with steps of particle lithography and immersion. A strategy for selectively backfilling irradiated sites provides nanoscale control for producing discrete small regions of MPTMS that can then be used with further biochemical steps to isolate and define surface sites for binding protein.
Changing the diameter of the mesoparticles used as masks for surface fabrication provides a way to control the surface coverage and density of the nanopatterns [68,69]. When using smaller diameters of silica mesoparticles, a greater surface density of nanopatterns can be generated. Further examples of AFM images of MPTMS nanopatterns prepared with different diameters of silica mesospheres are provided in the electronic supplementary material, figure S2.
3.2. Spatially selective attachment of fibrinogen to surface sites with MPTMS
Morphology changes are readily apparent for the sample surface after fibrinogen patterning (figure 3). The heights of the nanopatterns have increased, as shown for a representative 5 × 5 µm2 topography frame (figure 3a). For the most part, fibrinogen seems to bind primarily at the sites with MPTMS, and negligible adsorbates are located in areas between the taller protein clusters. The arrangement of nanopatterns indicates the sites of small clusters of protein, spaced at intervals of 500 nm. Further examples of fibrinogen nanopatterns with even smaller periodicities are shown in the electronic supplementary material, figure S3. A zoom-in view of a single protein nanodot is presented in figure 3b. The height measures 9 nm above the PEG-silane matrix at the tallest area in the centre of the nanopattern (figure 3c). As the MPTMS nanopatterns were 3.0 nm shorter than the surrounding matrix film, after protein attachment, the height increase measures an overall thickness of approximately 12 nm for the nanopattern.
Figure 3.
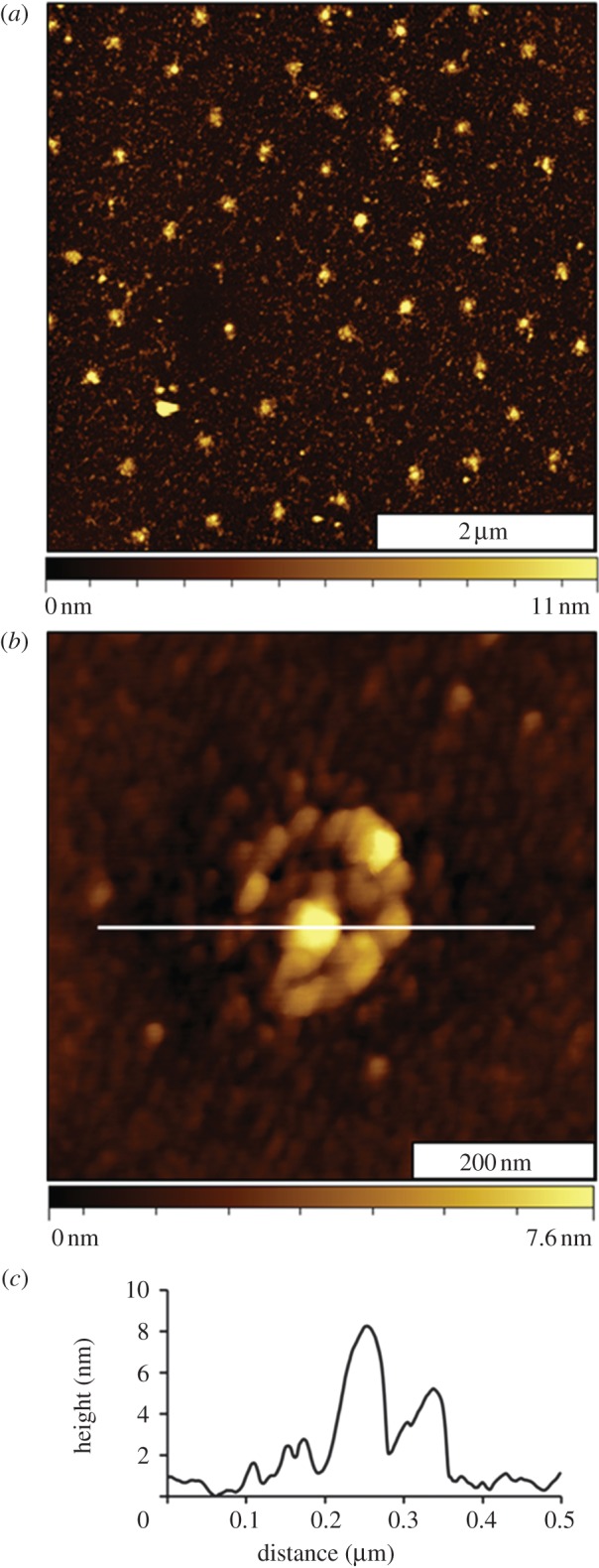
Surface changes after binding fibrinogen. (a) Fibrinogen attaches at the sites with MPTMS shown with an AFM topograph; (b) magnified view of a single nanostructure with fibrinogen; (c) corresponding height profile for the line in (b).
A sulfo-SMCC linker (0.83 nm) was used to couple fibrinogen to the surface of MPTMS nanopatterns; fibrinogen attached selectively at the regions patterned with sulfhydryl moieties (figure 3). When the chemically patterned samples were immersed in a sulfo-SMCC solution for 1 h, the maleimide groups of sulfo-SMCC reacted with thiol head groups at the interface. This produces an activated surface pattern with an NHS-ester that was available for binding protein. When the activated sample was immersed in a solution of fibrinogen, exposed lysine groups of the protein reacted with the NHS-ester to link the protein to the surface. The PEG-silane matrix furnished a protein-resistant background to surround and isolate discrete regions of bound protein.
After binding fibrinogen, the change in height of the protein nanopatterns ranged from 9 to 13 nm (average is 11.0 ± 1.1 nm) when including the depth of 3.0 nm from the MPTMS pores. There are multiple exposed lysine residues on each fibrinogen molecule for attaching to the surface. Thus, the protein may attach to the surface with different orientations. Studies with high-resolution AFM images of fibrinogen have previously been reviewed [43]. Trinodular molecules of fibrinogen were reported to measure lengths ranging from 48 to 60 nm, and the width measured from 8 to 28 nm. The values measured for the height of fibrinogen ranged from 1.2 to 4.1 nm. For the fibrinogen bound to MPTMS nanopatterns, the thickness of 11.0 ± 1.1 nm most likely corresponds to 3–5 layers of protein assuming a horizontal orientation. The multi-layer assembly for fibrinogen has previously been documented [70,71]. As fibrinogen adsorbs or covalently attaches to a surface, it spreads onto the surface and unfolds. It has been suggested that this unfolding exposes binding sites that initiate self-aggregation [72].
The surface density and coverage of protein nanopatterns can be controlled at the nanoscale by selecting the diameter of mesospheres used for particle lithography masks. Mesospheres with a smaller diameter generate a greater number density of smaller-sized nanopatterns; however, the overall surface coverage of protein increases. Analysis of AFM images from experiments with different sizes of mesospheres is summarized in table 1. Our studies show that the width of the patterns increased after protein attachment such that the proteins extend beyond the edges of the MPTMS nanodots. In the longest direction, fibrinogen was reported to measure 47 nm [50]; this corresponds to an orientation where the long direction is parallel to the sample surface. Measurements of the diameters of the nanopores before protein attachment suggest that two or three fibrinogen molecules can fit on a single nanopattern. Particle lithography using smaller spheres likewise generated nanopores with smaller diameters (table 1). Eventually, if the diameter of the nanopatterns can be matched to the size of a single protein, selective patterning of individual proteins on the surface will be achievable.
Table 1.
Surface coverage of MPTMS and fibrinogen as a function of mesosphere mask size.
| silica diameter (nm) | surface coverage MPTMS (%) | surface coverage fibrinogen (%) | pore diameter (nm) |
|---|---|---|---|
| 500 | 5.0 | 7.8 | 160 ± 20 |
| 250 | 7.7 | 9.8 | 134 ± 19 |
| 100 | 16 | 17 | 104 ± 18 |
3.3. Antigen–antibody binding studies
The final biochemical step for the studies with surface nanopatterning was to evaluate the activity of the immobilized fibrinogen for binding immunoglobulin G (IgG). Images were obtained after anti-fibrinogen was added to the sample ex situ (figure 4). The heights of the patterns have increased, as shown with topography views in figure 4. The periodic arrangement of nanopatterns is preserved, as shown in figure 4a; however, the nanopatterns are taller after binding anti-fibrinogen. Most of the antibody binding is localized at the central areas of the nanopatterns. A close-up view of a single nanostructure is shown in figure 4b. The surrounding areas of the matrix have also changed in appearance, indicating that there are adsorbates surrounding the nanopatterns. The nature of the adsorbates, which may be salts, contaminants or non-specifically bound protein or antibody, cannot be determined with AFM. The heights of the nanostructures after IgG attachment measured 17 ± 4 nm (including the 3 nm value for original depth of the nanopore). This value indicates an overall increase of 6 nm, which matches the approximate dimensions of a single layer of IgG with a side-on orientation.
Figure 4.
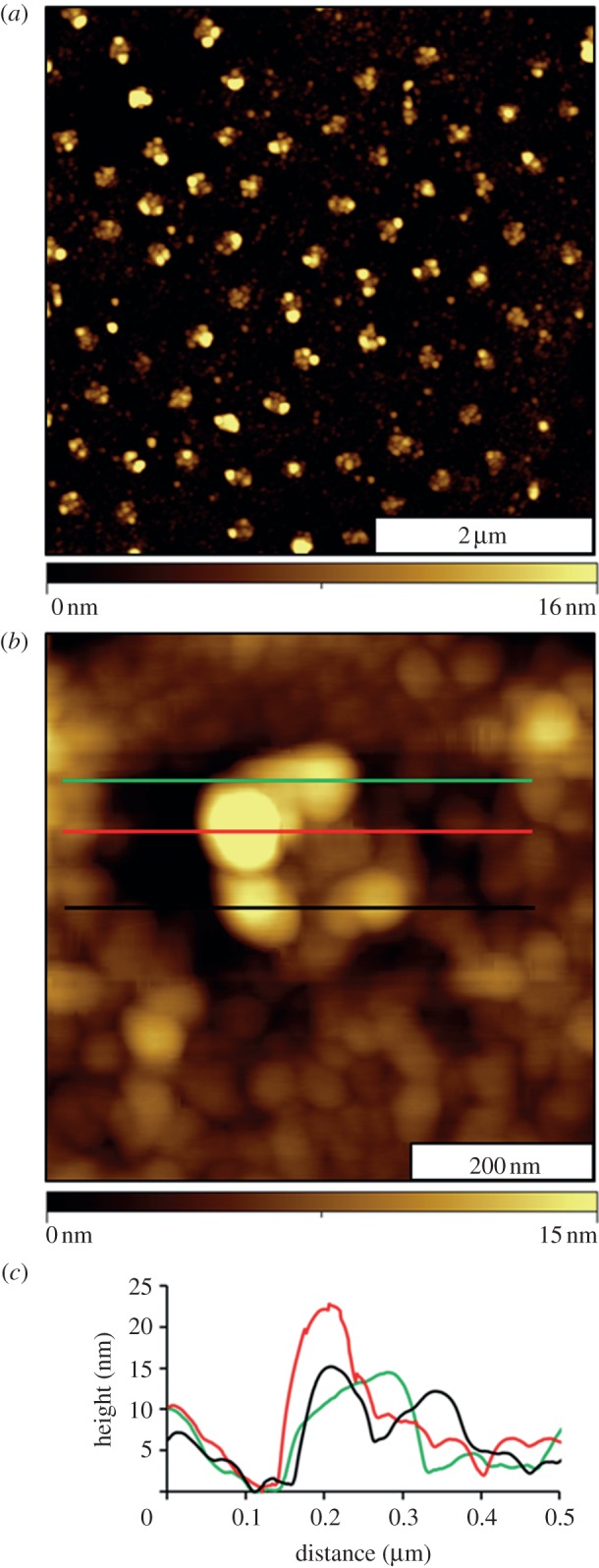
Changes of the nanostructures after binding antibody. (a) Topography image (5 × 5 µm2) acquired in air; (b) single nanostructure of fibrinogen–immunoglobulin G; (c) height profiles after antibody binding corresponding to the coloured lines in (b).
To view the overall changes taking place on the designed surface at different points of the nanofabrication and protein-binding processes, figure 5 presents a side-by-side comparison of AFM images during key steps of the reactions. The ex situ experiments disclose only very local areas of the surface; however, the frames are representative of multiple images acquired for different areas throughout the entire sample. The changes in height and lateral dimensions for snapshots at each step are shown in the top row of figure 5. A quantitative comparison of the changes in dimension is shown with representative cursor profiles drawn across the topography frames in the middle row. From left to right, the nanopores change from being holes, to forming isolated islands of fibrinogen, to taller protein–antibody clusters. At each step, the structures become both taller and slightly wider with the addition of fibrinogen and anti-fibrinogen molecules. Smaller regions measuring 1.5 × 1.5 µm2 are compared with three-dimensional views in the bottom row of figure 5. The progressive growth in height and width of the nanostructures is apparent, and surface selectivity for protein binding is mainly localized to the areas of MPTMS.
Figure 5.
Step-by-step changes of the surface during the process of nanofabrication and protein binding. Nanopores of PEG-silane: (a) topography, (b) cursor profile and (c) three-dimensional view. After coupling of fibrinogen to nanopores: (d) topography, (e) cursor profile and (f) three-dimensional representation. After binding anti-fibrinogen to nanopatterns: (g) topography; (h) corresponding cursor profile and (i) three-dimensional image.
A significant advantage for developing particle lithography-based approaches for patterning organosilanes is that high throughput at the scale of millions to billions of nanopatterns can be achieved with only basic steps of the chemical self-assembly. As organosilanes can be used with glass substrates, the procedures can be scaled to larger micron-scale dimensions for accomplishing protein-binding assays based on fluorescence detection. Future directions planned for AFM-based investigations with this surface protein sensor platform will be to study the interactions of small molecules or DNA with immobilized proteins and to develop more complex biochemical protocols for studies of protein bioactivity.
4. Conclusions
The reactivity of nanopatterns of organosilanes can be designed to spatially direct the immobilization of fibrinogen and IgG. Surface changes were monitored after steps of protein immobilization and antibody binding using ex situ AFM studies. Changes in the thickness of protein layers after immobilization were evaluated directly using height measurements with AFM, without the need for fluorescent or chemical labelling. Future work will investigate the size-dependent changes in the nanopattern size for the effectiveness of protein immobilization, protein activity for binding antibodies in situ, as well as the changes in the surface morphology within liquid media at different pH/ion concentrations.
Acknowledgements
This research was sponsored by funding from the National Science Foundation (Career and PECASE award, CHE-0847291) and by the Dreyfus Foundation, Camille Dreyfus Teacher-Scholar Award.
References
- 1.Blawas AS, Reichert WM. 1998. Protein patterning. Biomaterials 19, 595–609. 10.1016/S0142-9612(97)00218-4 ( 10.1016/S0142-9612(97)00218-4) [DOI] [PubMed] [Google Scholar]
- 2.Yang G, Garno JC, Liu G-Y. 2010. Scanning probe-based lithography for production of biological and organic nanostructures on surfaces. In Comprehensive nanoscience and technology (eds Andrews D, Scholes G, Wiederrecht G.). Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 3.Ngunjiri JN, Li J-R, Garno JC. 2006. Nanolithography: towards fabrication of nanodevices for life sciences. In Nanodevices for the life sciences (ed. Kumar CSSR.). Weinheim, Germany: Wiley-VCH. [Google Scholar]
- 4.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. 1999. Patterning proteins and cells using soft lithography. Biomaterials 20, 2363–2376. 10.1016/S0142-9612(99)00165-9 ( 10.1016/S0142-9612(99)00165-9) [DOI] [PubMed] [Google Scholar]
- 5.Renault JP, Bernard A, Bietsch A, Michel B, Bosshard HR, Delamarche E, Kreiter M, Hecht B, Wild UP. 2003. Fabricating arrays of single protein molecules on glass using microcontact printing. J. Phys. Chem. B 107, 703–711. 10.1021/jp0263424 ( 10.1021/jp0263424) [DOI] [Google Scholar]
- 6.Adams J, Tizazu G, Janusz S, Brueck SRJ, Lopez GP, Leggett GJ. 2010. Large-area nanopatterning of self-assembled monolayers of alkanethiolates by interferometric lithography. Langmuir 26, 13 600–13 606. 10.1021/la101876j ( 10.1021/la101876j) [DOI] [PubMed] [Google Scholar]
- 7.Dontha N, Nowall WB, Kuhr WG. 1997. Generation of biotin/avidin/enzyme nanostructures with maskless photolithography. Anal. Chem. 69, 2619–2625. 10.1021/ac9702094 ( 10.1021/ac9702094) [DOI] [PubMed] [Google Scholar]
- 8.Blawas AS, Oliver TF, Pirrung MC, Reichert WM. 1998. Step-and-repeat photopatterning of protein features using caged-biotin-BSA: characterization and resolution. Langmuir 14, 4243. 10.1021/la971231v ( 10.1021/la971231v) [DOI] [Google Scholar]
- 9.Chen XY, Su YD, Ajeti V, Chen SJ, Campagnola PJ. 2012. Cell adhesion on micro-structured fibronectin gradients fabricated by multiphoton excited photochemistry. Cell. Mol. Bioeng. 5, 307–319. 10.1007/s12195-012-0237-8 ( 10.1007/s12195-012-0237-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballav N, Thomas H, Winkler T, Terfort A, Zharnikov M. 2009. Making protein patterns by writing in a protein-repelling matrix. Angew. Chem. Int. Ed. 48, 5833–5836. 10.1002/anie.200900950 ( 10.1002/anie.200900950) [DOI] [PubMed] [Google Scholar]
- 11.Jeyachandran YL, Zharnikov M. 2012. Comprehensive analysis of the effect of electron irradiation on oligo(ethylene glycol) terminated self-assembled monolayers applicable for specific and nonspecific patterning of proteins. J. Phys. Chem. C 116, 14 950–14 959. 10.1021/jp303764h ( 10.1021/jp303764h) [DOI] [Google Scholar]
- 12.Kolodziej CM, Kim SH, Broyer RM, Saxer SS, Decker CG, Maynard HD. 2012. Combination of integrin-binding peptide and growth factor promotes cell adhesion on electron-beam-fabricated patterns. J. Am. Chem. Soc. 134, 247–255. 10.1021/ja205524x ( 10.1021/ja205524x) [DOI] [PubMed] [Google Scholar]
- 13.Delamarche E, Bernard A, Schmid H, Bietsch A, Michel B, Biebuyck H. 1998. Microfluidic networks for chemical patterning of substrate: design and application to bioassays. J. Am. Chem. Soc. 120, 500–508. 10.1021/ja973071f ( 10.1021/ja973071f) [DOI] [Google Scholar]
- 14.Patel N, Sanders GHW, Shakesheff KM, Cannizzaro SM, Davies MC, Langer R, Roberts CJ, Tendler SJB, Williams PM. 1999. Atomic force microscopic analysis of highly defined protein patterns formed by microfluidic networks. Langmuir 15, 7252–7257. 10.1021/la981268v ( 10.1021/la981268v) [DOI] [Google Scholar]
- 15.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. 2002. Protein nanoarrays generated by dip-pen nanolithography. Science 295, 1702–1705. 10.1126/science.1067172 ( 10.1126/science.1067172) [DOI] [PubMed] [Google Scholar]
- 16.Lee SW, Oh B-K, Sanedrin RG, Salaita K, Fujigaya T, Mirkin CA. 2006. Biologically active protein nanoarrays generated using parallel dip-pen nanolithography. Adv. Mater. 18, 1133–1136. 10.1002/adma.200600070 ( 10.1002/adma.200600070) [DOI] [Google Scholar]
- 17.Hyun J, Ahn SJ, Lee WK, Chilkoti A, Zauscher S. 2002. Molecular recognition-mediated fabrication of protein nanostructures by dip-pen lithography. Nano. Lett. 2, 1203–1207. 10.1021/nl0257364 ( 10.1021/nl0257364) [DOI] [Google Scholar]
- 18.Choi D-S, Yun S-H, An Y-C, Lee M-J, Kang D-G, Chang S-I, Kim H-K, Kim K-M, Lim J-H. 2007. Nanopatterning proteins with a stamp tip for dip-pen nanolithography. Biochip. J. 1, 200–203. [Google Scholar]
- 19.Lee KB, Lim J-H, Mirkin CA. 2003. Protein nanostructures formed via direct-write dip-pen nanolithography. J. Am. Chem. Soc. 125, 5588–5589. 10.1021/ja034236p ( 10.1021/ja034236p) [DOI] [PubMed] [Google Scholar]
- 20.Jianhua Gu CMY, Sha Li, Chengzhi Cai. 2004. Nanometric protein arrays on protein-resistant monolayers on silicon surfaces. J. Am. Chem. Soc. 126, 8098–8099. 10.1021/ja048405x ( 10.1021/ja048405x) [DOI] [PubMed] [Google Scholar]
- 21.Qin G, Gu J, Liu K, Xiao Z, Yam CM, Cai C. 2011. Conductive AFM patterning on oligo(ethylene glycol)-terminated alkyl monolayers on silicon substrates: proposed mechanism and fabrication of avidin patterns. Langmuir 27, 6987–6994. 10.1021/la1047358 ( 10.1021/la1047358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoff JD, Cheng LJ, Meyhofer E, Guo LJ, Hunt AJ. 2004. Nanoscale protein patterning by imprint lithography. Nano Lett. 4, 853–857. 10.1021/nl049758x ( 10.1021/nl049758x) [DOI] [Google Scholar]
- 23.Subramani C, Cengiz N, Saha K, Gevrek TN, Yu X, Jeong Y, Bajaj A, Sanyal A, Rotello VM. 2011. Direct fabrication of functional and biofunctional nanostructures through reactive imprinting. Adv. Mater. 23, 3165–3169. 10.1002/adma.201101292 ( 10.1002/adma.201101292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan YH, Liu M, Nolting B, Go JG, Gervay-Hague J, Liu GY. 2008. A nanoengineering approach for investigation and regulation of protein immobilization. ACS Nano 2, 2374–2384. 10.1021/nn800508f ( 10.1021/nn800508f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadu-Mesthrige K, Amro NA, Garno JC, Xu S, Liu G-Y. 2001. Fabrication of nanometer-sized protein patterns using atomic force microscopy and selective immobilization. Biophys. J. 80, 1891–1899. 10.1016/S0006-3495(01)76158-9 ( 10.1016/S0006-3495(01)76158-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M, Amro NA, Liu G-Y. 2008. Nanografting for surface physical chemistry. Annu. Rev. Phys. Chem. 59, 367–386. 10.1146/annurev.physchem.58.032806.104542 ( 10.1146/annurev.physchem.58.032806.104542) [DOI] [PubMed] [Google Scholar]
- 27.Ngunjiri JN, Stark DJ, Tian T, Briggman KA, Garno JC. 2012. Immobilization of proteins on carboxylic acid-functionalized nanopatterns. Anal. Bioanal. Chem. 405, 1985–1993. 10.1007/s00216-012-6621-3 ( 10.1007/s00216-012-6621-3) [DOI] [PubMed] [Google Scholar]
- 28.Garno JC, Amro NA, Wadu-Mesthrige K, Liu G-Y. 2002. Production of periodic arrays of protein nanostructures using particle lithography. Langmuir 18, 8186–8192. 10.1021/la020518b ( 10.1021/la020518b) [DOI] [Google Scholar]
- 29.Li J-R, Henry GC, Garno JC. 2006. Fabrication of nanopatterned films of bovine serum albumin and staphylococcal protein A using latex particle lithography. Analyst 131, 244–250. 10.1039/b511010f ( 10.1039/b511010f) [DOI] [PubMed] [Google Scholar]
- 30.Ngunjiri JN, Daniels SL, Li J-R, Serem WK, Garno JC. 2008. Controlling the surface coverage and arrangement of proteins using particle lithography. Nanomedicine 3, 529–541. 10.2217/17435889.3.4.529 ( 10.2217/17435889.3.4.529) [DOI] [PubMed] [Google Scholar]
- 31.Taylor ZR, Keay JC, Sanchez ES, Johnson MB, Schmidtke DW. 2012. Independently controlling protein dot size and spacing in particle lithography. Langmuir 28, 9656–9663. 10.1021/la300806m ( 10.1021/la300806m) [DOI] [PubMed] [Google Scholar]
- 32.Cai Y, Ocko BM. 2005. Large-scale fabrication of protein nanoarrays based on nanosphere lithography. Langmuir 21, 9274–9279. 10.1021/la051656e ( 10.1021/la051656e) [DOI] [PubMed] [Google Scholar]
- 33.Taylor ZR, Patel K, Spain TG, Keay JC, Jernigen JD, Sanchez ES, Grady BP, Johnson MB, Schmidtke DW. 2009. Fabrication of protein dot arrays via particle lithography. Langmuir 25, 10 932–10 938. 10.1021/la901512z ( 10.1021/la901512z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satriano C, Fragala ME, Aleeva Y. 2012. Ultrathin and nanostructured ZnO-based films for fluorescence biosensing applications. J. Colloid Interface Sci. 365, 90–96. 10.1016/j.jcis.2011.09.014 ( 10.1016/j.jcis.2011.09.014) [DOI] [PubMed] [Google Scholar]
- 35.Lord ST. 2007. Fibrinogen and fibrin: scaffold proteins in hemostasis. Curr. Opin. Hematol. 14, 236–241. 10.1097/MOH.0b013e3280dce58c ( 10.1097/MOH.0b013e3280dce58c) [DOI] [PubMed] [Google Scholar]
- 36.Roach P, Eglin D, Rohde K, Perry CC. 2007. Modern biomaterials: a review—bulk properties and implications of surface modifications. J. Mater. Sci. Mater. Med. 18, 1263–1277. 10.1007/s10856-006-0064-3 ( 10.1007/s10856-006-0064-3) [DOI] [PubMed] [Google Scholar]
- 37.Horbett TA. 1993. Principles underlying the role of adsorbed plasma-proteins in blood interactions with foreign materials. Cardiovasc. Pathol. 2, 137–148. 10.1016/1054-8807(93)90054-6 ( 10.1016/1054-8807(93)90054-6)25990608 [DOI] [Google Scholar]
- 38.Liu J, Hlady V. 1996. Chemical pattern on silica surface prepared by UV irradiation of 3-mercaptopropyltriethoxy silane layer: surface characterization and fibrinogen adsorption. Colloids Surf. B Biointerfaces 8, 25–37. 10.1016/S0927-7765(96)01298-2 ( 10.1016/S0927-7765(96)01298-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corum LE, Hlady V. 2012. The effect of upstream platelet–fibrinogen interactions on downstream adhesion and activation. Biomaterials 33, 1255–1260. 10.1016/j.biomaterials.2011.10.074 ( 10.1016/j.biomaterials.2011.10.074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soman P, Rice Z, Siedlecki CA. 2008. Immunological identification of fibrinogen in dual-component protein films by AFM imaging. Micron 39, 832–842. 10.1016/j.micron.2007.12.013 ( 10.1016/j.micron.2007.12.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okusa H, Kurihara K, Kunitake T. 1994. Chemical modification of molecularly smooth mica surface and protein attachment. Langmuir 10, 3577–3581. 10.1021/la00022a034 ( 10.1021/la00022a034) [DOI] [Google Scholar]
- 42.Kasemo B. 2002. Biological surface science. Surf. Sci. 500, 656–677. 10.1016/S0039-6028(01)01809-X ( 10.1016/S0039-6028(01)01809-X) [DOI] [Google Scholar]
- 43.Averett LE, Schoenfisch MH. 2010. Atomic force microscope studies of fibrinogen adsorption. Analyst 135, 1201–1209. 10.1039/b924814e ( 10.1039/b924814e) [DOI] [PubMed] [Google Scholar]
- 44.Lal R, John SA. 1994. Biological applications of atomic force microscopy. Am. J. Physiol. 266, C1–C21. [DOI] [PubMed] [Google Scholar]
- 45.Fotiadis D, Scheuring S, Muller SA, Engel A, Muller DJ. 2002. Imaging and manipulation of biological structures with the AFM. Micron 33, 385–397. 10.1016/S0968-4328(01)00026-9 ( 10.1016/S0968-4328(01)00026-9) [DOI] [PubMed] [Google Scholar]
- 46.Santos NC, Castanho M. 2004. An overview of the biophysical applications of atomic force microscopy. Biophys. Chem. 107, 133–149. 10.1016/j.bpc.2003.09.001 ( 10.1016/j.bpc.2003.09.001) [DOI] [PubMed] [Google Scholar]
- 47.Tunc S, Maitz MF, Steiner G, Vazquez L, Pham MT, Salzer R. 2005. In situ conformational analysis of fibrinogen adsorbed on Si surfaces. Colloids Surf. B Biointerfaces 42, 219–225. 10.1016/j.colsurfb.2005.03.004 ( 10.1016/j.colsurfb.2005.03.004) [DOI] [PubMed] [Google Scholar]
- 48.Agnihotri A, Siedlecki CA. 2004. Time-dependent conformational changes in fibrinogen measured by atomic force microscopy. Langmuir 20, 8846–8852. 10.1021/la049239+ ( 10.1021/la049239+) [DOI] [PubMed] [Google Scholar]
- 49.Xu LC, Siedlecki CA. 2009. Atomic force microscopy studies of the initial interactions between fibrinogen and surfaces. Langmuir 25, 3675–3681. 10.1021/la803258h ( 10.1021/la803258h) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yermolenko IS, Lishko VK, Ugarova TP, Magonov SN. 2011. High-resolution visualization of fibrinogen molecules and fibrin fibers with atomic force microscopy. Biomacromolecules 12, 370–379. 10.1021/bm101122g ( 10.1021/bm101122g) [DOI] [PubMed] [Google Scholar]
- 51.Saner CK, Lusker KL, LeJeune ZM, Serem WK, Garno JC. 2012. Self-assembly of octadecyltrichlorosilane: surface structures formed using different protocols of particle lithography. Beilstein J. Nanotechnol. 3, 114–122. 10.3762/bjnano.3.12 ( 10.3762/bjnano.3.12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J-R, Garno JC. 2008. Elucidating the role of surface hydrolysis in preparing organosilane nanostructures via particle lithography. Nano Lett. 8, 1916–1922. 10.1021/nl0806062 ( 10.1021/nl0806062) [DOI] [PubMed] [Google Scholar]
- 53.Lusker KL, Li J-R, Garno JC. 2011. Nanostructures of functionalized gold nanoparticles prepared by particle lithography with organosilanes. Langmuir 27, 13 269–13 275. 10.1021/la202816k ( 10.1021/la202816k) [DOI] [PubMed] [Google Scholar]
- 54.Lewandowski BR, Kelley AT, Singleton R, Li JR, Lowry M, Warner IM, Garno JC. 2009. Nanostructures of cysteine-coated CdS nanoparticles produced with ‘two-particle' lithography. J. Phys. Chem. C 113, 5933–5940. 10.1021/jp808056x ( 10.1021/jp808056x) [DOI] [Google Scholar]
- 55.Uosaki K, Quayum ME, Nihonyanagi S, Kondo T. 2004. Decomposition processes of an organic monolayer formed on Si(111) via a silicon-carbon bond induced by exposure to UV irradiation or ozone. Langmuir 20, 1207–1212. 10.1021/la030211s ( 10.1021/la030211s) [DOI] [PubMed] [Google Scholar]
- 56.Zhang XC, Kumar S, Chen JH, Teplyakov AV. 2009. Covalent attachment of shape-restricted DNA molecules on amine-functionalized Si(111) surface. Surf. Sci. 603, 2445–2457. 10.1016/j.susc.2009.05.023 ( 10.1016/j.susc.2009.05.023) [DOI] [Google Scholar]
- 57.Necas DK. 2007. Gwyddion 2.25. Czech Metrology Institute. See http://gwyddion.net/.
- 58.Wilxox C. 1996. ImageTool. San Antonio, TX: University of Texas Health Science Center (UTHSCSA). [Google Scholar]
- 59.Hu MH, Noda S, Okubo T, Yamaguchi Y, Komiyama H. 2001. Structure and morphology of self-assembled 3-mercaptopropyltrimethoxysilane layers on silicon oxide. Appl. Surf. Sci. 181, 307–316. 10.1016/S0169-4332(01)00399-3 ( 10.1016/S0169-4332(01)00399-3) [DOI] [Google Scholar]
- 60.Jo S, Park K. 2000. Surface modification using silanated poly(ethylene glycol)s. Biomaterials 21, 605–616. 10.1016/S0142-9612(99)00224-0 ( 10.1016/S0142-9612(99)00224-0) [DOI] [PubMed] [Google Scholar]
- 61.Prime KL, Whitesides GM. 1993. Adsorption of proteins onto surfaces containing end-attached oligo(ethylene oxide)—a model system using self-assembled monolayers. J. Am. Chem. Soc. 115, 10 714–10 721. 10.1021/ja00076a032 ( 10.1021/ja00076a032) [DOI] [Google Scholar]
- 62.Chapman RG, Ostuni E, Takayama S, Holmlin RE, Yan L, Whitesides GM. 2000. Surveying for surfaces that resist the adsorption of proteins. J. Am. Chem. Soc. 122, 8303–8304. 10.1021/ja000774f ( 10.1021/ja000774f) [DOI] [Google Scholar]
- 63.Ostuni E, Chapman RG, Liang MN, Meluleni G, Pier G, Ingber DE, Whitesides GM. 2001. Self-assembled monolayers that resist the adsorption of proteins and the adhesion of bacterial and mammalian cells. Langmuir 17, 6336–6343. 10.1021/la010552a ( 10.1021/la010552a) [DOI] [Google Scholar]
- 64.Holmlin RE, Chen X, Chapman RG, Takayama S, Whitesides GM. 2001. Zwitterionic SAMs that resist nonspecific adsorption of protein from aqueous buffer. Langmuir 17, 2841–2850. 10.1021/la0015258 ( 10.1021/la0015258) [DOI] [PubMed] [Google Scholar]
- 65.Luk Y-Y, Kato M, Mrksich M. 2000. Self-assembled monolayers of alkanethiolates presenting mannitol groups are inert to protein adsorption and cell attachment. Langmuir 16, 9604–9608. 10.1021/la0004653 ( 10.1021/la0004653) [DOI] [Google Scholar]
- 66.Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. 2001. A survey of structure–property relationships of surfaces that resist the adsorption of protein. Langmuir 17, 5605–5620. 10.1021/la010384m ( 10.1021/la010384m) [DOI] [PubMed] [Google Scholar]
- 67.Herrwerth S, Eck W, Reinhardt S, Grunze M. 2003. Factors that determine the protein resistance of oligoether self-assembled monolayers—internal hydrophilicity, terminal hydrophilicity, and lateral packing density. J. Am. Chem. Soc. 125, 9359–9366. 10.1021/ja034820y ( 10.1021/ja034820y) [DOI] [PubMed] [Google Scholar]
- 68.Li JR, Lusker KL, Yu JJ, Garno JC. 2009. Engineering the spatial selectivity of surfaces at the nanoscale using particle lithography combined with vapor deposition of organosilanes. ACS Nano 3, 2023–2035. 10.1021/nn9004796 ( 10.1021/nn9004796) [DOI] [PubMed] [Google Scholar]
- 69.Lusker KL, Yu JJ, Garno JC. 2011. Particle lithography with vapor deposition of organosilanes: a molecular toolkit for studying confined surface reactions in nanoscale liquid volumes. Thin Solid Films 519, 5223–5229. 10.1016/j.tsf.2011.01.164 ( 10.1016/j.tsf.2011.01.164) [DOI] [Google Scholar]
- 70.Yermolenko IS, Gorkun OV, Fuhrmann A, Podolnikova NP, Lishko VK, Oshkadyerov SP, Lord ST, Ros R, Ugarova TP. 2012. The assembly of nonadhesive fibrinogen matrices depends on the alpha C regions of the fibrinogen molecule. J. Biol. Chem. 287, 41 979–41 990. 10.1074/jbc.M112.410696 ( 10.1074/jbc.M112.410696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steiner G, Tunc S, Maitz M, Salzer R. 2007. Conformational changes during protein adsorption. FT-IR spectroscopic imaging of adsorbed fibrinogen layers. Anal. Chem. 79, 1311–1316. 10.1021/ac061341j ( 10.1021/ac061341j) [DOI] [PubMed] [Google Scholar]
- 72.Yermolenko IS, Fuhrmann A, Magonov SN, Lishko VK, Oshkadyerov SP, Ros R, Ugarova TP. 2010. Origin of the nonadhesive properties of fibrinogen matrices probed by force spectroscopy. Langmuir 26, 17 269–17 277. 10.1021/la101791r ( 10.1021/la101791r) [DOI] [PMC free article] [PubMed] [Google Scholar]