Abstract
Organohalide respiration is an anaerobic bacterial respiratory process that uses halogenated hydrocarbons as terminal electron acceptors during electron transport-based energy conservation. This dechlorination process has triggered considerable interest for detoxification of anthropogenic groundwater contaminants. Organohalide-respiring bacteria have been identified from multiple bacterial phyla, and can be categorized as obligate and non-obligate organohalide respirers. The majority of the currently known organohalide-respiring bacteria carry multiple reductive dehalogenase genes. Analysis of a curated set of reductive dehalogenases reveals that sequence similarity and substrate specificity are generally not correlated, making functional prediction from sequence information difficult. In this article, an orthologue-based classification system for the reductive dehalogenases is proposed to aid integration of new sequencing data and to unify terminology.
Keywords: organohalide respiration, reductive dehalogenase, phylogenetics
1. Organohalide respiration
Organohalide respiration is the preferred term for the energy-conserving respiratory process wherein a halogen–carbon bond is broken and the halogen atom is liberated as a halide. The removal of halogens from these compounds may reduce or eliminate toxicity or render the compound more biodegradable, making this process important for the remediation of contaminated sites [1]. Organohalide-respiring bacteria (OHRB) are micro-organisms capable of deriving energy for growth from dehalogenation of aromatic and/or aliphatic halogenated compounds. These bacteria are of environmental importance because they reduce anthropogenic halogenated compounds, many of which are significant contaminants in groundwater systems that pose hazards to human health and the environment [2,3]. Beyond bioremediation, the activities of OHRB are part of the global halogen cycle [4].
OHRB have been identified from diverse bacterial phyla, including the Proteobacteria, Firmicutes and Chloroflexi [5–8]. The known OHRB can be grouped as either obligate or non-obligate organohalide respirers [9]. The proteobacterial OHRB, including Geobacter, Desulfuromonas, Anaeromyxobacter and Sulfurospirillum, are all non-obligate organohalide respirers with versatile metabolisms encoded on relatively large genomes [9]. In contrast, the currently known organohalide-respiring Chloroflexi are all obligate organohalide respirers, meaning they are niche specialists with a very restricted metabolism. The Firmicutes contain non-obligate organohalide-respiring Desulfitobacterium spp. as well as metabolically restricted Dehalobacter OHRB [9]. It has recently been shown that some Dehalobacter spp. are able to ferment dichloromethane [10,11]. There is little to no correlation between phylogenetic affiliation and chlorinated substrate specificities: aliphatic and aromatic substrates are used by taxonomically diverse organisms [12].
Genome sequences exist for several OHRB, including five Dehalococcoides mccartyi strains [13–16] and Dehalogenimonas lykanthroporepellens strain BL-DC-9 [17] within the Chloroflexi; Geobacter lovleyi strain SZ [18] and Anaeromyxobacter dehalogenans strains 2CP-C and 2CP-1 [19] from the Proteobacteria; and Desulfitobacterium hafniense strains Y51 and DCB-2 [6,20], Desulfitobacterium dehalogenans (NC_018017), Desulfitobacterium dichloroeliminans (NZ_AGJE00000000) and most recently two Dehalobacter strains [21] from the Firmicutes. Given the low cost and broad availability of sequencing from pure or mixed cultures, the number of OHRB genomes will increase rapidly. Consistent with this expectation, a draft genome for Dehalobacter sp. E1 was recently elucidated based on metagenome analysis of a defined co-culture [22]. The availability of OHRB genomes has allowed comparative genomic studies within genera [15,23] as well as across phyla [6]. Genome sequences of the Dehalococcoides have revealed specific metabolic requirements, including corrinoid auxotrophy for the majority of sequenced strains [13–15].
The diversity of OHRB is not yet fully described. Moving forward, novel strains, species and even new genera with organohalide-respiring activity will probably be discovered, enriched and isolated from environments where suitable halogenated electron acceptors, either of natural or anthropogenic origin, are available.
Metagenomes of defined mixed cultures and entire microbial communities are currently being unravelled, allowing determination of metabolic interactions within organohalide-respiring consortia. Metagenomes were sequenced from consortia such as (i) the KB-1 culture, containing populations of Dehalococcoides, Geobacter, Methanosarcina, Spirochaeta and Sporomusa [24,25]; (ii) the ANAS tetrachloroethene (PCE)-dechlorinating mixed culture containing Dehalococcoides, fermentative bacteria and two methanogenic populations, among other organisms [26,27]; and (iii) a highly stable and efficient dechlorinating bioreactor community containing Dehalococcoides [26,28]. In addition, a defined coculture of Dehalobacter sp. E1 and Sedimentibacter sp. B4 that reductively dechlorinates beta-hexachlorocyclohexane has been sequenced [22].
2. Reductive dehalogenases
Organohalide respiration reactions are catalysed by reductive dehalogenases. The first reductive dehalogenase that was biochemically characterized was the 3-chlorobenzoate reductive dehalogenase of Desulfomonile tiedjei strain DCB-1 [29]. The first gene sequences encoding reductive dehalogenases were identified using classical reverse genetics approaches based on partial amino acid sequences of purified enzymes [30,31]. PCR and genome studies have since identified new sequences with high sequence identity to the characterized genes [6,13,14,32–42]. Reductive dehalogenase genes (or reductive dehalogenase homologous genes (rdh) if the encoded protein has not yet been biochemically characterized) typically comprise an operon containing rdhA, the gene for the catalytically active enzyme (RDase if characterized, otherwise RdhA), rdhB, a gene encoding a putative membrane-anchoring protein [34], and sometimes one or more members of rdhTKZECD, genes associated with reductive dehalogenase genes [43]. For some, but not all, of the predicted gene products, function in transcription regulation and maturation of the holoenzyme has been experimentally confirmed [44,45].
The nomenclature of the rdh genes as homologues is based on the presence of specific conserved motifs [46]. RdhA proteins have several conserved features, including two iron–sulfur cluster-binding motifs and a twin-arginine signal motif for translocation to or across the cell membrane [34]. Many, if not all, characterized RdhAs contain corrinoid co-factors (derivatives of vitamin B12), including the PCE dehalogenase from Sulfurospirillum multivorans (formerly Dehalospirillum), which was shown to contain the very specific, previously unknown corrinoid norpseudo-B12 [47]. Involvement of a Co(I) corrinoid in the catalytic activity of reductive dehalogenases has been demonstrated by the reversible inactivation of corrinoid using propyl iodides for a number of enzymes, and by the fact that the lack of the corrinoid cofactor resulted in loss of the PCE-dechlorination ability in S. multivorans and Dehalobacter restrictus PER-K23 [38,48]. Six chlorophenol and four PCE/trichloroethene (TCE) reductive dehalogenases have been purified from different Desulfitobacterium strains, and the majority of reductive dehalogenases that have so far been tested for the presence of a corrinoid prosthetic group contained such a group [49]. These corrinoid co-factors are thought to be essential for enzymatic function, and show some specificity: cocultures of Geobacter strains and Dehalococcoides mccartyi strains revealed cobamides produced by the OHRB Geobacter lovleyi strain SZ supported dechlorination by the Dehalococcoides strain, while cobamides produced by the non-OHRB Geobacter sulfurreducens did not [50].
Although the identity of several rdhA genes has been confirmed based on partial (N-terminal) amino acid sequences of purified enzymes with proven activity, the three-dimensional structure of an RdhA protein has not yet been determined, nor has an active site been identified. Furthermore, identification of most rdhA genes, especially those encountered in genome sequences of OHRB and other bacteria, has been based entirely on sequence similarity and the presence of the above-mentioned motifs. It is, in addition, not certain if all RdhA proteins are homologous (i.e. share a common evolutionary origin). To this end, we are maintaining the nomenclature of rdhA genes as homologues based on high sequence similarity along the entire length of the genes, conservation of shared motifs, and an absence of evidence of convergent evolution.
Reductive dehalogenase encoding genes have been identified in a wide variety of strictly anaerobic bacteria, including Sulfurospirillum [51], Desulfitobacterium [1,31,52,53], Dehalobacter [7] and Dehalococcoides [13–15,54], and in microaerophilic bacteria as in the case of Anaeromyxobacter [19], and others (figure 1) [18,58]. Only one archaeal putative reductive dehalogenase gene has been identified to date from a Ferroglobus species [59], but this organism has not been demonstrated to conserve energy via organohalide respiration. Most, but not all, of these rdhA gene-carrying organisms are known to reduce halogenated organic compounds. Within the known dehalogenating organisms, some do not use halogenated substrates for energy conservation in a respiratory process; for example, a strain of Dehalobacter has been shown to ferment dichloromethane [11].
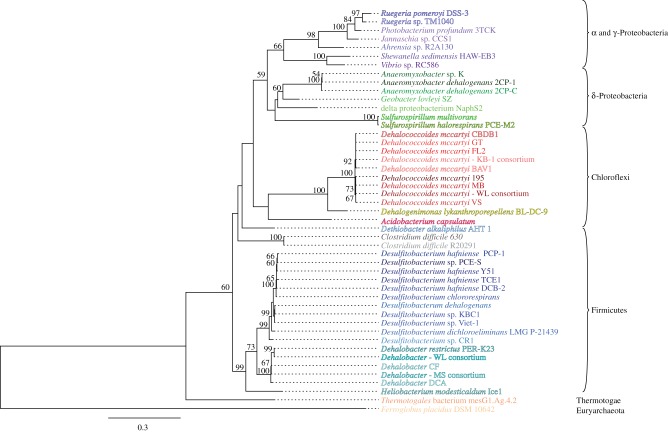
Figure 1.
Maximum-likelihood (ML) tree of 16S rRNA gene sequences from all known organisms containing a reductive dehalogenase homologous (rdhA) gene. For organisms with multiple 16S rRNA genes, one representative gene sequence was chosen for clarity (see the electronic supplementary material, table S2 for 16S rRNA gene sequence accessions). 16S rRNA gene sequences were mined from IMG-M and NCBI. The sequences were aligned using the greengenes NAST alignment algorithm [55], and manually curated and masked in Geneious [56] to a final alignment of 1117 unambiguously aligned positions. Ten ML trees were calculated using RAxML HPC v. 7.2.8 [57] under the GTR+γ model of nucleotide evolution, and the tree with the highest likelihood chosen. Organism names are coloured to correspond to the colours on the RdhA tree (figure 2).
The reductive dehalogenases present within an organism, enrichment culture or contaminated site dictate the range of halogenated electron acceptors used. Several molecular tools have been developed for examining a broad range of rdhA genes in a given sample (site, isolate or mixed culture), including both microarray-based methods and PCR-based protocols [60–63]. Identification of novel reductive dehalogenase genes has primarily come from PCR studies [32,33,35–37,39,40,42] and subsequently from genome and metagenome sequencing efforts [6,13,14,41,63], with each new organism or environment sampled increasing the known diversity of putative rdhA genes.
Analysis of genome sequences divides the OHRB into those whose genomes contain one or two rdhA genes and those containing several (more than two) different rdhA [5,64]. Genomes of Desulfitobacterium, Dehalococcoides, Dehalogenimonas and Dehalobacter strains contain multiple rdhA genes. The complete genome sequence of Desulfitobacterium hafniense strain DCB-2 contains seven putative rdhA genes [20]. From the Dehalococcoides mccartyi genomes, the current minimum number of rdhA genes per genome is 10 [15], with up to 36 rdhA genes present on a single genome [15]. The genome of Dehalococcoides mccartyi (formerly ethenogenes) strain 195 [65] harbours 17 different rdhA genes [14], while strain CBDB1′s genome contains 32 rdhAs [13]. The genome of Dehalogenimonas lykanthroporepellens strain BL-DC-9, the only sequenced Dehalogenimonas, contains 19 rdhA genes [17,66]. The presence of multiple rdhA genes is also a feature in the Dehalobacter genus, with up to 24 complete putative rdhA genes within sequenced Dehalobacter genomes [21,22,67].
(a). Substrate specificity of reductive dehalogenases
Only few reductive dehalogenases have been biochemically characterized. The enzymes are inactivated by molecular oxygen, and a genetically tractable system for expressing a specific reductive dehalogenase is not yet available. Both these factors have impeded extensive biochemical experimentation in the past. In the case of Dehalococcoides, Dehalogenimonas and some Dehalobacter, detailed studies on the biochemistry of reductive dehalogenases have been hampered by difficulties in obtaining sufficient biomass. Substrates for RDases have been identified using culture-based methods as well as by enzymatic assays with pure RDases. Much of our understanding of reductive dehalogenases has come from well-studied organisms, including Sulfurospirillum multivorans and Dehalobacter restrictus PER-K23 which dechlorinate PCE to cis-dichloroethene (DCE) [7,68–70]. PceAs, active on the chlorinated ethenes PCE and TCE, have been identified from Sulfurospirillum, Desulfitobacterium and Dehalobacter. The four known Desulfitobacterium PCE/TCE reductive dehalogenases were isolated from Desulfitobacterium hafniense strains PCE-S, TCE1 and Y51, and Desulfitobacterium sp. strain PCE1. The Sulfurospirillum and Desulfitobacterium PCE/TCE reductive dehalogenases cannot dehalogenate isomers of DCE. The substrate spectrum dehalogenated by the reductive dehalogenase of Desulfitobacterium hafniense strain Y51 is similar to the PCE reductive dehalogenase of Dehalobacter restrictus [35,38].
CprA and CrdA dehalogenases from Desulfitobacterium dehalogenans and Desulfitobacterium hafniense dechlorinate a variety of chlorinated phenols [31,37,39,71]. Substrate specificities towards ortho or meta and para dechlorination are observed for the different CprAs. CrdA is a unique chlorophenol reductive dehalogenase that is capable of ortho dechlorination of several polychlorinated phenols but not of 3-chloro-4-hydroxyphenylacetate, in contrast to the other chlorophenol reductive dehalogenases [37]. CrdA was isolated from Desulfitobacterium strain PCP-1 and the crdA gene is also present in Desulfitobacterium hafniense strains TCP-A, DP7, DCB-2, Y51 and TCE1, Desulfitobacterium dehalogenans, Desulfitobacterium chlororespirans and Desulfitobacterium sp. strain PCE-1 [72]. Another unique chlorophenol reductive dehalogenase is CprA5 found in Desulfitobacterium hafniense strains PCP-1, TCP-A and DCB-2, which has dechlorination activity against several chlorophenols at all three substituent (ortho, meta and para) positions [39,72].
A 1,2-dichloroethane (1,2-DCA)-specific reductive dehalogenase was identified in Desulfitobacterium dichloroeliminans strain DCA1 and also in the metagenome of a 1,2-DCA organohalide-respiring enrichment culture from a contaminated aquifer [73]. Sequence analysis of the catalytic subunit of these DcaA proteins showed specific sequence differences and signature motifs compared with the other known reductive dehalogenases, suggesting that these enzymes may have specifically adapted to 1,2-DCA reductive dechlorination [73].
Within the Dehalococcoides, five proteins involved in respiring chlorinated ethenes, ethanes and chlorinated benzenes have been partially characterized. PceA has been shown to dechlorinate PCE to TCE [54]. TceA has been shown to dechlorinate TCE to ethene, though was most active on TCE [34], and VcrA catalyses the dechlorination of TCE, DCE isomers and vinyl chloride (VC) to ethene [40,74]. The VcrA protein is additionally capable of dihaloelimination of 1,2-DCA to ethene [74]. Using native polyacrylamide gel electrophoresis (PAGE), CbrA from Dehalococcoides mccartyi strain CBDB1 was found to dechlorinate 1,2,3,4-tetrachlorobenzene and 1,2,3-trichlorobenzene [75]. Recent studies using blue native PAGE have confirmed that BvcA also catalyses dechlorination of TCE, DCE isomers, VC and 1,2-DCA and further expands the known substrate ranges for the enzymes BvcA, VcrA and TceA [76], indicating that at least these RDases, and probably many others, can dechlorinate multiple substrates.
Beyond these partially characterized reductive dehalogenases, many RdhA proteins have been assigned a putative function through transcriptional analyses in the presence of specific substrates, or through peptide sequencing of partially purified proteins produced during specific stages of dechlorination [40,61,77,78]. While these proteins often cannot be assigned substrate specificity with certainty, these experiments provide evidence for function and can be used to better illustrate the sequence similarity/substrate affinity relationship within this group of enzymes. These methods have been somewhat confounded in the case of Dehalococcoides, where multiple reductive dehalogenase genes are typically expressed in the presence of a single halogenated substrate [79,80].
The oxygen sensitivity of reductive dehalogenases, their association with the cell membrane, and the tendency for multiple rdhA genes to be transcribed have impeded determination of substrate specificities or tertiary structures for these enzymes. Blue Native PAGE is a promising approach for examining in vitro reductive dehalogenase activity [76,81]. The current frontier of biochemical research on the reductive dehalogenases involves attempts to heterologously express reductive dehalogenase encoding genes and to engineer an OHRB with desirable substrate preferences. Reductive dehalogenase genes have been successfully cloned, but efforts to produce an active enzyme have failed so far [30,82,83].
(b). Sequence analysis of reductive dehalogenases
This themed issue is the result of a meeting at the KAVLI centre, UK, on 4–5 July 2011. At the meeting, a need for a classification system and a consistent nomenclature was identified. To address this need, the diversity of reductive dehalogenases is examined based on sequence analysis on a curated protein dataset of RDases and RdhA proteins. The ability to predict an organism's dechlorination potential based on sequence similarity and the known functional information for RDases is examined, and orthologous groups for transparent nomenclature of the gene family are defined. The orthologue groups are intended to serve as a reference point for all subsequently identified RdhA proteins, and to facilitate comparisons between sequenced organohalide respirers. Orthologues were defined based on pairwise sequence identities greater than 90 per cent at the amino acid level, and orthologue groups constrained to this level of sequence identity. Additionally, orthologue groups were confirmed as highly supported clades in protein tree analyses.
A basic search for ‘reductive dehalogenase’ in the NCBI nucleotide database results in over 600 sequences as of June 2012. For the curated, comprehensive dataset of RdhA proteins discussed below, a reductive dehalogenase homologue had to have been sequenced from a known organism for which a 16S rRNA gene sequence was available. Based on this filter, environmental sequences from contaminated sites, ocean surveys and uncharacterized enrichment cultures were excluded. It was not required that the organism had demonstrated reductive dehalogenation activity. In the majority of cases, rdhA genes were mined from finished genome sequences, or from known dechlorinating isolates whose genomes have not yet been sequenced (e.g. Dehalococcoides mccartyi strain MB [60,61]). rdhA genes from consortia (KB-1, ACT-3 and WL) were included in cases where metagenome sequences or quantitative PCR studies allow connection of rdhA sequences with specific OHRB genera [24,84–86]. Additionally, the rdhA genes were required to be full-length or close to full-length (greater than 300 amino acids), which excluded many partial rdhA genes in the database. Only the catalytically active subunit (RdhA) was included in this analysis; the RdhB proteins were not considered. The final curated set contained 264 RdhA proteins from 44 micro-organisms (see the electronic supplementary material, table S1 for sequence names, accessions and classification identifiers). Sequence comparisons were conducted both including and excluding the twin-arginine translocation (Tat) signal sequences (approx. 50 amino acids, composed of the characteristic Tat-motif and a hydrophobic stretch) at the N-termini of the proteins, which may be expected to evolve at a different rate from motifs involved in catalysis [87]. For further comparison, phylogenetic analyses on the aligned TIGRFAM reductive dehalogenase domain (removing Tat signal sequences as well as C-terminal FeS cluster-binding motifs from the original alignment) were conducted.
The Tat signal sequence trimmed alignment gave the best-resolved tree in terms of strongest bootstrap support across the tree. In general, the relationships defined by the three trees (full-length proteins, Tat signal-trimmed and TIGRFAM domain only) were consistent, with the same overall topology, and with all orthologue groups recovered with high bootstrap support and posterior probabilities (greater than 90%, greater than 0.9, respectively). Many of the differences between the three trees were in the branching order of divergent, long-branch sequences, which may be the result of long-branch attraction artefacts. Other topological differences were the result of rearrangements of the backbone of the tree, which was poorly supported in all three trees, and as such, the correct topology in the tree backbone cannot be ascertained. The Tat signal-trimmed tree is depicted in figure 2, while all three trees are presented with bootstrap support values in the electronic supplementary material, figures S1–S3.
Figure 2.
ML tree of 264 reductive dehalogenases and translated rdhA genes based on an amino acid alignment of the TIGRFAM domain and FeS-binding domains. The protein sequences were aligned using the Geneious muscle plugin [56,88] and the alignment iteratively refined using Hmmer v. 2.3.2 until the iterations converged on a single solution [89]. The alignment was manually masked to remove ambiguously aligned positions, leaving a final alignment with 831 positions. The best model of amino acid substitution was determined using ProtTest v. 3 [90,91]. ML trees were generated using RAxML HPC v. 7.2.8 [57] under the LG+γ+F model of amino acid substitution with 100 bootstrap resampling trees conducted. Bayesian inference trees were generated using Mr. Bayes [92], with aamodelpr set to ‘mixed’, lset rates as ‘invgamma’, and default conditions otherwise. The analyses were continued until the standard deviation of split frequencies was below 0.1 (400 000 generations). Posterior probabilities were generated with a final burn-in of 150 000 and were mapped onto the nodes of the ML tree for display. Protein accessions are coloured by organism as in figure 1: red, Dehalococcoides; yellow, Dehalogenimonas; blue/grey, Firmicutes; purple, α and γ-Proteobacteria; green, δ-Proteobacteria; orange, Thermotogae and Archaea. See electronic supplementary material, figure S1 for a rectangular version of this phylogeny with scale bar and bootstrap support values for this tree. Reductive dehalogenases with known function on specific substrates, based on protein isolation, proteomics or transcriptomics are numbered as follows: (1) TceA (DET0079_tceA, Dehalococcoides mccartyi st. 195; TCE) [34]; (2) BvcA (BAV1_0847, Dehalococcoides mccartyi st. BAV1; VC, cis-DCE, 1,2-DCA) [40]; (3) VcrA (8658217VS, Dehalococcoides mccartyi st. VS; VC, TCE, cis-DCE) [74]; (4) MbrA (GU120391 (MB_mbrA on tree), Dehalococcoides mccartyi st. MB; TCE) [60]; (5) CbrA (cbdbA84, Dehalococcoides mccartyi st. CBDB1; 1,2,3,4-tetrachlorobenzene, 1,2,3-trichlorobenzene, pentachlorobenzene) [75]; (6) PceA (DET0318, Dehalococcoides mccartyi st. 195; PCE) [54]; (7) PceA (AJ439607, Dehalobacter restrictus PER-K23; PCE) [38]; (8) CprA (AY349165, Desulfitobacterium hafniense st. PCP-1; pentachlorophenol, 2,3,4,5-tetrachlorophenol, and 2,3,4-trichlorophenol) [71]; (9) CprA (AF259790, Desulfitobacterium dehalogenans; ortho-chlorophenols) [31]; 10-PrdA (AB194706, Desulfitobacterium sp. strain KBC1; PCE) [93]; (11) CfrA (ACT3_rdh02, Dehalobacter sp. strain CF; 1,1,1-trichloroethane, CF) [81]; (12) DcrA (ACT3_rdh01, Dehalobacter sp. strain DCA; 1,1-DCA) [81]; (13) DcrA (FJ010189 (AGWLrdh01 on tree), Dehalobacter sp. strain WL; 1,2-DCA) [94]; (14) DcaA (AM183918, Desulfitobacterium dichloroeliminans; 1,2-DCA) [73]; (15) PceA (AF022812; Sulfurospirillum multivorans; PCE) [51].
The RdhA protein tree is dominated by sequences from Dehalococcoides (red), Dehalogenimonas (yellow), Dehalobacter (light blue) and Desulfitobacterium (dark blue) (figure 2). The preponderance of these organisms is partially due to the intense research efforts concentrated on these bacterial groups based on their environmental significance as dechlorinating organisms, but is also due to the fact that genomes from these organisms typically contain multiple rdhA genes [20,32,80,94].
The Chloroflexi RdhA proteins, predominantly from the Dehalococcoides, cluster within a large clade. Within this large Dehalococcoidales clade, several sub-clades of RdhA identified in multiple Dehalococcoides mccartyi strains share branching order with the 16S rRNA gene tree. In several instances, orthologous RdhAs (greater than 90% pairwise amino acid identity (PID) over the full-length of the sequences and bidirectional best BLASTp hits) are present in only two or three strains of the six sequenced strains. This patchy distribution of rdhA genes points either to rapid gene loss within the Dehalococcoides, or to lateral acquisition of novel rdhA genes within subsets of Dehalococcoides mccartyi strains. Lateral gene transfer has been invoked for rdhA gene acquisition in several scenarios [95–99] and is potentially a factor in the RdhA family diversification. Sequence information from new strains and complete genome sequences from strains for which some rdhA genes have been identified will help clarify orthologue and paralogue relationships.
The only core RdhA present in all sequenced Dehalococcoides genomes (11 o'clock in figure 2) falls outside of the main Dehalococcoidales RdhA clade. This core RdhA clade matches the 16S rRNA phylogeny in terms of strain relationships, and shares synteny across Dehalococcoides genomes [15], indicating it may have diverged within Dehalococcoides from a shared ancestral gene. A second smaller clade of Dehalococcoides sequences is also present in the non-Dehalococcoides region of the tree (next to marked conserved syntenic clade in figure 2), though only three sequenced strains are represented (CBDB1, 195 and VS). These clades may represent distant orthologues to non-Dehalococcoides RdhA proteins, though the current analysis is not conclusive.
There are 15 RdhA proteins on the tree (see numbers in figure 2), for which functional information is available in the form of transcriptional data or biochemical characterization of purified proteins. These RDases are distributed across the RdhA tree, and are known to catalyse the dehalogenation of a broad range of substrates, including chlorinated ethenes (VcrA, BvcA, TceA, MbrA, PceA, PrdA [38,40,54,60,74,93,100]), chlorinated ethanes (CfrA, DcrA, DcaA [73,81,94]), and chlorinated aromatics (CbrA, CprA [31,71,75,101]). For most characterized reductive dehalogenases, the full substrate range is not known; instead activity on a specific substrate has been verified. It is clear that reductive dehalogenases that share a specific function do not necessarily share high sequence similarity: the four PCE-dehalogenating enzymes (nos. 6, 7, 10 and 15) are located at three distant positions on the tree. Similarly, the two chlorophenol reductive dehalogenases (nos. 8 and 9) are at opposite edges of the non-Dehalococcoides RdhA portion of the tree and have relatively low sequence similarity (28% PID). One note from this is that neither the structure nor the identities of the catalytic amino acid residues in the active centre of the reductive dehalogenase enzymes are known. It is possible that the dehalogenation of similar targets is conducted using different catalytic mechanisms, but it is also possible that shared residues and motifs between these divergent sequences with shared function will provide clues as to the critical residues for RdhA folding and function. When genetic manipulation of these targets becomes possible, those shared residues would be primary targets of interest for mutation experiments.
In a single example of sequence identity hinting at function, the chlorinated ethene reductive dehalogenases TceA (DET0079_tceA), BvcA (BAV1_0847) and VcrA (DhcVS_1291) in Dehalococcoides form a monophyletic group on the tree (with approx. 48% PID), and proteins in this group are associated with enrichment cultures capable of dechlorinating chlorinated ethenes and 1,2-DCA. In general, the RdhA phylogeny does not indicate that similarity analysis of reductive dehalogenases will allow confident prediction of substrate specificity for novel RdhA proteins identified. An example of this concerns the DcrA (ACT3_rdh01) and CfrA (ACT3_rdh02) RDases from Dehalobacter strains DCA and CF, respectively, from the ACT-3 culture, which share a high level of sequence similarity (95% PID), group closely together (nos. 11 and 12, at 8.30 in figure 2), but do not share overlapping substrate specificities [81]. Similarly, the Dehalobacter/Desulfitobacterium PceAs share 88–89% PID with the Desulfitobacterium dichloroeliminans DcaA but do not share overlapping substrate specificities [73]. These two examples present the reverse scenario to the four PceA proteins, where amino acid residues that differ between the two proteins are potentially important in substrate specificity.
(c). Classification system for reductive dehalogenases
The need for a classification system for the reductive dehalogenases is tied to the rapid expansion of the family through genome and metagenome sequencing. These enzymes and genes are being used extensively as biomarkers for monitoring active and passive bioremediation at contaminated sites. Greater clarity in nomenclature would facilitate this transfer of science to practice. Typically, comparisons of proteins encoded by novel rdhA genes focus on the characterized reductive dehalogenases. As discussed above, these provide only a partial picture of the sequence diversity of the protein family. In defining a classification system for the reductive dehalogenases, we hope to clarify the relationships within the family as well as provide a platform for more robust comparisons of new sequences to the existing family. Furthermore, such classification can guide the selection of representative genes and proteins for more detailed functional characterization.
Reductive dehalogenases are included in the TIGRFAM and PFAM databases of homologous proteins (TIGR02486 and PF13486), but are not recognized as a protein family by other major databases (PROSITE and SMART). In the absence of a crystal structure and information as to key residues for folding or the active site, it is difficult to implement many of the standard methods for classifying protein families. Instead, we have implemented a sequence identity-based classification of orthologues into groups, anchored by the tree of curated reductive dehalogenase protein sequences.
We defined orthologue groups based on a threshold of 90% PID in amino acid alignments between all members of a group that are additionally supported on the tree with bootstrap support values more than 90%. PID and similarity scores for all RdhAs within this curated amino acid dataset were generated using ClustalW [102,103]. The orthologue groups allow direct comparisons of RdhAs between current sequenced strains and facilitate placement of newly sequenced RdhAs in the protein family. Based on these criteria, we have identified 46 orthologue groups containing in total 176 RdhA proteins. The minimum number of proteins in a group is 2, while the current maximum is 10. A complete description of the defined groups, including organisms and gene names within each group, number of encoded proteins per group, and the respective PID ranges within the groups is presented in the electronic supplementary material, table S3, and the groups are highlighted in colour in the electronic supplementary material, figure S1.
In defining such a classification system for the reductive dehalogenase family, we endeavoured to create a flexible format that was informative, and which would not require constant revision as new RdhA proteins are described. To that end, all orthologue groups have been named sequentially based on the date of first publication of the earliest described sequence in the group. For example, the first sequenced RdhA was the tetrachloroethene reductive dehalogenase PceA from Sulfurospirillum multivorans in August 1998 [30], and as a result, the orthologue group containing this protein has been designated Reductive Dehalogenase Orthologue Group 1 (RD_OG1). In the case where multiple RdhA proteins were reported in the same article [13,14,32,40], group numbers were assigned based on locus tag or accession numbers, in ascending order. In one case, two RdhAs were presented in the same journal issue [39,74]; in this case, the lower group number was assigned based on alphabetical order of first author names.
An advantage to this classification method compared with large clade-based classification methods [9,15] is that the orthologue groups are not expected to change with identification of divergent sequences, while larger order relationships may shift with additional sequences. Given the lack of support for the backbone of the protein tree, the proposed classification system represents a conservative solution. The intention for this classification system moving forward is to allow the addition of new groups as novel RdhA proteins are discovered. A novel group would be created when a full-length or near full-length (greater than 75%) RdhA from a known organism was identified that had 90% or more PID with an RdhA not currently placed in an orthologue group. The group would be defined as containing that pair of sequences, and it would receive the next available group number. Such a classification system allows flexibility for adding novel sequences and provides a level of intuitive interpretation, where the lower the group number, the longer that particular RdhA group has been known or studied in multiple organisms.
(i). An example: the WBC-2 consortium
WBC-2 is a microbial consortium capable of dechlorinating chlorinated ethenes and ethanes [104]. This consortium contains Dehalobacter, Dehalococcoides and Dehalogenimonas strains [104]. Previous sequence data from this consortium have been limited to 16S rRNA gene clones, leaving the reductive dehalogenase complement unknown. A metagenome from a 1,1,2,2-tetrachloroethane-degrading WBC-2 subculture was sequenced and partially assembled, resulting in 144 846 contigs with total scaffold length of 68 536 230 bp, a maximum contig length of 334 963 and an N50 of 2192 bp. A total of 16 reductive dehalogenase genes were identified from the WBC-2 metagenome using tblastx with the curated gene set as queries. Of these, 13 were readily classified into existing orthologue groups as described above, while the remaining three shared greater than 90% PID with a single existing sequence. A protein tree placing the WBC-2 RdhA within the orthologue groups can be found in the electronic supplementary material, figure S4. As the reductive dehalogenases from the WBC-2 consortium cannot be ascribed to a specific OHRB, this example illustrates how novel sequences can be described in the context of the RD_OGs rather than how a new RD_OG could be generated and numbered.
The curated reductive dehalogenase dataset, a detailed method for incorporating novel reductive dehalogenase sequences into the RD_OG framework and for creating new RD_OGs, and a spreadsheet detailing the current RD_OGs are all available for public access and editing at the following link: docs.google.com/folder/d/0BwCzK8wzlz8ON1o2Z3FTbHFPYXc/edit. It is the hope of the authors that this allows simple adoption of this naming system for newly discovered reductive dehalogenase sequences, with iterations of the RD_OG framework easily generated and maintained.
3. Summary
Organohalide respiration is a unique metabolic process that is implicated in the global halogen cycle as well as being of environmental and societal significance for remediation efforts. The reductive dehalogenase family is diverse and growing: newly available genome sequences of organohalide-respiring organisms or gene surveys of natural and anthropogenic environments provide novel RdhA sequences, suggesting the diversity of the protein family is nowhere near exhausted. In this contribution, a curated database of RdhA proteins was generated and a phylogenetic comparison of the RdhA-encoding organisms and their respective RdhA proteins conducted. From this, a classification system for the RdhAs was proposed based on sequence identity. The intention for this classification is to facilitate placement of novel RdhAs within the context of the gene family, and to aid communication about and comparison of the multiple rdhA genes and their encoded proteins in different strains of organohalide-respiring bacteria.
Acknowledgements
The authors acknowledge the KAVLI centre meeting for generating the impetus for this themed issue. Support was provided to L.A.H. and E.A.E. by the Government of Canada through NSERC, Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405) and the Government of Ontario through the ORF-GL2 program. Support to L.A.H., E.A.E. and F.E.L. was provided by the United States Department of Defence through the Strategic Environmental Research and Development Program (SERDP) (project ER-1586). The Netherlands Genomics Initiative is acknowledged for support to F.M. and H.S. through the Ecogenomics and ECOLINC projects. L.A. and D.L. are supported by the European Research Council (ERC) and L.A. acknowledges further support by the German Research Foundation, DFG-FOR1530. We further acknowledge the helpful suggestions and comments from two anonymous reviewers.
References
- 1.Löffler FE, Sanford RA, Tiedje JM. 1996. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl. Environ. Microbiol. 62, 3809–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creech JL, Johnson MN. 1974. Angiosarcoma of liver in the manufacture of polyvinyl chloride. J. Occup. Med. 16, 150–151 [PubMed] [Google Scholar]
- 3.Jackson RE. 2004. Recognizing emerging environmental problems: the case of chlorinated solvents in groundwater. Technol. Cult. 45, 55–79 10.1353/tech.2004.0022 (doi:10.1353/tech.2004.0022) [DOI] [Google Scholar]
- 4.Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SCB, Novak PJ. 2012. Natural niche for organohalide-respiring Chloroflexi. Appl. Environ. Microbiol. 78, 393–401 10.1128/AEM.06510-11 (doi:10.1128/AEM.06510-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung Y, Fletcher KE, Ritalahti KM, Apkarian RP, Ramos-Hernández N, Sanford RA, Mesbah NM, Löffler FE. 2006. Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl. Environ. Microbiol. 72, 2775–2782 10.1128/AEM.72.4.2775-2782.2006 (doi:10.1128/AEM.72.4.2775-2782.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nonaka H, et al. 2006. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188, 2262–2274 10.1128/JB.188.6.2262-2274.2006 (doi:10.1128/JB.188.6.2262-2274.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJ. 1998. Dehalobacter restrictus gen. nov. and sp. nov, a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169, 313–321 10.1007/s002030050577 (doi:10.1007/s002030050577) [DOI] [PubMed] [Google Scholar]
- 8.Maymó-Gatell X, Chien Y, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276, 1568–1571 10.1126/science.276.5318.1568 (doi:10.1126/science.276.5318.1568) [DOI] [PubMed] [Google Scholar]
- 9.Maphosa F, de Vos WM, Smidt H. 2010. Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria. Trends Biotechnol. 28, 308–316 10.1016/j.tibtech.2010.03.005 (doi:10.1016/j.tibtech.2010.03.005) [DOI] [PubMed] [Google Scholar]
- 10.Lee M, Low A, Zemb O, Koenig J, Michaelsen A, Manefield M. 2012. Complete chloroform dechlorination by organochlorine respiration and fermentation. Environ. Microbiol. 14, 883–894 10.1111/j.1462-2920.2011.02656.x (doi:10.1111/j.1462-2920.2011.02656.x) [DOI] [PubMed] [Google Scholar]
- 11.Justicia-Leon SD, Ritalahti KM, Mack EE, Löffler FE. 2012. Dichloromethane fermentation by a Dehalobacter sp. in an enrichment culture derived from pristine river sediment. Appl. Environ. Microbiol. 78, 1288–1291 10.1128/AEM.07325-11 (doi:10.1128/AEM.07325-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smidt H, de Vos WM. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58, 43–73 10.1146/annurev.micro.58.030603.123600 (doi:10.1146/annurev.micro.58.030603.123600) [DOI] [PubMed] [Google Scholar]
- 13.Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273 10.1038/nbt1131 (doi:10.1038/nbt1131) [DOI] [PubMed] [Google Scholar]
- 14.Seshadri R, et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307, 105–108 10.1126/science.1102226 (doi:10.1126/science.1102226) [DOI] [PubMed] [Google Scholar]
- 15.McMurdie PJ, et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5, e1000714. 10.1371/journal.pgen.1000714 (doi:10.1371/journal.pgen.1000714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72, 1980–1987 10.1128/AEM.72.3.1980-1987.2006 (doi:10.1128/AEM.72.3.1980-1987.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddaramappa S, et al. 2012. Complete genome sequence of Dehalogenimonas lykanthroporepellens type strain (BL-DC-9T) and comparison to ‘Dehalococcoides’ strains. Stand. Genomic Sci. 6, 251–264 10.4056/sigs.2806097 (doi:10.4056/sigs.2806097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner DD, Hug LA, Hatt JK, Spitzmiller MA, Padilla-Crespo E, Ritalahti KM, Edwards EA, Konstantinidis KT, Löffler FE. 2012. Genomic determinants of organohalide-respiration in Geobacter lovleyi, an unusual member of the Geobacteraceae. BMC Genomics 13, 200. 10.1186/1471-2164-13-200 (doi:10.1186/1471-2164-13-200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas SH, Wagner RD, Arakaki AK, Skolnick J, Kirby JR, Shimkets LJ, Sanford RA, Löffler FE. 2008. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS ONE 3, e2103. 10.1371/journal.pone.0002103 (doi:10.1371/journal.pone.0002103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S-H, Harzman C, Davis JK, Hutcheson R, Broderick JB, Marsh TL, Tiedje JM. 2012. Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol. 12, 21. 10.1186/1471-2180-12-21 (doi:10.1186/1471-2180-12-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang S, Gong Y, Edwards EA. 2012. Semi-automatic in silico gap closure enabled de novo assembly of two Dehalobacter genomes from metagenomic data. PLoS ONE 7, e52038. 10.1371/journal.pone.0052038 (doi:10.1371/journal.pone.0052038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maphosa F, van Passel MWJ, de Vos WM, Smidt H. 2012. Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ. Microbiol. Rep. 4, 604–616 10.1111/j.1758-2229.2012.00376.x (doi:10.1111/j.1758-2229.2012.00376.x) [DOI] [PubMed] [Google Scholar]
- 23.Lee PKH, et al. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 5, 1014–1024 10.1038/ismej.2010.202 (doi:10.1038/ismej.2010.202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duhamel M, Edwards EA. 2007. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ. Sci. Technol. 41, 2303–2310 10.1021/es062010r (doi:10.1021/es062010r) [DOI] [PubMed] [Google Scholar]
- 25.Hug LA. 2012. A metagenome-based examination of dechlorinating enrichment cultures: Dehalococcoides and the role of the non-dechlorinating microorganisms, pp. 1–264 PhD thesis, Cell and Systems Biology, University of Toronto, Canada [Google Scholar]
- 26.Hug LA, Beiko RG, Rowe AR, Richardson RE, Edwards EA. 2012. Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: the role of the non-dechlorinating community. BMC Genomics 13, 327. 10.1186/1471-2164-13-327 (doi:10.1186/1471-2164-13-327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brisson VL, West KA, Lee PKH, Tringe SG, Brodie EL, Alvarez-Cohen L. 2012. Metagenomic analysis of a stable trichloroethene-degrading microbial community. ISME J. 6, 1702–1714 10.1038/ismej.2012.15 (doi:10.1038/ismej.2012.15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe AR, Lazar BJ, Morris RM, Richardson RE. 2008. Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc-associated ‘Dehalococcoides’ and Methanospirillum species. Appl. Environ. Microbiol. 74, 6709–6719 10.1128/AEM.00445-08 (doi:10.1128/AEM.00445-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni S, Fredrickson JK, Xun L. 1995. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J. Bacteriol. 177, 5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann A, Wohlfarth G, Diekert G. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180, 4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Pas BA, Smidt H, Hagen WR, van der Oost J, Schraa G, Stams AJ, de Vos WM. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274, 20 287–20 292 10.1074/jbc.274.29.20287 (doi:10.1074/jbc.274.29.20287) [DOI] [PubMed] [Google Scholar]
- 32.Hölscher T, Krajmalnik-Brown R, Ritalahti KM, von Wintzingerode F, Görisch H, Löffler FE, Adrian L. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70, 5290–5297 10.1128/AEM.70.9.5290-5297.2004 (doi:10.1128/AEM.70.9.5290-5297.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Wintzingerode F, Schlötelburg C, Hauck R, Hegemann W, Göbel UB. 2001. Development of primers for amplifying genes encoding CprA- and PceA-like reductive dehalogenases in anaerobic microbial consortia, dechlorinating trichlorobenzene and 1,2-dichloropropane. FEMS Microbiol. Ecol. 35, 189–196 10.1016/S0168-6496(01)00093-9 (doi:10.1016/S0168-6496(01)00093-9) [DOI] [PubMed] [Google Scholar]
- 34.Magnuson JK, Romine MF, Burris DR, Kingsley MT. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66, 5141–5147 10.1128/AEM.66.12.5141-5147.2000 (doi:10.1128/AEM.66.12.5141-5147.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suyama A, Yamashita M, Yoshino S, Furukawa K. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184, 3419–3425 10.1128/JB.184.13.3419-3425.2002 (doi:10.1128/JB.184.13.3419-3425.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee S-K, Fennell DE, Häggblom MM, Kerkhof LJ. 2003. Detection by PCR of reductive dehalogenase motifs in a sulfidogenic 2-bromophenol-degrading consortium enriched from estuarine sediment. FEMS Microbiol. Ecol. 43, 317–324 10.1111/j.1574-6941.2003.tb01072.x (doi:10.1111/j.1574-6941.2003.tb01072.x) [DOI] [PubMed] [Google Scholar]
- 37.Boyer A, Pagé-BéLanger R, Saucier M, Villemur R, Lépine F, Juteau P, Beaudet R. 2003. Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of 2,4,6-trichlorophenol from Desulfitobacterium frappieri PCP-1. Biochem. J. 373, 297–303 10.1042/BJ20021837 (doi:10.1042/BJ20021837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maillard J, Schumacher W, Vazquez F, Regeard C, Hagen WR, Holliger C. 2003. Characterization of the corrinoid iron–sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69, 4628–4638 10.1128/AEM.69.8.4628-4638.2003 (doi:10.1128/AEM.69.8.4628-4638.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thibodeau J, Gauthier A, Duguay M, Villemur R, Lépine F, Juteau P, Beaudet R. 2004. Purification, cloning, and sequencing of a 3,5-dichlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 70, 4532–4537 10.1128/AEM.70.8.4532-4537.2004 (doi:10.1128/AEM.70.8.4532-4537.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70, 6347–6351 10.1128/AEM.70.10.6347-6351.2004 (doi:10.1128/AEM.70.10.6347-6351.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Futagami T, Morono Y, Terada T, Kaksonen AH, Inagaki F. 2009. Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl. Environ. Microbiol. 75, 6905–6909 10.1128/AEM.01124-09 (doi:10.1128/AEM.01124-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regeard C, Maillard J, Holliger C. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56, 107–118 10.1016/j.mimet.2003.09.019 (doi:10.1016/j.mimet.2003.09.019) [DOI] [PubMed] [Google Scholar]
- 43.Smidt H, van Leest M, van der Oost J, de Vos WM. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182, 5683–5691 10.1128/JB.182.20.5683-5691.2000 (doi:10.1128/JB.182.20.5683-5691.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maillard J, Genevaux P, Holliger C. 2011. Redundancy and specificity of multiple trigger factor chaperones in Desulfitobacteria. Microbiology 157, 2410–2421 10.1099/mic.0.050880-0 (doi:10.1099/mic.0.050880-0) [DOI] [PubMed] [Google Scholar]
- 45.Gábor K, Veríssimo CS, Cyran BC, Ter Horst P, Meijer NP, Smidt H, de Vos WM, van der Oost J. 2006. Characterization of CprK1, a CRP/FNR-type transcriptional regulator of halorespiration from Desulfitobacterium hafniense. J. Bacteriol. 188, 2604–2613 10.1128/JB.188.7.2604-2613.2006 (doi:10.1128/JB.188.7.2604-2613.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Futagami T, Goto M, Furukawa K. 2008. Biochemical and genetic bases of dehalorespiration. Chem. Record 8, 1–12 10.1002/tcr.20134 (doi:10.1002/tcr.20134) [DOI] [PubMed] [Google Scholar]
- 47.Kräutler B, et al. 2003. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of a natural corrinoid. Helv. Chim. Acta 86, 3698–3716 10.1002/hlca.200390313 (doi:10.1002/hlca.200390313) [DOI] [Google Scholar]
- 48.Siebert A, Neumann A, Schubert T, Diekert G. 2002. A non-dechlorinating strain of Dehalospirillum multivorans: evidence for a key role of the corrinoid cofactor in the synthesis of an active tetrachloroethene dehalogenase. Arch. Microbiol. 178, 443–449 10.1007/s00203-002-0473-8 (doi:10.1007/s00203-002-0473-8) [DOI] [PubMed] [Google Scholar]
- 49.Villemur R, Lanthier M, Beaudet R, Lépine F. 2006. The Desulfitobacterium genus. FEMS Microbiol. Rev. 30, 706–733 10.1111/j.1574-6976.2006.00029.x (doi:10.1111/j.1574-6976.2006.00029.x) [DOI] [PubMed] [Google Scholar]
- 50.Yan J, Ritalahti KM, Wagner DD, Löffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78, 6630–6636 10.1128/AEM.01535-12 (doi:10.1128/AEM.01535-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neumann A, Scholz-Muramatsu H, Diekert G. 1994. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch. Microbiol. 162, 295–301 10.1007/BF00301854 (doi:10.1007/BF00301854) [DOI] [PubMed] [Google Scholar]
- 52.Christiansen N, Ahring BK, Wohlfarth G, Diekert G. 1998. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436, 159–162 10.1016/S0014-5793(98)01114-4 (doi:10.1016/S0014-5793(98)01114-4) [DOI] [PubMed] [Google Scholar]
- 53.Miller E, Wohlfarth G, Diekert G. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169, 497–502 10.1007/s002030050602 (doi:10.1007/s002030050602) [DOI] [PubMed] [Google Scholar]
- 54.Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64, 1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 10.1128/AEM.03006-05 (doi:10.1128/AEM.03006-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond A, et al. 2012. Geneious v5.6. Created by Biomatters. See http://www.geneious.com. [Google Scholar]
- 57.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 58.Selesi D, Jehmlich N, von Bergen M, Schmidt F, Rattei T, Tischler P, Lueders T, Meckenstock RU. 2010. Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47. J. Bacteriol. 192, 295–306 10.1128/JB.00874-09 (doi:10.1128/JB.00874-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hafenbradl D, Keller M, Dirmeier R, Rachel R, Rossnagel P, Burggraf S, Huber H, Stetter KO. 1996. Ferroglobus placidus gen. nov, sp. nov, a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch. Microbiol. 166, 308–314 10.1007/s002030050388 (doi:10.1007/s002030050388) [DOI] [PubMed] [Google Scholar]
- 60.Cheng D, He J. 2009. Isolation and characterization of ‘Dehalococcoides’ sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl. Environ. Microbiol. 75, 5910–5918 10.1128/AEM.00767-09 (doi:10.1128/AEM.00767-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chow WL, Cheng D, Wang S, He J. 2010. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 4, 1020–1030 10.1038/ismej.2010.27 (doi:10.1038/ismej.2010.27) [DOI] [PubMed] [Google Scholar]
- 62.Wagner A, Adrian L, Kleinsteuber S, Andreesen JR, Lechner U. 2009. Transcription analysis of genes encoding homologues of reductive dehalogenases in ‘Dehalococcoides’ sp. strain CBDB1 by using terminal restriction fragment length polymorphism and quantitative PCR. Appl. Environ. Microbiol. 75, 1876–1884 10.1128/AEM.01042-08 (doi:10.1128/AEM.01042-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.West KA, et al. 2008. Comparative genomics of ‘Dehalococcoides ethenogenes’ 195 and an enrichment culture containing unsequenced ‘Dehalococcoides’ strains. Appl. Environ. Microbiol. 74, 3533–3540 10.1128/AEM.01835-07 (doi:10.1128/AEM.01835-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Picardal F, Arnold RG, Huey BB. 1995. Effects of electron donor and acceptor conditions on reductive dehalogenation of tetrachloromethane by Shewanella putrefaciens 200. Appl. Environ. Microbiol. 61, 8–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Löffler FE, et al. 2012. Dehalococcoides mccartyi gen. nov., sp. nov., obligate organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidetes classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635 10.1099/ijs.0.034926-0 (doi:10.1099/ijs.0.034926-0) [DOI] [PubMed] [Google Scholar]
- 66.Moe WM, Yan J, Nobre MF, Da Costa MS, Rainey FA. 2009. Dehalogenimonas lykanthroporepellens gen. nov, sp. nov, a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 59, 2692–2697 10.1099/ijs.0.011502-0 (doi:10.1099/ijs.0.011502-0) [DOI] [PubMed] [Google Scholar]
- 67.Rupakula A, Kruse T, Boeren S, Holliger C, Smidt H, Maillard J. 2013. The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics. Phil. Trans. R. Soc. B 368, 20120325. 10.1098/rstb.2012.0325 (doi:10.1098/rstb.2012.0325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scholz-Muramatsu H, Neumann A, Meßmer M, Moore E, Diekert G. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov, sp. nov, a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163, 48–56 10.1007/BF00262203 (doi:10.1007/BF00262203) [DOI] [Google Scholar]
- 69.Luijten MLGC, de Weert J, Smidt H, Boschker HTS, de Vos WM, Schraa G, Stams AJM. 2003. Description of Sulfurospirillum halorespirans sp. nov, an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53, 787–793 10.1099/ijs.0.02417-0 (doi:10.1099/ijs.0.02417-0) [DOI] [PubMed] [Google Scholar]
- 70.Holliger C, Schraa G, Stams AJ, Zehnder AJ. 1993. A highly purified enrichment culture couples the reductive dechlorination of tetrachloroethene to growth. Appl. Environ. Microbiol. 59, 2991–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bisaillon A, Beaudet R, Lépine F, Déziel E, Villemur R. 2010. Identification and characterization of a novel CprA reductive dehalogenase specific to highly chlorinated phenols from Desulfitobacterium hafniense strain PCP-1. Appl. Environ. Microbiol. 76, 7536–7540 10.1128/AEM.01362-10 (doi:10.1128/AEM.01362-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gauthier A, Beaudet R, Lépine F, Juteau P, Villemur R. 2006. Occurrence and expression of crdA and cprA5 encoding chloroaromatic reductive dehalogenases in Desulfitobacterium strains. Can. J. Microbiol. 52, 47–55 10.1139/w05-111 (doi:10.1139/w05-111) [DOI] [PubMed] [Google Scholar]
- 73.Marzorati M, et al. 2007. A novel reductive dehalogenase, identified in a contaminated groundwater enrichment culture and in Desulfitobacterium dichloroeliminans strain DCA1, is linked to dehalogenation of 1,2-dichloroethane. Appl. Environ. Microbiol. 73, 2990–2999 10.1128/AEM.02748-06 (doi:10.1128/AEM.02748-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Müller JA, Rosner BM, von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain vs and its environmental distribution. Appl. Environ. Microbiol. 70, 4880–4888 10.1128/AEM.70.8.4880-4888.2004 (doi:10.1128/AEM.70.8.4880-4888.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adrian L, Rahnenführer J, Gobom J, Hölscher T. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73, 7717–7724 10.1128/AEM.01649-07 (doi:10.1128/AEM.01649-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang S, Chan WWM, Fletcher KE, Seifert J, Liang X, Löffler FE, Edwards EA, Adrian L. 2012. Functional characterization of reductive dehalogenases by using blue native polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 79, 974–981 10.1128/AEM.01873-12 (doi:10.1128/AEM.01873-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L. 2008. Temporal transcriptomic microarray analysis of ‘Dehalococcoides ethenogenes’ strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74, 2864–2872 10.1128/AEM.02208-07 (doi:10.1128/AEM.02208-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, Richardson RE. 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73, 320–326 10.1128/AEM.02129-06 (doi:10.1128/AEM.02129-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holmes VF, He J, Lee PKH, Alvarez-Cohen L. 2006. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl. Environ. Microbiol. 72, 5877–5883 10.1128/AEM.00516-06 (doi:10.1128/AEM.00516-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waller AS, Krajmalnik-Brown R, Löffler FE, Edwards EA. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71, 8257–8264 10.1128/AEM.71.12.8257-8264.2005 (doi:10.1128/AEM.71.12.8257-8264.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang S, Edwards EA. 2013. Identification of Dehalobacter reductive dehalogenases that catalyze dechlorination of chloroform, 1,1,1-trichloroethane and 1,1-dichloroethane. Phil. Trans. R. Soc. B 368, 20120318. 10.1098/rstb.2012.0318 (doi:10.1098/rstb.2012.0318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller E, Wohlfarth G, Diekert G. 1996. Studies on tetrachloroethene respiration in Dehalospirillum multivorans. Arch. Microbiol. 166, 379–387 10.1007/BF01682983 (doi:10.1007/BF01682983) [DOI] [PubMed] [Google Scholar]
- 83.Sjuts H, Fisher K, Dunstan MS, Rigby SE, Leys D. 2012. Heterologous expression, purification and cofactor reconstitution of the reductive dehalogenase PceA from Dehalobacter restrictus. Protein Expr. Purif. 85, 224–229 10.1016/j.pep.2012.08.007 (doi:10.1016/j.pep.2012.08.007) [DOI] [PubMed] [Google Scholar]
- 84.Duhamel M, Edwards EA. 2006. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol. Ecol. 58, 538–549 10.1111/j.1574-6941.2006.00191.x (doi:10.1111/j.1574-6941.2006.00191.x) [DOI] [PubMed] [Google Scholar]
- 85.Grostern A, Edwards EA. 2006. A 1,1,1-trichloroethane-degrading anaerobic mixed microbial culture enhances biotransformation of mixtures of chlorinated ethenes and ethanes. Appl. Environ. Microbiol. 72, 7849–7856 10.1128/AEM.01269-06 (doi:10.1128/AEM.01269-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grostern A, Edwards EA. 2006. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl. Environ. Microbiol. 72, 428–436 10.1128/AEM.72.1.428-436.2006 (doi:10.1128/AEM.72.1.428-436.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10, 483–496 10.1038/nrmicro2814 (doi:10.1038/nrmicro2814) [DOI] [PubMed] [Google Scholar]
- 88.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 10.1093/nar/gkr367 (doi:10.1093/nar/gkr367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 10.1093/bioinformatics/bti263 (doi:10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- 91.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 10.1093/bioinformatics/btr088 (doi:10.1093/bioinformatics/btr088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 93.Tsukagoshi N, Ezaki S, Uenaka T, Suzuki N, Kurane R. 2006. Isolation and transcriptional analysis of novel tetrachloroethene reductive dehalogenase gene from Desulfitobacterium sp. strain KBC1. Appl. Microbiol. Biotechnol. 69, 543–553 10.1007/s00253-005-0022-x (doi:10.1007/s00253-005-0022-x) [DOI] [PubMed] [Google Scholar]
- 94.Grostern A, Edwards EA. 2009. Characterization of a Dehalobacter coculture that dechlorinates 1,2-dichloroethane to ethene and identification of the putative reductive dehalogenase gene. Appl. Environ. Microbiol. 75, 2684–2693 10.1128/AEM.02037-08 (doi:10.1128/AEM.02037-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krajmalnik-Brown R, Sung Y, Ritalahti KM, Saunders FM, Löffler FE. 2007. Environmental distribution of the trichloroethene reductive dehalogenase gene (tceA) suggests lateral gene transfer among Dehalococcoides. FEMS Microbiol. Ecol. 59, 206–214 10.1111/j.1574-6941.2006.00243.x (doi:10.1111/j.1574-6941.2006.00243.x) [DOI] [PubMed] [Google Scholar]
- 96.Maillard J, Regeard C, Holliger C. 2005. Isolation and characterization of Tn-Dha1, a transposon containing the tetrachloroethene reductive dehalogenase of Desulfitobacterium hafniense strain TCE1. Environ. Microbiol. 7, 107–117 10.1111/j.1462-2920.2004.00671.x (doi:10.1111/j.1462-2920.2004.00671.x) [DOI] [PubMed] [Google Scholar]
- 97.McMurdie PJ, Hug LA, Edwards EA, Holmes S, Spormann AM. 2011. Site-specific mobilization of vinyl chloride respiration islands by a mechanism common in Dehalococcoides. BMC Genomics 12, 287. 10.1186/1471-2164-12-287 (doi:10.1186/1471-2164-12-287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McMurdie PJ, Behrens SF, Holmes S, Spormann AM. 2007. Unusual codon bias in vinyl chloride reductase genes of Dehalococcoides species. Appl. Environ. Microbiol. 73, 2744–2747 10.1128/AEM.02768-06 (doi:10.1128/AEM.02768-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Regeard C, Maillard J, Dufraigne C, Deschavanne P, Holliger C. 2005. Indications for acquisition of reductive dehalogenase genes through horizontal gene transfer by Dehalococcoides ethenogenes strain 195. Appl. Environ. Microbiol. 71, 2955–2961 10.1128/AEM.71.6.2955-2961.2005 (doi:10.1128/AEM.71.6.2955-2961.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakamura K, Mizumoto M, Ueno T, Ishida H. 2006. Cloning and analysis of trichloroethene reductive dehalogenase gene and its detection by quantitative real-time PCR. Environ. Eng. Res. 43, 119–125 [Google Scholar]
- 101.Krasotkina J, Walters T, Maruya KA, Ragsdale SW. 2001. Characterization of the B12- and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 276, 40 991–40 997 10.1074/jbc.M106217200 (doi:10.1074/jbc.M106217200) [DOI] [PubMed] [Google Scholar]
- 102.Higgins DG, Sharp PM. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73, 237–244 10.1016/0378-1119(88)90330-7 (doi:10.1016/0378-1119(88)90330-7) [DOI] [PubMed] [Google Scholar]
- 103.Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 (doi:10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 104.Manchester MJ, Hug LA, Zarek M, Zila A, Edwards EA. 2012. Discovery of a trans-dichloroethene respiring Dehalogenimonas in the 1,1,2,2-tetrachloroethane-dechlorinating WBC-2 consortium. Appl. Environ. Microbiol. 78, 5280–5287 10.1128/AEM.00384-12 (doi:10.1128/AEM.00384-12) [DOI] [PMC free article] [PubMed] [Google Scholar]