Abstract
Dehalococcoides mccartyi strains are strictly anaerobic organisms specialized to grow with halogenated compounds as electron acceptor via a respiratory process. Their genomes are among the smallest known for free-living organisms, and the embedded gene set reflects their strong specialization. Here, we briefly review main characteristics of published Dehalococcoides genomes and show how genome information together with cultivation and biochemical experiments have contributed to our understanding of Dehalococcoides physiology and biochemistry. We extend this approach by the detailed analysis of cofactor metabolism in Dehalococcoides strain CBDB1. Dehalococcoides genomes were screened for encoded proteins annotated to contain or interact with organic cofactors, and the expression of these proteins was analysed by shotgun proteomics to shed light on cofactor requirements. In parallel, cultivation experiments testing for vitamin requirements showed that cyanocobalamin (vitamin B12), thiamine and biotin were essential supplements and that cyanocobalamin could be substituted by dicyanocobinamide and dimethylbenzimidazole. Dehalococcoides genome analysis, detection of single enzymes by shotgun proteomics and inhibition studies confirmed the expression of the biosynthetic pathways for pyridoxal-5-phosphate, flavin nucleotides, folate, S-adenosylmethionine, pantothenate and nicotinic acids in strain CBDB1. Haem/cytochromes, quinones and lipoic acids were not necessary for cultivation or dechlorination activity and no biosynthetic pathways were identified in the genomes.
Keywords: proteomics, Dehalococcoides, comparative genomics, vitamin requirements, cofactors, biosynthesis
1. Introduction
(a). Dehalococcoides mccartyi
Dehalococcoides mccartyi strains are mesophilic, strictly anaerobic bacteria affiliated with the phylum Chloroflexi [1]. Several pure cultures have been isolated from anaerobic sludge or sediments, namely strains 195, CBDB1, FL2, BAV1, MB, VS, ANAS1, ANAS2 and DCMB5 [2–9]. Isolation strategies always exploited the ability of Dehalococcoides strains to grow by organohalide respiration, i.e. using halogenated aliphatic or aromatic compounds as a respiratory electron acceptor. None of the isolates has been shown to grow by respiration with non-halogenated electron acceptors or by fermentation. All described Dehalococcoides strains use hydrogen as an electron donor and, as shown in detail for strains 195 [10] and CBDB1 [11], acetate plus CO2/bicarbonate as carbon sources. Owing to these characteristics and to the fact that Dehalococcoides are regularly found in actively dechlorinating communities, Dehalococcoides are considered to be key organisms for remediation of soils and aquifers contaminated with halogenated ethenes or aromatics. Known Dehalococcoides cells have small disc-like morphologies and do not have a peptidoglycan cell wall. On the basis of their physiological, morphological and 16S rRNA gene similarity, all Dehalococcoides were recently described as members of one species, D. mccartyi [1]. With descriptions of Dehalogenimonas lykanthroporepellens [12,13] and ‘Dehalobium chlorocoercia’ strain DF-1 [14], two more obligate organohalide-respiring genera within the Chloroflexi have been identified. Similar to the Dehalococcoides, both are strictly anaerobic coccoid bacteria.
(b). Dehalococcoides genomics
Full genome sequences have been determined for the Dehalococcoides strains 195 [15], CBDB1 [16], BAV1, VS [17] and GT and for Dehalogenimonas lykanthroporepellens [18]. The genomes are relatively small with 1.34–1.47 Mbp for Dehalococcoides strains and 1.69 Mbp for Dehalogenimonas. While the genome of strain 195 contains a prophage, genomes of other Dehalococcoides strains do not. All Dehalococcoides genomes contain a variety of genes indicating genomic rearrangements and mobile elements, such as transposases, integrases or site-specific recombinases. Clustered regularly interspaced short palindromic repeats (CRISPR) elements are present in the genomes of strains CBDB1 and GT but not in the genomes of strains 195, BAV1, VS or Dehalogenimonas lykanthroporepellens. Whole genome comparison shows that most genes are strongly conserved between all sequenced Dehalococcoides strains and that extensive synteny exists throughout the genomes. Only two regions show massive gene variation [16]. These regions are located on both sides around the origin of replication (ORI), each starting about 50 000 bp away from the ORI, and were termed ‘high plasticity regions’ (HPRs) and analysed in detail [17]. These HPRs contain most of the reductive dehalogenase homologous genes (rdh genes), and associated genes predicted to be involved in the regulation of rdh transcription. All Dehalococcoides strains contain multiple rdh operons, amounting to numbers between 11 in strain BAV1 and 36 in strain VS. The high number of rdh genes close to the ORI, the strong bias for the location of rdh genes on the leading DNA strand, the high number of rdh paralogues in each strain and the close association with regulatory genes suggest a pivotal role of rdh genes in the metabolism of Dehalococcoides strains.
(c). Contribution of genome annotation to the understanding of physiology and biochemistry
The key enzyme, catalysing the reductive dehalogenation reaction in organohalide-respiring organisms, is a membrane-associated protein, RdhA. It contains a corrinoid in the active centre as indicated by inhibition studies [19] and biochemical studies with homologous proteins in bacteria of other genera (e.g. Sulfurospirillum multivorans [20]). In addition, RdhA proteins contain two iron–sulfur clusters as indicated by two iron–sulfur-cluster binding motifs in the protein sequence. The RdhA proteins in Dehalococcoides strains also contain a twin-arginine-transport signal peptide, indicating that the protein is transported through the membrane and is attached to the outside the cell, presumably via a small anchor protein, RdhB. Comparative genome annotations have revealed the presence of multiple non-redundant rdhA genes (more than 130 rdhA genes) in sequenced Dehalococcoides strains, and PCR analysis detected many hundreds more rdhA sequences related to Dehalococcoides rdhA genes in mixed cultures or complex environments, indicating the potential of Dehalococcoides to use a broad range of halogenated compounds as electron acceptors for respiration. Knowledge on reductive dehalogenases function is important to be able to use the sequences as biomarkers for field diagnosis in contaminated sites [21]. However, the substrate range of only five reductive dehalogenases from Dehalococcoides is currently known: a chlorobenzene reductive dehalogenase (CbrA) identified in strain CBDB1 [22]; tetrachloroethene (PCE) reductive dehalogenase (PceA) and trichloroethene (TCE) reductive dehalogenase (TceA) of strain 195 catalysing the reaction of PCE to TCE and TCE to ethene, respectively [23]; VcrA of strain VS with substrate specificity towards vinyl chloride [24,25]; and the PCE and TCE to trans-dichloroethene (DCE) reductive dehalogenase named MbrA in Dehalococcoides sp. MB [26]. The different dechlorination capacity among Dehalococcoides strains demonstrates that these strains are physiologically versatile although phylogenetically very close [8].
In contrast to the versatility with respect to halogenated electron acceptors, all Dehalococcoides isolates use exclusively hydrogen as electron donor. The importance of hydrogen in its physiology is exemplified by the presence of genes encoding five different hydrogenase complexes: one cytoplasmatic (Vhu) and four membrane-bound (Hup, Hyc, Ech, Hym) hydrogenases [15,16]. Among them, the Hup hydrogenase is of special interest because it is highly expressed at the transcriptional level in cultures containing PCE, whereas Ech and Hyc are hypothesized to generate low-potential electrons for biosynthesis rather than being involved with organohalide respiration [27]. Despite Dehalococcoides strains being strict hydrogenotrophs, genome sequences have revealed the presence of a gene coding for a predicted formate dehydrogenase (Fdh). Expression studies showed unexpected high levels of this gene in strain 195 and CBDB1 but also in Dehalococcoides-containing enrichment cultures under different growth conditions [27,28]. A closer look at the Fdh gene sequence revealed the presence of a serine residue at a key position in the active site of the enzyme instead of cysteine or selenocysteine coded in all known FdhA homologues, suggesting a novel function other than predicted for catabolic Fdh [28]. Regardless of its function, Fdh is an excellent candidate to serve as Dehalococcoides-specific indicator owing to its high expression and presence in all known Dehalococcoides strains [29].
An additional interesting finding from genome sequencing projects of Dehalococcoides isolates was the identification of a mannosylglycerate synthase (MGS), a bifunctional enzyme associated with osmotic stress response in thermophiles. By expressing the gene of Dehalococcoides strain 195, designated mgsD, in Escherichia coli and Saccharomyces cerevisiae, it was demonstrated that this enzyme catalyses MG synthesis at low temperatures [30]. The presence of MGS in strain 195 argues for a role in the osmotic adaptation of Dehalococcoides strains to salt stress and may reflect a horizontal gene transfer [30].
Energy and carbon metabolism is not interconnected in Dehalococcoides strains unlike in most other organisms as evidenced by physiological and genomic studies. The anabolism relies on acetate and carbonate/CO2 as carbon sources, whereas the catabolism is restricted to organohalide respiration with hydrogen and halogenated organics. The tricarboxylic acid (TCA) cycle is a central pathway in which anabolism and catabolism are normally coupled. Dehalococcoides strains, however, possess an incomplete TCA cycle with an oxidative half-cycle to 2-oxoglutarate and a reductive half-cycle to fumarate, which has been reported for many anaerobes [31,32]. Unlike aerobic organisms, the gene coding for a citrate synthase in Dehalococcoides is Re face-specific [10] and was recently identified in strain CBDB1 as cbdbA1708, previously annotated as homocitrate synthase [11]. The analysis of amino acid isotopologue distributions after feeding Dehalococcoides with 13C-labelled carbon sources, either by analysing free amino acids after total protein hydrolysis or by analysing full peptides after tryptic digest, has depicted amino acid biosynthetic pathways [10,33]. While uptake and incorporation of some amino acids into proteins was reported when growing strain 195 over several transfers on high concentrations of externally added amino acids in the medium [34], no such incorporation was detected with strain CBDB1 when amino acids were added at lower concentration for only one transfer [33].
(d). Cofactors and vitamins in Dehalococcoides strains
Organic cofactors play crucial roles in the catalysis of biochemical reactions in the metabolism of all living organisms. They are loosely or tightly (prosthetic groups) bound to enzymes and often participate directly in a reaction mechanism. In accordance with the multitude of reactions in which they participate, they are also structurally diverse. However, many cofactors are purine nucleotides (ATP, GTP) or contain them (FAD, NAD, CoA, MGD, S-adenosylmethionine, corrinoids), or purine nucleotides are involved in their biosynthesis (e.g. thiamine, tetrahydrofolate, molybdopterine, corrinoids, riboflavin). This has fuelled the concept that cofactors represent relicts of ancient biochemistry long before genomes and proteins had evolved [35].
To meet cofactor requirements, bacteria can catalyse de novo biosynthesis, take up stable precursor molecules or import the fully functional cofactors from the environment. For the cultivation of pure cultures, cofactors that are not de novo synthesized have to be supplied as vitamins with the medium, as fully functional cofactor or cofactor precursor. Here, we have analysed cofactor use and requirements in Dehalococcoides isolates by using five complementary approaches which have in common that they can be applied with cultures from which only low amounts of biomass can be recovered: (i) the comparative analysis of Dehalococcoides genome annotations for genes annotated to code for cofactor-containing/interacting proteins and genes involved in cofactor biosynthesis, (ii) expression analysis of cofactor-containing/interacting proteins and proteins involved in cofactor biosynthesis by nano-liquid chromatography–tandem mass spectrometry (nLC–MS/MS), (iii) cultivation experiments with or without selected vitamins, (iv) inhibition studies, targeting cofactor biosynthesis and (v) enzyme activity tests. All wet-laboratory experiments were carried out with Dehalococcoides strain CBDB1, which grows on purely synthetic mineral medium without the addition of any complex additions and therefore allows for precise cofactor requirement analysis.
2. Material and methods
(a). Chemicals
All chemicals were of analytical grade with the exception of Ti(III) citrate that was produced from technical grade Ti(III) chloride [6]. 5,6-Dimethylbenzimidazole (DMB) was obtained at 99 per cent purity from Alfa Aesar (Karlsruhe, Germany), dicyanocobinamide (DCC; 95% purity), trimethoprim and sulfamethoxazole were obtained from Sigma-Aldrich (Steinheim, Germany).
(b). Cultivation
Dehalococcoides mccartyi strain CBDB1 was grown under strictly anaerobic conditions in 100 ml glass serum bottles containing 60 ml of synthetic medium and 40 ml gas phase (80% N2/20% CO2). The basal medium contained Milli-Q water, minerals, trace elements (SL9 solution) and 5 mM acetate as a source of carbon. Hexachlorobenzene (HCB) in crystallized form (100 mg per serum bottle), 1,2,3 plus 1,2,4-trichlorobenzene, chlorophenols or bromobenzenes were added as electron acceptor as described previously [6,36], and the bottles were sterilized by autoclaving. The medium was then reduced with 1.6 mM Ti(III) citrate plus 30 µM Na2S, buffered to pH 7.0–7.2 with bicarbonate and amended with hydrogen as an electron donor (0.3 bar overpressure). The standard medium contained a vitamin solution with final concentrations of 40 nM cyanocobalamin, 150 nM thiamine chloride hydrochloride, 20 nM D(+)-biotin, 150 nM p-aminobenzoate, 410 nM nicotinic acid, 100 nM pantothenate and 360 nM pyridoxine hydrochloride, but this vitamin solution was substituted in the different experiments described in this study. When indicated, trimethoprim (14 µM), sulfamethoxazole (395 µM), folic acid (45 nM) or avidin (12 µM) were added. All cultures were set up in five identical parallels and inoculated to a final cell density of 1–5 × 106 cells ml−1. To remove unwanted compounds from the inoculum, cells from 200 ml of culture were harvested under strictly anaerobic conditions on a 0.2 µm filter, washed twice with 5 ml of supplement-free reduced medium and resuspended in 20 ml of fresh medium before injecting into the cultures. Cell numbers were determined by direct cell counting of SYBR-green labelled cells on agarose-covered slides using a highly standardized procedure and self-developed cell-counting software [37]. The counting allows a fast and reliable determination of cell numbers between 106 and 108 cells ml−1 with a standard deviation of about 10 per cent when cells are above 107 cells ml−1 as determined from counting dilution series. Dechlorination products from HCB were quantified by extracting 1 ml samples with 1 ml of hexane and analysing the extracts by gas chromatography and flame ionization detection (GC–FID) as described [6].
(c). Whole cell activity assay for reductive dechlorination
Reductive dehalogenase activity in whole cell suspensions of strain CBDB1 was measured as reported previously [19], using 1,2,3,4-tetrachlorobenzene (1,2,3,4-TeCB) as terminal electron acceptor. The assay was conducted in 100 mM potassium acetate (pH 5.8), 2 mM Ti(III) citrate, 500 µM 1,2,3,4-TeCB (added from 50 mM acetonic stock solution) and 1 mM electron donor. As electron donor either methyl viologen, menadione, menaquinone MK-4 (vitamin K2), ubiquinone-0 (Q-0), ubiquinone-4 (Q-4) or ubiquinone-10 (Q-10) was used from 100 mM acetonic stock solutions. The reaction was started by the addition of 200 µl of whole cell suspension and incubated for 1 h at 25°C. If necessary, cells were concentrated via filtration on a 0.2 µm filter. Dechlorination products were analysed by GC–FID [6]. Protein concentrations were determined using a fluorescamine protein assay (Thermo Fisher Scientific, Bremen, Germany).
(d). Protein expression analysis
Protein expression was analysed from 30 ml cultures of D. mccartyi strain CBDB1 with cell numbers between 5 and 8 × 107 cells ml−1. Cells were collected by centrifugation (5000g, 40 min, 4°C) and suspended in 50 mM ammonium bicarbonate buffer (pH 7.0). Cell lysis, protection of methionine oxidation with dithiothreitol, carbamidomethylation of cysteines with iodoacetamide, trypsin digestion, peptide purification on C18 ZipTip columns and analysis by nLC–MS/MS on an LTQ-Orbitrap (Thermo Fisher Scientific) equipped with a nanoUPLC system (nanoAcquity, Waters) was carried out as described previously [11]. A total of 30 cultures were analysed and included in the data processing.
Peptide identification was conducted by Mascot searches using the CBDB1 genome (NCBI taxonomy ID no. 255470) as a database as described [11]. The peptide identification threshold was set at a false-positive probability of 0.05 (probability-based ion score threshold of Aziz et al. [38]). Proteins were considered to be expressed if at least two peptides were identified. Protein coverage, peptide score and emPAI value [39] were calculated for each identified protein using the proteome discoverer software (Thermo Fisher Scientific). Further analyses were conducted on a Microsoft Excel spreadsheet. Proteins were ranked in each analysis, according to their emPAI values. The mean emPAI rank averaged over all identifications, and the absolute counts of identifications among the 30 samples were both taken as semi-quantitative measures to estimate the expression of a protein in the proteome of strain CBDB1.
(e). Bioinformatics
For genome comparisons, gene data from the NCBI (http://www.ncbi.nlm.nih.gov/genome/) or IMG database (http://img.jgi.doe.gov) were used. Automatic annotation was carried out with MaGe [40] and RAST [38]. Within these pipelines or standalone, the following tools and databases were applied: BLASTP [41], InterPro [42], KEGG pathways (http://www.genome.jp/kegg/pathway.html), Uniprot [43] and BRENDA [44]. Cofactor content of proteins was predicted using information from the UniProt and BRENDA databases.
3. Results
(a). Comparative genomics and expression analysis of Dehalococcoides proteins that contain or use organic cofactors
Screening through the annotations of all published Dehalococcoides genomes suggested the use of corrinoids, thiamine, biotin, pyrodoxal-5-phosphate, flavin nucleotides and molybdopterin as enzyme-bound organic cofactors and the soluble cofactors tetrahydrofolate, S-adenosylmethionine and pantothenate/CoA in Dehalococcoides strains. No genes encoding for enzymes containing haem or lipoic acid as cofactors were annotated in the genome. Freely soluble DNA/RNA-forming nucleotides and nicotinic acid derivates (NAD+, NADP+) are predicted to be abundant as in other organisms, whereas no indications were found for the use of archaeal cofactors (i.e. coenzyme F420, tetrahydromethanopterin, coenzyme B, coenzyme M, coenzyme F430, methanofuran). The electronic supplementary material, table S1 gives an overview of proteins predicted to contain one of the enzyme-bound cofactors or interact with tetrahydrofolate-derivates together with results of protein identification by nLC–MS/MS obtained from 30 cultures of strain CBDB1 grown on acetate and carbonate as carbon source, hydrogen as electron donor and a halogenated compound as electron acceptor.
Similar to other Dehalococcoides isolates, strain CBDB1 encodes corrinoid-dependent reductive dehalogenases playing a pivotal role in the reduction of many halogenated pollutants by the strain. The corrinoid is assumed to be located at the active site of the enzyme on the basis of the ability of cobalamins to catalyse dehalogenation in the presence of low-potential electron donors [45] and light-reversible inhibition by alkyl iodide [22], a characteristic of corrinoid-dependent reactions. Expression of the reductive dehalogenase CbdbA80 was detected in almost all tested cultures (29 of 30 tested cultures, in the following depicted as ‘29/30’), regardless of the type of electron acceptor that was added. This high frequency of detection is in accordance with previous studies suggesting the constitutive expression of CbdbA80 [22]. The chlorobenzene dechlorinating protein CbrA (CbdbA84) was found in 16 of 30 analyses together with CbdbA1588 (15/30), CbdbA1491 (8/30), CbdbA1598 (5/30), CbdbA1092 (3/30) and CbdbA88 (3/30). All these results suggest a strong need for corrinoids as a cofactor. Besides reductive dehalogenases, Dehalococcoides also encode cobalamin-dependent enzymes annotated as ribonucleoside-diphosphate reductases (Nrd) [46] for which expression of CbdbA287 was found in four of 30 cultures of strain CBDB1 (see the electronic supplementary material, table S1).
Genomes of Dehalococcoides strains contain several genes encoding proteins annotated to contain thiamine diphosphate as a cofactor. These proteins are predicted to be involved in a range of crucial metabolic reactions such as the pyruvate ferredoxin reductase (PFOR), transketolase (Tkt), acetolactate synthase for the biosynthesis of branched amino acids (IlvB) and 1-deoxy-d-xylulose-5-phosphate synthase for the biosynthesis of isoprenes (Dxs). These enzymes were not only encoded in Dehalococcoides strains but also expressed in strain CBDB1 as evidenced by nLC–MS/MS analysis. Among them, PFOR (27/30 for the thiamin-containing β-subunit PorB, cbdbA682) and acetolactate synthase (15/30) were most frequently detected and both were among the first 100 proteins in the mean emPAI rank list (see the electronic supplementary material, table S1).
According to the overall genome model, Dehalococcoides strains need biotin as cofactor in two central reactions: carboxylation of pyruvate to oxaloacetate and carboxylation of acetyl-CoA to malonyl-CoA for fatty acid biosynthesis. The two subunits of pyruvate carboxylase were 22 and 18 times detected in cultures of strain CBDB1, respectively, within the 30 data files, whereas acetyl-CoA carboxylase is not confidently annotated in strain CBDB1.
Pyridoxal-5-phosphate is predicted as a cofactor in several transferase and lyase reactions in Dehalococcoides strains (see the electronic supplementary material, table S1). Among the transferases, the most predominant proteins in the proteomic data were serine hydroxymethyltransferase (GlyA, CbdbA390, 24/30), ll-diaminopimelate aminotransferase (DapL, CbdbA714, 10/30), branched chain amino acid aminotransferase (IlvE, CbdbA11, 11/30), aspartate aminotransferase (AspC, CbdbA1292, 8/10) and histidinol-phosphate transaminase (HisC, CbdbA825, 12/30). Frequently detected lyases included tryptophan synthase (β-subunit, TrpB1, CbdbA1008, 27/30) and diaminopimelate carboxylase (LysA, CbdbA508, 12/30). All these detected proteins are involved in amino acid biosynthesis pathways.
Annotations indicate that Dehalococcoides strains need flavines in the form of flavin mononucleotide (FMN), e.g. for a HymB subunit [47] (cbdbA684, DET0729, DehalGT_0622, DehaBAV1_0661, DhcVS_635, Dehly_624) which is genetically linked with the Ech hydrogenase complex. This and a second HymB subunit also predicted to contain FMN were frequently detected in the proteomics data sets from strain CBDB1 cultures (22/30 and 10/30, respectively). Also, several proteins annotated to contain flavin adenine dinucleotide (FAD) are encoded in the genome of Dehalococcoides strains and some were identified in our shotgun proteomic data, for example a pyridine nucleotide-disulfide oxidoreductase that was by far the most frequently identified among them (CbdbA143, 25/30; see the electronic supplementary material, table S1).
Two membrane-bound protein complexes in Dehalococcoides strains are annotated to contain molybdopterin as cofactor, a formate dehydrogenase-like protein complex and a molybdopterin oxidoreductase complex without predicted substrate specificity. The latter complex was not found to be expressed in any of the 30 investigated CBDB1 cultures. By contrast, the major subunit of the formate dehydrogenase-like protein, CbdbA195, was found in all 30 samples at a mean emPAI rank of 6, indicating its very strong expression (see the electronic supplementary material, table S1). This is in accordance with previous studies showing a strong expression of a formate dehydrogenase-like protein in several Dehalococcoides strains [22,28]. However, its function is controversial because formate does not donate electrons for organohalide respiration in Dehalococcoides strains and cells of Dehalococcoides lack formate dehydrogenase activity [27]. Also crucial features in the active centre are different from known formate dehydrogenases [28,48]. Therefore, the need for molybdopterin in cells of CBDB1 is questionable.
Folate is a water-soluble, free-cycling cofactor comprising a pterin, a p-aminobenzoate and a glutamate moiety, and is transformed to its active forms dihydrofolate and tetrahydrofolate by two consecutive reductions. The metabolic function of tetrahydrofolate is the transfer of single-carbon functional groups such as methyl-, methylene- and formyl-groups. According to genome annotations, Dehalococcoides strains contain several crucial genes interacting with tetrahydrofolate or its C1-loaded forms, indicating the presence of tetrahydrofolate in Dehalococcoides. Analysis of nLC–MS/MS data from cultures of strain CBDB1 reveals strong expression of serine hydroxymethyltransferase (GlyA, CbdbA390, 24/30) and PurH (CbdbA1381, 23/30), a bifunctional purine biosynthesis protein using 10-formyl-tetrahydrofolate as C1 donor.
Dehalococcoides strains also encode many proteins predicted to use S-adenosylmethionine as a methyl-group donor, and many of these proteins were found in the nLC–MS/MS data from strain CBDB1 including many radical SAM proteins and S-adenosylmethionine-dependent methyl transferases.
CoA is a metabolic acyl group carrier and is formed from pantothenic acid in a five-step process. Strain CBDB1 encodes several enzymes that use CoA in essential metabolic pathways. Evidence of that is exemplified in the high expression found in strain CBDB1 of two acetyl-CoA synthetases (29/30 and 30/30) and the PFOR complex (PorA—23/30; PorB—27/30, PorD—2/30, PorG—23/30).
No cytochromes were annotated in any of the published Dehalococcoides genomes. Therefore, haem does not seem to play a role in Dehalococcoides metabolism. The absence of cytochrome b, a typical electron donor for quinones, also suggests that quinones are not involved in the respiratory electron transport chain in Dehalococcoides strains. Several proteins of strain CBDB1 are predicted to interact with ubiquinones and two are annotated to interact with menaquinones. However, the role of quinones is still not understood in Dehalococcoides because experimental data suggests that they cannot be used as electron carriers to support efficient anaerobic reductive dechlorination in this genus [49,50]. Similarly, an experiment testing quinones as electron donors for reductive dehalogenases in strain CBDB1 suggested that quinones do not function as natural electron donor. In this experiment, methyl viologen supported a much higher specific enzyme activity (203 nkat mg−1) than the tested quinones MK-4, Q-4 or Q-10 (below 0.15 nkat mg−1) or the quinone analogues menadione or Q-0 (no activity). By contrast, other authors have found indications for the presence of quinones in Dehalococcoides strains and speculated about a possible role of quinones in cell protection against free-radical species produced during reductive dehalogenation [51]. Several proteins predicted to interact with quinones were experimentally detected in the tested cultures of strain CBDB1, most importantly two subunits of the periplasmatic [Ni/Fe] hydrogenase Hym (22/30 and 10/30, respectively) and a cytoplasmatic [Fe] hydrogenase Vhu (23/30). However, it is not clear whether these subunits do interact with quinones or with other electron mediators in strain CBDB1.
No enzymes in Dehalococcoides strains are annotated to contain lipoic acid, a sulfur-containing cofactor often involved in large oxidoreductase complexes such as pyruvate or 2-oxoglutarate dehydrogenase, indicating that lipoic acid is not used by Dehalococcoides strains.
(b). Cultivation experiments with strain CBDB1 to identify vitamin requirements
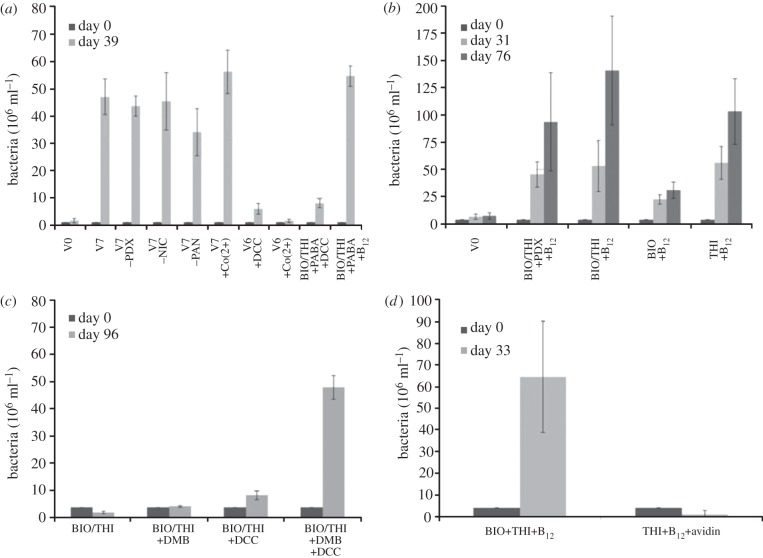
Strain CBDB1 has been cultured for many years over hundreds of transfers in a defined medium containing seven vitamins: cyanocobalamin, thiamine, biotin, p-aminobenzoate, pantothenate, nicotinate and pyridoxin hydrochloride. Riboflavin, folate, lipoic acid, naphthoquinone and haemin were not present in the medium, therefore demonstrating that these cofactors are either de novo synthesized or not needed at all. To elucidate the exact vitamin requirements in strain CBDB1, a first set of experiments was conducted investigating the need for pyridoxine hydrochloride, nicotinate and pantothenate (figure 1a). These three vitamins were investigated first because almost complete biosynthesis pathways were encoded in the genome (see below). The cultivation showed that cell numbers were not lower in cultures in which one or all of these three vitamins were omitted from the medium after 39 days of cultivation. The same results were obtained in a second cell counting after 63 days (results not shown). This demonstrated that pyridoxine hydrochloride, nicotinate and pantothenate were not required in the medium for growth. The removal of cyanocobalamin from the medium resulted in a strong decrease in the number of cells, confirming that vitamin B12 is essential for strain CBDB1. This growth inhibition could not be released by the addition of cobalt chloride or DCC (figure 1a). The cell numbers increased concurrently with the formation of dechlorination products from HCB (see the electronic supplementary material, figure S1). A second set of experiments were conducted to investigate the need for thiamine, biotin and p-aminobenzoate. Cell enumeration demonstrated that thiamine was required for growth, whereas p-aminobenzoate could be omitted without observing a decrease in the growth (figure 1b). Cultures that were not amended with biotin showed cell numbers and dechlorination rates similar to cultures amended with biotin (figure 1b). These results were unexpected owing to the lack of the required biosynthesis genes for biotin in Dehalococcoides. A feasible explanation for this result was the ubiquitous presence of low concentrations of biotin in the defined media or attached to the glassware used in the experiments, enough to sustain growth of strain CBDB1. Therefore, a new set of experiments was prepared to remove biotin from the media by adding the high-affinity biotin-binding protein avidin. As shown in figure 1d, the lack of available biotin in cultures containing avidin resulted in a negligible growth of Dehalococcoides, demonstrating the requirement for biotin as a vitamin (figure 1d).
Figure 1.
Cell concentration of strain CBDB1 after cultivation in the standard defined medium lacking specific cofactors. Bars show mean values of five parallel cultures, and standard deviations are indicated. The treatments depicted in the figures are as follows: V0, negative control without cofactors; V7, positive control containing the seven vitamins cyanocobalamin (B12), biotin (BIO), thiamine (THI), p-aminobenzoate (PABA), pantothenate (PAN), nicotinate (NIC) and pyridoxine hydrochloride (PDX); V6, V7 without B12. The symbol minus or plus preceding a vitamin abbreviation indicates that a specific cofactor was omitted or added, respectively. DCC, dicyanocobinamide; Co(2+), cobalt(II)chloride; DMB, dimethylbenzimidazole. (a) Vitamin requirements for pyridoxal-5-phosphate, nicotinate, pantothenate and cyanocobalamin; (b) vitamin requirements for thiamine and biotin; (c) cobalamin precursors; (d) effect of biotin removal with avidin.
Successively, cultures were regularly maintained in medium containing cyanocobalamin, thiamine and biotin as vitamins for over five transfers without diminishing the growth rate of CBDB1 cells, final cell numbers or dechlorination rates, demonstrating that the cultures are sustainable on this minimal medium. A more detailed analysis showed that cyanocobalamin could be replaced by a combination of DMB and DCC (figure 1c), both precursors for the synthesis of cobamins.
(c). Analysis of cofactor biosynthesis pathways in Dehalococcoides genomes and analysis of the expression of cofactor biosynthesis enzymes
Comparative genome analysis in Dehalococcoides genomes showed that cofactor biosynthesis pathways for DNA/RNA-forming nucleotides, nicotinamide nucleotides, pantothenate/CoA, pyridoxal-5-phosphate, S-adenosylmethionine and riboflavin are fully or almost fully encoded, whereas the biosynthesis of corrinoids, thiamine and biotin is incomplete (see the electronic supplementary material, table S2). Biosynthesis of p-aminobenzoate, folate and pterin is discussed below. The canonical pathways for lipoic acid biosynthesis, lipoic acid salvage and haemin biosynthesis are completely missing in all Dehalococcoides strains. While enzymes involved in amino acid, nucleic acids or lipid biosynthesis were abundant in the proteome of strain CBDB1 as judged by the frequency of detection and obtained emPAI values from the nLC–MS/MS data, cofactor biosynthesis proteins were generally much less abundantly expressed.
Cobalamin biosynthesis is incompletely encoded in all sequenced Dehalococcoides strains [15,16,52,53]. All genes for the de novo biosynthesis of the corrin ring system are missing, and also no gene was identified to encode for an enzyme catalysing cobalt incorporation into the corrin ring. By contrast, all proteins required for the uptake of corrinoids (ABC transporter) and the biosynthesis of cobalamin from cobyrinates or cobinamids (cobalamin salvage, cobA, -D, -U, -S, -C, -T and cbiB [54,55]) are encoded in Dehalococcoides genomes that includes the genes for the assembly of the nucleotide loop and the incorporation of DMB as a lower ligand, although the biosynthesis of DMB from FMN itself is not encoded. Also, the two amidases CbiA and CobQ are encoded in all Dehalococcoidetes. In strain CBDB1, several of the cobalamin biosynthesis enzymes were detected by mass spectrometry although the strain was cultivated in medium supplied with vitamin B12: CobA (cbdbA1141, 2/30), CobQ (cbdbA890, 5/30), CobT (cbdbA641, 16/30, medium emPAI rank 51) and CobC (cbdbA643, 3/30). CobT and CobC are involved in the transformation of DMB to α-ribazol to be used as the lower ligand in cobalamins, indicating the use of DMB as a lower ligand.
Although thiamine diphosphate containing enzymes are encoded in Dehalococcoides and expressed in strain CBDB1, the genomes do not encode a complete thiamine diphosphate biosynthesis pathway. Dehalococcoides strains encode thiamine biosynthesis protein ThiC (phosphomethylpyrimidine synthase; cbdbA749, DET0775, DehalGT0662, DehaBAV1_0701, DhcVS681, Dehly_0872), thiamine-phosphate diphosphorylase ThiE (cbdbA758, DET0782, DehalGT0669, DehaBAV1_0708, DhcVS688, Dehly_0766) and thiamine-phosphate kinase ThiL (cbdbA347, DET0397, DehalGT0340, DehaBAV1_0376, DhcVS340, Dehly_1128). While the synthesis of 4-amino-2-methyl-5-diphosphomethylpyrimidine might be catalysed by one or two not yet identified kinases, the synthesis of 4-methyl-5-(2-phosphoethyl)-thiazole seems not be encoded in Dehalococcoides strains, and several enzymes are missing. The genome annotations therefore hint to the necessity to take up thiamine or a precursor such as thiazole-phosphate or 3-methyl-4-amino-5-hydroxymethylpyrimidine diphosphate (HMP-PP). Degradation products of thiamine might also serve as a thiamine precursor in Dehalococcoides strains [56]. The protein encoded by cbdbA749 (thiC) was biochemically detected in one of the 30 expression analyses.
No indication for a biotin biosynthesis pathway in Dehalococcoides genomes was found. The only related enzyme is the biotin-(acetyl-CoA-carboxylase) ligase, BirA (cbdbA832, DET0849, DehalGT0729, DehaBAV1_0768, DhcVS753, Dehly_0846), which catalyses the ligation of biotin with a lysine residue in the active centre of acetyl-CoA-carboxylase. The enzyme was not detected in the expression analysis. By contrast, a putative biotin transporter (bioY—DET1184, cbdbA1098) was described in the genome [15], but no expression was found in strain CBDB1.
Pyridoxal-5-phosphate seems to be synthesized in Dehalococcoides strains via the deoxyxylulose 5-phosphate-independent pathway [57] with the two enzymes pyridoxal biosynthesis lyase PdxS (cbdbA327, DET0380, DehalGT0324, DehaBAV1_0362, DhcVS324, Dehly_0205) and glutamine amidotransferase subunit PdxT (cbdbA579, DET0598, DehalGT0535, DehaBAV1_0573, DhcVS538, Dehly_0745). The protein encoded by cbdbA327 (pdxS) was identified in 12 of 30 nLC–MS/MS analyses with an average emPAI rank of 97.
All enzymes for the biosynthesis of riboflavin and flavocofactors [58] are encoded in published Dehalococcoides genomes. Most of the enzymes were detected by mass spectrometry in cultures of strain CBDB1 (RibA: 11/30; RibD: 10/30, RibH: 7/30, RibF: 5/30), indicating a strong biosynthesis of riboflavin. RibE was not identified.
No genes coding for enzymes in the biosynthesis of molybdopterin or the molybdenum cofactor from GTP [59] are annotated in Dehalococcoides genomes. However, mobA (cbdbA115, DET0099, DehalGT_0130, DehaBAV1_0269, DhcVS_109, Dehly_0414), encoding an enzyme that connects the molybdenum cofactor and GTP to the functional MGD cofactor is present in all Dehalococcoides strains. This suggests that Dehalococcoides strains may take up the molybdenum cofactor and then assemble the functional MGD. However, it cannot be excluded that the organisms use an unknown molybdenum cofactor biosynthesis pathway or do not use molybdopterin cofactors at all. A candidate transporter protein would be cbdbA116 that forms an operon with mobA and genes for a molybdopterin-dependent oxidoreductase in strain CBDB1.
Although not all canonical enzymes of the folate biosynthesis pathway are encoded, the pathway appears to be functional considering the following parts (figure 2): (i) GTP cyclohydrolase (FolE) transforming GTP to 7,8-dihydroneopterin triphosphate, (ii) three proteins encoded by an operon of ptpS-III/folK/folP that is conserved and syntenic in all Dehalococcoides strains (cbdbA1696–1699, DET1603–1605, DehalGT1397–1399, DehaBAV1_1349–1351, DhcVS1485–1487, Dehly_006-007 and Dehly_1457), catalysing the biosynthesis of 7,8–dihydropteroate, (iii) FolC, producing dihydrofolate and (iv) a yet unidentified dihydrofolate reductase. The gene ptpS was shown to encode a type III 6-pyruvoyltetrahydropterin synthase, which directly produces 7,8-dihydropterin from the precursor 7,8-dihydroneopterin 3-triphosphate as present also in other anaerobic bacteria [60,61]. In the protein expression analysis, only CbdbA1120 (FolE) was detected in one of the analyses. Although p-aminobenzoate (PABA) is not needed as a vitamin for cultivation of Dehalococcoides strains, the required genes to synthesize PABA from chorismate, i.e. 4-amino-4-deoxychorismate synthase and aminodeoxychorismate lyase, are not annotated in the genomes. Because we can exclude that PABA is present in the medium and previous studies have shown that PABA is almost universally needed for tetrahydrofolate biosynthesis [62,63], our results suggest that the functions are either co-catalysed, e.g. by genes of the tryptophan biosynthesis pathways as shown for other organisms [64] or are catalysed by unidentified genes.
Figure 2.
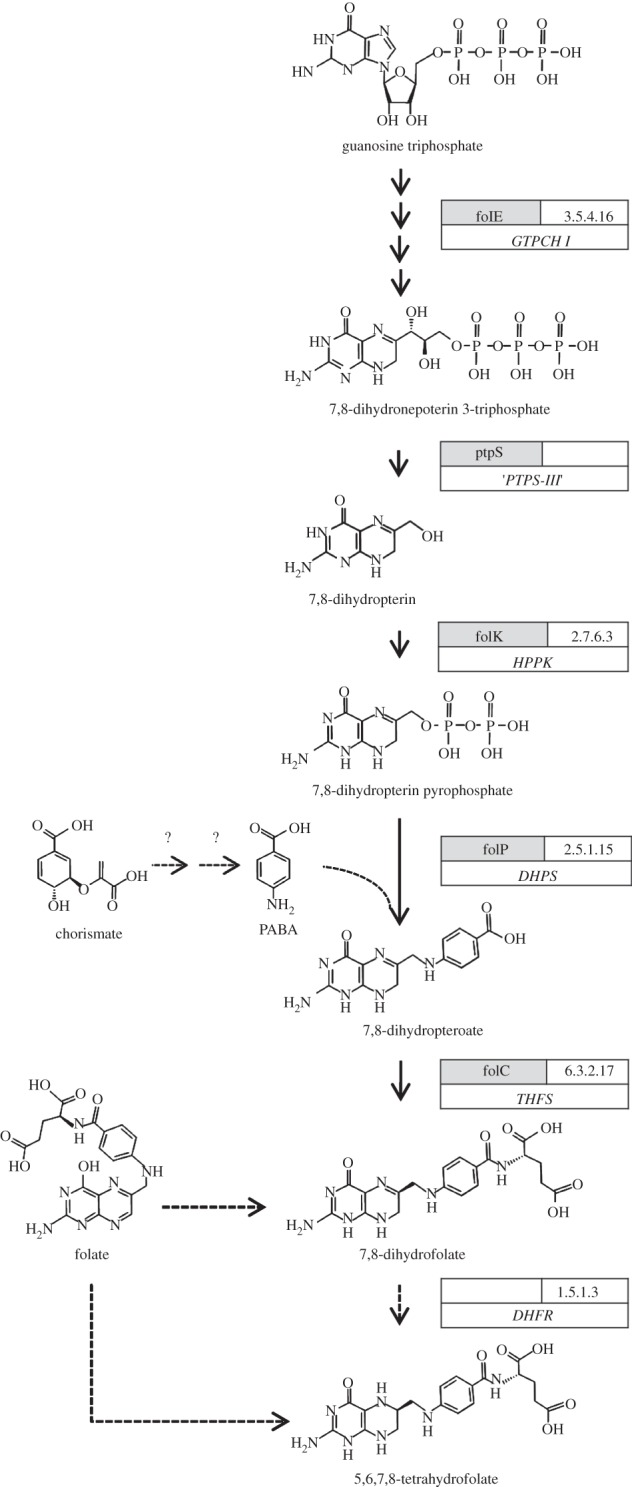
Folate biosynthesis in Dehalococcoides as inferred from cultivation, genome annotation, nLC–MS/MS and inhibition studies. Boxes show gene name, E.C. number of the encoded enzyme and a short name of the enzyme. PABA, p-aminobenzoate; GTPCH I, guanosine triphosphate cyclohydrolase I; PTPS-III, 6-pyruvoyltetrahydropterin synthase, type III [60]; HPPK, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase; DHPS, dihydropteroate synthase; THFS, dihydrofolate synthase/tetrahydrofolate synthase; DHFR, dihydrofolate reductase. Only FolE (CbdbA1120) was found in one of 30 nLC–MS/MS data sets from cultures of strain CBDB1.
The biosynthesis of S-adenosylmethionine is a one-step reaction catalysed by methionine adenosyl transferase from methionine and ATP. The enzyme is encoded in all Dehalococcoides genomes (cbdbA476, DET0512, DehalGT0451, DehaBAV1_0488, DhcVS453, Dehly_1062) and highly expressed in CBDB1 cultures (29/30 with a mean emPAI rank of 28), indicating that S-adenosylmethionine is an abundant cofactor in Dehalococcoides strains.
With the exception of 2-dehydropantoate 2-reductase (EC 1.1.1.169), all pantothenate and CoA biosynthesis genes are annotated in Dehalococcoides strains. The annotation of this 2-dehydropantoate 2-reductase, however, has also been difficult in many other bacteria, so that the pathway appears intact in Dehalococcoides strains. The first four enzymes in the pathway (IlvHBCD) could be detected by mass spectrometry with high abundance (11–24 times detected in 30 analyses with a mean emPAI rank near or below 100). However, these enzymes are also involved in the biosynthesis of branched amino acids, which explains their strong expression. All pantothenate-specific biosynthesis enzymes were not detected in our analyses from CBDB1 extracts with the exception of CoaE (cbdbA1268), which was detected once.
In addition, the biosynthesis of nicotinamide nucleotides is fully encoded in Dehalococcoides strains. Only in strain 195, the l-aspartate oxidase NadB/NabD seems to be disrupted, but this annotation might also be the result of a sequencing error. NadB, NadA and PncB were rarely detected in extracts of strain CBDB1 (1/30, 5/30, 3/30, respectively), whereas NadC was prominently found (mean emPAI rank of 46) in most analyses (26/30).
The deazaflavin cofactor F420 is used by many Archaea and some Bacteria, including the Chloroflexi species, as a cofactor of oxidoreductases [65]. However, in contrast to the genome of for example Chloroflexus aurianticus, Dehalococcoidia (Dehalococcoides and Dehalogenimonas), genomes do not encode any cofactor F420 biosynthesis genes indicating that this cofactor is not used. The biosynthesis of porphyrin rings such as haem and all haem-transforming enzymes are completely missing in all Dehalococcoides strains.
(d). Inhibition of the folate biosynthesis
Cultivation of strain CBDB1 with trimethoprim, an inhibitor of dihydrofolate reductase, or with sulfamethoxazole, an inhibitor of dihydropteroate reductase, reduced growth of strain CBDB1 by 42 per cent and 76 per cent, respectively, indicating that both enzymes are present and active in strain CBDB1 (figure 3). While dihydropteroate reductase is annotated, no dihydrofolate reductase is known in strain CBDB1. Addition of 20 µg ml−1 folic acid to the medium did not release inhibition, indicating that folic acid was not transported into the cells.
Figure 3.
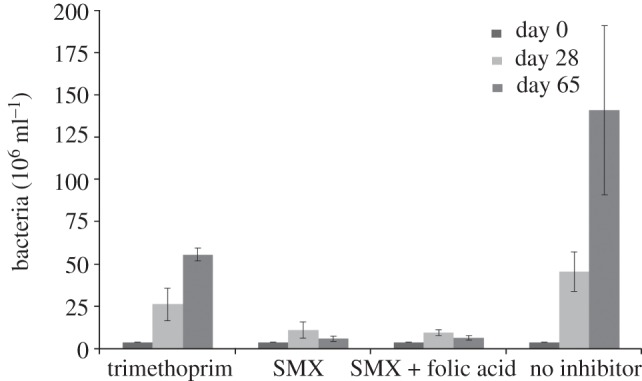
Cell numbers in cultures incubated with folic acid biosynthesis inhibitors. Cultures were inoculated at day 0 to a cell number of 4 × 106 cells ml−1. Trimethoprim is described to inhibit dihydrofolate reductase; sulfamethoxazole (SMX) inhibits dihydropteroate reductase. All cultures contained cyanocobalamin, p-aminobenzoate, biotin and thiamine. Concentrations: Trimethoprim, 4 mg l−1; sulfamethoxazole, 100 mg l−1; folic acid, 20 µg l−1. For each culture condition, five parallel cultures were set up and counted. Bars show the mean cell numbers and standard deviations are indicated.
4. Discussion
To date, Dehalococcoides isolates have always been cultivated with a mixture of different vitamins derived from classical formulations [66]. However, apart from work with cobamins, little biochemical evidence supports the link between cofactor requirements and biological activity. Here, we combined the rational prediction of cofactor requirements by genome annotation with experimental evidence coming from cultivation and proteomic analysis. With this approach, we obtained detailed insights into the cofactor requirements of Dehalococcoides strains. Our results allow the design of a minimal medium to grow Dehalococcoides cultures, which is an important aspect for rationally optimizing large-scale cultivation and application in the field at minimal costs and avoiding the introduction of unwanted or unnecessary chemicals into the environment. At the same time, the restriction of a medium to the inevitably needed vitamins can limit growth of competing organisms in a mixed culture or in the field.
(a). Organic cofactors that are not needed as vitamins
Several cofactors were not needed as vitamins for cultivation. This can either be due to the fact that de novo biosynthesis was catalysed or that the cofactor was not used in the metabolism. Dehalococcoides genomes harbour genes encoding the complete biosynthetic pathway of pyridoxal-5-phosphate, S-adenosylmethionine, pantothenic acid, flavines and nicotinamides. The folate biosynthesis pathway is almost complete but it is not clear which enzyme catalyses the reduction of 7,8-dihydrofolate to 5,6,7,8-tetrahydrofolate. We performed inhibition studies demonstrating that indeed the folate pathway was intact. At the same time, our experiments showed that folate was not taken up as a vitamin, corroborating that folic acid is synthesized de novo in Dehalococcoides strains. While the encoded biosynthesis pathways for pyridoxal-5-phosphate, S-adenosylmethionine, pantothenic acid, flavines, nicotinamides and folic acid were functional, the cultivation experiments showed that the cells did not need these cofactors as vitamins in the medium confirming that they were synthesized. These results are also in accordance with the nLC–MS/MS data from cultures of strain CBDB1 indicating a wide use of these cofactors in the metabolism (see the electronic supplementary material, table S1).
In contrast to the identification of cofactor-containing/interacting proteins, the approach to identify proteins that are involved in cofactor biosynthesis was less successful (see the electronic supplementary material, table S2). This was in accordance with the expectations at least for the enzyme-bound cofactor biosynthesis enzymes because such cofactor biosynthesis enzymes are needed in much lower amounts than enzymes involved in energy conservation, central pathways or the biosynthesis of amino acids, lipids or sugars. Still, the detection of several enzymes in cofactor biosynthesis pathways corroborates the de novo synthesis of the respective cofactors.
(b). Organic cofactors involved in respiration
A special interest lies in the analysis of cofactors involved in respiratory processes, because the energy metabolism of Dehalococcoides strains relies exclusively on them. In addition, the organohalide respiratory capacity makes Dehalococcoides strains important for remediation approaches. In other organohalide-respiring organisms such as Sulfurospirillum multivorans [67], Desulfitobacterium strain PCE1 [68], Desulfomonile tiedjei DCB-1 [69] or Dehalobacter restrictus PER-K23 [70], quinones play an important role in the electron transport through the membrane. Also for Dehalococcoides strains, the presence of quinones has been reported [51]; however, the biosynthesis pathways for ubiquinone and menaquinone are almost absent in all known Dehalococcoides genomes although the biosynthesis of the terpenoid backbone and even the octaprenyl-transferase MenA is encoded. We have not found any indications for the need to supplement CBDB1 cultures with quinones or quinone precursors and did not detect significant enzymatic activity with quinones as electron donor in an activity test for reductive dehalogenation. It is therefore a challenge for future work to either confirm the presence of quinones in Dehalococcoides and to determine the biosynthetic pathway or to identify functional substitutes to mediate electron transport between hydrogenases and reductive dehalogenases in Dehalococcoides strains. While the initial work on organohalide respiration in Desulfomonile tiedjei identified the involvement of cytochromes [71], the electron transport in other organohalide respirers was independent from cytochromes. In Dehalococcoides strains, haem clearly does not play any role as shown by several lines of evidence. By contrast, cobamin is an essential cofactor for all currently characterized reductive dehalogenases in Dehalococcoides and other organohalide respirers and a positive correlation between vitamin B12 concentration in the medium, dechlorination and cellular growth has been established for strain 195 [72]. Recently, it was shown for strain 195 that dehalogenation of TCE was catalysed by reductive dehalogenases containing DMB or 5-methyl benzimidazole as the lower ligand of the cobamide cofactor and that other benzimidazol derivatives could be modified and also incorporated into the cobamide to produce active reductive dehalogenases [53]. Corrinoid proteins make up a larger part of the total proteome in Dehalococcoides strains than in any other analysed bacterial group [46]. Because the de novo biosynthesis of corrinoids is incomplete, Dehalococcoides strains rely on other organisms in their environment for the supply of these corrinoids [52] and undergo large changes in the metabolism in response to corrinoid supplies [73]. We have confirmed this pivotal role of corrinoids for growth of strain CBDB1 and that cyanocobalamin can be substituted by DMB plus DCC. As judged from genome annotations, Dehalococcoides seem to be specialized to take up different precursors with corrinoid ring structure and to modify the upper and lower ligands according to their special needs. For corrinoid remodelling, the genome of strain CBDB1 contains three copies of cobA to catalyse adenosyl transfers as the upper ligand, and eight copies of cbiZ, previously annotated as RdhF [16], predicted to encode amidolyases, cleaving off non-functional lower ligands. The close association of cbiZ genes with rdh clusters [16] underlines the importance of corrinoid salvage and remodelling for reductive dehalogenase functioning.
Molybdopterin-dependent oxidoreductases are also often involved in respiratory electron chains. The identification of molybdopterin-containing proteins could therefore help to explain respiratory energy conservation in Dehalococcoides cells. However, although two MGD-containing complexes are predicted from annotation, the bacteria clearly do not need molybdopterin additions to the medium for growth and do not seem to be able to synthesize it de novo. One explanation for the observed data is that the molybdopterin-oxidoreductase might not be expressed when molybdopterin is unavailable. Therefore, the addition of molybdopterin could possibly extend the catabolic capacity of Dehalococcoides cells. The formate dehydrogenase-like protein, by contrast, is highly expressed. It has to be investigated in more detail if this protein indeed contains molybdopterin, if it is active without molybdopterin cofactor or if possibly the enzyme is inactive owing to the absence of the cofactor.
(c). Essential vitamins involved in central anabolism
The requirement to amend Dehalococcoides CBDB1 cultures with thiamine was clearly shown by several lines of evidence, including cultivation, lack of the required biosynthesis genes in the genome and the detection of several proteins predicted to contain thiamine. These essential enzymes catalyse some of the most central anabolic functions of Dehalococcoides strains and include pyruvate–ferredoxin–oxidoreductase through which a large part of the fixed carbon flows, transketolase which is needed for the synthesis of pentoses for protein, DNA, RNA and cofactor biosynthesis and acetolactate synthase involved in branched amino acid biosynthesis. As described for cobalamin-dependent functions in the metabolism, these functions are all crucial for the metabolism and it might surprise why the biosynthesis of thiamine is not encoded in the genome. The conclusion can only be, as described and experimentally shown for corrinoids [52], that thiamine is supplied in excess in anaerobic environments by syntrophic organisms and that the biosynthesis genes are therefore dispensable. The same seems true for biotin, for which a biosynthesis pathway is missing, although biotin is predicted to play a major role in the central metabolism and also biotin-dependent enzymes were detected in high abundance. Our experiments including the biotin-binding protein avidin in the cultures to remove traces of biotin confirmed the essentiality of biotin for Dehalococcoides.
5. Conclusion
With the analysis of cofactor biosynthesis and use in Dehalococcoides strains, not only can a rational approach for cultivation and large-scale production of Dehalococcoides cells be advanced, but also the basic knowledge on central metabolic functions can be extended. Dehalococcoides cells are excellent candidates for the analysis of basic and probably ancient core metabolic functions owing to (i) the very small size of the genome and accordingly the low number of encoded proteins, (ii) the highly restricted mode of energy conservation, (iii) the clear separation of anabolic and catabolic functions and (iv) the apparent lack of complex regulatory networks. Therefore, Dehalococcoides strains also represent model organisms for much slower growing anaerobic bacteria in subsurface environments, including the widely distributed and highly abundant Dehalococcoides-related Chloroflexi [74].
Acknowledgements
We acknowledge the expert technical help of Benjamin Scheer. Funding was provided by the ERC project ‘Microflex’ and the DFG (FOR1530) to L.A. E.M.-U. acknowledges the support from the Catalan Government (Generalitat de Catalunya) for the mobility grant no. BE-DGR (2011 BE1 00321).
References
- 1.Löffler FE, et al. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligate organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635 10.1099/ijs.0.034926-0 (doi:10.1099/ijs.0.034926-0) [DOI] [PubMed] [Google Scholar]
- 2.Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276, 1568–1571 10.1126/science.276.5318.1568 (doi:10.1126/science.276.5318.1568) [DOI] [PubMed] [Google Scholar]
- 3.Cheng D, He J. 2009. Isolation and characterization of ‘Dehalococcoides’ sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl. Environ. Microbiol. 75, 5910–5918 10.1128/aem.00767-09 (doi:10.1128/aem.00767-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7, 1442–1450 10.1111/j.1462-2920.2005.00830.x (doi:10.1111/j.1462-2920.2005.00830.x) [DOI] [PubMed] [Google Scholar]
- 5.He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424, 62–65 10.1038/nature01717 (doi:10.1038/nature01717) [DOI] [PubMed] [Google Scholar]
- 6.Adrian L, Szewzyk U, Wecke J, Görisch H. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408, 580–583 10.1038/35046063 (doi:10.1038/35046063) [DOI] [PubMed] [Google Scholar]
- 7.Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72, 1980–1987 10.1128/AEM.72.3.1980-1987.2006 (doi:10.1128/AEM.72.3.1980-1987.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee PKH, et al. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 5, 1014–1024 10.1038/ismej.2010.202 (doi:10.1038/ismej.2010.202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunge M, Wagner A, Fischer M, Andreesen JR, Lechner U. 2008. Enrichment of a dioxin-dehalogenating Dehalococcoides species in two-liquid phase cultures. Environ. Microbiol. 10, 2670–2683 10.1111/j.1462-2920.2008.01688.x (doi:10.1111/j.1462-2920.2008.01688.x) [DOI] [PubMed] [Google Scholar]
- 10.Tang YJ, Yi S, Zhuang W-Q, Zinder SH, Keasling JD, Alvarez-Cohen L. 2009. Investigation of carbon metabolism in ‘Dehalococcoides ethenogenes’ strain 195 by use of isotopomer and transcriptomic analyses. J. Bacteriol. 191, 5224–5231 10.1128/jb.00085-09 (doi:10.1128/jb.00085-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marco-Urrea E, Paul S, Khodaverdi V, Seifert J, von Bergen M, Kretzschmar U, Adrian L. 2011. Identification and characterization of a Re-citrate synthase in Dehalococcoides strain CBDB1. J. Bacteriol. 193, 5171–5178 10.1128/JB.05120-11 (doi:10.1128/JB.05120-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J, Rash BA, Rainey FA, Moe WM. 2009. Isolation of novel bacteria within the Chloroflexi capable of reductive dechlorination of 1,2,3-trichloropropane. Environ. Microbiol. 11, 833–843 10.1111/j.1462-2920.2008.01804.x (doi:10.1111/j.1462-2920.2008.01804.x) [DOI] [PubMed] [Google Scholar]
- 13.Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA. 2009. Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 59, 2692–2697 10.1099/ijs.0.011502-0 (doi:10.1099/ijs.0.011502-0) [DOI] [PubMed] [Google Scholar]
- 14.May HD, Miller GS, Kjellerup BV, Sowers KR. 2008. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl. Environ. Microbiol. 74, 2089–2094 10.1128/AEM.01450-07 (doi:10.1128/AEM.01450-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seshadri R, et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307, 105–108 10.1126/science.1102226 (doi:10.1126/science.1102226) [DOI] [PubMed] [Google Scholar]
- 16.Kube M, Beck A, Zinder S, Kuhl H, Reinhardt R, Adrian L. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273 10.1038/nbt1131 (doi:10.1038/nbt1131) [DOI] [PubMed] [Google Scholar]
- 17.McMurdie PJ, et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5, e1000714. 10.1371/journal.pgen.1000714 (doi:10.1371/journal.pgen.1000714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddaramappa S, et al. 2012. Complete genome sequence of Dehalogenimonas lykanthroporepellens type strain (BL-DC-9(T)) and comparison to ‘Dehalococcoides’ strains. Stand. Genomic Sci. 6, 251–264 10.4056/sigs.2806097 (doi:10.4056/sigs.2806097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hölscher T, Görisch H, Adrian L. 2003. Reductive dehalogenation of chlorobenzene congeners in cell extracts of Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 69, 2999–3001 10.1128/AEM.69.5.2999-3001.2003 (doi:10.1128/AEM.69.5.2999-3001.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kräutler B, et al. 2003. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of a natural corrinoid. Helvetica Chim. Acta 86, 3698–3716 10.1002/hlca.200390313 (doi:10.1002/hlca.200390313) [DOI] [Google Scholar]
- 21.Lee PKH, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 72, 6161–6168 10.1128/aem.01070-06 (doi:10.1128/aem.01070-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adrian L, Rahnenführer J, Gobom J, Hölscher T. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73, 7717–7724 10.1128/AEM.01649-07 (doi:10.1128/AEM.01649-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64, 1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70, 6347–6351 10.1128/AEM.70.10.6347-6351.2004 (doi:10.1128/AEM.70.10.6347-6351.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller JA, Rosner BM, von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70, 4880–4888 10.1128/AEM.70.8.4880-4888.2004 (doi:10.1128/AEM.70.8.4880-4888.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow WL, Cheng D, Wang S, He J. 2010. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 4, 1020–1030 10.1038/ismej.2010.27 (doi:10.1038/ismej.2010.27) [DOI] [PubMed] [Google Scholar]
- 27.Morris RM, Sowell S, Barofsky D, Zinder S, Richardson R. 2006. Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ. Microbiol. 8, 1499–1509 10.1111/j.1462-2920.2006.01090.x (doi:10.1111/j.1462-2920.2006.01090.x) [DOI] [PubMed] [Google Scholar]
- 28.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, Richardson RE. 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73, 320–326 10.1128/AEM.02129-06 (doi:10.1128/AEM.02129-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahm B, Richardson R. 2008. Correlation of respiratory gene expression levels and pseudo-steady-state PCE respiration rates in Dehalococcoides ethenogenes. Environ. Sci. Technol. 42, 416–421 10.1021/es071455s (doi:10.1021/es071455s) [DOI] [PubMed] [Google Scholar]
- 30.Empadinhas N, Albuquerque L, Costa J, Zinder SH, Santos MAS, Santos H, da Costa MS. 2004. A gene from the mesophilic bacterium Dehalococcoides ethenogenes encodes a novel mannosylglycerate synthase. J. Bacteriol. 186, 4075–4084 10.1128/JB.186.13.4075-4084.2004 (doi:10.1128/JB.186.13.4075-4084.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahn U, Huber H, Eisenreich W, Hügler M, Fuchs G. 2007. Insights into the autotrophic CO2 fixation pathway of the archaeon Ignicoccus hospitalis: Comprehensive analysis of the central carbon metabolism. J. Bacteriol. 189, 4108–4119 10.1128/jb.00047-07 (doi:10.1128/jb.00047-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Hagemeier CH, Seedorf H, Gottschalk G, Thauer RK. 2007. Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and isopropylmalate synthase rather than to Si-citrate synthase. J. Bacteriol. 189, 4299–4304 10.1128/jb.00198-07 (doi:10.1128/jb.00198-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marco-Urrea E, Seifert J, von Bergen M, Adrian L. 2012. Stable isotope peptide mass spectrometry to decipher amino acid metabolism in Dehalococcoides strain CBDB1. J. Bacteriol. 194, 4169–4177 10.1128/jb.00049-12 (doi:10.1128/jb.00049-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang W-Q, Yi S, Feng X, Zinder SH, Tang YJ, Alvarez-Cohen L. 2011. Selective utilization of exogenous amino acids by Dehalococcoides ethenogenes strain 195 and the effects on growth and dechlorination activity. Appl. Environ. Microbiol. 77, 7797–7803 10.1128/aem.05676-11 (doi:10.1128/aem.05676-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White HB. 1976. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 7, 101–104 10.1007/bf01732468 (doi:10.1007/bf01732468) [DOI] [PubMed] [Google Scholar]
- 36.Jayachandran G, Görisch H, Adrian L. 2003. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180, 411–416 10.1007/s00203-003-0607-7 (doi:10.1007/s00203-003-0607-7) [DOI] [PubMed] [Google Scholar]
- 37.Adrian L, Hansen SK, Fung JM, Görisch H, Zinder SH. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ. Sci. Technol. 41, 2318–2323 10.1021/Es062076m (doi:10.1021/Es062076m) [DOI] [PubMed] [Google Scholar]
- 38.Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. 10.1186/1471-2164-9-75 (doi:10.1186/1471-2164-9-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 10.1074/mcp.M500061-MCP200 (doi:10.1074/mcp.M500061-MCP200) [DOI] [PubMed] [Google Scholar]
- 40.Vallenet D, et al. 2009. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009, bap021. 10.1093/database/bap021 (doi:10.1093/database/bap021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter S, et al. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 10.1093/nar/gkr948 (doi:10.1093/nar/gkr948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu CH, et al. 2006. The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34, D187–D191 10.1093/nar/gkj161 (doi:10.1093/nar/gkj161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheer M, et al. 2011. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 39, D670–676 10.1093/nar/gkq1089 (doi:10.1093/nar/gkq1089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gantzer CJ, Wackett LP. 1991. Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ. Sci. Technol. 25, 715–722 10.1021/es00016a017 (doi:10.1021/es00016a017) [DOI] [Google Scholar]
- 46.Zhang Y, Rodionov D, Gelfand M, Gladyshev V. 2009. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10, 78. 10.1186/1471-2164-10-78 (doi:10.1186/1471-2164-10-78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graentzdoerffer A, Rauh D, Pich A, Andreesen JR. 2003. Molecular and biochemical characterization of two tungsten- and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase. Arch. Microbiol. 179, 116–130 10.1007/s00203-002-0508-1 (doi:10.1007/s00203-002-0508-1) [DOI] [PubMed] [Google Scholar]
- 48.Soboh B, Pinske C, Kuhns M, Waclawek M, Ihling C, Trchounian K, Trchounian A, Sinz A, Sawers G. 2011. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen: benzyl viologen oxidoreductase activity. BMC Microbiol. 11, 173. 10.1186/1471-2180-11-173 (doi:10.1186/1471-2180-11-173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nijenhuis I, Zinder SH. 2005. Characterization of hydrogenase and reductive dehalogenase activities of Dehalococcoides ethenogenes strain 195. Appl. Environ. Microbiol. 71, 1664–1667 10.1128/AEM.71.3.1664-1667.2005 (doi:10.1128/AEM.71.3.1664-1667.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jayachandran G, Görisch H, Adrian L. 2004. Studies on hydrogenase activity and chlorobenzene respiration in Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 182, 498–504 10.1007/s00203-004-0734-9 (doi:10.1007/s00203-004-0734-9) [DOI] [PubMed] [Google Scholar]
- 51.White DC, Geyer R, Peacock AD, Hedrick DB, Koenigsberg SS, Sung Y, He J, Löffler FE. 2005. Phospholipid furan fatty acids and ubiquinone-8: lipid biomarkers that may protect Dehalococcoides strains from free radicals. Appl. Environ. Microbiol. 71, 8426–8433 10.1128/AEM.71.12.8426-8433.2005 (doi:10.1128/AEM.71.12.8426-8433.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan J, Ritalahti KM, Wagner DD, Löffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78, 6630–6636 10.1128/aem.01535-12 (doi:10.1128/aem.01535-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi S, Seth EC, Men Y-J, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports the corrinoid-dependent metabolism of Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78, 7745–7752 10.1128/aem.02150-12 (doi:10.1128/aem.02150-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Escalante-Semerena JC. 2007. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J. Bacteriol. 189, 4555–4560 10.1128/JB.00503-07 (doi:10.1128/JB.00503-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gray MJ, Tavares NK, Escalante-Semerena JC. 2008. The genome of Rhodobacter sphaeroides strain 2.4.1 encodes functional cobinamide salvaging systems of archaeal and bacterial origins. Mol. Microbiol. 70, 824–836 10.1111/j.1365-2958.2008.06437.x (doi:10.1111/j.1365-2958.2008.06437.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. 2007. A new thiamin salvage pathway. Nat. Chem. Biol. 3, 492–497 10.1038/nchembio.2007.13 (doi:10.1038/nchembio.2007.13) [DOI] [PubMed] [Google Scholar]
- 57.Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T. 2007. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem. J. 407, 1–13 10.1042/BJ20070765 (doi:10.1042/BJ20070765) [DOI] [PubMed] [Google Scholar]
- 58.Fischer M, Bacher A. 2005. Biosynthesis of flavocoenzymes. Nat. Product Rep. 22, 324–350 10.1039/b210142b (doi:10.1039/b210142b) [DOI] [PubMed] [Google Scholar]
- 59.Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. 2008. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 68, 918–932 10.1111/j.1365-2958.2008.06208.x (doi:10.1111/j.1365-2958.2008.06208.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pribat A, Jeanguenin L, Lara-Núñez A, Ziemak MJ, Hyde JE, de Crécy-Lagard V, Hanson AD. 2009. 6-Pyruvoyltetrahydropterin synthase paralogs replace the folate synthesis enzyme dihydroneopterin aldolase in diverse bacteria. J. Bacteriol. 191, 4158–4165 10.1128/jb.00416-09 (doi:10.1128/jb.00416-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dittrich S, Mitchell SL, Blagborough AM, Wang Q, Wang P, Sims PFG, Hyde JE. 2008. An atypical orthologue of 6-pyruvoyltetrahydropterin synthase can provide the missing link in the folate biosynthesis pathway of malaria parasites. Mol. Microbiol. 67, 609–618 10.1111/j.1365-2958.2007.06073.x (doi:10.1111/j.1365-2958.2007.06073.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. 2004. Conservation of mechanism in three chorismate-utilizing enzymes. J. Am. Chem. Soc. 126, 2378–2385 10.1021/ja0389927 (doi:10.1021/ja0389927) [DOI] [PubMed] [Google Scholar]
- 63.Kuratsu M, Hamano Y, Dairi T. 2010. Analysis of the Lactobacillus metabolic pathway. Appl. Environ. Microbiol. 76, 7299–7301 10.1128/aem.01514-10 (doi:10.1128/aem.01514-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slock J, Stahly DP, Han CY, Six EW, Crawford IP. 1990. An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J. Bacteriol. 172, 7211–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selengut JD, Haft DH. 2010. Unexpected abundance of coenzyme F420-dependent enzymes in Mycobacterium tuberculosis and other Actinobacteria. J. Bacteriol. 192, 5788–5798 10.1128/JB.00425-10 (doi:10.1128/JB.00425-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolin EA, Wolin MJ, Wolfe RS. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238, 2882–2886 [PubMed] [Google Scholar]
- 67.Scholz-Muramatsu H, Neumann A, Messmer M, Moore E, Diekert G. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163, 48–56 10.1007/BF00262203 (doi:10.1007/BF00262203) [DOI] [Google Scholar]
- 68.Gerritse J, Renard V, Pedro G, Lawson PA, Collins MD, Gottschal JC. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165, 132–140 10.1007/s002030050308 (doi:10.1007/s002030050308) [DOI] [PubMed] [Google Scholar]
- 69.Louie TM, Mohn WW. 1999. Evidence of a chemiosmotic model of dehalorespiration in Desulfomonile tiedjei DCB-1. J. Bacteriol. 181, 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holliger C, Hahn D, Harmsen H, Ludwig W, Tindall B, Vazquez F, Weiss N, Zehnder AJB. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169, 313–321 10.1007/s002030050577 (doi:10.1007/s002030050577) [DOI] [PubMed] [Google Scholar]
- 71.Louie TM, Ni S, Xun L, Mohn WW. 1997. Purification and gene sequence analysis of a novel cytochrome c co-induced with reductive dechlorination activity in Desulfomonile tiedjei DCB-1. Arch. Microbiol. 168, 520–527 10.1007/s002030050530 (doi:10.1007/s002030050530) [DOI] [PubMed] [Google Scholar]
- 72.He J, Holmes VF, Lee PKH, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73, 2847–2853 10.1128/aem.02574-06 (doi:10.1128/aem.02574-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson DR, Nemir A, Andersen GL, Zinder SH, Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol. Lett. 294, 198–206 10.1111/j.1574-6968.2009.01569.x (doi:10.1111/j.1574-6968.2009.01569.x) [DOI] [PubMed] [Google Scholar]
- 74.Adrian L. 2009. ERC-group Microflex: Microbiology of Dehalococcoides-like Chloroflexi. Rev. Environ. Sci. Biotechnol. 8, 225–229 10.1007/s11157-009-9166-y (doi:10.1007/s11157-009-9166-y) [DOI] [Google Scholar]