Abstract
Chlorinated pollutants are hardly biodegradable under oxic conditions, but they can often be metabolized by anaerobic bacteria through organohalide respiration reactions. In an attempt to identify bottlenecks limiting aerobic catabolism of 1,3-dichloroprop-1-ene (1,3-DCP; a widely used organohalide) in Pseudomonas pavonaceae, the possible physiological restrictions for this process were surveyed. Flow cytometry and a bioluminescence reporter of metabolic state revealed that cells treated with 1,3-DCP experienced an intense stress that could be traced to the endogenous production of reactive oxygen species (ROS) during the metabolism of the compound. Cells exposed to 1,3-DCP also manifested increased levels of d-glucose-6-P 1-dehydrogenase activity (G6PDH, an enzyme key to the synthesis of reduced NADPH), observed under both glycolytic and gluconeogenic growth regimes. The increase in G6PDH activity, as well as cellular hydroperoxide levels, correlated with the generation of ROS. Additionally, the high G6PDH activity was paralleled by the accumulation of d-glucose-6-P, suggesting a metabolic flux shift that favours the production of NADPH. Thus, G6PDH and its cognate substrate seem to play an important role in P. pavonaceae under redox stress caused by 1,3-DCP, probably by increasing the rate of NADPH turnover. The data suggest that oxidative stress associated with the biodegradation of 1,3-DCP reflects a significant barrier for the evolution of aerobic pathways for chlorinated compounds, thereby allowing for the emergence of anaerobic counterparts.
Keywords: Pseudomonas pavonaceae, biodegradation, redox balance, stress, NADPH, organochloride
1. Introduction
Chloro-organic compounds rank among the most persistent environmental pollutants [1]. The chemical stability of the C–Cl bond is a major problem, which often causes polychlorinated compounds to be minimally degradable by most environmental micro-organisms [2–4]. Some aerobic bacteria metabolize highly chlorinated organic compounds, including polychlorinated biphenyls [5], haloalkanes [6–8], chlorobenzenes [9,10] and chlorophenols [11,12]. The removal of the halogen substituent occurs through the action of specific dehalogenases [13,14] or as part of a downstream reaction of Cl-containing metabolic intermediates [15]. While Gram-negative aerobic bacteria, in particular pseudomonads, are favourite hosts for designing new biodegradation pathways, attempts to improve the biodegradation of chlorinated compounds through genetic engineering have not had a high degree of success [16,17]. In recent years, however, the number of cases of anaerobic bacteria that can, to some degree, degrade recalcitrant polychlorinated chemicals has increased [18–21]. In these cases, dechlorination of the target molecules occurs through the action of dedicated anoxic dehalogenases [22,23] or as a side effect of organohalide respiration [24], for example, the dehalogenation process in Dehalococcoides mccartyi [25,26]. It seems that the natural evolution of aerobic routes for chloro-organic molecules endures more difficulties than the evolution of their anoxic counterparts; however, the precise nature of such bottlenecks has not been clearly documented.
While aerobic metabolism of many organohalides may be biochemically feasible [27,28], their in vivo use is often impossible because of high toxicity. The specifics of the toxicity and the mechanism of the limits on biodegradation in the presence of molecular O2 are largely unknown. We have examined the physiological regimes experienced by Pseudomonas pavonaceae 170 (formerly known as Pseudomonas cichorii) when exposed to the highly toxic nematocide 1,3-dichloroprop-1-ene (1,3-DCP). This strain is a Gram-negative soil bacterium that can use some organohalides as a sole carbon and energy source [29]. Because of its carcinogenic properties and widespread occurrence in groundwater supplies, 1,3-DCP is an important environmental pollutant [30]. The key biodegradation steps of this compound involve a hydrolytic dehalogenation reaction executed by the haloalkane dehalogenase DhlA (figure 1). During the process, one of the halogen substituents of the molecule is replaced in a nucleophilic substitution reaction by an H2O-derived hydroxyl group. Although the biodegradation of 1,3-DCP can proceed to completion, the compound is highly toxic for cells [31,32]. Environmental bacteria thus face the challenge of performing an efficient biodegradation process while enduring the stress associated with the target compound itself and its metabolism.
Figure 1.
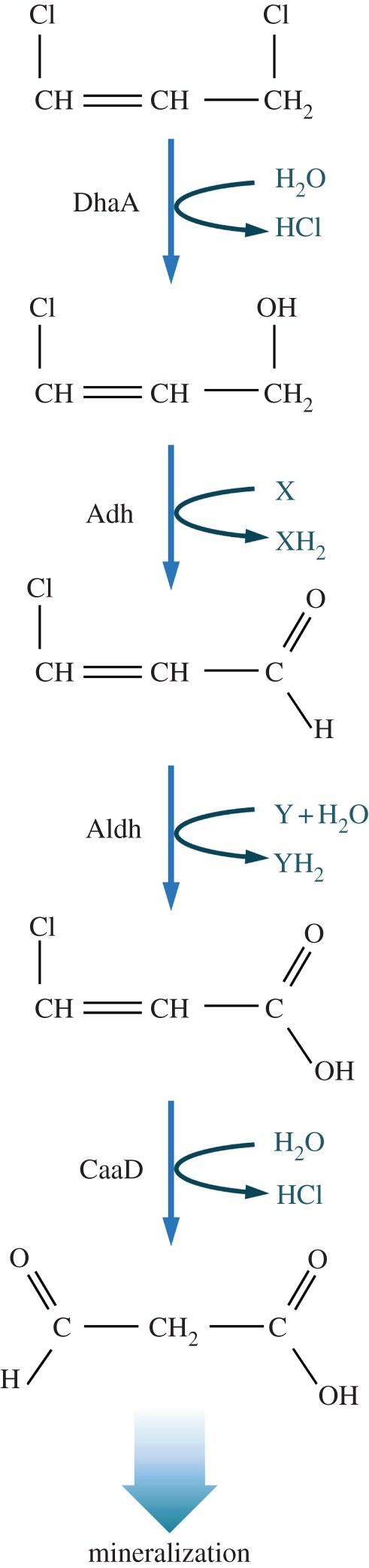
Biochemical pathway proposed for 1,3-dichloroprop-1-ene biodegradation in the Gram-negative bacterium Pseudomonas pavonaceae 170. The first step in the biodegradation of this organohalide is catalysed by a hydrolytic haloalkane dehalogenase (DhaA, E.C. 3.8.1.5) that has broad substrate specificity. After two sequential oxidation steps catalysed by an alcohol dehydrogenase (Adh) and an aldehyde dehydrogenase (Aldh), the first intermediate metabolite (3-chloro-2-propene-1-ol) is converted into 3-chloro-2-propenal and finally into 3-chloropropenoic acid. The latter compound is the substrate for a 3-chloroacrylic acid dehalogenase (CaaD, E.C. 3.8.1; composed by CaaD1, the α subunit, and CaaD2, the β subunit) that yields 3-oxopropanoate (malonate semialdehyde). This metabolite finally enters into the central metabolic pathways upon decarboxylation by a malonate semialdehyde decarboxylase. Components X and Y represent electron acceptors which have not been identified yet. (Online version in colour.)
The bacteria of the genus Pseudomonas, as the majority of the heterotrophic bacteria, oxidize organic carbon sources through the Entner–Doudoroff pathway and the tricarboxylic acid cycle [33–35]. d-Glucose is transported through a dedicated ABC uptake system into the cytoplasm, where it is phosphorylated to G6P and d-gluconate-6-P. d-Glucose can diffuse into the periplasm and be converted to d-gluconate and then to 2-keto-d-gluconate; these intermediates are transported into the cytoplasm to be phosphorylated to d-gluconate-6-P and 2-keto-d-gluconate-6-P. These upstream intermediates converge into d-gluconate-6-P, which can enter into the Entner–Doudoroff or the pentose phosphate pathways. Several of these oxidative steps are coupled to the reduction of NADP+ to NADPH, used to generate anabolic precursors needed for bacterial growth and as an antioxidant under stressful conditions (both environmental and endogenous) [36,37]. G6PDH catalyses the NADP+-dependent oxidation of G6P and is a key step in the generation of precursor metabolites for the Entner–Doudoroff and the pentose phosphate pathways [38]. The connection between these central catabolic pathways and biodegradation has not been explored in P. pavonaceae.
Antioxidant responses in cells exposed to xenobiotics vary widely in the specificity and mechanism of action, but the majority of them rely on the generation of reducing power that the cells use to counteract reactive oxygen species (ROS) production. The appropriate balance of NADPH/NADP+ is a central feature to the continued performance of virtually any aerobic micro-organism [39]. A reductive intracellular environment mediated by an appropriate supply of NADPH enables growth and provides the reducing power needed by the cell to counteract the accumulation of ROS generated as a direct consequence of oxidative phosphorylation (a process inherent to the generation of ATP), as well as the ROS derived from the environmental stresses mentioned before [16]. The effectiveness of the vast majority of ROS-detoxifying enzymes, such as superoxide dismutase, catalase and glutathione peroxidase, largely depends on the intracellular availability of NADPH.
The data below document the effects of 1,3-DCP on the cell physiology and oxidative stress responses of P. pavonaceae. Specifically, we observed that this micro-organism undergoes a severe metabolic stress when exposed to 1,3-DCP and that its ability to defeat such a condition arises, at least partially, from the modulation of the G6PDH activity. Accumulation of d-glucose-6-P (G6P) as a source of electrons was also observed under stressful conditions. These results expose some of the difficulties of degrading chlorinated compounds aerobically and provide a rationale for the emergence of alternative anaerobic pathways.
2. Material and methods
(a). Bacterial strains, culture media and growth conditions
The isolation and some phenotypic properties of the 1,3-DCP degrader P. pavonaceae 170 were described by Verhagen et al. [40]. This strain was used throughout this study. Escherichia coli DH5α (Φ80lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169) [41] and CC118 (araD139 Δ(ara-leu)7697 ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB (rifampicinR) argE(Am) recA1) [42] were used for routine cloning procedures and plasmid maintenance. Escherichia coli BW25113 (F− λ− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) rph-1 Δ(rhaD-rhaB)568 hsdR514) [43] was used in some metabolomic determinations. During genetic manipulations and for inocula propagation, P. pavonaceae and E. coli strains were grown under oxic conditions at 30°C and 37°C, respectively. The bacteria were grown in LB medium (containing 10 g l−1 tryptone, 10 g l−1 NaCl and 5 g l−1 yeast extract) in Erlenmeyer flasks filled with medium up to one-fifth of their nominal volume and with agitation at 170 rpm. Pseudomonas pavonaceae was grown batchwise in a semi-synthetic culture medium (MMY), containing 6 g l−1 Na2HPO4, 3 g l−1 KH2PO4, 1.5 g l−1 (NH4)2SO4, 0.3 g l−1 K2SO4, 0.2 g l−1 MgSO4·7H2O and 0.5 g l−1 yeast extract (Becton–Dickinson Co., Sparks, MD, USA) was amended as a nutritional additive, predominantly because of its high content of soluble vitamins. d-Glucose, sodium d-gluconate, sodium citrate or sodium succinate were separately added at 30 mM as filter-sterilized solutions to the MMY medium, as the sole carbon source (which approximately corresponds to the molar concentration of 5 g l−1 d-glucose). The seed cultures for all experiments were prepared by dispersing a loopful of cells from a fresh LB agar plate (containing the appropriate antibiotics if needed) into 5 ml of MMY medium and incubating the resulting suspension at the corresponding growth temperature for 18 h. Growth was estimated by measuring the optical density at 600 nm (OD600) of the cultures and measured using an Ultrospec 3000 pro UV/visible spectrophotometer (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). The solid medium contained 15 g l−1 agar, and 50 μg ml−1 kanamycin was added as a filter-sterilized solution when appropriate. In some experiments, oxidative stress conditions were imposed by adding either 0.5 mM diamide (1,1′-azo-bis(N,N-dimethylformamide)) or 0.15 mM menadione (2-methylnaphthalene-1,4-dione) to exponentially growing cells. Either drug was added to the cultures from concentrated solutions freshly prepared in dimethyl sulfoxide, and an appropriate volume of dimethyl sulfoxide was added to the control cultures.
Commercial glass vials with teflon-stoppered metallic screw caps (Sigma-Aldrich Co., St Louis, MO, USA) were used for some cultivations involving 1,3-DCP. These vials are designed for headspace gas chromatography (GC), and we used this system because it ensures that no organochloride is lost by evaporation (the system is tightly closed from the point at which 1,3-DCP is added to the culture onwards) [44]. Vials, filled up to one-fifth of their nominal volume, were incubated at 30°C, with rotary agitation to prevent biomass sedimentation, and one vial was used per time point for further analysis. For enzymatic determinations, the cells were immediately used in the biochemical assays. For headspace GC coupled to mass spectrometry (MS) analysis, the cells were rapidly inactivated by incubating the vials at 85°C for 15 min. Vials were cooled to room temperature and stored at −20°C until analysis.
In the experiments to determine the growth kinetic properties of P. pavonaceae under different culture conditions, the OD600 of each culture condition was determined using a SpectraMax Plus384 microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 15 min intervals during 24–36 h. Clear polystyrene 96-well microtitre plates (Nunc A/S, Roskilde, Denmark), containing 200 μl of the corresponding culture medium initially inoculated at an OD600 of approximately 0.05 units, were shaken for 30 s before each measurement; homogeneous conditions were ensured during the cultivation period. Turbidity measurements were computed during exponential growth, and the specific growth rate (μ) was calculated for each condition as μ = [ln(OD600 at t1) − ln(OD600 at t0)]/(t1 − t0). Normalized growth coefficients were calculated as 100 × (μexperimental/μcontrol), where μexperimental and μcontrol are the specific growth rates under stressful and control (i.e. in the absence of the stressor) growth conditions, respectively. The extension of the lag phase in each case was analytically obtained as detailed by Dalgaard & Koutsoumanis [45].
(b). Construction of a lux-based reporter for in vivo evaluation of the metabolic state of the cells
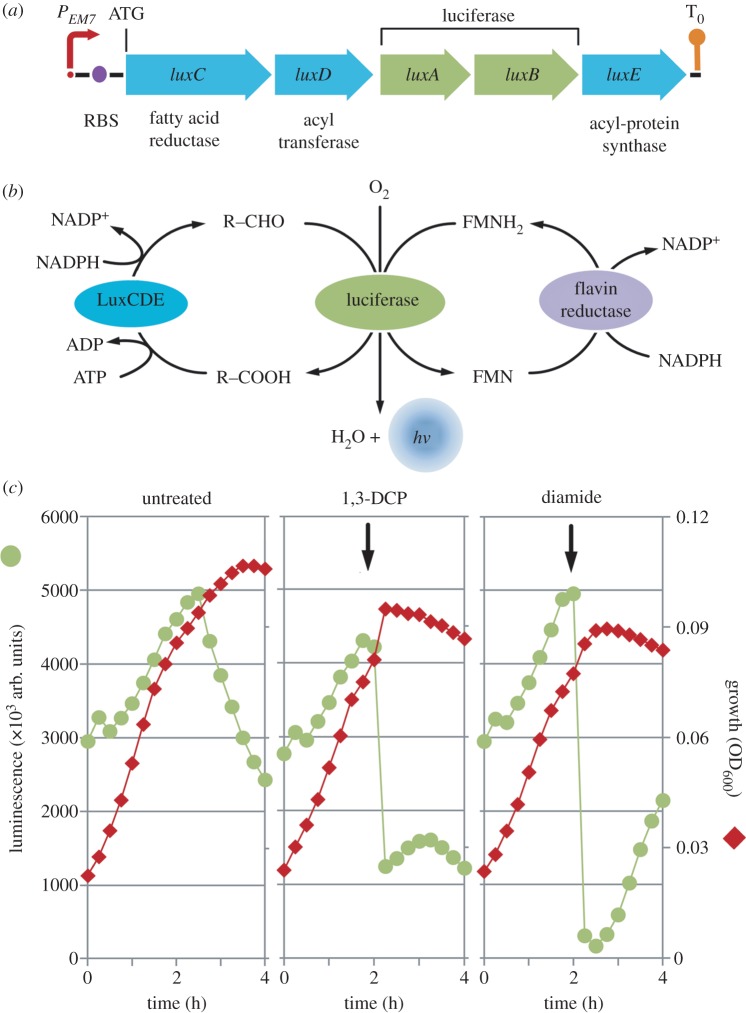
The pentacistronic lux operon from Photorhabdus luminescens consists of a luxAB component, which encodes a luciferase (a mixed-function oxidase composed of two subunits: α and β) that oxidizes a reduced flavin mononucleotide molecule (FMNH2) and a long-chain aliphatic aldehyde, in the presence of molecular O2, to yield a characteristic 490-nm optical signature [46,47]. The aldehyde is subsequently regenerated by a multi-enzyme fatty acid reductase encoded by luxC, luxD and luxE [48]. A broad-host-range plasmid carrying the lux-based reporter was constructed by applying standard molecular biology methods [49]. In the first step, oligonucleotides PEM7-F1 (5′-AGT GAG CTC GTT GAC AAT TAA TCA TCG GCA TAG TAT ATC GGC ATA GTA TAA TAC GAC AAG GTG AGG AAC TAA ACC ACT AGT CAG-3′) and PEM7-R1 (5′-CTG ACT AGT GGT TTA GTT CCT CAC CTT GTC GTA TTA TAC TAT GCC GAT ATA CTA TGC CGA TGA TTA ATT GTC AAC GAG CTC ACT-3′) were annealed to generate a 84-bp double-stranded DNA fragment that spans the EM7 promoter sequence, bracketed by recognition sites for SacI and SpeI (underlined in the corresponding sequences). This DNA fragment was digested with SacI and SpeI and ligated into pUC18-mini-Tn7T-Gm-lux [50], restricted with the same enzymes. This intermediate plasmid was used as the template in a PCR amplification with oligonucleotides PEM7-F2 (5′-TAG CGA ATT CAG TGA GCT CGT TGA CAA TTA-3′, EcoRI site underlined) and PEM7-R2 (5′-CTG AGG ATC CTG ACT AGT GGT TTA GTT CC-3′, BamHI site underlined). The resulting PCR product was digested with EcoRI and BamHI and ligated into pSEVA226, restricted with the same enzymes. Plasmid pSEVA226 contains luxCDABE from P. luminescens, with an optimized ribosome binding site (RBS; 5′-AGG AGG AAA AAC AT-3′), a RK2 replication origin and a kanamycin resistance determinant [51]. These procedures resulted in pSEVA226E, in which the expression of luxCDABE is under the control of the constitutive EM7 promoter. This promoter possesses medium strength in Pseudomonas species (D. Pérez-Pantoja et al. 2012, unpublished data). Plasmid pSEVA226E was introduced by electroporation into P. pavonaceae cells rendered electrocompetent by washing the biomass with 0.3 M sucrose, as described by Choi et al. [52].
To assess emission of light in P. pavonaceae carrying pSEVA226E, cultures were first pre-grown overnight at 30°C, in M9 medium with d-glucose as the carbon source. The cultures were diluted in fresh medium to an OD600 of approximately 0.05 units, and 200 μl aliquots were disposed, in triplicate, in black polystyrene 96-well microtitre plates (Nunc A/S). Where indicated, 1,3-DCP or diamide was added, and the incubation was continued at 30°C, with shaking at 200 rpm every 15 min. Growth and light emission were recorded in a Wallac VICTOR2 1420 multi-label counter and plate reader (PerkinElmer, Inc., Waltham, MA, USA) and analysed using the WorkOut applications data analysis software (Wallac Oy, Turku, Finland).
(c). Preparation of cell-free extracts and in vitro determination of enzyme activities and intracellular metabolites concentration
Cell-free extracts were prepared from cells grown to the mid-exponential phase (OD600 of approx. 0.4–0.6 units) and harvested by centrifugation from an appropriate culture volume (usually between 5 and 25 ml) at 4000g at 4°C for 10 min. The pellets were suspended in one volume of 10 mM sodium phosphate buffer (pH = 7.5, previously refrigerated) containing 10 mM 2-mercaptoethanol and centrifuged as described earlier. The cells were resuspended in a 0.3–0.5 volume of the same buffer at approximately 0.5 g wet cells ml−1 and sonicated intermittently for 3–5 min with successive sonication (1 min) and cooling (1 min) cycles. A similar procedure was used to prepare cell-free extracts for haloalkane dehalogenase activity determination, but the cells (harvested at different time points) were washed and resuspended in 0.3–0.5 volume of a buffer composed of 10 mM Tris·H2SO4 (pH = 8.2), 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.02% (w/v) NaN3 and 10% (v/v) glycerol. In both cases, the sonicated cells preparations were centrifuged at 7500g at 4°C for 30 min to remove cell debris. The protein concentration in the cell-free extracts was measured using the Bradford method [53] (with Coomassie brilliant blue G-250), using a kit from Sigma-Aldrich Co. and with crystalline bovine serum albumin as the standard.
The G6PDH activity was determined by following the rate of NADP+ reduction at 340 nm at 30°C in a reaction mixture (1 ml final volume) that contained 50 mM phosphate buffer (pH = 7.5), 10 mM MgSO4, 0.75 mM NADP+, 2 mM G6P and 8–100 μl of the cell-free extract. The reaction was started by the addition of G6P, and an extinction coefficient (ɛNADH) of 6.22 mM−1 cm−1, representing the difference between the extinction coefficients of NADPH and NADP+, was used to calculate the specific enzymatic activity. One unit of G6PDH activity was defined as the quantity of enzyme that catalysed the formation of 1 μmol of product for 1 min at 30°C. Haloalkane dehalogenase assays were performed by incubating an appropriate amount of cell extract with 3 ml of a 5 mM substrate (either 1,3-DCP or 1-chlorobutane) solution in 50 mM Tris·H2SO4 buffer (pH = 8.2) at 30°C. Halide liberation was monitored colourimetrically, using a modification of the method described by Bergmann & Sanik [54] as described by Nikel & de Lorenzo [44], and corrected in each experimental condition by the signal obtained using un-inoculated MMY medium. One unit of activity was defined as the quantity of enzyme that catalysed the formation of 1 μmol of halide during 1 min at 30°C. Under the assay conditions described in this study, one unit of G6PDH activity≡1 unit of haloalkane dehalogenase activity≡16.67 nkat.
For the determination of intracellular metabolites, cell lysates were cleared by centrifugation at 13 000g at 4°C for 15 min to remove cell debris and to reduce the NAD(P)+/H background in the extract. The resulting cell-free extracts were boiled for 10 min, chilled in an ice bath and centrifuged at 13 000g at 4°C for 15 min, and the supernatants were immediately used for determination. Appropriate cell-free extract samples (50–350 μl) were incubated at 30°C in a reaction mixture that contained 50 mM Tris HCl (pH = 7.4), 15 mM MgCl2, 0.75 mM NADP+ and 0.5 U ml−1 G6PDH from Saccharomyces cerevisiae (Sigma-Aldrich Co.). G6P concentration was spectrophotometrically assessed at 340 nm. d-Gluconate-6-P concentration was determined by applying the procedure as for G6P, except that d-gluconate-6-P dehydrogenase (0.025 U ml−1) was used instead of G6PDH. Authentic G6P and d-gluconate-6-P standards (>99% purity, Sigma-Aldrich Co.) were used at concentrations ranging from 0.005 to 0.25 mM to calibrate the assay, and some experimental samples were spiked with known amounts of metabolites to confirm that boiling did not affect their stability in the heat-treated cell-free extracts. Metabolite contents, expressed as nmol mg protein−1, were calculated from the corresponding ΔA340, the extinction coefficients of NADPH and NADH, and the protein concentration in the cell-free extracts.
(d). In vitro determination of the total hydroperoxide content
Hydroperoxides can oxidize Fe2+ to Fe3+ in acidic solution. The latter ions, in the presence of xylenol orange [3,3′-bis[N,N-bis(carboxymethyl)aminomethyl]-o-cresolsulfonephthalein disodium salt], form a stable Fe3+–xylenol orange complex that absorbs at 560 nm. We used a modification of the procedure described by Jiang et al. [55] to estimate the total hydroperoxide content in the cell-free extracts. The FOXII reagent, which consisted of 0.15 mM xylenol orange, 0.25 mM (NH4)2Fe(SO4)2·6H2O, 4 mM 2,6-bis(1,1-dimethylethyl)-4-methylphenol and 25 mM H2SO4 (in 90% (v/v) CH3OH), was freshly prepared for each set of determinations and stored at −20°C for no longer than 24 h. For measurements, 75–150 μl of cell-free extract (obtained as described in the preceding section, and containing a fixed amount of soluble proteins) was pipetted into a 1.5-ml microtube and, when necessary, made up to 150 μl with CH3OH, to which 1350 μl of FOXII reagent was added. The reaction mixtures were thoroughly mixed by vortexing and incubated in the dark at room temperature for 45 min. Absorbances were read at 560 nm against a blank of 150 μl CH3OH and 1350 μl FOX2 reagent. A calibration curve was run with each set of experimental determination with tert-butyl hydroperoxide (Sigma-Aldrich Co.) as the standard. An extinction coefficient (εFOXII) of 4.52 mM−1 cm−1, representing the intrinsic absorbance of the Fe3+–xylenol orange complex, was used to calculate the content of hydroperoxides in the samples, and the results are expressed as molar equivalents of tert-butyl hydroperoxide.
(e). In vivo determination of oxidative stress by flow cytometry
Fluorescence-activated cell sorter cytometry analysis was performed using the ROS-activated green fluorescent dye, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA; Sigma-Aldrich Co.). Pseudomonas pavonaceae was grown in M9 minimal medium containing the corresponding carbon source at 30°C to the mid-exponential phase, when the cell suspension was split into two cultures. One culture was a control, and the other was amended with 0.5 mM 1,3-DCP. Both cultures were incubated, as described earlier, for 1.5 h, and the cells from 0.5–1.5 ml culture aliquots were pelleted by centrifugation at 8000g during 5 min in a benchtop microfuge (Eppendorf centrifuge 5415D; Eppendorf North America, Inc., Hauppauge, NY, USA), washed once with PBS, resuspended in PBS containing 20 μM H2DCF-DA (added from a fresh 4 mM stock solution in dimethyl sulfoxide), and incubated in the dark for 15 min at 30°C. The cell suspensions were diluted 1 : 500–1 : 1000 in PBS, as needed, just prior to the measurements. Flow cytometry analysis of the fluorescence levels was performed in a Gallios flow cytometer (Beckman Coulter Inc., Indianapolis, IN, USA), equipped with an argon ion laser of 15 mW at 488 nm as the excitation source. The H2DCF-DA fluorescence emission at 530 nm was detected using a 530/30-nm band pass filter array. Size-related forward scatter signals gathered by the cytometer were used by the Cyflogic v. 1.2.1 software (CyFlo Ltd., Turku, Finland) to gate fluorescence data from bacteria in the stream, thus avoiding the mixing of data from the bacteria with data from smaller, non-living particles in the suspension. Data for 25 000 cells per experimental condition were collected, and the Cyflogic v. 1.2.1 software was used to calculate the geometric mean of fluorescence per bacterial cell and the percentage of H2DCF-DA+-cells in each sample.
(f). Analytical procedures
Headspace GC–MS analysis of 1,3-DCP was performed using a Varian3000 gas chromatograph and a 4000MS ion trap mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany), equipped with a Combi PAL auto sampler (Altmann Analytik GmbH & Co. KG, Munich, Germany). The sample volumes of 100 μl were injected at 250°C (injector temperature) without split. The gas chromatograph was equipped with a 30 m × 0.25 mm × 0.25 μm FactorFour 5 ms column (low polarity phase, 5% phenyl and 95% dimethylpolysiloxane, Bruker Daltonik GmbH). The interface temperature was adjusted to 230°C, and the ion source was set to 200°C. The helium carrier gas was set to a constant flow of 0.6 ml min−1. After 6 min of constant heating at 35°C, the oven temperature was raised in a temperature ramp of 10°C min−1 to 250°C. To equilibrate the system for the next injection, the temperature was set to 35°C for 10 min. Mass spectra were recorded at two scans per second, with a scanning range of m/z = 50–301. The evaluation of the chromatograms was performed with the Varian Star Workstation v. 6.2 software (Bruker Daltonik GmbH). The metabolites were identified by comparison with the NIST atomic spectra database v. 4 (National Institute of Standards and Technology, Gaithersburg, MD, USA) and purified standards. The automatic peak quantification of the selected metabolites was implemented in the processing setup of the Varian Star Workstation v. 6.2 software. Virtual fragmentations were performed separately with the Mass Frontier v. 2.0 software (Thermo Fisher Scientific Inc.). d-Glucose concentration in culture supernatants was determined using an enzymatic kit based on the reactions sequentially catalysed by hexokinase and G6PDH (from S. cerevisiae), according to the manufacturer's instructions (R-Biopharm AG, Darmstadt, Germany).
(g). Statistical analysis
The reported experiments were independently repeated at least twice (as indicated in the corresponding figure legend), and the mean value of the corresponding parameter ± s.d. is presented. The statistical significance between multiple comparisons was obtained by the analysis of variance (ANOVA) followed by a Bonferroni post-test using, if necessary, transformed data. For the flow cytometry experiments, the median value is reported in box plots with the first and third quartiles. In these experiments, the statistical significance was analysed with the Mann–Whitney U-test. In all cases, data were considered statistically significant when p < 0.05.
3. Results
(a). Characterization of growth parameters and haloalkane dehalogenase activity in batch cultures of Pseudomonas pavonaceae containing d-glucose as the primary carbon source
Pseudomonas pavonaceae 170 was isolated from soil samples heavily contaminated with 1,3-DCP and 1,2-dichloropropane [40]. This Gram-negative micro-organism has been classified as Pseudomonas cichorii, using a phenotypic characterization strategy [29], and the genes encoding the haloalkane dehalogenase activity that enables growth on 1,3-DCP were traced to a self-transmissible plasmid that also confers resistance to Hg2+ salts [40]. Strain 170 also grows on 3-chloroallyl alcohol, 3-chloroacrylic acid and several 1-halo-n-alkanes [56,57]. The structural and mechanistic characteristics of haloalkane dehalogenase enzymes helped to elucidate the 1,3-DCP biodegradation pathway (figure 1). The first step in 1,3-DCP biodegradation in P. pavonaceae 170 is catalysed by a hydrolytic haloalkane dehalogenase (DhaA, E.C. 3.8.1.5) with broad substrate specificity [22]. After two sequential oxidation steps catalysed by an alcohol dehydrogenase and an aldehyde dehydrogenase, the first intermediate metabolite (3-chloro-2-propene-1-ol) is converted into 3-chloro-2-propenal, and finally into 3-chloro-2-propenoic acid. The latter compound, also termed 3-chloroacrylic acid, is the substrate for a specific 3-chloroacrylic acid dehalogenase (CaaD, E.C. 3.8.1; composed by CaaD1, the α subunit and CaaD2, the β subunit), which yields 3-oxopropanoate (malonate semialdehyde). Neither DhaA nor CaaD seem to need cofactors for the dechlorination reactions that they catalyse, and the cofactors for Aldh and Adh have not been identified. In the last step of 1,3-DCP biodegradation and mineralization, 3-oxopropanoate can enter into the central metabolic pathways as acetaldehyde upon decarboxylation by a malonate semialdehyde decarboxylase [58,59].
Little is known about the physiological responses elicited in native organochloride-degrader micro-organisms when exposed to 1,3-DCP. We evaluated the growth, consumption of the primary carbon substrate and organohalide biodegradation in batch cultures of P. pavonaceae developed in MMY, a semi-synthetic culture medium containing 30 mM d-glucose as the primary carbon source. Microbial growth was completely inhibited when the 1,3-DCP concentration was higher than 0.5 mM, regardless of the primary carbon source used (data not shown), and we routinely used this concentration in the experiments involving addition of 1,3-DCP. It was proposed that growth inhibition is primarily caused by the toxic effect of 1,3-DCP itself, rather than by the intermediate metabolites derived, because the corresponding 3-chloroallyl alcohols could serve as growth substrates at concentrations up to 5 mM [57]. In cultures containing d-glucose as the primary carbon source, cells started to grow after a lag phase of 2.8 ± 0.4 h with μ = 0.27 ± 0.04 h−1 and reached the stationary phase after approximately 20 h (figure 2a). The OD600 of these cultures increased to 1.38 ± 0.16 units between 20 and 48 h (an OD600 value approx. 1.6-fold higher than that at 24 h), but at a much slower rate. The kinetics of 1,3-DCP biodegradation followed a distinguishable behaviour throughout the cultivation period. We detected a lag phase during which the organohalide substrate was not consumed. The residual 1,3-DCP concentration did not noticeably change over the first 6 h (probably as a consequence of the low cell density within this timeframe) and started to trend sharply downwards thereafter (figure 2a). The highest rate of 1,3-DCP degradation was registered between 12 and 48 h, and at the end of this cultivation period, the residual organohalide concentration reached 0.154 ± 0.042 mM, indicating that more than 72 per cent of the initial 1,3-DCP was consumed with a specific consumption rate of 85 ± 3 μmol OD600−1 h−1. During exponential growth, the specific rate of d-glucose consumption was 6.8 ± 0.4 mmol OD600−1 h−1, and after 18–20 h, most of the primary carbon source was consumed (>90%, data not shown). By contrast, the specific rate of d-glucose consumption in control cultures, in which no 1,3-DCP had been added, reached 5.1 ± 0.2 mmol OD600−1 h−1. These results suggest that 1,3-DCP can be used with d-glucose as a carbon source and that the presence of an organochloride in the culture medium stimulates d-glucose utilization.
Figure 2.
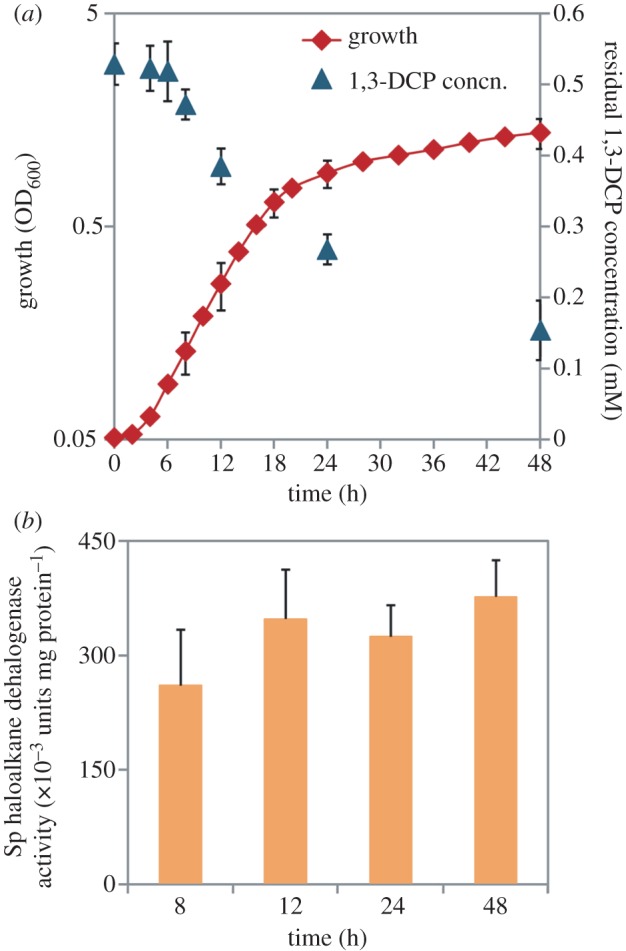
Characterization of growth, 1,3-dichloroprop-1-ene (1,3-DCP) biodegradation, and haloalkane dehalogenase activity in P. pavonaceae cells grown in batch cultures developed on MMY medium containing 30 mM d-glucose as the primary carbon source. (a) The cell density (estimated as the optical density measured at 600 nm (OD600) of appropriate culture dilutions) was evaluated along with the kinetics of organohalide biodegradation, expressed as the residual concentration of the compound and assessed by means of headspace GC coupled to MS. See §2 for further experimental details. (b) In vitro quantification of the specific (Sp) haloalkane dehalogenase activity at selected points during the experiment. Each data point or bar represents the mean value of the corresponding parameter ± s.d. of duplicate measurements from at least three independent experiments. (Online version in colour.)
To test whether the uncoupling between growth and 1,3-DCP consumption in P. pavonaceae may arise from a carbon catabolite repression phenomenon mediated by d-glucose, we measured the specific haloalkane dehalogenase activity during the cultivation period (figure 2b). This type of global regulation is typical in biodegradation pathways of environmental bacteria, and the expression of the pWW0-encoded TOL functions for aromatic catabolism in P. putida mt-2 is a paradigmatic example [60]. We observed that the enzymatic activity did not vary significantly between 8 and 48 h (p > 0.1), peaking at (469 ± 75) × 10−3 units mg protein−1 at the end of the cultivation period. When the specific haloalkane dehalogenase activity was quantified in the cell-free extracts obtained from exponential cultures developed on d-glucose or citrate (i.e. no 1,3-DCP added), values similar to those in cultures amended with the organohalide were obtained ((279 ± 12) and (381 ± 43) × 10−3 units mg protein−1, respectively). These experimental values are within the range reported in the literature for cultures of P. pavonaceae, developed on several organohalide compounds as the carbon and energy sources [29,57], and they indicate that haloalkane dehalogenase activity is constitutively present in this micro-organism under both glycolytic and gluconeogenic metabolic regimes.
(b). Addition of 1,3-dichloroprop-1-ene to Pseudomonas pavonaceae cultures has a different effect on growth parameters, depending on the primary carbon source used for growth
We investigated the consistent lag phase in growth and 1,3-DCP consumption, which occurs even when the haloalkane dehalogenase activity is constitutively present in P. pavonaceae. The cells were grown batchwise on two glycolytic substrates (i.e. d-glucose or d-gluconate) or two gluconeogenic substrates (i.e. citrate or succinate) as the primary carbon sources, and the extent of the lag phase and the normalized growth coefficient were determined in each condition (figure 3). Glycolytic carbon sources supported microbial growth, at specific growth rates very close to those observed in the absence of 1,3-DCP, with normalized growth coefficients of 92 ± 3% and 85 ± 5% for d-glucose and d-gluconate, respectively. The lag phase before exponential growth of the cells was less than 4 h for both carbon sources. By contrast, cells grown on citrate or succinate exhibited a lag phase 2.5- and 2.1-fold longer, respectively, than that observed on d-glucose (p < 0.05). Judging by the growth kinetics in these batch cultures, cells grown on gluconeogenic carbon sources were also more affected by the presence of 1,3-DCP than those grown under a glycolytic regime, resulting in normalized growth coefficients of 62 ± 3% and 71 ± 4% for citrate and succinate, respectively (p < 0.01, when compared with the normalized growth coefficient observed for d-glucose cultures). As an additional control of the differences on the kinetic parameters caused by using different carbon sources, we compared the normalized growth coefficients for d-gluconate, citrate and succinate, when compared with d-glucose cultures. The values obtained ranged between 85 per cent and 108 per cent, suggesting that the reduction in the specific growth rate in the presence of 1,3-DCP can be predominantly attributed to the organochloride presence in the culture medium.
Figure 3.
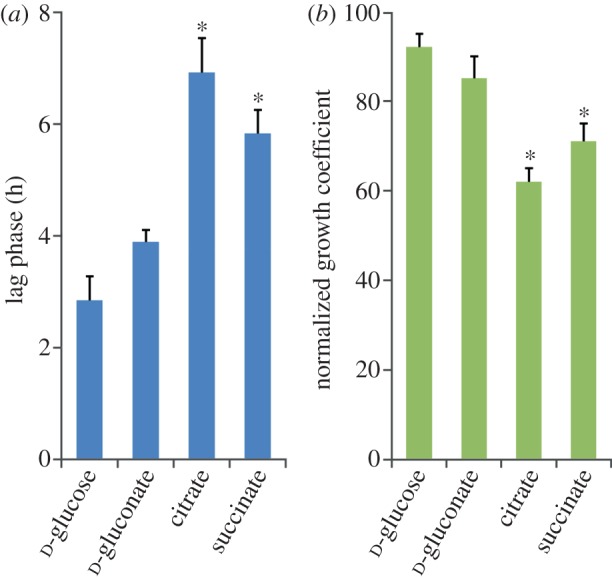
Evaluation of the effects of glycolytic and gluconeogenic metabolic regimes imposed by the primary carbon source on growth parameters of P. pavonaceae. Batch cultures were developed on MMY medium containing 30 mM of the corresponding carbon source and 0.5 mM 1,3-dichloroprop-1-ene. (a) The extension of the lag phase was quantified analytically from growth parameters as described by Dalgaard & Koutsoumanis [45]. (b) Normalized growth coefficients represent the fraction of the specific growth rate attained by the cells in the presence of the organohalide when compared with that computed under control conditions. See §2 for further experimental details. Each bar represents the mean value of the corresponding parameter ± s.d. of quadruplicate measurements from at least ten independent experiments. The asterisk denotes significant differences in the corresponding parameter at the 95% level when compared with the control culture (d-glucose), as evaluated with ANOVA. (Online version in colour.)
(c). 1,3-Dichloroprop-1-ene elicits stressful conditions in Pseudomonas pavonaceae under both glycolytic and gluconeogenic regimes
In addition to the metabolic state of the cells, we considered the possibility that the addition of 1,3-DCP might impose stressful conditions on P. pavonaceae. To test this scenario, we quantified the percentage of cells that gave a positive signal for H2DCF-DA (a molecular probe highly sensitive to intracellular ROS) using flow cytometry, as an estimation of the stress level (figure 4a) upon exposure of the cells to the organochloride. The total percentage of H2DCF-DA+ cells increased under glycolytic and gluconeogenic conditions when 1,3-DCP was added to the cultures. The increase was significantly greater in cells growing on citrate as the primary carbon source (a 6.1-fold increase in the median value of H2DCF-DA+ cells), compared with that in cells growing on d-glucose (a 4.2-fold increase in the median value of H2DCF-DA+ cells). A stressed subpopulation of cells, representing a median value of 59 per cent of the cell count, was detected after the addition of 1,3-DCP under gluconeogenic growth conditions, as opposed to 32 per cent under glycolytic conditions.
Figure 4.
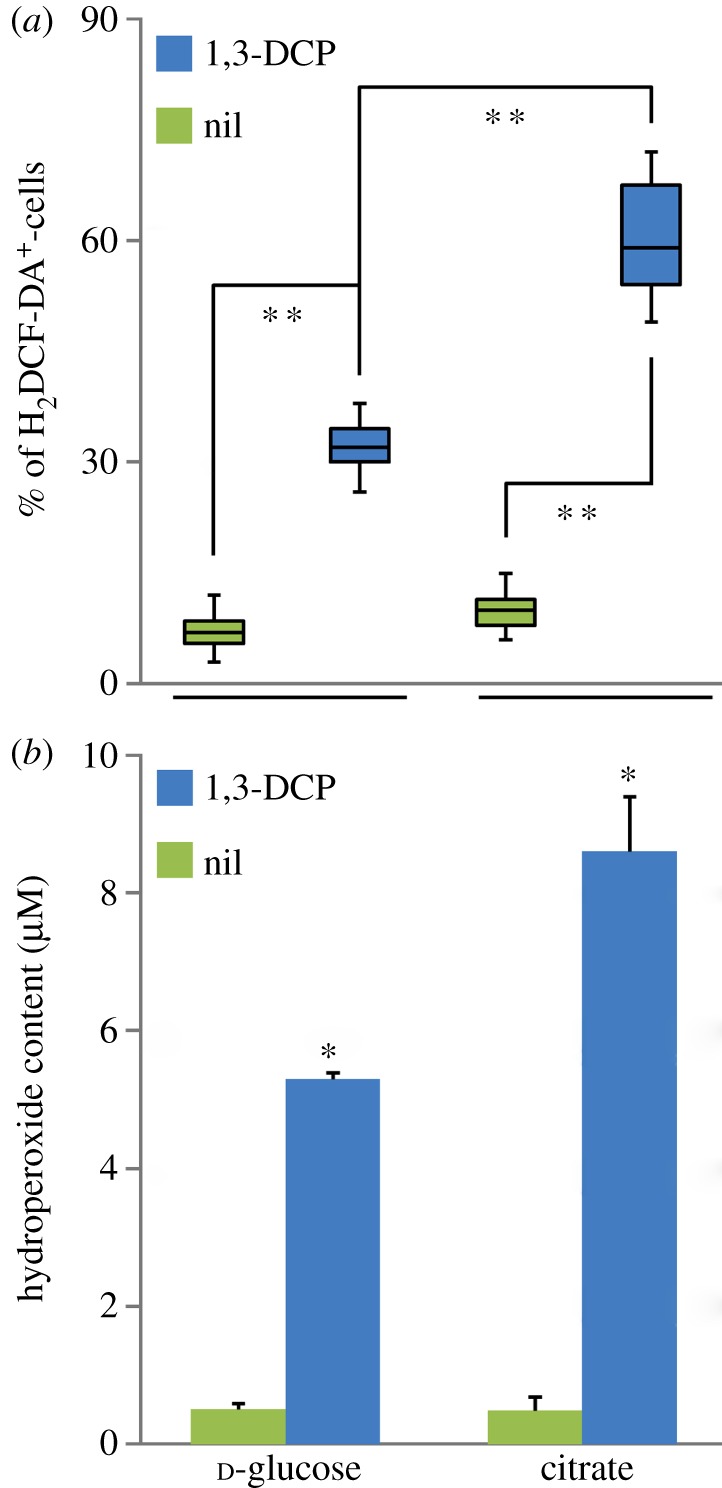
Evaluation of stress parameters in P. pavonaceae cells growing exponentially on MMY medium containing either 30 mM d-glucose or citrate (i.e. glycolytic and gluconeogenic metabolic regimes) under control conditions (nil) and in the presence of 0.5 mM 1,3-dichloroprop-1-ene (1,3-DCP). (a) Fluorescence-activated cell sorter cytometry analysis was performed to identify stressed bacterial subpopulations in vivo using the fluorescent dye 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA), which is activated upon exposure to ROS. Box plots represent the median value and the first and third quartiles from at least five independent experiments performed in triplicates, and the asterisk denotes significant differences among treatments at the 99% level as evaluated with the Mann–Whitney U-test. (b) In vitro determination of the total hydroperoxide content in the cells by means of the FOXII reagent, based on the formation of a stable Fe3+–xylenol orange complex. Each bar represents the mean value of the hydroperoxide content (expressed as equivalents of tert-butyl hydroperoxide) ± s.d. of duplicate measurements from at least three independent experiments. The asterisk denotes significant differences in the hydroperoxide content at the 95% level when compared with the control culture (without 1,3-DCP), as evaluated with ANOVA.
The next question was whether this stressed state translates into damage to the cells when exposed to 1,3-DCP. The level of oxidation of unsaturated lipids, which is a process that involves an allylic hydrogen atom abstraction, an insertion of molecular O2, and a subsequent reduction of the resulting hydroperoxyl radical to a free hydroperoxide, is widely used as an estimation of the cellular damage imposed by compounds that elicit oxidative conditions in vivo [61,62]. Considering the differences in the content of ROS determined at the single-cell level, we quantified spectrophotometrically the content of hydroperoxides in cultures of P. pavoneaceae, growing under glycolytic and gluconeogenic conditions, as a consequence of 1,3-DCP addition (figure 4b). Consistent with the changes in stressed populations, as analysed by flow cytometry, we observed a significant increase in the hydroperoxide content in cells exposed to 1,3-DCP, compared with that in untreated cultures. While the hydroperoxide content increased 10.4-fold in d-glucose-grown cells upon addition of 1,3-DCP, the difference between 1,3-DCP-amended and untreated cultures developed on citrate reached 17.6-fold. These results show that cells growing under the metabolic regimes tested have different abilities to use 1,3-DCP as a secondary substrate and to withstand the stressful conditions imposed by the organochloride substrate.
(d). Bacterial luciferase activity correlates with an altered redox state upon exposure of the cells to 1,3-dichloroprop-1-ene
Light emission by bacterial luciferase has been used in a variety of applications, including biosensors and reporters for gene expression, which resulted in useful whole cell-based assays for high-throughput screening and environmental monitoring [47,63]. Galluzzi & Karp [64] observed that light emission is strongly dependent on the redox state of cells (specifically to NADPH availability), a characteristic that to a degree limited the wide application of lux-based reporters. FMNH2 should continually be supplied in the bacterial cell for bioluminescence to remain at a constant level [65]. Both ATP and NADPH are directly involved in the formation of the luciferase substrates, although they do not directly participate in the light-emitting bioreaction. Therefore, bioluminescence is not an isolated process inside the cells because it is connected in the central biochemical network at the redox and energy homeostasis levels. It is also directly linked with other important pathways, including the tricarboxylic acid cycle, the electron transport chain and the synthesis of fatty acids and riboflavin. In particular, the relationship between the bioluminescence pathway and NADPH availability occurs at different points (figure 5b). NADPH is required to reduce an acyl group, previously transferred from the synthetase to the reductase component for the formation of the aldehyde that serves as the luciferase substrate. Reducing power is also required to regenerate FMNH2 from its oxidized counterpart (FMN). In most Gram-negative bacteria, this reductive pathway is based on the activity of a flavin reductase enzyme, which uses NADPH as the cofactor [66].
Figure 5.
Construction and analysis of the expression of a reporter tailored for monitoring the overall metabolic state of cells exposed to different stress agents. (a) Schematic of the synthetic reporter in which luxCDABE from P. luminescens, preceded by an optimized RBS, are placed under the control of a constitutive PEM7 element as a single transcriptional unit. The transcriptional terminator of the bacteriophage λ is depicted as T0. The elements in this outline borne by plasmid pSEVA226E, are not drawn to scale. (b) Scheme of the relationship between the biochemical luciferase reaction and the redox and energy state of the cells. A long-chain aldehyde (R-CHO) and a reduced flavine mononucleotide molecule (FMNH2) are used as the substrates in an O2-dependent reaction that produces H2O and light. Note that the regeneration of the aldehyde from the cognate carboxylic acid (R-COOH) as well as the reduction of FMN are biochemical processes that consume NADPH as reducing power. (c) Growth and bioluminescence trajectories of P. pavonaceae cells carrying pSEVA226E. Cultures were developed on MMY medium containing 30 mM d-glucose as the primary carbon source, and either 0.5 mM 1,3-dichloroprop-1-ene (1,3-DCP) or 0.5 mM diamide were added during exponential growth at the time indicated by the vertical arrow. Each data point represents the mean value of the corresponding parameter of triplicate measurements from at least two independent experiments, and standard deviation bars (consistently below 15% of the corresponding mean value for all experimental conditions) are omitted in the graphics for the sake of clarity.
We used the sensitivity of the lux-dependent bioluminescence to monitor the redox state of the cells exposed to 1,3-DCP. Pseudomonas pavonaceae was transformed with a reporter plasmid in which the expression of luxCDABE is driven by the EM7 promoter, a synthetic derivative of the T7 promoter that has constitutive and moderate activity in Pseudomonas species. When cells were grown on d-glucose alone, the bioluminescence steadily increased as growth progressed and decreased as the cells transitioned into the stationary phase, as expected for the physiological conditions under which NADPH (as well as O2 and ATP generation) becomes limiting. By contrast, when 1,3-DCP was added to cells growing exponentially, the bioluminescence dropped to approximately 25 per cent of the initial value almost immediately and remained at very low levels. In a separate experiment, we evaluated the effect of diamide addition on bioluminescence generation. Diamide is a strong thiol-oxidant agent known to deplete NADPH and interfere with the balance between reduced and oxidized glutathione in vivo [67]. As expected for an oxidative stress-inducing agent, we observed a strong reduction in bioluminescence levels with the addition of diamide, and the bioluminescence values reached values near zero, which were more apparent than those observed for the 1,3-DCP addition. The bioluminescence began to increase approximately 1 h after the diamide addition, suggesting that cells have a detoxification mechanism(s) that allows them to respond to the oxidative stress caused by the toxicant. These results strongly suggest that 1,3-DCP alters the redox homeostasis in P. pavonaceae.
(e). d-Glucose-6-P dehydrogenase is a key metabolic step that enables Pseudomonas pavonaceae cells to withstand stressful conditions imposed by 1,3-dichloroprop-1-ene addition
G6PDH is a major dehydrogenase involved in NADPH generation in the biochemical network, and we evaluated its activity in P. pavonaceae under different growth conditions to investigate the relationship between enzymatic activity and the stressful conditions elicited by 1,3-DCP. As the complete genome sequence of P. pavonaceae is not available, we first explored the presence of a zwf gene (encoding G6PDH) in this micro-organism. The amplification of a 460-bp product by PCR, using primers based on published sequences (details not shown), indicated that P. pavonaceae has a zwf gene which exhibits a 93 per cent identity to that of P. fluorescens Pf0-1 (Pfl01_2587) [68], its closer orthologue. When the G6PDH activity was evaluated in cell-free extracts (figure 6a), approximately half of the activity observed in d-glucose-grown cells was present in cells growing under a gluconeogenic regime on citrate (38.2 ± 2.4 nmol min−1 mg protein−1), suggesting that this metabolic step contributes to replenish the pentose pool (and generates reducing equivalents) when hexoses are not available as growth substrates. When 1,3-DCP was added to these cultures, the specific G6PDH activity increased under the glycolytic and gluconeogenic regimes, but the increase in the cells grown on citrate was higher than that observed on d-glucose (1.9- and 1.2-fold, respectively, p < 0.05). In both cases, the differences in enzymatic activity are expected to be accompanied by higher rates of NADPH turnover than under untreated conditions, which is consistent with the sharper stressful effect exerted by 1,3-DCP on citrate-growing cells than on d-glucose-growing cells. As a further evidence of the role of G6PDH on NADPH generation under stressful conditions, we quantified its activity in cells exposed to 0.15 mM menadione (a naphtoquinone known to mediate the generation of superoxide radical [69]), and obtained a pattern similar to the one observed for the cells treated with 1,3-DCP. Addition of menadione to the cells grown on citrate provoked a 4.7-fold increase in G6PDH, compared with a 1.6-fold increase attained in the cells grown on d-glucose. These results demonstrate that the metabolic state of the cells is determinant in their ability to withstand stressful conditions.
Figure 6.
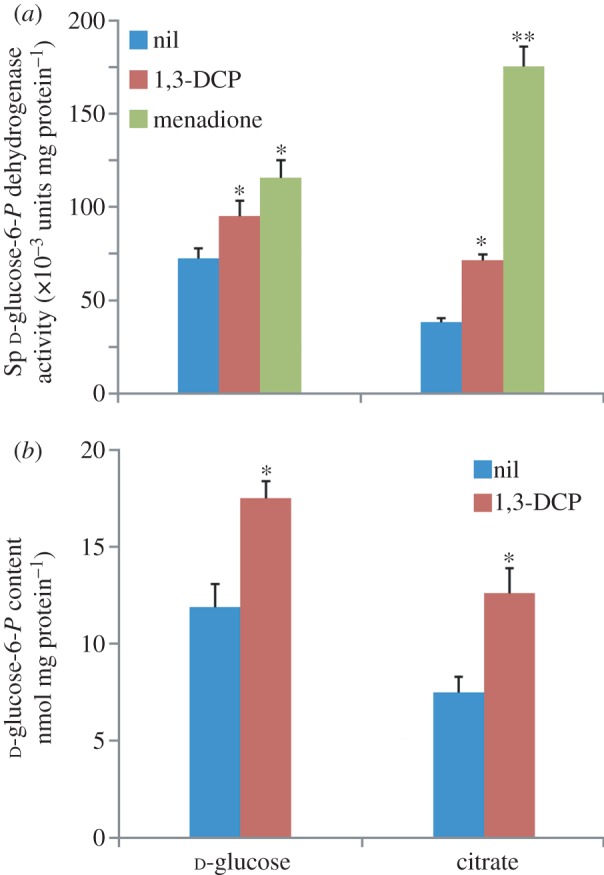
Evaluation of biochemical and metabolomic parameters in P. pavonaceae cells growing exponentially on MMY medium containing either 30 mM d-glucose or citrate (i.e. glycolytic and gluconeogenic metabolic regimes) under control conditions (nil) and in the presence of 0.5 mM 1,3-dichloroprop-1-ene (1,3-DCP). (a) In vitro quantification of the specific (Sp) d-glucose-6-P dehydrogenase activity. Note that the superoxide-generating naphtoquinone menadione was added at 0.15 mM to some cultures to impose oxidative stress conditions. (b) Determination of the intracellular d-glucose-6-P concentration. Appropriate aliquots of clarified cell-free extracts were boiled and centrifuged, and supernatants were spectrophotometrically analysed for d-glucose-6-P as detailed in §2. Each bar represents the mean value of the corresponding parameter ± s.d. of duplicate measurements from at least three independent experiments. In both cases, the asterisk marks denote significant differences in the corresponding parameter at either the 95% level (single asterisk) or the 99% level (double asterisks) when compared with the control culture (without additives), as evaluated with ANOVA.
(f). Addition of 1,3-dichloroprop-1-ene stimulates d-glucose-6-P accumulation in Pseudomonas pavonaceae
We questioned whether the differences observed in specific G6PDH activity among the conditions assayed result in different contents of G6P. The level of G6P in pseudomonads is thought to be determined by the tight metabolic balance that exists between the catabolic and anabolic reactions in the metabolic pathways, including the Entner–Doudoroff pathway, gluconeogenesis and the pentose phosphate pathway [70]. Depending on the carbon source and cellular needs, G6P can be (i) directly derived from d-glucose, (ii) biosynthesized from precursors in the lower metabolism through gluconeogenesis, (iii) oxidized by glycolysis to generate ATP (unlikely in this case because most Pseudomonas species lack the 6-phosphofructokinase activity needed to catalyse this transformation [35]) and/or (iv) oxidized through the pentose phosphate pathway (by means of the sequential activity of G6PDH and d-gluconate-6-P dehydrogenase) to generate NADPH. These biochemical steps can occur either separately or in combination, allowing the cells to satisfy the need of metabolic intermediates and to maintain the redox homeostasis.
We monitored the content of this phosphosugar among different metabolic regimes and culture conditions (figure 6b). Under control conditions, the content of G6P reached 11.9 ± 1.2 nmol mg protein−1 in cells grown on d-glucose. The value for cells grown on citrate was lower, as expected for gluconeogenic conditions. It is interesting to note that E. coli BW25113, grown in the same culture medium containing 30 mM d-glucose as carbon source, had a G6P content of 4.2 ± 0.3 nmol mg protein−1, which suggests that G6P might have a distinctive metabolic role in P. pavonaceae different than that in Enterobacteria. Upon addition of 1,3-DCP, the content of G6P in P. pavonaceae increased significantly under both glycolytic and gluconeogenic regimes, mirroring the behaviour observed for G6PDH activity. The intracellular G6P concentration had a 1.4- and 1.8-fold increase in cells grown on d-glucose and citrate, respectively (p < 0.05). These figures indicate that under biodegradation conditions, G6P is replenished to meet the demand determined by high G6PDH activities. This result implies that under a vigorous glycolytic regime of growth, the phosphorylation of d-glucose should be higher in the presence of 1,3-DCP, compared with untreated conditions in which cells are grown on d-glucose alone. As mentioned in a preceding section, the specific rate of d-glucose consumption increased approximately 1.4-fold when 1,3-DCP was added as a co-substrate, which is consistent with the notion that more G6P is produced by direct hexose phosphorylation. In cells growing on citrate, a high gluconeogenic activity (in particular, that of D-fructose-1,6-bisphosphatase) is expected to account for an elevated G6P content. In cells grown on d-glucose, d-gluconate-6-P was accumulated at 8.3 ± 0.6 nmol mg protein−1, and at 3.9 ± 0.2 nmol mg protein−1 when citrate was used as the carbon source. d-Gluconate-6-P levels changed upon the addition of 1,3-DCP in a similar fashion to that observed for G6P, which might be an indication of high levels of both G6PDH and d-gluconate-6-P dehydrogenase activity (which feeds the pentose phosphate pool) under stressed conditions. These results show that the high levels of G6P detected in P. pavonaceae play a crucial role as a potential source of reducing power, the availability of which is adjusted by different levels of G6PDH activity according to the cellular requirements.
4. Discussion
The synthetic halogenated aliphatic 1,3-DCP was introduced as a nematocidic soil fumigant in the 1950s, and since then has been used in large quantities [32,71]. The organohalides are recalcitrant to biodegradation for several reasons: (i) they are highly toxic, (ii) most complex organohalides are unnatural compounds recently introduced into the environment by human activities, and micro-organisms are unlikely to possess complete catabolic pathways for their mineralization, and (iii) if degradation is initiated by a dehalogenation reaction catalysed by an existing dehalogenase or oxidase, the lack of enzymes for the rapid conversion of the resulting halogenated alcohols or aldehydes would lead to the accumulation of toxic and/or highly reactive metabolic intermediates. Besides, the chemical bonds in these compounds are extraordinarily stable, making them very persistent in most environments. The slow biodegradation of 1,3-DCP observed in contaminated soils leads to the theory that some micro-organisms have assembled their catabolic pathways recently, possibly by recruiting (pre-)existing enzymes [22,72,73]. The constitutive expression of the haloalkane dehalogenase activity in P. pavonaceae grown on different carbon sources could indicate that the enzymes involved are part of a recently assembled catabolic pathway, which lacks an active regulatory system to tightly regulate the expression of the cognate genes. Pseudomonas pavonaceae thus affords an exceptional experimental system to examine how still-evolving environmental bacteria manage the interplay between different metabolic and stress programmes because some of the growth substrates used by this micro-organism constitute acute physiological stressors. In general, our data reveal that P. pavonaceae cells undergo stressful conditions when facing (and degrading) 1,3-DCP.
Three major types of response to stressful conditions imposed by xenobiotics have been recognized [16]. Metabolic programmes consist of the tuning (both at the gene transcription and regulation of activity levels) of enzyme sets required for the catabolism or anabolism of nutrients and intermediates; stress-response programmes for adaptation to suboptimal growth conditions; and morphological programmes related to shape, transport and surface chemistry of the bacterial cell. These three programmes are closely connected, and their functioning largely determines the survival of a given population in specific niches or its displacement by a fitter organism when facing adverse conditions.
In this study, we focused on the interplay between metabolic and stress-response programmes elicited by 1,3-DCP in P. pavonaceae. As a result of our findings, we hypothesized that the metabolic state of the cells influences their ability to face (and metabolize) the organochloride as an alternative carbon and energy source. The homeostasis conditions within metabolic networks are orchestrated by fine-tuning mechanisms that prevent drastic responses to nutritional or environmental perturbations. Instead of the typical all-or-none behaviour of most bacterial transcriptome constituents, biochemical networks are regulated by devices that tune (rather than switch) metabolic regimes, according to the overall physiological conditions. These processes allow the cells to generate the appropriate ratios of biosynthetic precursors for balanced growth [74]. As the haloalkane dehalogenase activity is constitutively present in P. pavonaceae, the biochemical network in this micro-organism is expected to respond to the presence of 1,3-DCP by adjusting metabolic fluxes to obtain the relevant metabolites and cofactors needed for biodegradation and growth. Our data suggest that in conditions that foster metabolic stress, under both glycolytic and gluconeogenic regimes, G6P is accumulated at high levels and possibly maintained as a reservoir of reducing power. The G6P amassed under such conditions is most likely used as a substrate of G6PDH to generate NADPH. The strong activity of gluconeogenic pathways, needed to fulfil G6P needs when cells are grown on citrate, supports the theory that fluxes throughout the biochemical network are adapted to meet the demand of metabolic precursors under stressful conditions. In addition to these core bioreactions, the need for a fine-tuned balancing of different redox cofactors (i.e. NADH and NADPH) is partially accounted for in several bacteria by the presence of transhydrogenases, which are enzymes that catalyse the electron transfer reactions between NADH and NADPH to equilibrate the respective pools thus meeting metabolic demands [38]. Although not investigated in this study, transhydrogenases could contribute to the homeostasis of NADH and NADPH together with the major dehydrogenases of the central catabolic pathways.
A possible generalization of the data presented in this study is that xenobiotic-mediated stress sets a limit to the possibilities of evolving aerobic pathways for the degradation of organochloride compounds. Bacteria must evolve adequate biodegradation pathways for the compound at stake, and they will also encounter the metabolic stress imposed by the potential substrate during an oxic metabolic regime. In general, anaerobic metabolisms are less prone to generate oxidative stress [75], and the chemical and thermodynamic itinerary for assembling new biodegradation routes could be less constrained than in aerobiosis. On the other hand, the strong electronegativity of multiple chlorine substituents contributes to make an oxidative attack on organochlorides more unlikely than a reductive attack. These results might explain, at least partially, the observed capacity of anaerobes to perform organohalide respiration for processing compounds that are virtually impossible to metabolize in the presence of oxygen. Finally, the results obtained in this study open new avenues for the rational design of biocatalysts tailored for the efficient degradation of organohalides [44].
Acknowledgements
The authors thank Prof. Dick B. Janssen (Institute for Biomolecular Sciences and Biotechnology, Groningen, The Netherlands) for the kind gift of Pseudomonas pavonaceae 170. This study was supported by the BIO and FEDER CONSOLIDER-INGENIO programme of the Spanish Ministry of Science and Innovation, the BACSINE, MICROME, and ST-FLOW Contracts of the EU and the PROMT Project of the CAM. P.I.N. is a researcher from the Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina) and holds an EMBO long-term fellowship (ALTF 13-2010); and D.P.P. is the holder of a Marie Curie grant of the EC for visiting Scholars.
References
- 1.Häggblom MM, Bossert ID. 2003. Halogenated organic compounds: a global perspective. In Dehalogenation: microbial processes and environmental applications (eds Häggblom MM, Bossert ID.), pp. 3–29 New York, NY: Kluwer Academic Publishers [Google Scholar]
- 2.Copley SD. 1997. Diverse mechanistic approaches to difficult chemical transformations: microbial dehalogenation of chlorinated aromatic compounds. Chem. Biol. 4, 169–174 10.1016/s1074-5521(97)90285-4 (doi:10.1016/s1074-5521(97)90285-4) [DOI] [PubMed] [Google Scholar]
- 3.Lee MD, Odom JM, Buchanan RJ., Jr 1998. New perspectives on microbial dehalogenation of chlorinated solvents: insights from the field. Annu. Rev. Microbiol. 52, 423–452 10.1146/annurev.micro.52.1.423 (doi:10.1146/annurev.micro.52.1.423) [DOI] [PubMed] [Google Scholar]
- 4.Futagami T, Goto M, Furukawa K. 2008. Biochemical and genetic bases of dehalorespiration. Chem. Rec. 8, 1–12 10.1002/tcr.20134 (doi:10.1002/tcr.20134) [DOI] [PubMed] [Google Scholar]
- 5.Field JA, Sierra-Alvarez R. 2008. Microbial transformation and degradation of polychlorinated biphenyls. Environ. Pollut. 155, 1–12 10.1016/j.envpol.2007.10.016 (doi:10.1016/j.envpol.2007.10.016) [DOI] [PubMed] [Google Scholar]
- 6.Hardman DJ. 1991. Biotransformation of halogenated compounds. Crit. Rev. Biotechnol. 11, 1–40 10.3109/07388559109069182 (doi:10.3109/07388559109069182) [DOI] [PubMed] [Google Scholar]
- 7.Janssen DB, Jager D, Witholt B. 1987. Degradation of n-haloalkanes and α,ω-dihaloalkanes by wild-type and mutants of Acinetobacter sp. strain GJ70. Appl. Environ. Microbiol. 53, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkin S. 1992. Biodegradation of haloalkanes. Biodegradation 3, 299–313 10.1007/bf00129090 (doi:10.1007/bf00129090) [DOI] [Google Scholar]
- 9.Guerin TF. 2008. Ex situ bioremediation of chlorobenzenes in soil. J. Hazard Mater. 154, 9–20 10.1016/j.jhazmat.2007.09.094 (doi:10.1016/j.jhazmat.2007.09.094) [DOI] [PubMed] [Google Scholar]
- 10.Goulding C, Gillen CJ, Bolton E. 1988. Biodegradation of substituted benzenes. J. Appl. Bacteriol. 65, 1–5 10.1111/j.1365-2672.1988.tb04310.x (doi:10.1111/j.1365-2672.1988.tb04310.x) [DOI] [PubMed] [Google Scholar]
- 11.Olaniran AO, Igbinosa EO. 2011. Chlorophenols and other related derivatives of environmental concern: properties, distribution and microbial degradation processes. Chemosphere 83, 1297–1306 10.1016/j.chemosphere.2011.04.009 (doi:10.1016/j.chemosphere.2011.04.009) [DOI] [PubMed] [Google Scholar]
- 12.Solyanikova IP, Golovleva LA. 2004. Bacterial degradation of chlorophenols: pathways, biochemica, and genetic aspects. J. Environ. Sci. Health B 39, 333–351 10.1081/pfc-120035921 (doi:10.1081/pfc-120035921) [DOI] [PubMed] [Google Scholar]
- 13.de Jong RM, Dijkstra BW. 2003. Structure and mechanism of bacterial dehalogenases: different ways to cleave a carbon-halogen bond. Curr. Opin. Struct. Biol. 13, 722–730 10.1016/j.sbi.2003.10.009 (doi:10.1016/j.sbi.2003.10.009) [DOI] [PubMed] [Google Scholar]
- 14.Kurihara T, Esaki N. 2008. Bacterial hydrolytic dehalogenases and related enzymes: occurrences, reaction mechanisms, and applications. Chem. Rec. 8, 67–74 10.1002/tcr.20141 (doi:10.1002/tcr.20141) [DOI] [PubMed] [Google Scholar]
- 15.Janssen DB, Oppentocht JE, Poelarends GJ. 2001. Microbial dehalogenation. Curr. Opin. Biotechnol. 12, 254–258 10.1016/s0958-1669(00)00208-1 (doi:10.1016/s0958-1669(00)00208-1) [DOI] [PubMed] [Google Scholar]
- 16.Loza-Tavera H, de Lorenzo V. 2011. Microbial bioremediation of chemical pollutants: how bacteria cope with multi-stress environmental scenarios? In Bacterial stress responses (eds Storz G, Hengge R.), pp. 481–492 Washington, DC: ASM Press [Google Scholar]
- 17.Haro MA, de Lorenzo V. 2001. Metabolic engineering of bacteria for environmental applications: construction of Pseudomonas strains for biodegradation of 2-chlorotoluene. J. Biotechnol. 85, 103–113 10.1016/s0168-1656(00)00367-9 (doi:10.1016/s0168-1656(00)00367-9) [DOI] [PubMed] [Google Scholar]
- 18.Bedard DL. 2008. A case study for microbial biodegradation: anaerobic bacterial reductive dechlorination of polychlorinated biphenyls: from sediment to defined medium. Annu. Rev. Microbiol. 62, 253–270 10.1146/annurev.micro.62.081307.162733 (doi:10.1146/annurev.micro.62.081307.162733) [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Bennett GN. 2005. Biodegradation of xenobiotics by anaerobic bacteria. Appl. Microbiol. Biotechnol. 67, 600–618 10.1007/s00253-004-1864-3 (doi:10.1007/s00253-004-1864-3) [DOI] [PubMed] [Google Scholar]
- 20.Mogensen AS, Dolfing J, Haagensen F, Ahring BK. 2003. Potential for anaerobic conversion of xenobiotics. Adv. Biochem. Eng. Biotechnol. 82, 69–134 10.1007/3-540-45838-7_3 (doi:10.1007/3-540-45838-7_3) [DOI] [PubMed] [Google Scholar]
- 21.Häggblom MM, Knight VK, Kerkhof LJ. 2000. Anaerobic decomposition of halogenated aromatic compounds. Environ. Pollut. 107, 199–207 10.1016/s0269-7491(99)00138-4 (doi:10.1016/s0269-7491(99)00138-4) [DOI] [PubMed] [Google Scholar]
- 22.Janssen DB. 2004. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 8, 150–159 10.1016/j.cbpa.2004.02.012 (doi:10.1016/j.cbpa.2004.02.012) [DOI] [PubMed] [Google Scholar]
- 23.Wohlfarth G, Diekert G. 1997. Anaerobic dehalogenases. Curr. Opin. Biotechnol. 8, 290–295 10.1016/s0958-1669(97)80006-7 (doi:10.1016/s0958-1669(97)80006-7) [DOI] [PubMed] [Google Scholar]
- 24.El Fantroussi S, Naveau H, Agathos SN. 1998. Anaerobic dechlorinating bacteria. Biotechnol. Prog. 14, 167–188 10.1021/bp980011k (doi:10.1021/bp980011k) [DOI] [PubMed] [Google Scholar]
- 25.Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Häggblom MM. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38, 2075–2081 10.1021/es034989b (doi:10.1021/es034989b) [DOI] [PubMed] [Google Scholar]
- 26.Nijenhuis I, Zinder SH. 2005. Characterization of hydrogenase and reductive dehalogenase activities of Dehalococcoides ethenogenes strain 195. Appl. Environ. Microbiol. 71, 1664–1667 10.1128/aem.71.3.1664-1667.2005 (doi:10.1128/aem.71.3.1664-1667.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhry GR, Chapalamadugu S. 1991. Biodegradation of halogenated organic compounds. Microbiol. Rev. 55, 59–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Commandeur LC, Parsons JR. 1990. Degradation of halogenated aromatic compounds. Biodegradation 1, 207–220 10.1007/bf00058837 (doi:10.1007/bf00058837) [DOI] [PubMed] [Google Scholar]
- 29.Poelarends GJ, Wilkens M, Larkin MJ, van Elsas JD, Janssen DB. 1998. Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl. Environ. Microbiol. 64, 2931–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xuan R, Yates SR, Ashworth DJ, Luo L. 2012. Mitigating 1,3-dichloropropene, chloropicrin, and methyl iodide emissions from fumigated soil with reactive film. Environ. Sci. Technol. 46, 6143–6149 10.1021/es300388r (doi:10.1021/es300388r) [DOI] [PubMed] [Google Scholar]
- 31.Ibekwe AM, Papiernik SK, Yates SR, Crowley DE, Yang CH. 2001. Microcosm enrichment of 1,3-dichloropropene-degrading soil microbial communities in a compost-amended soil. J. Appl. Microbiol. 91, 668–676 10.1046/j.1365-2672.2001.01431.x (doi:10.1046/j.1365-2672.2001.01431.x) [DOI] [PubMed] [Google Scholar]
- 32.van Dijk H. 1974. Degradation of 1,3-dichloropropenes in the soil. AgroEcosystems 1, 193–204 10.1016/0304-3746(74)90026-2 (doi:10.1016/0304-3746(74)90026-2) [DOI] [Google Scholar]
- 33.Fuhrer T, Fischer E, Sauer U. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590 10.1128/jb.187.5.1581-1590.2005 (doi:10.1128/jb.187.5.1581-1590.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavarría M, Kleijn RJ, Sauer U, Pflüger-Grau K, de Lorenzo V. 2012. Regulatory tasks of the phosphoenolpyruvate-phosphotransferase system of Pseudomonas putida in central carbon metabolism. mBio 3, e00028-00012. 10.1128/mbio.00028-12 (doi:10.1128/mbio.00028-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nogales J, Palsson BØ, Thiele I. 2008. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst. Biol. 2, 79. 10.1186/1752-0509-2-79 (doi:10.1186/1752-0509-2-79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Jeon CO, Park W. 2008. Dual regulation of zwf-1 by both 2-keto-3-deoxy-6-phosphogluconate and oxidative stress in Pseudomonas putida. Microbiology 154, 3905–3916 10.1099/mic.0.2008/020362-0 (doi:10.1099/mic.0.2008/020362-0) [DOI] [PubMed] [Google Scholar]
- 37.Sandoval JM, Arenas FA, Vásquez CC. 2011. Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS ONE 6, e25573. 10.1371/journal.pone.0025573 (doi:10.1371/journal.pone.0025573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuhrer T, Sauer U. 2009. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 191, 2112–2121 10.1128/jb.01523-08 (doi:10.1128/jb.01523-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach. Sunderland, MA: Sinauer Associates [Google Scholar]
- 40.Verhagen C, Smit E, Janssen DB, van Elsas JD. 1995. Bacterial dichloropropene degradation in soil; screening of soils and involvement of plasmids carrying the dhlA gene. Soil Biol. Biochem. 27, 1547–1557 10.1016/0038-0717(95)00105-n (doi:10.1016/0038-0717(95)00105-n) [DOI] [Google Scholar]
- 41.Hanahan D, Meselson M. 1983. Plasmid screening at high colony density. Methods Enzymol. 100, 333–342 10.1016/0076-6879(83)00066-x (doi:10.1016/0076-6879(83)00066-x) [DOI] [PubMed] [Google Scholar]
- 42.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 172, 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 0008. 10.1038/msb4100050 (doi:10.1038/msb4100050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikel PI, de Lorenzo V. 2012. Engineering an anaerobic metabolic regime in Pseudomonas putida KT2440 for the anoxic biodegradation of 1,3-dichloroprop-1-ene. Metab. Eng. 15, 98–112 10.1016/j.ymben.2012.09.006 (doi:10.1016/j.ymben.2012.09.006) [DOI] [PubMed] [Google Scholar]
- 45.Dalgaard P, Koutsoumanis K. 2001. Comparison of maximum specific growth rates and lag times estimated from absorbance and viable count data by different mathematical models. J. Microbiol. Methods 43, 183–196 10.1016/s0167-7012(00)00219-0 (doi:10.1016/s0167-7012(00)00219-0) [DOI] [PubMed] [Google Scholar]
- 46.Wilson T, Hastings JW. 1998. Bioluminescence. Annu. Rev. Cell Dev. Biol. 14, 197–230 10.1146/annurev.cellbio.14.1.197 (doi:10.1146/annurev.cellbio.14.1.197) [DOI] [PubMed] [Google Scholar]
- 47.Waidmann MS, Bleichrodt FS, Laslo T, Riedel CU. 2011. Bacterial luciferase reporters: the Swiss army knife of molecular biology. Bioeng. Bugs 2, 8–16 10.4161/bbug.2.1.13566 (doi:10.4161/bbug.2.1.13566) [DOI] [PubMed] [Google Scholar]
- 48.Meighen EA. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55, 123–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- 50.Choi KH, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protocols 1, 153–161 10.1038/nprot.2006.24 (doi:10.1038/nprot.2006.24) [DOI] [PubMed] [Google Scholar]
- 51.Silva-Rocha, et al. 2013. The standard European vector architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675 10.1093/nar/gks1119 (doi:10.1093/nar/gks1119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 10.1016/j.mimet.2005.06.001 (doi:10.1016/j.mimet.2005.06.001) [DOI] [PubMed] [Google Scholar]
- 53.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 (doi:10.1016/0003-2697(76)90527-3) [DOI] [PubMed] [Google Scholar]
- 54.Bergmann JG, Sanik J. 1957. Determination of trace amounts of chlorine in naphtha. Anal. Chem. 29, 241–243 10.1021/ac60122a018 (doi:10.1021/ac60122a018) [DOI] [Google Scholar]
- 55.Jiang ZY, Hunt JV, Wolff SP. 1992. Ferrous ion oxidation in the presence of xylenol orange for detection of lipid hydroperoxide in low density lipoprotein. Anal. Biochem. 202, 384–389 10.1016/0003-2697(92)90122-N (doi:10.1016/0003-2697(92)90122-N) [DOI] [PubMed] [Google Scholar]
- 56.Poelarends GJ, Kulakov LA, Larkin MJ, van Hylckama Vlieg JET, Janssen DB. 2000. Roles of horizontal gene transfer and gene integration in evolution of 1,3-dichloropropene- and 1,2-dibromoethane-degradative pathways. J. Bacteriol. 182, 2191–2199 10.128/jb.182.8.2191-2199.2000 (doi:10.128/jb.182.8.2191-2199.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poelarends GJ, Saunier R, Janssen DB. 2001. trans-3-Chloroacrylic acid dehalogenase from Pseudomonas pavonaceae 170 shares structural and mechanistic similarities with 4-oxalocrotonate tautomerase. J. Bacteriol. 183, 4269–4277 10.1128/jb.183.14.4269-4277.2001 (doi:10.1128/jb.183.14.4269-4277.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almrud JJ, Poelarends GJ, Johnson WH, Jr, Serrano H, Hackert ML, Whitman CP. 2005. Crystal structures of the wild-type, P1A mutant, and inactivated malonate semialdehyde decarboxylase: a structural basis for the decarboxylase and hydratase activities. Biochemistry 44, 14 818–14 827 10.1021/bi051383m (doi:10.1021/bi051383m) [DOI] [PubMed] [Google Scholar]
- 59.Poelarends GJ, Johnson WH, Jr, Murzin AG, Whitman CP. 2003. Mechanistic characterization of a bacterial malonate semialdehyde decarboxylase: identification of a new activity on the tautomerase superfamily. J. Biol. Chem. 278, 48 674–48 683 10.1074/jbc.m306706200 (doi:10.1074/jbc.m306706200) [DOI] [PubMed] [Google Scholar]
- 60.Ramos JL, Marqués S, Timmis KN. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51, 341–373 10.1146/annurev.micro.51.1.341 (doi:10.1146/annurev.micro.51.1.341) [DOI] [PubMed] [Google Scholar]
- 61.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8 [PubMed] [Google Scholar]
- 62.Imlay JA. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418 10.1146/annurev.micro.57.030502.090938 (doi:10.1146/annurev.micro.57.030502.090938) [DOI] [PubMed] [Google Scholar]
- 63.Heitzer A, Applegate B, Kehrmeyer S, Pinkart H, Webb OF, Phelps TJ, White DC, Sayler GS. 1998. Physiological considerations of environmental applications of lux reporter fusions. J. Microbiol. Methods 33, 45–57 10.1016/S0167-7012(98)00043-8 (doi:10.1016/S0167-7012(98)00043-8) [DOI] [Google Scholar]
- 64.Galluzzi L, Karp M. 2007. Intracellular redox equilibrium and growth phase affect the performance of luciferase-based biosensors. J. Biotechnol. 127, 188–198 10.1016/j.jbiotec.2006.06.019 (doi:10.1016/j.jbiotec.2006.06.019) [DOI] [PubMed] [Google Scholar]
- 65.Koga K, Harada T, Shimizu H, Tanaka K. 2005. Bacterial luciferase activity and the intracellular redox pool in Escherichia coli. Mol. Genet. Genomics 274, 180–188 10.1007/s00438-005-0008-5 (doi:10.1007/s00438-005-0008-5) [DOI] [PubMed] [Google Scholar]
- 66.Fieschi F, Nivière V, Frier C, Décout JL, Fontecave M. 1995. The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli. J. Biol. Chem. 270, 30 392–30 400 10.1074/jbc.270.51.30392 (doi:10.1074/jbc.270.51.30392) [DOI] [PubMed] [Google Scholar]
- 67.Kosower NS, Kosower EM. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251, 123–133 10.1016/0076-6879(95)51116-4 (doi:10.1016/0076-6879(95)51116-4) [DOI] [PubMed] [Google Scholar]
- 68.Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock REW, Brinkman FSL. 2011. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 10.1093/nar/gkq869 (doi:10.1093/nar/gkq869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hassan HM, Fridovich I. 1979. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch. Biochem. Biophys. 196, 385–395 10.1016/0003-9861(79)90289-3 (doi:10.1016/0003-9861(79)90289-3) [DOI] [PubMed] [Google Scholar]
- 70.Blank LM, Ionidis G, Ebert BE, Bühler B, Schmid A. 2008. Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS J. 275, 5173–5190 10.1111/j.1742-4658.2008.06648.x (doi:10.1111/j.1742-4658.2008.06648.x) [DOI] [PubMed] [Google Scholar]
- 71.Roberts TR, Stoydin G. 1976. The degradation of (Z)- and (E)-1,3-dichloropropenes and 1,2-dichloropropane in soil. Pestic. Sci. 7, 325–335 10.1002/ps.2780070402 (doi:10.1002/ps.2780070402) [DOI] [Google Scholar]
- 72.Janssen DB, Dinkla IJT, Poelarends GJ, Terpstra P. 2005. Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ. Microbiol. 7, 1868–1882 10.1111/j.1462-2920.2005.00966.x (doi:10.1111/j.1462-2920.2005.00966.x) [DOI] [PubMed] [Google Scholar]
- 73.Poelarends GJ, Whitman CP. 2004. Evolution of enzymatic activity in the tautomerase superfamily: mechanistic and structural studies of the 1,3-dichloropropene catabolic enzymes. Bioorg. Chem. 32, 376–392 10.1016/j.bioorg.2004.05.006 (doi:10.1016/j.bioorg.2004.05.006) [DOI] [PubMed] [Google Scholar]
- 74.Blank LM, Ebert BE, Buehler K, Bühler B. 2010. Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis. Antioxid. Redox Signal 13, 349–394 10.1089/ars.2009.2931 (doi:10.1089/ars.2009.2931) [DOI] [PubMed] [Google Scholar]
- 75.Imlay JA. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microbial Physiol. 46, 111–153 10.1016/s0065-2911(02)46003-1 (doi:10.1016/s0065-2911(02)46003-1) [DOI] [PubMed] [Google Scholar]