Abstract
The remarkable capacity of the genus Dehalococcoides to dechlorinate a multitude of different chlorinated organic compounds reflects the number and diversity of genes in the genomes of Dehalococcoides species encoding reductive dehalogenase homologues (rdh). Most of these genes are located in the vicinity of genes encoding multiple antibiotic resistance regulator (MarR)-type or two-component system regulators. Here, the transcriptional response of rdhA genes (coding for the catalytic subunit) to 2,3- and 1,3-dichlorodibenzo-p-dioxin (DCDD) was studied in Dehalococcoides mccartyi strain CBDB1. Almost all rdhA genes were transcribed in the presence of 2,3-DCDD, albeit at different levels as shown for the transcripts of cbrA, cbdbA1453, cbdbA1624 and cbdbA1588. By contrast, 1,3-DCDD did not induce rdhA transcription. The putative MarR CbdbA1625 was heterologously produced and its ability to bind in vitro to the overlapping promoter regions of the genes cbdbA1624 and cbdbA1625 was demonstrated. To analyse regulation in vivo, single-copy transcriptional promoter–lacZ fusions of different rdhA genes and of cbdbA1625 were constructed and introduced into the heterologous host Escherichia coli, and expression levels of the fusions were measured. The cbdbA1625 gene was cloned into a vector allowing a regulation of expression by arabinose and it was transformed into the strains containing the rdh-promoter–lacZ fusion derivatives. CbdbA1625 was shown to downregulate transcription from its own promoter resulting in a 40–50% reduction in the β-galactosidase activity, giving the first hint that it acts as a repressor.
Keywords: Dehalococcoides, reductive dehalogenase, transcriptional regulation, multiple antibiotic resistance regulator, dichlorodibenzo-p-dioxin, trichlorobenzene
1. Introduction
The obligately organohalide-respiring Dehalococcoides mccartyi strains isolated so far are able to dehalogenate a variety of organohalides ranging from chlorinated aliphatic to chlorinated aromatic compounds. These include notoriously recalcitrant compounds such as polychlorinated biphenyls or dibenzo-p-dioxins. From an evolutionary standpoint, this broad substrate specificity is logical and because nature harbours a multitude of halogenated compounds [1]; however, each of these is present at relatively low concentrations compared with the abundance of electron acceptors such as sulfate or carbonate that provide the basis for other anaerobic respiratory processes. Recent results [2] demonstrated a positive correlation between the number of Dehalococcoides-like Chloroflexi and the natural organochlorine content in forest soils. It is thought that the high number of up to 36 non-identical genes encoding putative reductive dehalogenases (rdh) in the D. mccartyi genomes [3–5] forms the genetic basis for the capability to attack chlorinated compounds with a broad range of electronic and steric properties. Reductive dehalogenases consist of a catalytic cobalamin- and [Fe–S] cluster-containing subunit facing the outer side of the cytoplasmic membrane and a putative membrane anchor, encoded by the rdhA and rdhB genes, respectively. Owing to the large number of rdhAB operons, it can be assumed that their synthesis is controlled by regulatory events.
This assumption is supported by data obtained with another group of organohalide-respiring bacteria, the chlorophenol-dechlorinating members of the Gram-positive genus Desulfitobacterium [6]. In contrast to Dehalococcoides, these bacteria are facultative organohalide respirers, and regulation must confer an adequate response to the presence of alternative electron acceptors such as nitrate or chlorinated compounds in the environment or even to fermentative growth conditions. The o-chlorophenol reductive dehalogenase-encoding genes cprBA were specifically induced in the presence of chlorinated phenols by high affinity binding of the CRP/FNR-type transcriptional activator CprK to target sequences (dehaloboxes) in the promoter regions within the cpr gene cluster [7]. CprK is activated by an allosteric mechanism. Binding of o-chlorophenols leads to conformational changes, which are required for specific DNA binding [8]. An examination of the genome sequences of D. mccartyi and relatives for the presence of putative regulators of the CRP/FNR-type revealed that they are present in the genomes of D. mccartyi strains 195 and CBDB1 and of Dehalogenimonas lykanthroporepellens BL-DC-9, but no orthologues were annotated in the genomes of the D. mccartyi strains GT, VS and BAV-1, ruling out a general role in D. mccartyi organohalide respiration.
In the vcrAB operon, encoding a vinyl chloride (VC) reductive dehalogenase, in D. mccartyi strain VS a gene is present coding for a putative regulator of the NosR/NirI-type [9]. It shows some similarity to CprC, which is also encoded in the cpr gene cluster of the o-chlorophenol-respiring Desulfitobacterium dehalogenans [10]. Although closely related orthologues (94–95% identity) are found in the genomes of the two D. mccartyi strains 195 and CBDB1, these are not associated with rdh genes. Interestingly, two other types of regulators are frequently encoded in the vicinity of rdhAB genes in all described D. mccartyi genomes: two-component system (TCS) regulators and MarR-type regulators (table 1).
Table 1.
Number of rdhAB genes and of genes encoding putative MarR-type and two-component system regulators in the genomes of D. mccartyi strains.a
| genes encoding TCS regulators |
genes encoding MarR-regulators |
||||
|---|---|---|---|---|---|
| strain | rdhAB genes | total | adjacent to rdhA genes | total | adjacent to rdhA genes |
| CBDB1 | 32 | 24 | 14 | 16 | 10 |
| 195 | 17 | 19 | 10 | 8 | 3 |
| BAV1 | 11 | 13 | 5 | 4 | 1 |
| GT | 20 | 3 | 1 | 12 | 7 |
| VS | 36 | 21 | 11 | 14 | 10 |
TCS regulators typically consist of a histidine kinase (HK) and a response regulator (RR). HKs possess a modular architecture with diverse input domains linked to a conserved catalytic core, which allows the coupling of a variety of input signals to output responses through a conserved autophosphorylation and phosphotransfer pathway [12]. Localization of the sensory domains on extracytoplasmic or membrane-spanning regions allows direct signal perception at the cell periphery; however, the 14 putative HKs encoded in the vicinity of rdh genes in strain CBDB1 have no transmembrane helices and are predicted to be cytoplasmic proteins [3]. For 13 of these HKs, at least one Per–Arndt–Sim (PAS) fold is predicted as part of the input domain. PAS domains can sense a variety of signals such as oxygen, light, redox potential or the presence of small molecules [13] and can bind cofactors such as haem or flavin. In general, upon signal recognition, a histidine residue in the HK is autophosphorylated, and the phosphoryl group is transferred to an aspartate of the RR. The RR proteins in D. mccartyi predicted to be involved in organohalide respiration possess a signal receiver domain with a conserved aspartate and a C-terminal effector domain forming a DNA-binding winged helix, which could mediate activation of transcription of target genes [14]. The range of signals sensed by the cognate sensor kinase can be extended by the so-called TCS connector proteins, transmitting signals from other regulatory circuits to TCS regulators by protein–protein interaction [15,16]. Therefore, the TCS regulators encoded in the genome of D. mccartyi might represent important target molecules for the integration of different signals such as the redox state of the cell or the presence of organochlorines.
The multiple antibiotic resistance regulator (MarR) protein family is distributed throughout the bacterial and archaeal domains and mediates cellular responses to changing environmental conditions, such as antibiotic or peroxide stress and adaptation to the catabolism of aromatic compounds [17]. The latter fact is interesting in the light of the capability of strains of D. mccartyi and its close relatives to use chlorinated aromatics for organohalide respiration. In general, the MarRs of aromatic compound metabolism act as repressors. Binding of the aromatic ligand releases the repressor from the promoter and leads to the induction of the regulated gene(s). Structurally, the reported aromatic ligands range from salicylate (MarR) [18], 3-chlorobenzoate [19], 2-methylhydroquinone [20] to flavonoids [21]. The genes encoding MarR proteins are generally part of the gene cluster that they regulate and are divergently oriented to them. This is also the case for most rdhA gene-associated marR genes in D. mccartyi genomes.
The typical structural element of MarR proteins is the winged helix–turn–helix DNA recognition fold. Some structures of MarRs have been resolved with and without bound ligand, providing evidence for the displacement of the DNA-binding helix upon ligand binding [22]. Interestingly, the rdh-associated regulators form a separate branch in the phylogenetic tree of MarRs (see the electronic supplementary material, figure S1). It is also interesting that putative sensory and catalytic proteins in organohalide respiration seem to have co-evolved, as indicated by the conservation of gene clusters comprising a specific rdh gene and its associated regulatory gene in different D. mccartyi genomes.
First indications of a differential expression of rdhA genes in response to specific compounds came from transcription analyses of D. mccartyi in pure and mixed cultures that dechlorinate chlorinated ethenes. During VC respiration by strain BAV1, the gene bvcA was highly expressed, suggesting its gene product functions as a VC reductase, and indeed, it was detected in several VC-dechlorinating mixed cultures [23,24]. Another gene encoding a functional VC reductase was also induced by VC [9]. In addition, albeit at lower levels, transcripts of further rdhA genes were formed, suggesting that multiple rdhA genes are induced by a single chlorinated ethene [24]. Studies with trichloroethene (TCE)- and tetrachloroethene (PCE)-grown cells of strain 195 indicated that the genes encoding the PCE and TCE reductive dehalogenases (PceA, TceA) were transcribed to the highest levels among all 17 rdhA genes [25], and the corresponding proteins were identified with a high coverage in the proteome of the respective cells [26]. Furthermore, rdhA gene expression levels depended on the growth phase and the PCE respiration rate [27–29]. Consequently, they were used as biomarkers for active PCE or TCE dechlorination in technical and groundwater systems [30,31].
The D. mccartyi strain CBDB1 is adapted to organohalide respiration with chlorinated aromatic compounds such as different chlorinated benzenes, phenols, biphenyls and even chlorinated dibenzo-p-dioxins [32–35]. Figure 1 shows three selected chloroaromatic compounds and the dechlorination route observed with strain CBDB1. With regard to the dechlorination of polychlorinated dibenzo-p-dioxins, strain CBDB1 can even dechlorinate the most toxic congeners 1,2,3,7,8-penta- and 2,3,7,8-tetrachlorodibenzo-p-dioxin to 2,7- or 2,8-dichlorodibenzo-p-dioxin (DCDD), resulting in a strong reduction in the toxicity [33]. The transcriptional response of all 32 rdhA genes of strain CBDB1 during dechlorination of 1,2,3- and 1,2,4-trichlorobenzenes (TCBs) was investigated using a combined reverse transcription (RT)-PCR/terminal restriction fragment length polymorphism analysis (t-RFLP) approach [36]. This approach enabled the simultaneous detection of 29 of 32 rdhA transcripts. The transcription of the remaining three rdhA genes was also detected by reverse transcription-quantitative PCR (RT-qPCR). In summary, the upregulation of all 32 rdhA genes could be observed following the addition of 1,2,3- or 1,2,4-TCB. However, a differential upregulation of at least two rdhA genes, cbdbA1453 and cbdbA1624, in response to 1,2,3- and 1,2,4-TCB, respectively, was suggested by the results. RT-qPCR revealed that the transcript levels of ten analysed rdhA genes differed by several orders of magnitude. As expected, the rdhA gene cbrA, encoding a chlorobenzene reductase [37], was transcribed at the highest level in the presence of both TCBs [36]. Although cbrA is co-localized with genes encoding a TCS system, cbdbA1624 and cbdbA1453 are preceded by marR genes suggesting that different regulatory principles might be involved in controlling the dechlorination of 1,2,3-TCB and 1,2,4-TCB.
Figure 1.
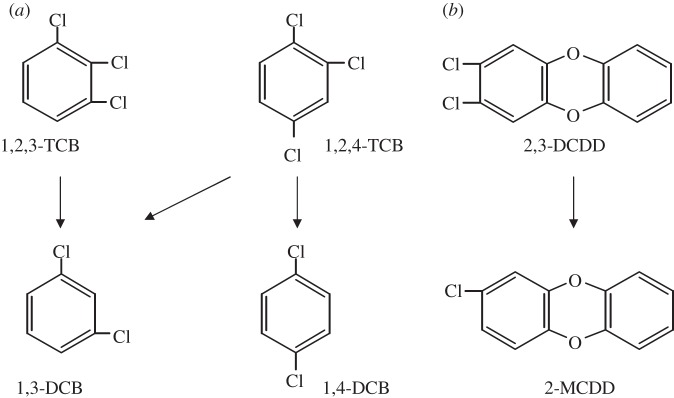
Scheme of (a) reductive dechlorination of 1,2,3- and 1,2,4-trichlorobenzene (TCB) to 1,3- and 1,4-dichlorobenzene (DCB) [32] and (b) 2,3-dichlorodibenzo-p-dioxin (DCDD) to 2-monochlorodibenzo-p-dioxin (MCDD) [33].
In this study, we report the results of transcription analyses of rdhA genes in response to two chlorinated dibenzo-p-dioxins. The two congeners 1,3- and 2,3-DCDD were chosen because they are dechlorinated by strain CBDB1 in one step to 2-monochlorodibenzo-p-dioxin (MCDD) (shown for 2,3-DCDD in figure 1). We also addressed the possible role of MarR-type regulators in organohalide respiration and focused on cbdbA1625, a marR gene preceding the rdhA gene cbdbA1624, which shows a specific transcriptional response to 1,2,4-TCB [36]. The putative MarR CbdbA1625 was heterologously produced in Escherichia coli and analysed in vitro for its ability to interact with the intergenic region between cbdbA1624 and cbdbA1625. Furthermore, promoter–reporter gene fusions were developed to analyse the functionality of certain marR and rdhA gene promoters in the E. coli host and to investigate the regulation of the system in vivo by the MarR CbdbA1625.
2. Material and methods
(a). Bacterial strains, plasmids and culture conditions
Dehalococcoides mccartyi strain CBDB1 was grown in Ti(III) citrate-reduced, carbonate-buffered synthetic medium with hydrogen as electron donor and 5 mM acetate as carbon source as described previously [36]. Two-liquid phase cultures (50 ml) supplemented with 1,2,3-TCB (200 mM, with a nominal concentration of 10 mM) dissolved in hexadecane served as inoculum. 1,3- and 2,3-DCDD (AccuStandard, New Haven, CT, USA) were added from 300 µM stock solutions in acetone to several replicates of empty 100 ml serum bottles and in parallel to 20 ml gas chromatography (GC) vials at final concentrations of 15–40 µM. The acetone was evaporated in a stream of filter-sterilized N2 gas before the bottles and vials were closed with Teflon-coated butyl rubber stoppers, filled with 50 ml and 3 ml, respectively, of sterile, anaerobic medium and inoculated to 10% (v/v) with the pre-culture resulting in 5 × 107 cells ml–1 as determined by qPCR targeting the 16S rRNA gene [36]. Cultures were statically incubated at 30°C.
The E. coli strains, plasmids and phages used in this study are given in table 2. The E. coli strains were grown at 37°C in nutrient or lysogeny broth (LB) supplemented with the appropriate antibiotics, diagnostic substrates or inducers according to standard procedures [42]. BL21(DE3)-CodonPlus-RIL served as host for the cbdbA1625 expression vectors derived from pASK-IBA5 and pASK-IBA3 (IBA, Göttingen, Germany). Escherichia coli XL1-Blue MRF′ (Stratagene, Amsterdam, The Netherlands) was used as host for the cloning vectors pGEM T-Easy and pRS551. The λ-phage-sensitive E. coli strain MC1061 was used to produce a heterogeneous phage lysate for the subsequent transfer of the promoter–reporter gene constructs into the chromosome of E. coli MC4100.
Table 2.
Escherichia coli strains, plasmids and phages used in this study. The plasmids were derived by standard cloning procedures from vectors.
| E. coli strain, plasmid, or phage | description/genotype (insert size) | reference or source |
|---|---|---|
| strains | ||
| MC1061 | F− Δ(ara-leu)7697 [araD139]B/r Δ(codB-lacI)3 galK16 galE15 λ− e14− mcrA0 relA1 rpsL150(strR) spoT1 mcrB1 hsdR2(r−m+) | [38] |
| MC4100 | F− araD 139 Δ (argF-lac) U 169 ptsF25 deoCI selA1 fibB530 rpsL 150λ− | [39] |
| XL1-Blue MRF` | Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene, Amsterdam, The Netherlands |
| BL21(DE3)-CodonPlus-RIL | B F− ompT hsdS(rb− mB−) dcm+ Tetr gal λ (DE3) endA Hte [argU ileYleuW Cmr] | Statagene, Amsterdam, The Netherlands |
| LS4 | MC4100 λ RS_pRS551, carrying the promoter-less lacZ of pRS551 | this study |
| LS5 | MC4100 λ RS_P1625, carrying a chromosomal P1625–lacZ promoter fusion | this study |
| LS11 | MC4100 λ RS_P1455, carrying a chromosomal P1455–lacZ promoter fusion | this study |
| LS13 | MC4100 λ RS_P1624, carrying a chromosomal P1624–lacZ promoter fusion | this study |
| plasmids | ||
| pP1–P2a | AmpR, comprising complete intergenic region of cbdbA1624/1625 PCR-amplified with primer pair P1/P2 (269 bp) | this study |
| pP1–P3a | AmpR fragment of intergenic region of cbdbA1624/1625 PCR-amplified with primer pair P1/P3 (97 bp) | this study |
| pP4–P5a | AmpR fragment of intergenic region of cbdbA1624/1625 PCR-amplified with primer pair P4/P5 (106 bp) | this study |
| pIR-CBDBA84a | AmpR fragment of intergenic region of cbdbA83/84 PCR-amplified with primer pair CbdbA84-for/rev (350 bp) | this study |
| pIR-CBDBA1453a | AmpR fragment of intergenic region of cbdbA1454/1453 PCR-amplified with primer pair CbdbA1453-for/rev (475 bp) | this study |
| pCBDBA1625-IBA5b | AmpR, CmR, PCR-amplified cbdbA1625 cloned into BsaI site of pASK-IBA5 (482 bp) | this study |
| pCBDBA1625-IBA3b | AmpR, CmR, PCR-amplified cbdbA1625 cloned into BsaI site of pASK-IBA3 (485 bp) | this study |
| pRS551_P1624::lacZc | KanR, complete cbdbA1625–cbdbA1624 intergenic region (247 bp), cbdbA1624–lacZ promoter fusion | this study |
| pRS551_P1625::lacZc | KanR cbdbA1624–cbdbA1625 intergenic region, truncated 5′ by 14 bp (233 bp), cbdbA1625–lacZ promoter fusion | this study |
| pRS551_P1455::lacZc | KanR complete cbdbA1456–cbdbA1455 intergenic region (251 bp), cbdbA1455–lacZ promoter fusion | this study |
| pBAD1625d | AmpR, marR gene cbdbA1625 cloned into EcoRI/ HindIII restriction sites in front of arabinose inducible pBAD promoter | this study |
| phages | ||
| λ RS_lacZe | λ RS45 (lacZ) | this study |
| λ RS_P1624e | λ RS45 (P1624::lacZ), cbdbA1624–lacZ promoter fusion | this study |
| λ RS_P1625e | λ RS45 (P1625::lacZ), cbdbA1625–lacZ promoter fusion | this study |
| λ RS_P1455e | λ RS45 (P1455::lacZ), cbdbA1455–lacZ promoter fusion | this study |
The plasmids were derived by standard cloning procedures from vectors apGEM T-Easy (Promega), b661 pASK-IBA 3 and 5, respectively (IBA), cpRS551 [40] and dpBAD30 [41].
eThe phage variants were obtained by homologous recombination using λ RS45 [40].
(b). Measurement of chlorinated dibenzo-p-dioxins
The dibenzo-p-dioxins were analysed by GC with a Shimadzu 14A gas chromatograph and a DB608-megabore-capillary column (30 m, 0.331 mm i.d., 0.5 µm film thickness, J&W Scientific, Folsom, CA, USA). 2-MCDD was quantitatively determined by solid-phase microextraction from the headspace of 3-ml cultures as described before [43]. Subsequently, the dichlorodibenzo-p-dioxins were extracted from the 3-ml cultures by the addition of 3 ml of hexane and shaking (250 r.p.m.) at 30°C for 12 h. The extraction was repeated twice. The combined hexane extracts were supplemented with 50 µl of the internal standard 5,6-dibromoacenaphthene (0.25 mM) dissolved in 2,2,4,4,6,8,8-heptamethylnonane (HMN), concentrated to the HMN phase in a stream of nitrogen and diluted with 200 µl of hexane. The samples were analysed by GC–flame ionization detector, at a split ratio of 1 : 10 and injector and detector temperatures of 250°C and 280°C, respectively. The oven temperature programme was as follows: 3 min at 170°C, 1°C per min to 175°C, 5°C per min to 290°C, hold for 5 min.
(c). Transcription experiments, nucleic acids extraction and reverse transcription
For each transcription experiment, several replicate 50 ml cultures and 3 ml cultures were inoculated by 10% (v/v) with cells from one selected pre-culture. Duplicate 50 ml cultures were removed immediately after inoculation and after 24, 48, 72 and 168 h and completely harvested for nucleic acid extraction. The concentration of chlorinated dibenzo-p-dioxins was followed in the 3 ml cultures, which were incubated in parallel, extracted and analysed in duplicate at each time point. Cultures (50 ml) without the addition of the chlorinated electron acceptors served as controls. Chromosomal DNA of Dehalococcoides sp. strain CBDB1 was extracted from 1 ml of the 50 ml cultures using the NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany) according to the instructions of the manufacturer. Forty-five millilitres of the liquid culture was harvested by low-speed centrifugation and subjected to RNA isolation according to the procedure described previously [36], including the addition of Coleoptera luciferase mRNA (Promega, Mannheim, Germany), which served as an internal standard for normalization owing to losses during mRNA preparation and reverse transcription inefficiencies. Contaminating DNA was removed using a DNaseI kit (Fermentas, St Leon-Rot, Germany) with a treatment period of 3 h. RNA was stored at −80°C until cDNA synthesis was performed. Within each experiment, equal amounts of sample RNA in the range of 0.1–1 µg were subjected to reverse transcription using random hexamer primers and the RevertAid H minus first strand cDNA synthesis kit (Fermentas), according to the manufacturer's recommendations.
(d). PCR amplification of rdhA targets from DNA or cDNA and t-RFLP analysis
Thirteen degenerate primer pairs were used to amplify fragments of all 32 rdhA sequences encoded in the genome of strain CBDB1 using exactly the same primers and procedure as described previously [36]. Each primer pair was designed to recognize clusters of up to five specific rdhA genes or transcripts. The forward primers were labelled with 6-carboxyfluorescein to allow a subsequent t-RFLP analysis as described previously [36]. Briefly, the amplicons obtained with each degenerate primer pair were divided into aliquots, which were subjected to separate digestions with at least two appropriate restriction enzymes (MspI, RsaI, AluI, MboI, FnuDII, PstI or BsuRI; Fermentas). The labelled terminal restriction fragments were separated on a genetic analyser 3100 (PE Applied Biosystems, Langen, Germany), and sizes were determined by comparison with an internal size standard (ROX Genescan 500). The fragments were assigned to specific rdhA transcripts on the basis of t-RFLP profiles obtained with the same set of PCR primers and restriction enzymes from genomic DNA of strain CBDB1 and compared with computational digests of the respective rdhA gene sequences.
(e). Quantification of genes and transcripts
The copy number of the rdhA genes cbrA, cbdbA1624, cbdbA1453, cbdbA1588 and of the respective transcripts, of the 16S rRNA gene and of luciferase mRNA were measured by qPCR using total DNA or cDNA as template and the QuantiTect SYBR green kit (Qiagen, Hildesheim, Germany), the primers, reaction mixtures, cycling conditions and external standards for quantification as described previously [36]. Samples and external standards were analysed in duplicate. The transcription rate of the rdhA genes was calculated as the ratio of the copy number of the rdhA transcript, which was normalized for the recovered luciferase transcript number, and the copy number of the corresponding rdhA gene.
(f). Mapping the transcriptional start sites
Total RNA was extracted from cells of strain CBDB1 grown for 94 h in the presence of 50 µM 1,2,4-TCB and from E. coli strains LS5 and LS13 grown to an OD600 of 0.25. Primer extension analyses were carried out using the 5′-rapid amplification of cDNA ends according to the manufacturer's recommendations (Roche, Mannheim, Germany). Total RNA (50 ng from strain CBDB1 and 1 µg from E. coli strains) was subjected to cDNA synthesis using the cbdbA1624- and pRS551-specific SP1 primers, respectively. The 3′-end of cDNA was tailed with poly-dATP and subjected to nested PCR with the corresponding SP2 and SP3 primers (see the electronic supplementary material, table S1) and sequencing.
(g). Construction of plasmids and promoter–lacZ transcriptional fusions
Standard procedures were used for plasmid preparations, restriction enzyme digestions, ligations, transformations, phage transduction and gel electrophoresis [42]. The desired DNA fragments were amplified from genomic DNA of strain CBDB1 using the primers indicated in the electronic supplementary material, table S1, digested with restriction endonucleases as appropriate, ligated into linearized cloning vectors pGEM T-Easy or pRS551 (promoter fragments) or expression vectors pASK-IBA3/5 (cbdbA1625 gene). The plasmids and primers used in this study are listed in table 2 and the electronic supplementary material, table S1, respectively. The promoter–lacZ fusions were constructed by cloning the PCR-amplified intergenic regions into the EcoRI site of pRS551. Fusions with the correct orientation were then transformed into MC1061 cells, which were subsequently infected by λ-RS45. The resulting lysates of recombinant phages (table 2) were used to transduce MC4100 cells to a kanamycin (Kan)-resistant phenotype as described by Sawers & Böck [44]. The single-copy integration of the promoter–lacZ fusions and of the promoter-less lacZ gene (control) into the genomes of strains LS5, LS11, LS13 and LS4, respectively, was examined by PCR and sequencing.
(h). In vivo promoter probe assay
Strains harbouring single-copy promoter–lacZ transcriptional fusions (LS5, LS11, LS13) or the promoter-less lacZ gene (LS4) were inoculated into completely filled anaerobic Hungate tubes containing LB medium and were incubated statically at 37°C. To study the effect of the putative MarR-type regulator on gene expression, the appropriate strains were transformed with plasmids pBAD1625 (table 2) or pBAD30 [41] (vector control) and cultivated in LB supplemented with ampicillin (125 µg ml−1). For the induction of CbdbA1625 synthesis, 0.02% (w/v) of filter-sterilized arabinose was added. The strains were grown in triplicate to an optical density (600 nm) of 0.4–0.6. To study the gene expression, 100 µl of cell suspensions were added to a 96-well plate, and β-galactosidase activity was measured as described [42] by recording the absorbance at 415 nm using a microplate reader (Bio-Rad Laboratories, München, Germany). The rate of o-nitrophenol formation was normalized to the optical density of the respective culture, and the specific activity was given in Miller units. In addition, strains were grown on MacConkey-lactose agar (Roth, Karlsruhe, Germany), and β-galactosidase activity of colonies was qualitatively indicated by the formation of a pink colour.
(i). Overproduction and purification of CbdbA1625–Strep tag fusion proteins
The constructed vectors pCBDBA1625-IBA3 and -IBA5 encoding a C- and N-terminal Strep-tag II fusion of CbdbA1625 (CbdbA1625-StrepC and -StrepN, respectively) were transformed into E. coli BL21(DE3)-CodonPlus-RIL (table 2). The heterologous production of CbdbA1625 was induced by the addition of anhydrotetracycline (final concentration 0.2 µg ml−1), when the cultures had reached an optical density of 0.5–0.8 at 600 nm. Cells were harvested after 3 h by centrifugation at 4000g at 4°C for 10 min, and the pellet was resuspended in 1 ml lysis buffer (100 mM Tris/HCl, pH 7.6, 1 mM EDTA, 2 mM dithiothreitol (DTT)). The cells were lysed using a French press (SLM Instruments) at 1260 psi after prior treatment with DNase I (5 µg ml−1) and lysozyme (1 mg ml−1). The protein was purified by chromatography on StrepTactin–Sepharose columns following the recommendations of the manufacturer (IBA). Protein concentration was measured photometrically at 280 nm using a NanoDrop (Thermo Scientific, Schwerte, Germany). Purified protein was either used immediately or stored on ice for maximally 2–3 days prior to use.
(j). Electrophoretic mobility shift assay
The templates for the electrophoretic mobility shift assay (EMSA) were produced by re-amplification from plasmids pP1–P2, pP1–P3, pP4–P5, pIR-CBDBA84 and pIR-CBDBA1453, which contained the complete intergenic regions, or portions thereof (table 2), using the indicated primers (see the electronic supplementary material, table S1). The PCR products were purified using the QIAquick PCR purification kit. Different concentrations (10, 3.3, 1.1, 0.4, 0.1 µM) of both N- and C-terminally tagged CbdbA1625-Strep protein were incubated with 13–18 nM of the target DNA in 1× reaction buffer (20% (w/v) glycerol, 50 mM Tris/HCl pH 7.5, 5 mM MgCl2, 2.5 mM EDTA, 250 mM NaCl, 3 mM DTT, 1 µg µl−1 poly(dI -dC) for 30 min at 24°C in a reaction volume of 200 µl. The effect of TCBs was examined by the addition of 1,2,3-, 1,2,4- or 1,3,5-TCB each at a final concentration of 40 µM. Twenty microlitres of the reaction mixtures was separated by electrophoresis on a non-denaturating 8% (w/v) polyacrylamide gel in 0.5 × TBE (1.78 mM Tris, 1.78 mM boric acid, 0.04 mM EDTA; pH 8.0), and the DNA was visualized by GelRed nucleic acid gel stain (Biotrend, Köln, Germany).
3. Results and discussion
(a). Transcription of rdhA genes in response to dichlorodibenzo-p-dioxins
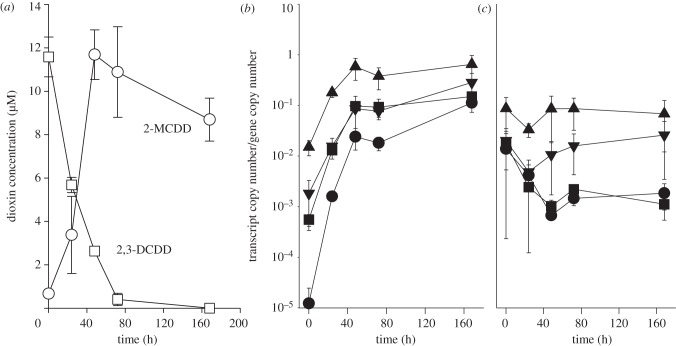
The transcriptional response of the rdhA genes in strain CBDB1 to chlorinated dibenzo-p-dioxins was studied using the two congeners 2,3-DCDD and 1,3-DCDD. 2,3-DCDD was dechlorinated to 2-MCDD within 48 h (figure 2a). The transcription of clusters of rdhA genes was analysed by RT-PCR using 13 pairs of degenerate primers [36]. Almost no rdhA transcript was detectable at time-point 0 and in the control cultures without added dioxin. However, after 24 h of incubation with 2,3-DCDD, products were obtained, with 12 of the 13 primer pairs demonstrating a 2,3-DCDD-dependent induction of transcription (see the electronic supplementary material, figure S2). The abundance of products increased up to 48 h, when reductive dechlorination was complete, but remained relatively stable until the end of the experiment (168 h). t-RFLP was used to identify individual rdhA transcripts within the cluster-specific amplicons. As indicated in the electronic supplementary material, figure S2, altogether 29 individual rdhA transcripts were detected. The three rdhA transcripts of cbdbA80, cbdbA88 and cbdbA243 targeted by the cluster-3 primers were not detected previously in TCB-grown cells, presumably owing to their very low abundance [36]. In summary, the general transcription profile of rdhA genes strongly resembled the situation as reported for 1,2,3-TCB-grown cells [36]. Four rdhA genes belonging to different PCR-amplified clusters were selected for a quantitative transcription analysis: the three rdhA genes cbrA, cbdbA1453 and cbdbA1624, which previously had been shown to exhibit a strong response to TCBs [36], and cbdbA1588 encoding an orthologue of PceA, identified as a bifunctional PCE and 2,3-dichlorophenol reductive dehalogenase in strain 195 [25]. The 2,3-DCDD-dependent induction of all four genes could be demonstrated (figure 2b), whereas in the control without dioxin no induction of transcription was observed (figure 2c). It is noteworthy that after 48 h cbrA transcripts attained the highest level of the four genes followed by cbdbA1453, whereas the level of cbdbA1624 was one to two orders of magnitude lower, again agreeing with the transcription levels for 1,2,3-TCB-grown cells reported earlier [36]. Interestingly, cbdbA1588, which was expressed at a lower level in 1,2,3-TCB-grown cells [36], attained the same transcription level as cbdbA1453 in the presence of 2,3-DCDD. The transcript copy number of roughly one per cell determined for cbrA was very low. This might be due to the low concentration and the low water solubility (59 nM) [45] of 2,3-DCDD. However, the results emphasize a possible role of CbrA in reductive dechlorination of dioxins. It is likely, however, that other reductive dehalogenases are also involved, as suggested by the fact that so far only D. mccartyi strain DCMB5, which contains a close orthologue of cbrA (unpublished data, 2012), is not able to dechlorinate 2,3-DCDD [43].
Figure 2.
(a) Kinetics of reductive dechlorination of 2,3-dichloro (DCDD)- to 2-monochlorodibenzo-p-dioxin (MCDD) by strain CBDB1. Two 3 ml cultures were completely extracted for the analyses of chlorinated dibenzo-p-dioxins at each time-point shown. Quantitative transcription analysis of the four rdhA genes cbrA (filled triangles), cbdbA1624 (filled circles), cbdbA1453 (filled inverted triangles) and cbdbA1588 (filled squares) of strain CBDB1 in the presence of 2,3-DCDD (b), and (c) in the control not supplemented with an electron acceptor is shown. Mean values from duplicate cultures and s.d. are shown.
The other congener, 1,3-DCDD, has been shown previously to be a transient intermediate in the dechlorination of 1,2,4-trichlorodibenzo-p-dioxin to 2-MCDD [33]. When we applied it directly to the culture, it was slowly dechlorinated only in the primary culture and surprisingly was not dechlorinated in a subculture which was inoculated to 10% (v/v) with the previous culture (data not shown). In addition, transcription of rdhA genes was not detectable in the primary culture by RT-PCR and RT-qPCR (data not shown), indicating that 1,3-DCDD was not an inducer of rdhA gene transcription. Thus, dechlorination of 1,3-DCDD might be a co-metabolic process similar to the co-metabolic conversion of VC to ethene by D. mccartyi strain 195 [46].
The transcription analyses of rdhA genes in D. mccartyi strain CBDB1 exposed to two dichlorinated dibenzo-p-dioxins revealed two findings. First, it confirmed for another class of chloroaromatic compounds that a single chlorinated compound induces a general transcriptional response. This possibly enables Dehalococcoides to recognize further chlorinated compounds present in its environment, also extending the spectrum of substrates to non-inducing organochlorines such as 1,3-DCDD. Second, the genes are expressed to different levels, strongly suggesting the occurrence of specific regulatory events. These might include a specific activation of target and/or repression of non-target genes.
(b). In vitro study of MarR interaction with the intergenic region of cbdbA1624 and cbdbA1625
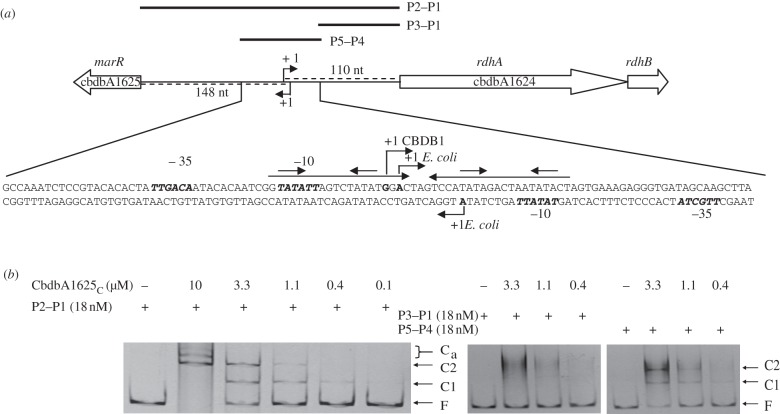
The gene cbdbA1625 encoding a putative MarR-type regulator is oppositely oriented to the rdhA gene cbdbA1624 (figure 3), which is specifically regulated in the presence of 1,2,4-TCB [36]. To study whether CbdbA1625 exerts a regulatory function on the expression of both genes, the cbdbA1625 gene was cloned into the expression vectors pASK-IBA5 and pASK-IBA3 and Strep-CbdbA1625N and -C were purified as N- and C-terminally Strep-tag II-fusion proteins, respectively, from E. coli BL21(DE3)-CodonPlus-RIL. The complete intergenic region between cbdbA1624 and cbdbA1625 (P1–P2) and two sub-fragments (P1–P3 and P4–P5) were used in EMSA. After the incubation of the purified P1–P2 PCR product with different concentrations of either Strep-CbdbA1625c or -N, a retardation of the DNA band was observed (shown for CbdbA1625C in figure 3b). Regardless of whether N- or C-terminally Strep-tagged CbdbA1625 was used, two protein–DNA complexes (C1, C2) appeared at protein concentrations between 0.4 and 3.3 µM, suggesting two specific binding sites for CbdbA1625 in the intergenic region. Additional discrete bands of lower electrophoretic mobility (Ca) appeared when the ratio of Strep-CbdbA1625 to the operator was further increased. Binding of Strep-CbdbA1625 to the cbdbA1624–cbdbA1625 intergenic region was shown to be specific, because no gel shift was observed when the amplified intergenic regions upstream of cbdbA1453 and cbrA were used in the gel retardation assay (data not shown). To locate the binding site of CbdbA1625 within the intergenic region, two sub-fragments comprising the 3′-region (P1–P3) and the putative promoter region (P4–P5) (figure 3a) were used in an EMSA. Addition of both the C- and N-terminally Strep-tagged CbdbA1625 fusion proteins resulted in mobility shifts of the P4–P5 fragment (exemplarily shown for CbdbA1625c in figure 3b), which were comparable to the shift observed with the complete intergenic region. By contrast, no discrete shifted bands were identified when the P1–P3 fragment was used (figure 3b). Despite several attempts, disruption of the protein–DNA complexes could not be achieved by the addition of 1,2,4-, 1,2,3- or 1,3,5-TCB, suggesting that specific conditions or further regulatory components are required for signal perception (data not shown).
Figure 3.
(a) Schematic of the intergenic region of cbdbA1624–cbdbA1625. The DNA sequence shown depicts the P5–P4 region. The long horizontal arrows above the sequence indicate inverted repeat sequences in the promoter region; the angled arrows indicate the experimentally determined transcriptional start sites; the short arrows above the inverted repeat sequences represent the putative recognition sequences of the regulator; bold and italic letters depict the consensus sequences of the −10 and −35 region predicted by the BPROM software (http://linux1.softberry.com/berry.phtml). The complete intergenic region (246 bp; P2–P1) and sub-fragments thereof (P3–P1/P5–P4) were amplified by PCR from total DNA of strain CBDB1 with primers binding in, or flanking, the intergenic region for EMSA. (b) Interaction of Strep-CbdbA1625C with the complete intergenic region (P2–P1) and the sub-fragments (P3–P1/P5–P4). EMSA was carried out in the presence (plus symbols) and absence (minus symbols) of Strep-CbdbA1625C protein. Free DNA (F) and retarded DNA–protein complexes (C1, C2, Ca) were visualized by staining with GelRed (Biotium, Hayward, CA, USA).
Analysis of the DNA sequence of the P4–P5 region (figure 3a) indicated that it contained a perfect 40 bp palindrome, which comprised short inverted repeats in its half sites, the typical binding motif of MarRs [17]. A primer extension analysis using RNA extracted from 1,2,4-TCB-grown cells of strain CBDB1 mapped the transcriptional start site of cbdbA1624 to a position within this palindrome 110 bp upstream of the translational initiation codon (figure 3a). Primer extension analysis of the marR transcript cbdbA1625 failed to identify a signal, possibly indicating transcript levels below the detection limit of the assay.
(c). In vivo study of rdhA and marR transcription and interaction with MarR in a heterologous system
Transcriptional promoter–lacZ fusions were constructed for the two rdhA genes (cbdbA1624 and cbdbA1455) and the marR gene (cbdbA1625), and these were introduced in single copy into the genome of E. coli strain MC4100. Each DNA fragment resulted in detectable β-galactosidase enzyme activity, which varied in the range between 40 and 2800 Miller units (see the electronic supplementary material, figure S3) depending on the promoter region. This finding suggested that the putative promoters within these DNA sequences were recognized by the E. coli RNA polymerase. To provide further evidence for this proposal, we determined the transcriptional start sites of the transcriptional fusions P1624-lacZ (strain LS13) and P1625-lacZ (strain LS5) by primer extension. For P1624, the transcription initiation site was mapped to a position almost identical (2 bp difference) to the transcriptional start of cbdbA1624 in the native host CBDB1, strongly suggesting that the E. coli RNA polymerase indeed recognized the Dehalococcoides promoter (figure 3a). The signal intensity was low perhaps owing to the unusually short distance between the putative −10 and −35 regions [47,48]. For P1625-lacZ, the promoter was mapped 149 bp upstream of the putative translational start codon of cbdbA1625 (figure 3a). The transcriptional start and the predicted −10 region are also within the palindrome suggesting that the promoter regions of cbdbA1624 and cbdbA1625 overlap and are both under the control of the MarR CbdbA1625.
We selected the P1625–lacZ construct (strain LS5) because of its high β-galactosidase activity for a first analysis of in vivo regulation by CbdbA1625. The native MarR CbdbA1625 was cloned into the pBAD30 vector and transformed into strain LS5. After inducing expression of the cbdbA1625 gene on the plasmid in anaerobically grown cells of E. coli by adding 0.02% (w/v) arabinose, a 40–50% reduction in β-galactosidase enzyme activity was observed, whereas no significant reduction in the β-galactosidase activity occurred in a vector control (table 3). In addition, the interaction of CbdbA1625 was specific: no reduction in the β-galactosidase activity was observed for the cbdbA1455 promoter–lacZ fusion strain (table 3). These results demonstrate for the first time that a MarR of D. mccartyi is functional in a heterologous host and suggest that CbdbA1625 acts as repressor and auto-regulates its own transcription.
Table 3.
Influence of the MarR CbdbA1625 on the expression of cbdbA1625–lacZ and cbdbA1455–lacZ transcriptional fusions in the heterologous host E. coli. The strains LS5 and LS11 were transformed with pBAD1625, which carries the marR gene cbdbA1625 under the control of the PBAD promoter, or pBAD30 (vector control) and grown to an OD600nm of 0.4–0.6 in the presence or absence of 0.02% (w/v) of arabinose. The strains were analysed in biological triplicates. One out of three repeated experiments is shown with standard deviations.
| pBAD1625 |
pBAD30 (vector control) |
||||
|---|---|---|---|---|---|
| promoter activity |
promoter activity |
||||
| strains (transcriptional fusion) | addition of arabinose | (Miller units) | (%) | (Miller units) | (%) |
| LS5 (P1625-lacZ) | — | 1023.5 ± 145.1 | 100 | 1217.2 ± 164.4 | 100 |
| + | 606.7 ± 103.5 | 59 | 1054.5 ± 185.7 | 87 | |
| LS11 (P1455-lacZ) | — | 937.5 ± 172.4 | 100 | 549.9 ± 108.9 | 100 |
| + | 904.5 ± 156.3 | 97 | 535.5 ± 120.9 | 97 | |
4. Concluding remarks
Our investigation of rdh gene expression in Dehalococcoides mccartyi strain CBDB1 demonstrated a general response to 2,3-dichlorodibenzo-p-dioxin. The levels of rdhA transcripts varied over several orders of magnitude, which was similar to the response to chlorinated benzenes. The fact that 1,3-DCDD did not induce rdh gene expression, despite being a substrate for reductive dehalogenases, highlights the importance of regulatory processes for the functionality of organohalide respiration and demonstrates specificity of regulation. As inferred from the genomic context of rdh-associated genes, MarR-type regulators might play specific roles in the regulation of a subset of the rdhA genes. This possibility was addressed by performing in vitro interaction studies involving the MarR CbdbA1625 and the respective rdhA and marR promoter regions. In addition, results of in vivo promoter probe assays suggest that CbdbA1625 functions as repressor. Given this is a typical regulatory mechanism of MarRs, we assume that rdhA genes regulated by MarR-type regulators undergo repression control and this may be the rule rather than the exception in the environmental life of Dehalococcoides. If halogenated compounds become available, a de-repression might enable specific induction of rdhA transcription. Negative auto-regulation would ensure a tightly regulated transcriptional response. However, to understand the regulation in more detail, future efforts must be directed towards elucidating the signal to which the regulator responds.
Acknowledgements
This study was supported by Deutsche Forschungsgemeinschaft (graduate college 416, LE 780/4-1) and a grant of FEMS to A.W. We thank Jan R. Andreesen for continued support and providing critical comments on the manuscript.
References
- 1.Gribble GW. 2003. The diversity of naturally produced organohalogens. Chemosphere 52, 289–297 10.1016/S0045-6535(03)00207-8 (doi:10.1016/S0045-6535(03)00207-8) [DOI] [PubMed] [Google Scholar]
- 2.Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SC, Novak PJ. 2012. Natural niche for organohalide-respiring Chloroflexi. Appl. Environ. Microbiol. 78, 393–401 10.1128/AEM.06510-11 (doi:10.1128/AEM.06510-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273 10.1038/nbt1131 (doi:10.1038/nbt1131) [DOI] [PubMed] [Google Scholar]
- 4.Seshadri R, et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307, 105–108 10.1126/science.1102226 (doi:10.1126/science.1102226) [DOI] [PubMed] [Google Scholar]
- 5.McMurdie PJ, et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5, e1000714. 10.1371/journal.pgen.1000714 (doi:10.1371/journal.pgen.1000714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisaillon A, Beaudet R, Lepine F, Villemur R. 2011. Quantitative analysis of the relative transcript levels of four chlorophenol reductive dehalogenase genes in Desulfitobacterium hafniense PCP-1 exposed to chlorophenols. Appl. Environ. Microbiol. 77, 6261–6264 10.1128/AEM.00390-11 (doi:10.1128/AEM.00390-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabor K, Verissimo CS, Cyran BC, Ter Horst P, Meijer NP, Smidt H, de Vos WM, van der Oost J. 2006. Characterization of CprK1, a CRP/FNR-type transcriptional regulator of halorespiration from Desulfitobacterium hafniense. J. Bacteriol. 188, 2604–2613 10.1128/JB.188.7.2604-2613.2006 (doi:10.1128/JB.188.7.2604-2613.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy C, Pike K, Heyes DJ, Joyce MG, Gabor K, Smidt H, van der Oost J, Leys D. 2008. Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol. Microbiol. 70, 151–167 10.1111/j.1365-2958.2008.06399.x (doi:10.1111/j.1365-2958.2008.06399.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller JA, Rosner BM, Von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70, 4880–4888 10.1128/AEM.70.8.4880-4888.2004 (doi:10.1128/AEM.70.8.4880-4888.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smidt H, van Leest M, van der Oost J, de Vos WM. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182, 5683–5691 10.1128/JB.182.20.5683-5691.2000 (doi:10.1128/JB.182.20.5683-5691.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caspi R, et al. 2012. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40, D742–D753 10.1093/nar/gkr1014 (doi:10.1093/nar/gkr1014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154 10.1146/annurev.micro.091208.073214 (doi:10.1146/annurev.micro.091208.073214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hefti MH, Francoijs KJ, de Vries SC, Dixon R, Vervoort J. 2004. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 271, 1198–1208 10.1111/j.1432-1033.2004.04023.x (doi:10.1111/j.1432-1033.2004.04023.x) [DOI] [PubMed] [Google Scholar]
- 14.Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 10.1128/JB.01887-05 (doi:10.1128/JB.01887-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611 10.1101/gad.1700308 (doi:10.1101/gad.1700308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 15, 118–124 10.1016/j.mib.2011.11.009 (doi:10.1016/j.mib.2011.11.009) [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 8, 51–62 [PubMed] [Google Scholar]
- 18.Cohen SP, Levy SB, Foulds J, Rosner JL. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175, 7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Providenti MA, Wyndham RC. 2001. Identification and functional characterization of CbaR, a MarR-like modulator of the cbaABC-encoded chlorobenzoate catabolism pathway. Appl. Environ. Microbiol. 67, 3530–3541 10.1128/AEM.67.8.3530-3541.2001 (doi:10.1128/AEM.67.8.3530-3541.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Töwe S, Leelakriangsak M, Kobayashi K, Van Duy N, Hecker M, Zuber P, Antelmann H. 2007. The MarR-type repressor MhqR (YkvE) regulates multiple dioxygenases/glyoxalases and an azoreductase which confer resistance to 2-methylhydroquinone and catechol in Bacillus subtilis. Mol. Microbiol. 66, 40–54 10.1111/j.1365-2958.2007.05891.x (doi:10.1111/j.1365-2958.2007.05891.x) [DOI] [PubMed] [Google Scholar]
- 21.Hirooka K, Danjo Y, Hanano Y, Kunikane S, Matsuoka H, Tojo S, Fujita Y. 2009. Regulation of the Bacillus subtilis divergent yetL and yetM genes by a transcriptional repressor, YetL, in response to flavonoids. J. Bacteriol. 191, 3685–3697 10.1128/JB.00202-09 (doi:10.1128/JB.00202-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2, 243–254 10.1093/jmcb/mjq021 (doi:10.1093/jmcb/mjq021) [DOI] [PubMed] [Google Scholar]
- 23.Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70, 6347–6351 10.1128/AEM.70.10.6347-6351.2004 (doi:10.1128/AEM.70.10.6347-6351.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waller AS, Krajmalnik-Brown R, Löffler FE, Edwards EA. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71, 8257–8264 10.1128/AEM.71.12.8257-8264.2005 (doi:10.1128/AEM.71.12.8257-8264.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fung JM, Morris RM, Adrian L, Zinder SH. 2007. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl. Environ. Microbiol. 73, 4439–4445 10.1128/AEM.00215-07 (doi:10.1128/AEM.00215-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, Richardson RE. 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73, 320–326 10.1128/AEM.02129-06 (doi:10.1128/AEM.02129-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahm BG, Morris RM, Richardson RE. 2006. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 72, 5486–5491 10.1128/AEM.00855-06 (doi:10.1128/AEM.00855-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahm BG, Richardson RE. 2008. Correlation of respiratory gene expression levels and pseudo-steady-state PCE respiration rates in Dehalococcoides ethenogenes. Environ. Sci. Technol. 42, 416–421 10.1021/es071455s (doi:10.1021/es071455s) [DOI] [PubMed] [Google Scholar]
- 29.Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L. 2008. Temporal transcriptomic microarray analysis of ‘Dehalococcoides ethenogenes’ strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74, 2864–2872 10.1128/AEM.02208-07 (doi:10.1128/AEM.02208-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behrens S, Azizian MF, McMurdie PJ, Sabalowsky A, Dolan ME, Semprini L, Spormann AM. 2008. Monitoring abundance and expression of ‘Dehalococcoides’ species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl. Environ. Microbiol. 74, 5695–5703 10.1128/AEM.00926-08 (doi:10.1128/AEM.00926-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PK, Macbeth TW, Sorenson KS, Jr, Deeb RA, Alvarez-Cohen L. 2008. Quantifying genes and transcripts to assess the in situ physiology of Dehalococcoides spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 74, 2728–2739 10.1128/AEM.02199-07 (doi:10.1128/AEM.02199-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adrian L, Szewzyk U, Wecke J, Görisch H. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408, 580–583 10.1038/35046063 (doi:10.1038/35046063) [DOI] [PubMed] [Google Scholar]
- 33.Bunge M, Adrian L, Kraus A, Opel M, Lorenz WG, Andreesen JR, Görisch H, Lechner U. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421, 357–360 10.1038/nature01237 (doi:10.1038/nature01237) [DOI] [PubMed] [Google Scholar]
- 34.Adrian L, Hansen SK, Fung JM, Görisch H, Zinder SH. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ. Sci. Technol. 41, 2318–2323 10.1021/es062076m (doi:10.1021/es062076m) [DOI] [PubMed] [Google Scholar]
- 35.Adrian L, Dudkova V, Demnerova K, Bedard DL. 2009. ‘Dehalococcoides’ sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl. Environ. Microbiol. 75, 4516–4524 10.1128/AEM.00102-09 (doi:10.1128/AEM.00102-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner A, Adrian L, Kleinsteuber S, Andreesen JR, Lechner U. 2009. Transcription analysis of genes encoding homologues of reductive dehalogenases in ‘Dehalococcoides’ sp. strain CBDB1 by using terminal restriction fragment length polymorphism and quantitative PCR. Appl. Environ. Microbiol. 75, 1876–1884 10.1128/AEM.01042-08 (doi:10.1128/AEM.01042-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adrian L, Rahnenführer J, Gobom J, Hölscher T. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73, 7717–7724 10.1128/AEM.01649-07 (doi:10.1128/AEM.01649-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138, 179–207 10.1016/0022-2836(80)90283-1 (doi:10.1016/0022-2836(80)90283-1) [DOI] [PubMed] [Google Scholar]
- 39.Casadaban MJ, Cohen SN. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: In vivo probe for transcriptional control sequences. Proc. Natl Acad. Sci. USA 76, 4530–4533 10.1073/pnas.76.9.4530 (doi:10.1073/pnas.76.9.4530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96 10.1016/0378-1119(87)90095-3 (doi:10.1016/0378-1119(87)90095-3) [DOI] [PubMed] [Google Scholar]
- 41.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrock J, Russell DW. 2001. Molecular cloning, 3rd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 43.Ewald EM, Wagner A, Nijenhuis I, Richnow HH, Lechner U. 2007. Microbial dehalogenation of trichlorinated dibenzo-p-dioxins by a Dehalococcoides-containing mixed culture is coupled to carbon isotope fractionation. Environ. Sci. Technol. 41, 7744–7751 10.1021/es070935g (doi:10.1021/es070935g) [DOI] [PubMed] [Google Scholar]
- 44.Sawers G, Böck A. 1988. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J. Bacteriol. 170, 5330–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiu WY, Doucette W, Gobas FAPC, Andren A, Mackay D. 1988. Physical-chemical properties of chlorinated dibenzo-p-dioxins. Environ. Sci. Technol. 22, 651–658 10.1021/es00171a006 (doi:10.1021/es00171a006) [DOI] [Google Scholar]
- 46.Maymó-Gatell X, Anguish T, Zinder SH. 1999. Reductive dechlorination of chlorinated ethenes and 1, 2-dichloroethane by ‘Dehalococcoides ethenogenes’ 195. Appl. Environ. Microbiol. 65, 3108–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dombroski AJ, Johnson BD, Lonetto M, Gross CA. 1996. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc. Natl Acad. Sci. USA 93, 8858–8862 10.1073/pnas.93.17.8858 (doi:10.1073/pnas.93.17.8858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawers G. 2001. A novel mechanism controls anaerobic and catabolite regulation of the Escherichia coli tdc operon. Mol. Microbiol. 39, 1285–1298 10.1111/j.1365-2958.2001.02316.x (doi:10.1111/j.1365-2958.2001.02316.x) [DOI] [PubMed] [Google Scholar]