Abstract
Dehalococcoides mccartyi strains are corrinoid-auxotrophic Bacteria and axenic cultures that require vitamin B12 (CN-Cbl) to conserve energy via organohalide respiration. Cultures of D. mccartyi strains BAV1, GT and FL2 grown with limiting amounts of 1 µg l−1 CN-Cbl quickly depleted CN-Cbl, and reductive dechlorination of polychlorinated ethenes was incomplete leading to vinyl chloride (VC) accumulation. In contrast, the same cultures amended with 25 µg l−1 CN-Cbl exhibited up to 2.3-fold higher dechlorination rates, 2.8–9.1-fold increased growth yields, and completely consumed growth-supporting chlorinated ethenes. To explore whether known cobamide-producing microbes supply Dehalococcoides with the required corrinoid cofactor, co-culture experiments were performed with the methanogen Methanosarcina barkeri strain Fusaro and two acetogens, Sporomusa ovata and Sporomusa sp. strain KB-1, as Dehalococcoides partner populations. During growth with H2/CO2, M. barkeri axenic cultures produced 4.2 ± 0.1 µg l−1 extracellular cobamide (factor III), whereas the Sporomusa cultures produced phenolyl- and p-cresolyl-cobamides. Neither factor III nor the phenolic cobamides supported Dehalococcoides reductive dechlorination activity suggesting that M. barkeri and the Sporomusa sp. cannot fulfil Dehalococcoides' nutritional requirements. Dehalococcoides dechlorination activity and growth occurred in M. barkeri and Sporomusa sp. co-cultures amended with 10 µM 5′,6′-dimethylbenzimidazole (DMB), indicating that a cobalamin is a preferred corrinoid cofactor of strains BAV1, GT and FL2 when grown with chlorinated ethenes as electron acceptors. Even though the methanogen and acetogen populations tested did not produce cobalamin, the addition of DMB enabled guided biosynthesis and generated a cobalamin that supported Dehalococcoides' activity and growth. Guided cobalamin biosynthesis may offer opportunities to sustain and enhance Dehalococcoides activity in contaminated subsurface environments.
Keywords: reductive dechlorination, organohalide respiration, vitamin B12, Dehalococcoides
1. Introduction
Chlorinated compounds, including tetrachloroethene (PCE) and trichloroethene (TCE), are widespread and toxic environmental pollutants [1,2]. Strains of the organohalide-respiring species Dehalococcoides mccartyi couple growth to the reductive dechlorination of a variety of chlorinated priority pollutants including polychlorinated biphenyls (PCBs), chlorobenzenes and chlorinated solvents. Some D. mccartyi strains dechlorinate PCE, TCE, dichloroethenes (DCEs) and vinyl chloride (VC) to non-toxic ethene, and thus are of great interest to restore aquifers contaminated with chlorinated ethenes [2–4].
Dehalococcoides mccartyi strains grow in completely synthetic, defined media, but cannot de novo synthesize the corrin ring and require exogenous vitamin B12 (cyanocobalamin, CN-Cbl) [2]. Corrinoids, such as CN-Cbl, contain a heterocyclic tetrapyrrole ring with a central cobalt atom [5]. Some Archaea and Bacteria synthesize corrinoids with an upper β-ligand moiety and a lower α-ligand moiety, so-called cobamides, which participate in important biological processes such as methanogenesis, acetogenesis and organohalide respiration [5–7]. Cobamides with 5′,6′-dimethylbenzimidazole (DMB) as the lower α-ligand are cobalamins such as adenosylcobalamin, hydroxocobalamin and cyanocobalamin (CN-Cbl). A cobalamin or a cobamide serve as cofactors for the PCE and TCE reductive dehalogenases (RDases) of several organohalide-respiring genera such as Dehalobacter, Desulfitobacterium and Sulfurospirillum, and exemplify the crucial roles these corrinoids play in the energy metabolism of organohalide-respiring Bacteria [7–10].
Genome sequence analysis demonstrated that D. mccartyi strains are corrinoid auxotrophs and lack the ability for de novo corrinoid biosynthesis [11,12]. A previous survey demonstrated that only about half (209 out of 410) CN-Cbl-requiring Bacteria possessed genes encoding corrinoid biosynthesis pathways [11]. Genes encoding the BtuBFCD type CN-Cbl/cobamide transport system were found in most (more than 90%) of the corrinoid-auxotrophic Bacteria [11], indicating external CN-Cbl/cobamide uptake rather than de novo biosynthesis is a common strategy of microorganisms to acquire this essential cofactor. Thus, it is not surprising that Dehalococcoides also possess BtuBFCD gene homologues as well as cobinamide (i.e. a cobamide lacking the lower α-ligand) salvage genes (cobU and cbiZ) to save the energy cost associated with adenosylcobalamin (Ado-Cbl) de novo biosynthesis [11].
Enhanced Dehalococcoides growth and faster dechlorination rates were observed in co- or tri-cultures of D. mccartyi strain 195 and corrinoid-producers such as Acetobacterium woodii, Desulfovibrio desulfuricans and Desulfovibrio vulgaris Hildenborough, suggesting that strain 195 benefited from the corrinoids produced in the co-cultures [13,14]. Recently, interspecies cobalamin transfer from Geobacter lovleyi to D. mccartyi strains BAV1 and FL2 was demonstrated in CN-Cbl-free co-cultures [15]. Geobacter sulfurreducens did not support Dehalococcoides activity unless DMB was supplied to the growth medium suggesting that only the cobamides with specific types of lower ligands, such as DMB, meet Dehalococcoides' nutritional requirements.
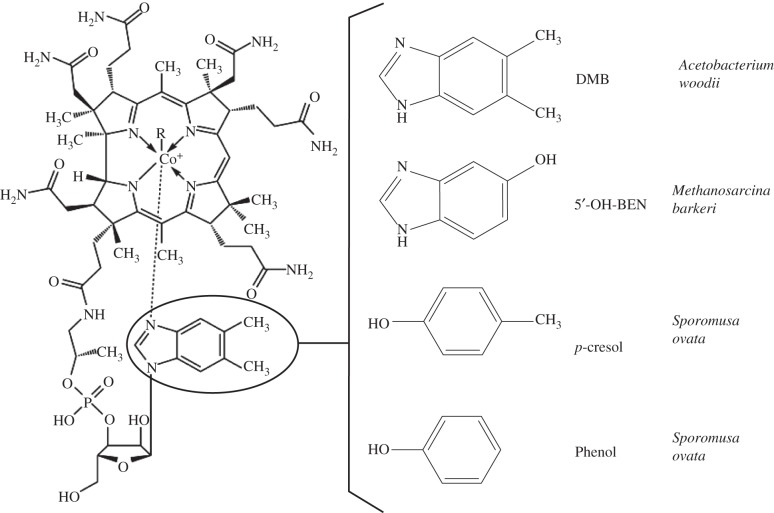
In anoxic subsurface environments, Dehalococcoides are members of diverse microbial communities, and some populations synthesize cobamides with different lower α-ligands [16,17]. Cobalamin and the naturally occurring cobamides tested in this study are shown in figure 1. For example, 5′-hydroxybenzimidazolyl-cobamide (factor III) or adenyl-cobamide (pseudo vitamin B12) are typically produced by methanogenic Archaea as the cofactors for methyltransferases involved in methane formation [18]. Acetogenic Bacteria synthesize benzimidazolyl- or phenolyl-type cobamides required for specific enzymes of the Wood–Ljungdahl pathway [18]. Cobalamin (5′,6′-dimethylbenzimidazolyl-cobamide) was identified as the corrinoid produced by A. woodii, phenolyl- and p-cresolyl-cobamide were purified from S. ovata, and 5′-methylbenzimidazolyl-cobamide was isolated from sulfate-reducers, such as Desulfobacterium autotrophicum and Desulfobulbus propionicus [19,20].
Figure 1.
Structures of cobalamin and the lower α-ligands of several characterized cobamides. In vitamin B12 (cyanocobalamin, CN-Cbl), a cyano group is the upper β-ligand and DMB is the lower α-ligand (circled). The structures were redrawn from [16,17]. DMB, 5′,6′-dimethylbenzimidazole; 5′-OH-BEN, 5′-hydroxybenzimidazole.
In this study, we investigated Dehalococcoides reductive dechlorination activity and growth during the co-cultivation with corrinoid-producing methanogenic Archaea and acetogenic Bacteria. Co-cultures established in CN-Cbl-free medium demonstrated interspecies cobamide transfer and identified the specific type(s) of corrinoid that Dehalococcoides can use. Knowledge of how Dehalococcoides' nutritional requirements are met in natural communities will reveal the ecophysiology of strictly organohalide-respiring, corrinoid-auxotrophic Chloroflexi. Understanding the bottlenecks (i.e. controls) limiting the activity of organohalide-respiring Chloroflexi may lead to innovative engineering approaches that enhance reductive dechlorination rates and extents and enable more efficient bioremediation applications.
2. Methods
(a). Pure cultures and growth conditions
Dehalococcoides mccartyi strain BAV1 (ATCC BAA-2100), strain GT (ATCC BAA-2099) and strain FL2 (ATCC BAA-2098) were grown in 160 ml serum bottles containing 100 ml reduced, bicarbonate-buffered, defined mineral salts medium amended with acetate (5 mM) and a N2–CO2 (80/20, v/v) headspace [21–23]. Following autoclaving, the bottles were amended with 10 ml hydrogen and 25 µg l−1 CN-Cbl as described [15]. Neat TCE (5 µl, 99%, Fisher, Pittsburgh, PA, USA) and cis-DCE (5 µl, 99.5%, Sigma-Aldrich-Fluka, St Louis, MO, USA) were added with a microlitre syringe (Hamilton, Reno, NV, USA). Following equilibration, the bottles received 3 per cent (v/v) inocula from dechlorinating cultures maintained under the same conditions. In addition, culture bottles amended with 1 µg l−1 CN-Cbl were maintained to evaluate the effects of low CN-Cbl concentrations on Dehalococcoides growth yields and dechlorination activities. Pure cultures of M. barkeri strain Fusaro (DSM 804), S. ovata (DSM 2662) and Sporomusa sp. strain KB-1 (16S rRNA gene GenBank accession no. AY780559.1) were maintained in the same defined mineral salts medium except that acetate, CN-Cbl and chlorinated solvent additions were omitted. All culture vessels were incubated stationary, with the stoppers down, at 30°C in the dark.
(b). Co-cultures
Triplicate co-cultures containing one of the three D. mccartyi strains and either M. barkeri or one of the Sporomusa sp. were established in 160 ml vessels inside an anoxic chamber as described [15]. The vessels were amended with 5 mM acetate and contained a H2–CO2 (80/20, v/v) headspace. To avoid CN-Cbl carry-over, cells for inoculation were collected by centrifugation at 14 000g for 10 min at room temperature, and suspended in CN-Cbl-free medium prior to inoculation. The positive control inocula were prepared in the same way and inoculated into culture vessels amended with 25 µg l−1 CN-Cbl. Dehalococcoides mccartyi pure cultures without CN-Cbl served as negative controls. Additional co-culture replicates received 10 µM DMB (more than 99% purity, Sigma-Aldrich-Fluka).
(c). DNA extraction and 16S rRNA gene quantification
Cells from Dehalococcoides pure cultures and co-cultures were harvested from 1 ml culture suspension as described [15,24]. Briefly, total genomic DNA was extracted using the MO BIO Soil DNA Isolation kit (MO BIO, Carlsbad, CA, USA), and a high efficiency bead ruptor homogenizer (Omni International, Kennesaw, GA, USA) following the manufacturer's protocols. Samples were processed with the bead ruptor homogenizer at a speed 3.25 m s−1 for 5 min. Quantitative real-time PCR (qPCR) to enumerate Dehalococcoides 16S rRNA gene copies followed established protocols using primer set Dhc1200F/Dhc1271R and probe FAM-BHQ1 Dhc1240 [24].
(d). Corrinoid measurements
(i). Microbiological B12 assay
Quantification of extracellular cobamides in pure and co-cultures was performed using a microbiological assay with Lactobacillus delbrueckii subsp. lactis (ATCC 7830) as the cobamide-auxotroph test organism following described procedures [15]. To detect intracellular cobamides, whole cells from 1.5 ml S. ovata or Sporomusa sp. strain KB-1 cultures were collected by centrifugation at 14 000 g for 2 min, suspended in sterile water, and broken either by heating at 95°C for 15 min or by sonicating with a Branson 250 Sonifier Analog Cell Disruptor (Branson Ultrasonics Corp., Danbury, CT, USA) for 15 min at 50 per cent duty cycle at an output of 60 W.
(ii). Total cobamide extraction and purification
Total intracellular cobamides were extracted and purified using an established protocol with the following modifications [25]. Sporomusa ovata or Sporomusa sp. strain KB-1 cultures (1 litre each) grown with H2/CO2 were centrifuged at 15 000g for 15 min at 4°C. Following removal of the supernatants, each cell pellet was suspended in 5 ml deionized water, the pH was adjusted to 5–6 with 3 per cent (v/v) glacial acetic acid, and KCN (100 mM stock solution) was added to reach a final concentration of 10 mM. The suspensions were vigorously shaken, each transferred to sterile 50 ml plastic tubes and incubated for 20 min in a boiling water bath. Following centrifugation at 15 000g for 15 min, the supernatants were collected and the pellets were extracted a second time. The combined supernatants were mixed with 0.01 volumes of a 3 per cent acetic acid solution prior to loading onto a Sep-Pak C18 cartridge (Waters Corp, Milford, MA, USA), which had been equilibrated with 2 ml 100 per cent methanol and 40 ml deionized water. Following sample loading, the cartridge was washed with 10 ml deionized water (20 interstitial volumes) and 7.5 ml 10 per cent methanol (15 interstitial volumes) prior to the final elution step with 10 ml 50 per cent methanol (20 interstitial volumes). The slightly pink-coloured solution obtained from the final elution step was vacuum dried and the residues were suspended in 0.5 ml sterile, deionized water.
(iii). HPLC analysis of cyanocobamides
Cyanocobamides were analysed using an Agilent 1200 series HPLC system equipped with an Eclipse XDB-C18 column (5 µm, 4.6 × 150 mm) and a diode array detector at a detection wavelength of 361 nm. Samples (20 µl each) were injected and separated at 30°C at a flow rate of 1.0 ml min−1 using 0.1 per cent acetic acid in water (eluent A) and 0.1 per cent acetic acid in methanol (eluent B) as mobile phases. The initial ratio of 90 per cent A/10 per cent B was linearly decreased to 35 per cent A/65 per cent B over a 12 min time window followed by a 4 min hold, before equilibration with 90 per cent A. CN-Cbl was identified by comparing retention times and absorption spectra with authentic CN-Cbl standards (more than 98% purity, Fisher Scientific, Pittsburgh, PA, USA) dissolved in deionized water.
(e). Other analytical methods
Methane, ethene and chlorinated ethenes were quantified using an Agilent 7890 gas chromatograph equipped with a flame ionization detector and a DB-624 capillary column (60 m × 0.32 mm × 1.8 µm) as described [22]. Acetate was analysed using an Agilent 1200 series HPLC system equipped with an Aminex HPX-87H column (Bio-Rad, Hercules, CA, USA). Samples were acidified with 1 M H2SO4 in a ratio of 19 : 1 (v/v) and separated with 4 mM aqueous H2SO4 as the mobile phase at a flow rate of 0.6 ml min−1 and quantified using a UV detector set to 210 nm.
3. Results
(a). CN-Cbl-dependent reductive dechlorination and growth of D. mccartyi strains
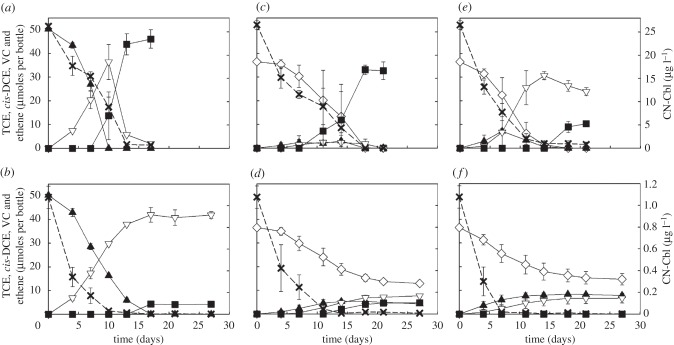
In defined medium amended with 25 µg l−1 CN-Cbl, strain BAV1 and strain GT cultures produced stoichiometric amounts of ethene from 50.3 ± 0.4 µmoles of cis-DCE (figure 2a) and 36.5 ± 0.9 µmoles of TCE (figure 2c) provided as electron acceptor, respectively. Strain FL2 cultures produced a mixture of VC and ethene (23.9 ± 1.7 and 10.3 ± 0.9 µmoles, respectively) during a 3-week incubation period (figure 2e). Cultures that received 1 µg l−1 CN-Cbl exhibited lower dechlorination rates and incomplete dechlorination. Strain BAV1 cultures dechlorinated cis-DCE (50.3 ± 0.4 µmoles) to VC (42.0 ± 3.1 µmoles) and small amounts of ethene (4.3 ± 0.4 µmoles) by day 17. VC remained the major dechlorination product (90.8 ± 0.1%) after 27 days and ethene accounted for 9.2 ± 0.1% (figure 2b). In strain GT cultures, more than one third of the initial TCE amount (36.5 ± 0.9 µmoles) remained after 27 days of incubation, and cis-DCE (4.3 ± 0.2 µmoles), VC (7.7 ± 0.7 µmoles) and ethene (4.7 ± 1.1 µmoles) were detected (figure 2d). TCE was dechlorinated at a rate of 9.0 ± 1.1 µmoles l−1 d−1 or about 2.2-fold slower compared with the TCE dechlorination rates (19.6 ± 0.2 µmoles l−1 d−1) observed in strain GT cultures that received 25 µg l−1 CN-Cbl (figures 2c,d). Similarly, strain FL2 cultures dechlorinated TCE at lower rates (i.e. 9.8 ± 2.3 versus 22.2 ± 0.8 µmoles l−1 d−1 in medium amended with 25 µg l−1 CN-Cbl), and after a 4-week incubation period, 40.3 ± 6.8% of the initial TCE was still present with cis-DCE (7.9 ± 0.4 µmoles) and VC (6.8 ± 2.1 µmoles) as the dechlorination products but no ethene was formed (figure 2f).
Figure 2.
Dechlorination of TCE or cis-DCE by Dehalococcoides under CN-Cbl limiting and non-limiting conditions. Strain BAV1 cultures were supplied with cis-DCE as electron acceptor and (a) 25 µg l−1 CN-Cbl or (b) 1 µg l−1 CN-Cbl. Strain GT cultures were supplied with TCE as electron acceptor and (c) 25 µg l−1 CN-Cbl or (d) 1 µg l−1 CN-Cbl. Strain FL2 cultures were supplied with TCE as electron acceptor and (e) 25 µg l−1 CN-Cbl or (f) 1 µg l−1 CN-Cbl. Open diamond, TCE; filled triangle, cis-DCE; open inverted triangle, vinyl chloride; filled square, ethene; cross symbol, CN-Cbl. Error bars represent the standard deviations of triplicate samples and are not shown if they are smaller than the symbols.
CN-Cbl concentrations in the medium decreased concomitantly with reductive dechlorination and Dehalococcoides growth, and the initial CN-Cbl concentration of 1 µg l−1 declined to below 20 ng l−1 during the 18-day incubation period (figure 2b,d,f). By day 27, reductive dechlorination of the remaining chlorinated ethenes ceased, and the CN-Cbl concentrations in strain GT, strain FL2 and strain BAV1 cultures were 7.7 ± 2.0 ng l−1, below 5 ng l−1 (i.e. below the quantification limit of the microbiological B12 assay), and below 2 ng l−1 (i.e. below the detection limit), respectively. CN-Cbl was depleted at higher rates in the 25 µg l−1 CN-Cbl-amended Dehalococcoides cultures. At day 17, when BAV1 cultures had completely dechlorinated cis-DCE to ethene, CN-Cbl concentrations dropped to 0.7 ± 0.1 µg l−1, averaging an uptake rate of 1500 ng l−1 d−1 versus 63 ng l−1 d−1 in the BAV1 cultures amended with 1 µg l−1 CN-Cbl (figure 2a,b). Strain GT and strain FL2 cultures depleted CN-Cbl from an initial amount of 26.5 ± 1.0 µg l−1 to 0.03 ± 0.03 and 1.0 ± 0.7 µg l−1, respectively, at 25- and 24-fold higher rates compared with those rates observed in the corresponding cultures amended with 1 µg l−1 CN-Cbl (figure 2c–f).
qPCR analysis demonstrated that limiting amounts of 1 µg l−1 CN-Cbl affected Dehalococcoides growth yields, which were calculated as described [22]. For example, 1.4 ± 0.8 × 107 BAV1 cells per µmole of Cl− released were obtained in cultures amended with 1 µg l−1 CN-Cbl, whereas 1.3 ± 0.1 × 108 BAV1 cells per µmole of Cl− released (a 9.1-fold increase) were measured in cultures amended with 25 µg l−1 CN-Cbl (table 1). Similar results were observed in strain GT cultures with an average 2.8-fold higher growth yield in medium amended with 25 µg l−1 CN-Cbl (table 1).
Table 1.
Comparison of Dehalococcoides mccartyi growth yields in axenic and in co-cultures amended with DMB.
| culture | CN-Cbl (µg l−1) | DMBa |
D. mccartyi 16S rRNA gene copies per ml |
growth yield (cells per µmole Cl− released) | ||
|---|---|---|---|---|---|---|
| inoculum | final | fold increasee | ||||
| axenic cultures | ||||||
| BAV1 | 1 | − | 6.3 ± 2.0 × 106 | 1.3 ± 0.4 × 107b | 2.1 | 1.4 ± 0.8 × 107 |
| 25 | − | 6.3 ± 2.0 × 106 | 1.3 ± 0.2 × 108c | 20.3 | 1.3 ± 0.1 × 108 | |
| GT | 1 | − | 4.0 ± 0.5 × 106 | 2.1 ± 0.3 × 107b | 5.4 | 5.3 ± 1.3 × 107 |
| 25 | − | 4.0 ± 0.5 × 106 | 1.5 ± 0.1 × 108c | 36.5 | 1.5 ± 0.3 × 108 | |
| FL2 | 1 | − | 2.4 ± 0.3 × 106 | 2.3 ± 0.7 × 106b | 1.0 | —f |
| 25 | − | 2.4 ± 0.3 × 106 | 3.7 ± 1.1 × 107c | 15.3 | 4.4 ± 1.5 × 107 | |
| co-cultures | ||||||
| BAV1/KB-1 | − | + | 6.3 ± 2.0 × 106 | 1.9 ± 0.1 × 108d | 29.8 | 2.1 ± 0.2 × 108 |
| BAV1/Fusaro | − | + | 6.3 ± 2.0 × 106 | 2.0 ± 0.1 × 108c | 30.9 | 2.1 ± 0.2 × 108 |
| GT/KB-1 | − | + | 4.0 ± 0.5 × 106 | 2.1 ± 0.3 × 108d | 51.5 | 2.1 ± 0.3 × 108 |
| GT/Fusaro | − | + | 4.0 ± 0.5 × 106 | 1.6 ± 0 × 108c | 41.0 | 1.8 ± 0.1 × 108 |
| FL2/KB-1 | − | + | 2.4 ± 0.3 × 106 | 9.5 ± 1.6 × 107d | 39.6 | 9.0 ± 1.4 × 107 |
| FL2/Fusaro | − | + | 2.4 ± 0.3 × 106 | 9.0 ± 2.7 × 107c | 37.3 | 9.8 ± 2.1 × 107 |
aDMB supplied at 10 µM.
bBiomass collected at day 27.
cBiomass collected at day 21.
dBiomass collected at day 17.
eData averaged from triplicates.
fNo growth occurred.
(b). Cobamide production in methanogenic and acetogenic cultures
During a 43-day incubation period, extracellular cobamide concentrations gradually increased to 4.2 ± 0.1 µg l−1 in M. barkeri strain Fusaro cultures concomitantly with methane formation (figure 3a). No extracellular cobamides were detected in the acetate-producing S. ovata (figure 3b) and Sporomusa sp. strain KB-1 cultures, which was unexpected because Sporomusa spp. are known cobamide producers when grown via H2/CO2 reductive acetogenesis. Cobamides were also not detected in Sporomusa cell lysates suggesting that Sporomusa produced cobamide(s), to which the indicator organism L. delbrueckii subsp. lactis did not respond (i.e. the cobamide cannot be detected and quantified with the microbiological B12 assay).
Figure 3.
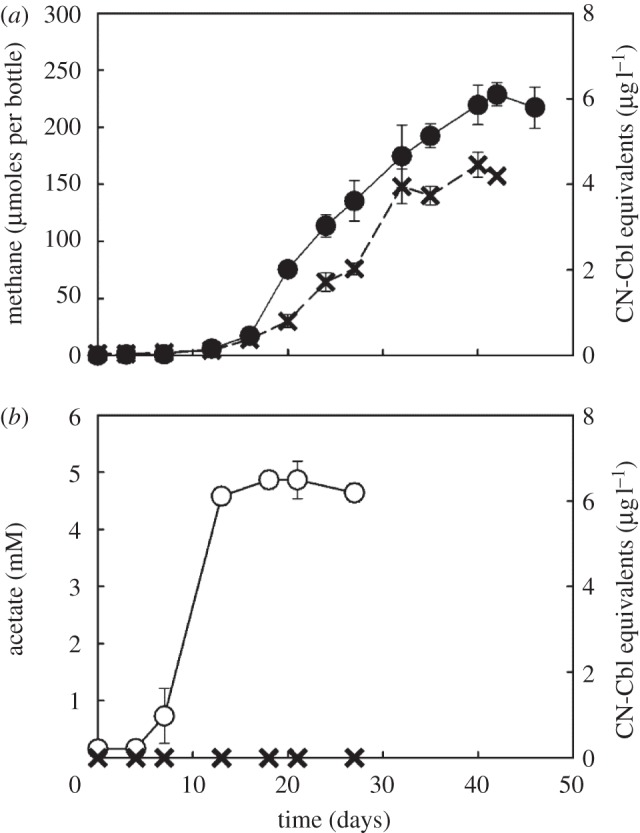
Extracellular cobamide production by Methanosarcina barkeri strain Fusaro and Sporomusa ovata during autotrophic growth with H2/CO2. (a) Methane and cobamide production by Methanosarcina barkeri strain Fusaro and (b) acetate production and lack of measureable cobamide(s) in Sporomusa ovata cultures. Filled circle, methane; open circle, acetate, cross symbol, cobamide(s) reported as CN-Cbl equivalents. Error bars represent the standard deviations of triplicate samples and are not shown if they are smaller than the symbols.
(c). Reductive dechlorination in co-cultures
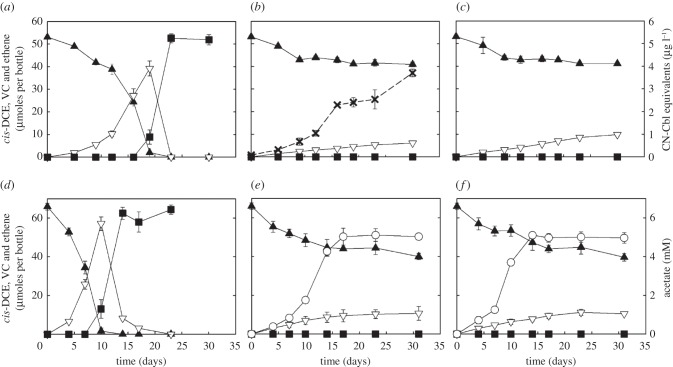
In D. mccartyi strain BAV1 and M. barkeri strain Fusaro co-cultures amended with 25 µg l−1 CN-Cbl, reductive dechlorination and methane formation occurred concomitantly. Strain BAV1 dechlorinated 53.1 ± 0.6 µmoles of cis-DCE to stoichiometric amounts of ethene with concomitant methane formation (86.9 ± 15.9 µmoles) by strain Fusaro within 25 days (figure 4a). In BAV1/Fusaro co-cultures not amended with CN-Cbl, methane was formed (225.2 ± 26.2 µmoles per bottle) and maximum amounts of 3.7 ± 0.2 µg l−1 cobamide were measured as CN-Cbl equivalents using the microbiological B12 assay in the culture suspensions after 30 days (figure 4b). Only negligible reductive dechlorination activity occurred in these co-cultures, and less than 12 per cent of the initial amount of cis-DCE was dechlorinated to VC (6.1 ± 0.1 µmoles). Negative controls (i.e. strain BAV1 cultures without CN-Cbl) produced 9.8 ± 0.9 µmoles of VC (figure 4c).
Figure 4.
Dechlorination and extracellular cobamide production in Dehalococcoides mccartyi strain BAV1/Methanosarcina barkeri strain Fusaro and strain BAV1/Sporomusa ovata co-cultures. (a) Strain BAV1/Methanosarcina barkeri strain Fusaro co-culture amended with 25 µg l−1 CN-Cbl (positive control); (b) strain BAV1/strain Fusaro co-culture without CN-Cbl addition; (c) strain BAV1 culture without CN-Cbl (negative control); (d) Strain BAV1/Sporomusa ovata co-culture amended with 25 µg l−1 CN-Cbl (positive control); (e) strain BAV1/Sporomusa ovata co-culture without CN-Cbl addition; (f) strain BAV1/Sporomusa sp. strain KB-1 co-culture without CN-Cbl addition. Filled triangle, cis-DCE; open inverted triangle, vinyl chloride; filled square, ethene; open circle, acetate; cross symbol, cobamide(s) reported as CN-Cbl equivalents. Error bars represent the standard deviations of triplicate samples and are not shown if they are smaller than the symbols.
Dehalococcoides/S. ovata co-cultures amended with 25 µg l−1 CN-Cbl exhibited acetate formation concomitantly with complete reductive dechlorination of 65.9 ± 1.7 µmoles of cis-DCE to ethene (figure 4d), indicating growth of both populations. Both S. ovata and Sporomusa sp. strain KB-1 grew in co-cultures lacking CN-Cbl but extracellular cobamides were not detected using the microbiological B12 assay, and only 10.6 ± 3.6 and 10.4 ± 0.5 µmoles of VC were produced in BAV1/S. ovata and BAV1/Sporomusa sp. strain KB-1 co-cultures, respectively (figure 4e,f). These findings demonstrate that Sporomusa sp. were unable to support BAV1 reductive dechlorination activity. Activities of D. mccartyi strains GT and FL2 were also tested in the co-cultures with M. barkeri strain Fusaro, S. ovata and Sporomusa sp. strain KB-1. Only insignificant dechlorination of TCE to cis-DCE (less than 0.5% of the initial amount of TCE) occurred in these co-cultures (data not shown).
(d). Guided biosynthesis of cobalamin in Sporomusa
The CN-Cbl standard (5 mg l−1) eluted at a retention time of 6.94 min (figure 5a). HPLC separation of cyanocobamides extracted from Sporomusa sp. strain KB-1 (figure 5b) and S. ovata (data not shown) cells collected from cultures not amended with DMB yielded two major peaks with retention times of 9.80 and 11.25 min, matching previous reports that two types of cobamides, phenolyl-cobamide and p-cresolyl-cobamide, were synthesized by S. ovata [26]. The addition of 10 µM DMB to the culture medium did not affect growth, and Sporomusa sp. strain KB-1 cultures reached similar OD 600 nm values of 0.040–0.041 (data not shown). Distinct cyanocobamide patterns were observed in Sporomusa sp. strain KB-1 cultures amended with DMB. The peak areas corresponding to phenolyl-cobamide and p-cresolyl-cobamide both decreased by about 80 per cent, and a major peak with a retention time of 6.94 min, identical to that of authentic CN-Cbl, emerged (figure 5c). These findings provide evidence for guided biosynthesis [27], and cyanocobalamin was the major cobamide when 10 µM DMB was added to the growth medium.
Figure 5.
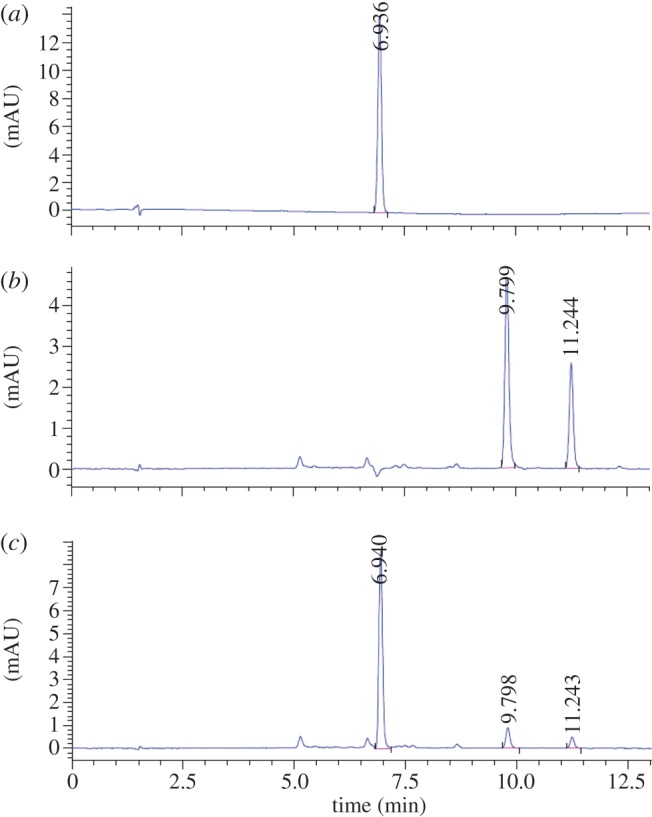
Demonstration of guided cobalamin biosynthesis. HPLC chromatograms of (a) authentic CN-Cbl standard (5 mg l−1 dissolved in water); (b) cyano-phenolyl and cyano-p-cresolyl cobamides extracted from 1 l of Sporomusa sp. strain KB-1 culture grown autotrophically with H2/CO2; and (c) cyanocobalamin extracted from 1 litre of Sporomusa sp. KB-1 culture grown autotrophically with H2/CO2 and the addition of 10 µM DMB. mAU = milliabsorbance units. (Online version in colour.)
(e). Recovery of Dehalococcoides dechlorination activity in DMB-amended co-cultures
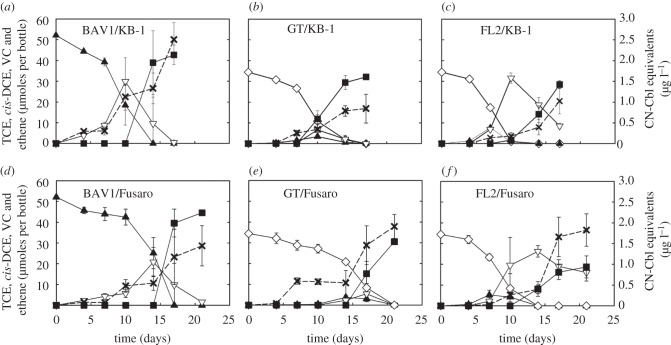
The addition of 10 µM DMB to non-dechlorinating Dehalococcoides/Sporomusa sp. strain KB-1 co-cultures stimulated reductive dechlorination activity. DMB-amended BAV1/KB-1 and GT/KB-1 co-cultures completely dechlorinated cis-DCE (52.1 ± 0.5 µmoles) and TCE (34.4 ± 0.7 µmoles) to ethene, respectively (figure 6a,b). FL2/KB-1 co-cultures reduced the initial TCE amount to VC (8.5 ± 1.2 µmoles) and ethene (28.5 ± 2.0 µmoles) (figure 6c). Concomitant with cis-DCE or TCE dechlorination, extracellular cobamides measured using the microbiological B12 assay after 17 days of incubation gradually increased from below the detection limit (i.e. less than 2 ng l−1) to 2.5 ± 0.4, 0.9 ± 0.4 and 1.0 ± 0.3 µg l−1 in strain BAV1/strain KB-1, strain GT/strain KB-1 and strain FL2/strain KB-1 co-cultures, respectively (figure 6a–c).
Figure 6.
Reductive dechlorination activity and extracellular cobamide production in Dehalococcoides/Sporomusa sp. strain KB-1 and Dehalococcoides/Methanosarcina barkeri strain Fusaro co-cultures amended with 10 µM DMB. (a) BAV1/KB-1 co-cultures; (b) GT/KB-1 co-cultures; (c) FL2/KB-1 co-cultures; (d) BAV1/Fusaro co-cultures; (e) GT/ Fusaro co-cultures and (f) FL2/ Fusaro co-cultures. Open diamond, TCE; filled triangle, cis-DCE; open inverted triangle, vinyl chloride; filled square, ethene; cross symbol, cobamide(s) reported as CN-Cbl equivalents. Error bars represent the standard deviations of triplicate samples and are not shown if they are smaller than the symbols.
Guided cobalamin biosynthesis through lower ligand exchange with DMB was also observed in Dehalococcoides/M. barkeri strain Fusaro co-cultures (figure 6d–f). Methane formation occurred and extracellular cobamide concentrations increased to 1.4–1.9 µg l−1. In addition, cis-DCE and TCE were completely dechlorinated to ethene in the strain BAV1 and strain GT co-cultures, respectively, and strain FL2 co-cultures dechlorinated the initial TCE amount (34.4 ± 0.7 µmoles) of VC (16.0 ± 4.1 µmoles) and ethene (18.8 ± 5.3 µmoles).
Negligible Dehalococcoides growth was observed in the co-cultures without DMB. In contrast, Dehalococcoides cell densities increased 29.8- to 51.5-fold in the DMB-amended co-cultures with Sporomusa sp. strain KB-1. Strain BAV1 cells increased from 6.3 ± 2.0 × 106 cells ml−1 (cells introduced with the inoculum) to 1.9 ± 0.1 × 108 cells ml−1, strain GT cells increased from 4.0 ± 0.5 × 106 to 2.1 ± 0.3 × 108 cells ml−1, and strain FL2 cells increased from 2.4 ± 0.3 × 106 to 9.5 ± 1.6 × 107 cells ml−1. In the DMB-amended M. barkeri strain Fusaro co-cultures, D. mccartyi strain BAV1, strain GT and strain FL2 cell densities increased 31-, 41- and 37-fold to reach 2.0 ± 0.1 × 108, 1.6 ± 0.0 × 108 and 9.0 ± 2.7 × 107 cells ml−1, respectively (table 1).
4. Discussion
CN-Cbl is a required cofactor for the characterized D. mccartyi strains grown in axenic culture [2]. The responses of D. mccartyi strain 195 to different CN-Cbl concentrations in the medium revealed that maximum TCE dechlorination rates and growth yields required CN-Cbl of 25 µg l−1 or higher concentrations [13]. In cultures of D. mccartyi strains BAV1, GT and FL2 amended with 1 µg l−1 CN-Cbl, CN-Cbl was quickly depleted to below 10 ng l−1, and TCE and cis-DCE dechlorination was incomplete. Ethene formation, about 2-fold higher dechlorination rates, and 2.8–9.1-fold increases in Dehalococcoides growth yields were observed in cultures that received 25 µg l−1 CN-Cbl. These findings demonstrate that Dehalococcoides' requirement for this essential cofactor must be understood to achieve and sustain desirable reductive dechlorination rates and endpoints at sites impacted with chloroorganic contaminants. Reductive dechlorination and growth of D. mccartyi strain BAV1 and strain FL2 were sustained in co-cultures with the PCE-to-cis-DCE dechlorinating species G. lovleyi strain SZ, which possesses the complete cobamide biosynthesis pathway [15]. The cobamide concentrations in these dechlorinating co-cultures ranged from 10 to 30 ng l−1, suggesting that the cobamide flux may be the relevant factor to sustain Dehalococcoides' dechlorination activity in microbial communities. Genes encoding the prokaryotic BtuBFCD corrinoid uptake system, which belongs to the ABC-type (ATP-binding cassette) transporter family, were identified in all sequenced Dehalococcoides genomes [11]. In vitro experiments demonstrated that the outer membrane corrinoid receptor BtuB of Escherichia coli had a high affinity for CN-Cbl with a half-saturation concentration of 0.5 nM (0.68 µg l−1) [28]. Dehalococcoides reductive dechlorination activity was sustained in D. mccartyi/G. lovleyi co-cultures with cobamide concentrations of 10–30 ng l−1, suggesting that D. mccartyi strains also possess high affinity cobamide uptake systems.
A crucial question is the source of the corrinoid cofactor to sustain Dehalococcoides reductive dechlorination activity in the environment, particularly in chlorinated solvent-contaminated aquifers. The Dehalococcoides-containing consortium KB-1 used for bioaugmentation contains methanogens and CO2-reducing acetogens [29]. Both methanogenic Archaea and acetogenic Bacteria are known to synthesize cobamides with different lower ligands (figure 1). 5′-Hydroxybenzimidazolyl-cobamide (factor III) and adenyl-cobamide (pseudo vitamin B12) are the predominant cobamides isolated from methanogenic Archaea [30], and factor III was identified in M. barkeri cultures [31]. In Dehalococcoides co-cultures with M. barkeri strain Fusaro, more than 4 µg l−1 CN-Cbl equivalents, presumably factor III, were quantified in culture suspensions; however, only negligible reductive dechlorination activity, similar to negative control cultures, was observed, indicating that factor III cannot fulfil Dehalococcoides' corrinoid requirement. These findings are supported by a previous study that reported that D. mccartyi strain 195 reductive dechlorination activity was not enhanced in co-cultures with Methanobacterium congolense [14]. Pure culture studies with D. mccartyi strain 195 corroborated that purified factor III and phenolic cobamides did not support reductive dechlorination, and growth only occurred when cobamides containing benzimidazolyl types of lower ligands were available [32]. Although the specific cobamides produced by Methanobacterium congolense and many other methanogens have not been thoroughly explored, the data reported here confirm that at least the 5′-hydroxybenzimidazolyl-cobamide produced by some methanogens cannot fulfil the D. mccartyi corrinoid cofactor requirement.
Cobamides containing a phenolic-type lower ligand, i.e. phenolyl- and p-cresolyl-cobamide, were first purified from S. ovata, and were found in other Bacteria capable of H2/CO2 reductive acetogenesis [26]. Sporomusa sp. strain KB-1 was isolated from the dechlorinating KB-1 consortium and shares 98.5 per cent sequence identity with the 16S rRNA gene of S. ovata (AJ279800.1). Both Sporomusa isolates produced phenolyl- and p-cresolyl-cobamide, suggesting that they represent the characteristic cobamides of the genus Sporomusa.
The test organism for the microbiological B12 assay, L. delbrueckii, did not respond to S. ovata or Sporomusa sp. strain KB-1 culture suspensions or cell lysates, indicating that the cobamides with phenolic lower ligands cannot fulfil the corrinoid cofactor requirement of L. delbrueckii. Many prokaryotes, including D. mccartyi strains and L. delbrueckii, use adenosyl-cobalamin-dependent class II ribonucleotide reductases (RNRs), which catalyse the conversion of ribonucleotides to deoxyribonucleotides [11]. Since D. mccartyi genomes harbour multiple copies of class II RNR genes, it is possible that the lack of dechlorination activity and Dehalococcoides growth in the Sporomusa co-cultures was not only caused by non-functional RDases but also because of non-functional RNRs.
Interestingly, Dehalococcoides dechlorination activity and growth occurred in the DMB-amended co-cultures. In the presence of 10 µM DMB, CN-Cbl was the predominant corrinoid extracted from Sporomusa sp. strain KB-1 cultures, indicating that guided biosynthesis allows Sporomusa sp. to generate a corrinoid cofactor that fulfils Dehalococcoides' nutritional requirements. Similar observations were made in D. mccartyi/G. sulfurreducens co-cultures that were amended with DMB [15], suggesting that guided biosynthesis may be a relevant process for supplying D. mccartyi strains with the required cobalamin cofactor.
In DMB-amended Sporomusa sp. strain KB-1 co-cultures with each of the D. mccartyi strains BAV1, GT and FL2, the dechlorinators attained population sizes of 2.1 ± 0.2 × 108, 2.1 ± 0.3 × 108 and 9.0 ± 1.4 × 107 cells per µmole of chloride released, respectively. These cell numbers were 1.7-, 1.4- and 2.2-fold higher compared with axenic cultures that received 25 µg l−1 CN-Cbl. Dehalococcoides mccartyi strains are known to grow better in mixed cultures, presumably because community members supply Dehalococcoides with unknown growth factors. Hence, it is possible that M. barkeri strain Fusaro and the Sporomusa isolates provided the D. mccartyi strains with not yet identified compounds that enhance growth yields. Another possible explanation for the lower growth yields observed with CN-Cbl is the cell's energetic burden associated with the replacement of the upper cyanide ligand [33].
The microbial synthesis of non-native cobamides in the presence of excess non-native lower ligand is known as guided biosynthesis [17,27,34]. Guided biosynthesis has been demonstrated with model organisms such as Propionibacterium shermanii, E. coli and Streptomyces griseus to obtain desired cobamides [17]. Factor III synthesized by Methanobacterium thermoautotrophicum was replaced by a cobalamin in medium amended with DMB without affecting methane formation and growth [35]. Synthesis of a cobalamin in the presence of DMB has also been demonstrated in M. barkeri cultures [36]. The co-culture experiments with D. mccartyi strains demonstrated that the addition of DMB to the medium enabled guided biosynthesis and the formation of a cobalamin by M. barkeri strain Fusaro and Sporomusa sp. strain KB-1. Sporomusa sp. strain KB-1 replaced the phenolic cobamides with a cobalamin without apparent effects on its own growth suggesting that cobalamin can replace the phenolyl- and p-cresolyl-cobamides during growth with H2/CO2.
Survival of corrinoid auxotrophs such as Dehalococcoides relies on cobamides and corrinoid precursors (e.g. cobinamides) produced and released by corrinoid-synthesizing Archaea and Bacteria. It is currently unclear to what extent and under what conditions microorganisms capable of de novo corrinoid synthesis produce and release cobamides and/or corrin ring precursors to the surrounding medium. While methanogens and acetogens readily release cobamides into the medium, at least under laboratory cultivation conditions, the formation of cobalamin requires DMB. Key questions are the origin of DMB in anoxic environments (i.e. who is synthesizing DMB?), and whether the availability of DMB limits cobalamin biosynthesis and Dehalococcoides activity. The biosynthesis pathway(s) of DMB under anoxic conditions has remained elusive, and the flux of DMB in subsurface environments has not been evaluated. While our experiments have demonstrated cobalamin production through guided biosynthesis, the relevance of this process in natural microbial communities has yet to be demonstrated. All D. mccartyi strains harbour cobT, cobC and cobS genes implicated in the cobalamin assembly from DMB and cobinamide [37], suggesting Dehalococcoides has the ability to assemble cobalamin if the precursors are available. Independently of whether Dehalococcoides or other microbes perform the final cobalamin assembly, the sources and fluxes of its precursors, in particular DMB, need to be understood in natural and disturbed microbial communities to identify Dehalococcoides' nutritional limitations and potentially predict and manipulate reductive dechlorination activity. Limited dechlorination activity of Dehalococcoides and related corrinoid-auxotroph, organohalide-respiring Chloroflexi (e.g. Dehalogenimonas [38,39], ‘Dehalobium’ [40]) is regarded as a major constraint to achieve detoxification at sites contaminated with chlorinated contaminants. Hence, elucidating the nutritional requirements and ecophysiologies of organohalide-respiring Chloroflexi in natural and contaminated environments will help develop predictive understanding and new strategies for stimulating in situ reductive dechlorination activity.
Acknowledgements
We thank Dr Kevin Sowers, University of Maryland, for providing Methanosarcina barkeri strain Fusaro, and Dr Kelly Nevin, University of Massachusetts, for providing Sporomusa ovata. We are grateful to Drs Elizabeth Edwards and Laura Hug, University of Toronto, for providing the Sporomusa sp. strain KB-1 culture. This work was supported by the Strategic Environmental Research and Development Program (SERDP) and the National Science Foundation (NSF).
References
- 1.Abelson PH. 1990. Inefficient remediation of ground-water pollution. Science 250, 733. 10.1126/science.2237418 (doi:10.1126/science.2237418) [DOI] [PubMed] [Google Scholar]
- 2.Löffler FE, et al. 2012. Dehalococcoides mccartyi gen. nov., sp. nov., obligate organohalide-respiring anaerobic bacteria, relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidetes classis nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. In Press 10.1099/ijs.0.034926-0 (doi:10.1099/ijs.0.034926-0) [DOI] [PubMed] [Google Scholar]
- 3.Maymó-Gatell X, Chien Y-t, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276, 1568–1571 10.1126/science.276.5318.1568 (doi:10.1126/science.276.5318.1568) [DOI] [PubMed] [Google Scholar]
- 4.Adrian L, Szewzyk U, Wecke J, Görisch H. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408, 580–583 10.1038/35046063 (doi:10.1038/35046063) [DOI] [PubMed] [Google Scholar]
- 5.Roth JR, Lawrence JG, Bobik TA. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181 10.1146/annurev.micro.50.1.137 (doi:10.1146/annurev.micro.50.1.137) [DOI] [PubMed] [Google Scholar]
- 6.Marsh ENG. 1999. Coenzyme B12 (cobalamin)-dependent enzymes. Essays Biochem. 34, 139–154 [DOI] [PubMed] [Google Scholar]
- 7.Maillard J, Schumacher W, Vazquez F, Regeard C, Hagen WR, Holliger C. 2003. Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69, 4628–4638 10.1128/AEM.69.8.4628-4638.2003 (doi:10.1128/AEM.69.8.4628-4638.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller E, Wohlfarth G, Diekert G. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169, 497–502 10.1007/s002030050602 (doi:10.1007/s002030050602) [DOI] [PubMed] [Google Scholar]
- 9.Neumann A, Wohlfarth G, Diekert G. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271, 16 515–16 519 10.1074/jbc.271.28.16515 (doi:10.1074/jbc.271.28.16515) [DOI] [PubMed] [Google Scholar]
- 10.Kräutler B, et al. 2003. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of natural corrinoid. Helv. Chim. Acta 86, 3698–3716 10.1002/hlca.200390313 (doi:10.1002/hlca.200390313) [DOI] [Google Scholar]
- 11.Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. 2009. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genom. 10, 78. 10.1186/1471-2164-10-78 (doi:10.1186/1471-2164-10-78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DR, Nemir A, Andersen GL, Zinder SH, Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol. Lett. 294, 198–206 10.1111/j.1574-6968.2009.01569.x (doi:10.1111/j.1574-6968.2009.01569.x) [DOI] [PubMed] [Google Scholar]
- 13.He J, Holmes VF, Lee PKH, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73, 2847–2853 10.1128/AEM.02574-06 (doi:10.1128/AEM.02574-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Men Y, et al. 2012. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J. 6, 410–421 10.1038/ismej.2011.111 (doi:10.1038/ismej.2011.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J, Ritalahti KM, Wagner DD, Löffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78, 6630–6636 10.1128/AEM.01535-12 (doi:10.1128/AEM.01535-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan CH, Escalante-Semerena JC. 2011. ArsAB, a novel enzyme for Sporomusa ovata activates phenolic bases for adenosylcobamide biosynthesis. Mol. Micro. 81, 952–967 10.1111/j.1365-2958.2011.07741.x (doi:10.1111/j.1365-2958.2011.07741.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider Z, Stroiński A. 1987. Comprehensive B12: chemistry, biochemistry, nutrition, ecology, medicine. Berlin, Germany: Walter de Gruyter & Co [Google Scholar]
- 18.Stupperich E, Eisinger HJ, Schurr S. 1990. Corrinoids in anaerobic bacteria. FEMS Microbiol. Rev. 87, 355–360 10.1111/j.1574-6968.1990.tb04936.x (doi:10.1111/j.1574-6968.1990.tb04936.x) [DOI] [Google Scholar]
- 19.Stupperich E, Eisinger HJ, Kräutler B. 1988. Diversity of corrinoids in acetogenic bacteria: p-cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicoaceticum and vitamin B12 from Acetobacterium woodii. Eur. J. Biochem. 172, 459–464 10.1111/j.1432-1033.1988.tb13910.x (doi:10.1111/j.1432-1033.1988.tb13910.x) [DOI] [PubMed] [Google Scholar]
- 20.Kräutler B, Kohler HPE, Stupperich E. 1988. 5′-Methylbenzimidazolyl-cobamides are the corrinoids from some sulfate-reducing and sulfur-metabolizing bacteria. Eur. J. Biochem. 176, 461–469 10.1111/j.1432-1033.1988.tb14303.x (doi:10.1111/j.1432-1033.1988.tb14303.x) [DOI] [PubMed] [Google Scholar]
- 21.He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424, 62–65 10.1038/nature01717 (doi:10.1038/nature01717) [DOI] [PubMed] [Google Scholar]
- 22.Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72, 1980–1987 10.1128/AEM.72.3.1980-1987.2006 (doi:10.1128/AEM.72.3.1980-1987.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7, 1442–1450 10.1111/j.1462-2920.2005.00830.x (doi:10.1111/j.1462-2920.2005.00830.x) [DOI] [PubMed] [Google Scholar]
- 24.Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Löffler FE. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72, 2765–2774 10.1128/AEM.72.4.2765-2774.2006 (doi:10.1128/AEM.72.4.2765-2774.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupperich E, Steiner I, Rühlemann M. 1986. Isolation and analysis of bacterial cobamides by high-performance liquid chromatography. Anal. Biochem. 155, 365–370 10.1016/0003-2697(86)90447-1 (doi:10.1016/0003-2697(86)90447-1) [DOI] [PubMed] [Google Scholar]
- 26.Stupperich E, Eisinger HJ, Kräutler B. 1989. Identification of phenolyl cobamide from the homoacetogenic bacterium Sporomusa ovata. Eur. J. Biochem. 186, 657–661 10.1111/j.1432-1033.1989.tb15256.x (doi:10.1111/j.1432-1033.1989.tb15256.x) [DOI] [PubMed] [Google Scholar]
- 27.Chandra T, Brown KL. 2008. Vitamin B12 and α-ribonucleosides. Tetrahedron 64, 9–38 10.1016/j.tet.2007.08.061 (doi:10.1016/j.tet.2007.08.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradbeer C, Kenley JS, Di Masi DR, Leighton M. 1978. Transport of vitamin B12 in Escherichia coli. Corrinoid specificities of the periplasmic B12-binding protein and of energy-dependent B12 transport. J. Biol. Chem. 253, 1347–1352 [PubMed] [Google Scholar]
- 29.Duhamel M, Mo K, Edwards EA. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70, 5538–5545 10.1128/AEM.70.9.5538-5545.2004 (doi:10.1128/AEM.70.9.5538-5545.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stupperich E, Kräutler B. 1988. Pseudo vitamin B12 or 5-hydroxybenzimidazolyl-cobamide are the corrinoids found in methanogenic bacteria. Arch. Microbiol. 149, 268–271 10.1007/BF00422016 (doi:10.1007/BF00422016) [DOI] [Google Scholar]
- 31.Pol A, van der Drift C, Vogels GD. 1982. Corrinoids from Methanosarcina barkeri: structure of the α-ligand. Biochem. Biophys. Res. Commun. 108, 731–737 10.1016/0006-291X(82)90890-7 (doi:10.1016/0006-291X(82)90890-7) [DOI] [PubMed] [Google Scholar]
- 32.Yi S, Seth EC, Men Y-J, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78, 7745–7752 10.1128/AEM.02150-12 (doi:10.1128/AEM.02150-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Gherasim C, Banerjee R. 2008. Decyanation of vitamin B12 by a trafficking chaperone. Proc. Natl Acad. Sci. USA 38, 14 551–14 554 10.1073/pnas.0805989105 (doi:10.1073/pnas.0805989105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen RH, Stabler SP. 2008. Identification and quantification of cobalamin and cobalamin analogues in human feces. Am. J. Clin. Nutr. 87, 1324–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stupperich E, Steiner I, Eisinger HJ. 1987. Substitution of Co-α(5-hydroxybenzimidazolyl) cobamide (factor III) by vitamin B12 in Methanobacterium thermoautotrophicum. J. Bacteriol. 169, 3076–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryzhkova EP, Brukhanov AL. 2009. Effect of a corrinoid on Methanosarcina barkeri DNA synthesis. Mikrobiologiya 78, 5–11 10.1134/S0026261709010019 (doi:10.1134/S0026261709010019) [DOI] [PubMed] [Google Scholar]
- 37.Scott AI, Roessner CA. 2002. Biosynthesis of cobalamin (vitamin B12). Biochem. Soc. Trans. 4, 613–620 10.1042/bst0300613 (doi:10.1042/bst0300613) [DOI] [PubMed] [Google Scholar]
- 38.Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA. 2009. Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 59, 2692–2697 10.1099/ijs.0.011502-0 (doi:10.1099/ijs.0.011502-0) [DOI] [PubMed] [Google Scholar]
- 39.Yan J, Rash BA, Rainey FA, Moe WM. 2009. Isolation of novel bacteria within the Chloroflexi capable of reductive dechlorination of 1,2,3-trichloropropane. Environ. Microbiol. 11, 833–843 10.1111/j.1462-2920.2008.01804.x (doi:10.1111/j.1462-2920.2008.01804.x) [DOI] [PubMed] [Google Scholar]
- 40.Wu Q, Watts JEM, Sowers KR, May H. 2002. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl. Environ. Microbiol. 68, 807–812 10.1128/AEM.68.2.807-812.2002 (doi:10.1128/AEM.68.2.807-812.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]