Abstract
The respiratory system comprises several scales of biological complexity: the genes, cells and tissues that work in concert to generate resultant function. Malfunctions of the structure or function of components at any spatial scale can result in diseases, to the detriment of gas exchange, right heart function and patient quality of life. Vast amounts of data emerge from studies across each of the biological scales; however, the question remains: how can we integrate and interpret these data in a meaningful way? Respiratory disease presents a huge health and economic burden, with the diseases asthma and chronic obstructive pulmonary disease (COPD) affecting over 500 million people worldwide. Current therapies are inadequate owing to our incomplete understanding of the disease pathophysiology and our lack of recognition of the enormous disease heterogeneity: we need to characterize this heterogeneity on a patient-specific basis to advance healthcare. In an effort to achieve this goal, the AirPROM consortium (Airway disease Predicting Outcomes through patient-specific computational Modelling) brings together a multi-disciplinary team and a wealth of clinical data. Together we are developing an integrated multi-scale model of the airways in order to unravel the complex pathophysiological mechanisms occurring in the diseases asthma and COPD.
Keywords: asthma, chronic obstructive pulmonary disease, computational modelling
1. The need for multi-scale models of the respiratory system
The diseases asthma and chronic obstructive pulmonary disease (COPD) are common chronic diseases associated with significant disability and societal burden [1]. These diseases affect over 500 million people worldwide [2] and related costs exceed €56 billion per year in the European Union (EU). Asthma and COPD are complex airway diseases encompassing several underlying pathological conditions that involve a variety of gene–cell–environment interactions giving rise to various clinical phenotypes. The diseases are characterized by an underlying inflammatory response that induces airway remodelling, airflow limitation and increased ventilation–perfusion (V/Q) mismatch resulting in diminished lung function. Current therapies are inadequate owing to our incomplete understanding of the pathophysiology of these diseases and our lack of recognition of the enormous disease heterogeneity, complexity and multi-scale nature of these diseases: we need to characterize this heterogeneity on a patient-specific basis to advance healthcare. In an effort to achieve this goal, the AirPROM consortium (Airway disease Predicting Outcomes through patient-specific computational Modelling; www.airprom.eu) brings together existing clinical consortia with expertise in physiology, radiology, image analysis, bioengineering, data harmonization (the process of synchronizing and bringing together different datasets), data security and ethics, computational modelling and systems biology. Together we are developing an integrated multi-scale model of the airways in order to unravel the complex pathophysiological mechanisms occurring in the diseases asthma and COPD.
It is estimated that COPD, typically associated with cigarette smoking but also related to biomass/environmental exposure, will be the third largest cause of global mortality by the year 2030. Few existing therapies have an impact upon the natural history or mortality in COPD [3]. COPD is associated with obstruction and obliteration of small (less than 2 mm in diameter) conducting airways (bronchiolitis) and alveolar airspace enlargement (emphysema). Both of these processes contribute towards respiratory disability in COPD [4], and can occur to differential degrees in patients with the same level of lung function loss. The pathogenesis of bronchiolitis and emphysema in COPD is poorly understood. However, these two processes are likely to be governed by a diverse array of cellular-scale processes, including airway inflammation, adaptive and innate immunity to cigarette smoking and microbiota, autoimmunity to external or self-peptides, accelerated senescence and dysregulated repair (reviewed in [5]). Finally, a variety of candidate genes have recently been reported in genome-wide association studies [6]. The focus to date has been upon single scales of the disease, and how airway inflammation and persistent infection are a consequence of genetic susceptibility and result in airway remodelling and dysfunction is uncertain.
Although asthma deaths appear to be decreasing owing to the consistent use of inhaled corticosteroids and wider recognition of markers associated with adverse risk [7], many patients with asthma continue to have persistence and poor control of symptoms [8]. A substantial proportion of the EU health budgets assigned to airway diseases is used by patients who experience asthma exacerbations. The pathogenesis of exacerbations remains poorly understood [9]. However, recognition that asthma is associated with multiple inflammatory phenotypes [10] and targeting these different phenotypes in specific ways using existing or novel asthma therapies [11] has been shown to reduce asthma exacerbations. The mechanism of how eosinophilic disease may modulate exacerbations in asthma remains unknown. Furthermore, non-eosinophilic phenotypes of asthma are common in the general population and new therapies targeting non-type 2 helper T-cell pathways (reviewed in [12]) and bronchial smooth muscle lability (bronchial thermoplasty; [13]) are emerging. These therapies are unlikely to show efficacy in every patient with asthma and will probably need to be personalized by patient phenotype informed by an understanding of the interactions between different scales of the disease.
It is clear from the descriptions above that both diseases encompass pathophysiological changes across a range of biological—spatial and time—scales: from gene to cell, tissue and whole organ. Understanding how the changes within these isolated components impact on whole organ (system) function requires a systems biology approach. Here we present an overview of the AirPROM project, which aims to do exactly this. The project brings together various computational tools and techniques and a wealth of patient data sampled from the respiratory system. The huge challenge we face in AirPROM is the integration of these tools and the application of the clinical and biological data to both inform and validate the models to provide an understanding of the multi-scale pathophysiological changes occurring in asthma and COPD, striving towards the realization of personalized medicine.
2. The approach
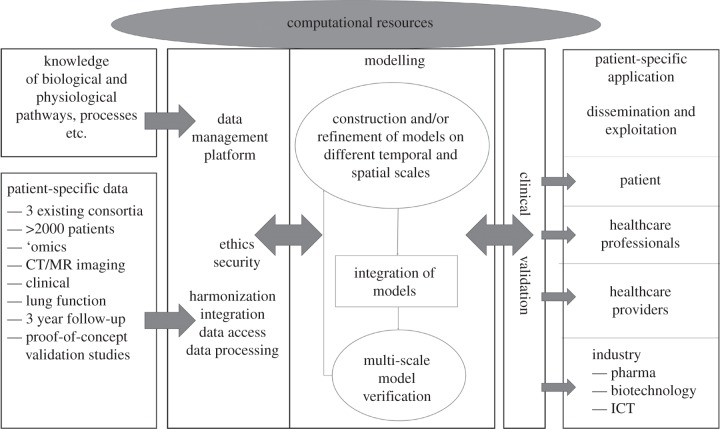
The workflow includes collection and analysis of patient data, extraction of structural information from medical images, the construction of computational meshes, three-dimensional computational fluid dynamics (CFD) and the development of a multi-scale model—capturing information from the gene–cell–tissue level—that predicts clinically relevant outcomes (figure 1).
Figure 1.
Schematic diagram illustrating the workflow structure within the AirPROM project.
2.1. Patient data
We are working towards integrating and extending existing clinical databases from three clinical consortia (U-BIOPRED IMI [14], EvA FP7 [15] and BTS severe asthma [16]). These data include extensive genomic, transcriptomic and proteomic profiles, detailed lung function with novel small airway physiological measures, bronchial challenge studies, imaging data from computed tomography (CT) imaging and hyperpolarized gas magnetic resonance imaging (HP MRI), and patient-reported outcomes. The clinical measurements provide a wealth of both cross-sectional and longitudinal follow-up data. Proof-of-concept clinical trials with standard and novel interventions will also contribute to the huge patient dataset available. These patient-specific data together with known biological pathway data from public databases are being integrated into a ‘knowledge management’ platform (see §2.2) that will feed into the computational modelling framework.
2.2. Knowledge management system
Leveraging the clinical utility of systems biology techniques requires the integrative assessment of complex, heterogeneous data and information on multiple scales. Over the last 15 years, a large number of methods and software tools have been developed to integrate aspects of biological knowledge, such as signalling pathways, with experimental data [17,18]. However, it has proved extremely difficult to couple true semantic integration across all information types relevant in a life science project [19]. The inherent challenges in knowledge management in the life sciences relate to primary data (e.g. natural text, omics and clinical datasets), to technically distributed data sources and to knowledge (e.g. objects, entity relationships, subnetworks and experimental procedures), the corresponding complexity in developing and changing data formats, lack of connectedness and missing semantic descriptions.
In order to address this challenge, Biomax Informatics AG has developed the BioXM knowledge management environment [20], which is widely used in European system biology and medicine projects.1 Within AirPROM BioXM establishes a comprehensive respiratory disease knowledge base to facilitate the information flow between all (approx. 120) consortium participants. The BioXM system provides a secure and sustainable infrastructure that semantically integrates the newly produced clinical and experimental data with existing biomedical knowledge from allied consortia and public databases. This resource is available on Web Apps and by APIs for analysis and modelling, facilitating sharing, collaboration and publication within AirPROM and beyond. The AirPROM Respiratory Disease Knowledge Base represents knowledge as a network of nodes (e.g. concepts such as ‘organ’, ‘disease’ and ‘protein’), their instantiations (e.g. ‘lung’, ‘asthma’ and ‘interleukins’) and their relationships (e.g. ‘is affected by’ and ‘produces’) as edges. The network is based on semantic standardization using ontologies [21,22]. Semantic mapping links the knowledge network with patient data, mathematical models, public resources and molecular concepts.
While molecular-level mathematical models can be represented in SBML or CellML [23,24] and their concepts referenced based on MIRIAM [25], no corresponding standard is currently available for supra-molecular models. Therefore, we developed a new concept which coincidentally arrived at the same conclusions as the FP7-VPH RICORDO project [26]. The AirPROM knowledge management system now represents the first (to our knowledge) independent implementation and validation of the RICORDO semantic model and data representation concept. In this concept, individual model and physiological parameters are semantically described based on terms independently collected from a set of controlled vocabularies. The resulting term feature vector is used in a network search to identify semantically ‘similar’ parameters [27]. This ‘identity of meaning’ is subsequently verified manually and is used to integrate models across scales and to connect between models and data.
3. Component parts of the modelling toolkit
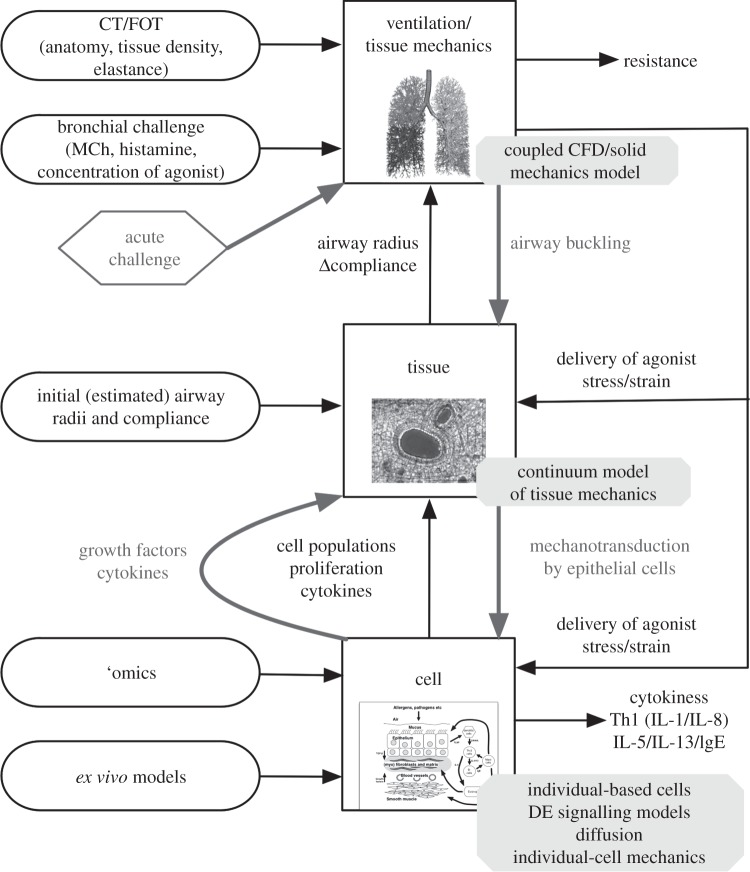
The primary function of the lungs is gas exchange; this process is optimized by bringing air and blood into contact over an extremely large alveolar surface area. All respiratory diseases result in a reduction in ventilation and/or perfusion. One of the major causes of the decline in gas exchange function in asthma and COPD is airflow obstruction. The main focus of the modelling within AirPROM is ventilation and the impact of pathophysiological changes on resultant ventilatory and gas exchange function. Because changes occur throughout the airway network—within both large and small airways—and in the parenchymal tissue, there are several components we need to include in an organ-scale model to understand the system as a whole. We optimize and bring together the existing techniques in three-dimensional CFD (see §3.1), small airway measurements and models (see §3.2), one-dimensional flow modelling, organ-level continuum mechanics and statistical models (see §3.4) to capture the structure and function of a whole lung. Sub-organ-level components also need to be integrated with the organ model to enable an understanding of the multi-scale nature of these diseases. Here, we again optimize and bring together existing tools and techniques using agent-based modelling to predict emergent tissue properties from gene–cell interactions (see §3.3). The framework for a multi-scale model was established at the outset of the project; a schematic illustration representing some of the model components required to represent an asthmatic patient is illustrated in figure 2.
Figure 2.
Schematic of the multi-scale model being developed within the AirPROM project; this illustration represents some of the model components required to represent an asthmatic patient. CFD, computational fluid dynamics; CT, computed tomography; DE, differential equation; FOT, forced oscillation technique; IgE, immunoglobulin E; IL, interleukin; MCh, methacholine; Th1, type 1 helper T cells.
3.1. Functional respiratory imaging
Classic pulmonary function tests and in particular the forced expiratory volume in 1 second are commonly used but are poor predictors for clinical symptoms, exercise tolerance and response to bronchodilators in chronic lung disease [28,29]. The combination of CT imaging and CFD, here termed functional respiratory imaging (FRI), yields novel, more sensitive outcome parameters enabling an improved prediction of clinically relevant changes in the respiratory system [30].
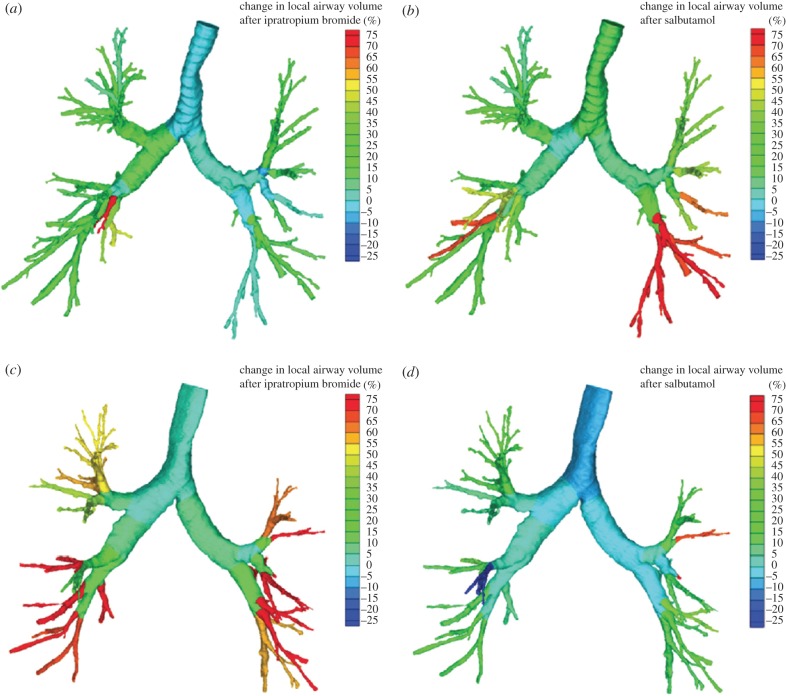
Structural information on the central airways (for example, airway volume, length and wall thickness) and lobes can be derived from CT images. Subsequently, solving the Navier–Stokes equations adds a functional element to the static images by providing an indication of the regional pressure, resistance, flow and wall shear stress. Following the changes in these image-based outcome parameters provides a strong indication of the efficiency and response to therapies (figure 3). In previous studies, this concept has been applied to long-acting β2-agonists (LABAs) [32], a combination of a LABA and an inhaled corticosteroid [30], a short-acting anticholinergic, a short-acting β2-agonist [31] and non-invasive ventilation [33]. The concept has been validated using gamma scintigraphy [34], single-photon emission computed tomography [35] and HP MRI [36].
Figure 3.
Changes in airway volume based on segmented airways from CT scans in a patient (a) mainly responsive to salbutamol and (b) less to ipratropium bromide, and in a patient (c) more responsive to ipratropium bromide and (d) less responsive to salbutamol [31]. Colour spectrums illustrate the change in local airway volume after treatment (%).
Within AirPROM, state-of-the-art software (Mimics, Materialise NV, Belgium; Airways, Institut Telecom, France) is being developed to enable automatic extraction of the morphological properties of airways and lobes from patient CT data. High-resolution computational meshes of the central airways and lung surface are generated for use in three-dimensional CFD (FRI) simulation studies using the Ansys software. These procedures will form part of a high-throughput semi-automated framework for modelling structure and function within the large airways (illustrated for a single subject in figure 4), applied so far in 20 asthmatic subjects and nine controls within AirPROM. Application of the FRI technique further within AirPROM will focus on the phenotyping and treatment of the central and distal airways. The alveolar and small airway regions are assessed indirectly via CT-based lobar expansion from functional residual capacity to total lung capacity, and applied as boundary conditions in the CFD simulations. The AirPROM consortium aims to correlate these image-based markers with other outcome predictors of small airway diseases such as measurements from the forced oscillation technique and ventilation imaging from HP MRI [9].
Figure 4.
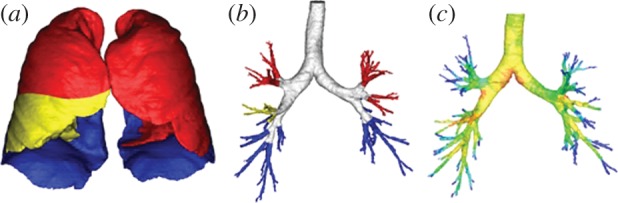
Computational meshes of the (a) lobar surfaces and (b) central airways using subject data for an asthmatic subject within the AirPROM database. (c) Illustrates the pressure solution within the central airways (red, high pressure; dark blue, lowest pressure) obtained using the FRI approach.
3.2. Small airway measurements and models
Severe asthma is characterized by disease within the small airways with an absence of alveolar pathology or emphysema contrasting that in COPD. Within AirPROM, small airway diseases are being characterized using multiple breath washout, impulse oscillometry, measurement of exhaled volatile organic compounds and HP MRI [9]. HP MRI provides a safe and repeatable technique for imaging multiple aspects of regional lung physiology with sensitivity to lung structures and function over multiple spatial and temporal length scales. The non-reliance on ionizing radiation also lends the technique to repeat studies in therapy follow-up (figure 5a), when modelling the regional effects of a personalized intervention in a given patient with lung disease. These functional measures cannot be accessed with other structural imaging modalities such as CT.
Figure 5.
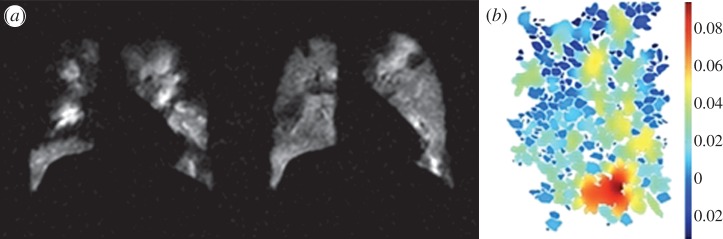
(a) HP MRI images for a patient with asthma before and after bronchodilator therapy. (b) Simulation of 3He diffusion in two-dimensional micro-sections of alveolar ducts and sacs. Colour spectrum represents apparent diffusion coefficients (cm2 s−1).
A combined HP MRI measurement and modelling approach [37,38] is being used to assess the structure and function of small airways in asthma and COPD. Models of HP MRI measurements are being developed to provide regional functional data upon which computational models of gas diffusion, washout, exchange and flow in the major airways can be based. For example, models of 3He diffusion within the lung micro-structure are being developed to understand changes in alveolar dimensions related to emphysema and to understand acinar gas exchange (figure 5b).
3.3. Cell–tissue-level modelling
Several previous studies have focused on acute bronchoconstrictive airway response, incorporating models of sub-cellular contraction (a nice review is given in [39]). In this paper, we focus more on the longer time-scale airway remodelling and how the composition of cells in the airway walls changes. Structural cells that make-up the tissue in the airway are composed of epithelial cells, airway smooth muscle cells and extracellular matrix components. In chronic asthma, in response to repeated inflammatory and bronchoconstrictive episodes, irreversible airway remodelling occurs through recruitment and proliferation of the various cell types [40–43]. These changes culminate in the increased thickness of the lamina propria, an increase in the amount of mucus and debris in the lumen and a reduction in the diameter of the airway lumen. Although airway remodelling is assumed to be damaging to the airways, there is also evidence that some of the structural changes could help to limit airway narrowing, thereby reducing the severity of an asthma attack [44,45].
We are applying the technique of individual-based modelling/agent-based modelling (ABM) to capture the changes occurring at the cell level. ABM has been most widely used in ecology [46], but also in finance, cell and crowd modelling. The rationale for using ABM is that the collective behaviour of a set of individuals is a result of their interaction, and this collective behaviour cannot be predicted from the individual behaviour. In ABM, individual entities (in this case, cells) are represented by software agents which interact within some framework. The interactions may be physical (forces) or signals (messages). The agents change their state (e.g. position in the cell cycle) depending on the interaction of a set of internal functions, information stored in memory (which could represent the phenotype) and external signals. A function could be a mathematical model of a signalling system, a statistical model of behaviour or a rule (a logical expression) [47,48]. The flexible large-scale agent modelling environment (http://www.flame.ac.uk/; [47]) enables the agents to interact with other modelling paradigms, for instance differential equation representations of signalling systems or diffusion models, to understand the behaviour of individual cells in the airway wall in response to stimulation by irritants or agonists.
For the tissue modelling perspective, we think of the airway wall as a ‘composite material’ composed of different cell types with different volume fractions. We are modelling, within a continuum framework, the corresponding changes in volume fraction of the structural components that potentially vary with time. This will in turn cause changes to the elastic properties of the tissue (airway), and hence compliance and contractile characteristics. The ultimate aim, at the tissue level, is to develop a model, for a single airway, that can predict changes in effective compliance and airway calibre as the airway wall remodels, extending from the study of Brook et al. [49]. However, at the cellular level, inflammatory mediators and growth factors released in response to inflammation or bronchoconstriction interact with structural cells within a complex biological network. In order to determine how the airway wall remodels, we therefore need to couple the change in volume fraction of the different cell types in the airway wall to the biological mechanisms at the cellular level. ABMs being developed alongside the simpler tissue models incorporate many of these mechanisms. The aim here is to enable parameterization of the simpler tissue model with outputs from ABMs, so that we may simplify the coupling of the cell- and tissue-level mechanisms and reduce computational and mathematical complexity.
3.4. Organ-level modelling
Multi-scale biophysically based computational models of the lung have been developed and applied to the understanding of several areas of respiratory physiology, including airflow and particle deposition, bronchoconstriction, airway closure [50] and airway hyper-responsiveness in asthmatic airways [51,52], pulmonary blood flow [53] with application to pulmonary embolism [54] and lung perfusion MRI [55] (for a more detailed review refer to [39,56]). These types of models have provided some understanding of structure–function relationships in the lung; however, to date, these types of mechanistic models have seldom (if at all) been applied to clinical medicine. By contrast, statistical modelling techniques already present a useful tool in clinical medicine. They describe how random variables (e.g. clinical measurements) are related to other random variables. Statistical models can be used to understand relationships between variables within a dataset and, for example, used to predict patient phenotypes based on previous population analysis of symptoms. Here we describe biophysically based models and statistical modelling approaches at the whole organ scale being brought together within the framework of AirPROM.
In order to combine the range of model components spanning multiple scales into a predictive whole organ model, several simplifications are required. Computational restrictions limit the use of three-dimensional CFD to around seven to nine airway branching generations; therefore, we are developing a one-dimensional model of ventilation within the distal conducting airways enabling inclusion of flow properties right down to the gas exchange units (approx. 16th generation). The geometry of the airway tree is created using a combination of information extracted from CT (central airways and lobar geometries) and a volume-filling branching algorithm [57]. The ventilation model developed within AirPROM is based on work by Swan et al. [58], and can predict the dynamic distribution of ventilation during breathing. This model is being used to understand the consequences of airway remodelling on ventilation. Ventilation is driven into the lung via deformation of the lung parenchymal tissue. To calculate these deformations and the stresses within the tissue acting on the airways and vessels, we are using a continuum finite deformation approach [59]. Properties of airway remodelling from the cell–tissue-level models will be coupled into both the ventilation and tissue mechanic models at the organ scale. These techniques are being developed within the Chaste (cancer, heart and soft tissue environment; www.cs.ox.ac.uk/chaste), which is a general purpose open-source multi-scale modelling platform.
The statistical modelling develops classifiers and predictors based on univariate and multivariate statistical analyses and clustering of data collected through clinical measurements. Different aspects of the diseases can be measured and quantified using factor analysis. Factor variables, which are unobserved variables inferred from the statistical modelling, can be defined which describe the underlying properties of a dataset using a smaller dimensional space than the full dataset. The factors can be thought of as representatives of processes or aspects of disease which correlate with variables associated with that process. Factors most commonly obtained for asthma include measurements associated with lung volumes, airway obstruction, symptoms, allergy, inflammation, airway hyper-responsiveness and lung physiology changes [10]. For COPD, these can include symptoms, lung volume and airway obstruction, inflammation, lung physiology changes, exercise capacity and gas exchange information [60]. Structural equation models, which can be used to describe the causal relationship between variables, can be converted to stochastic models that can connect computational and statistical modelling. Initial results for the BTS severe asthma study are already available. Integrating all clinical groups regardless of current diagnosis (asthma or COPD) will allow for the unbiased identification of subgroups defined by their genotype–phenotype without disease boundaries.
4. Where to from here?
We are working on a multi-scale model of the airways to investigate the natural history and progression of the airway diseases asthma and COPD. The airway model, following the ambitions of the VPH Initiative, will incorporate data and models spanning multiple levels of biological complexity, from genes–cells–tissue to whole organ-level structure and function. The models will be informed by an extensive cohort of medical imaging data and physiological and molecular measurements across the scales. Regarding asthma, we will focus on the impact of cellular remodelling within the airway walls occurring over longer time scales (chronic disease) on declining lung function, incorporating feedback from the change in ventilation and deformation patterns at the cell level. For COPD as well as the airway remodelling (in a similar way to asthma), we will also investigate the changes in parenchymal tethering, occurring because of emphysema on lung function.
Current clinical classification of asthma and COPD is imprecise owing to the range and overlap of symptoms. Patient phenotyping is of vital clinical significance, potentially helping to reveal the underlying pathophysiology, risk factors, natural history and treatment responses of the specific phenotypes—with the hope of enabling customized treatment regimes for individuals [61]. One of the major underlying goals within AirPROM is the improvement of patient phenotyping. This is where statistical modelling can play a major role. In addition, the biophysically based models will be used to search for new biomarkers of disease and indicators of patient outcomes and exacerbations. Further multi-scale phenotyping of patients will be done via genomics and the development of cell models. Cascading down the scales from the organ to the cell level allows investigation of several hypotheses resulting in a better understanding of the disease across multiple scales of biology and the subsequent interaction of therapies to alter the progression of the, often chronic, disease. Gaining insights into these pathways is a crucial step to bring personalized medicine closer to the patient.
Model validation brings together disparate aspects of the AirPROM project, in particular the modelling outcomes and patient data. Validation of computational models is a vital yet exceedingly difficult task in a project of this type; however, the range of patient data makes this challenge feasible. Patient-specific data will both inform and validate the models. For example, HP MRI has been shown to provide insight into ventilation dynamics in human lungs. We are developing quantitative time-resolved methods for measuring gas flow velocity to provide additional validation for CFD and one-dimensional ventilation predictions. Pioneering multi-material additive layer manufacturing will be used to produce CT-based models that are not only anatomically realistic, but also mimic the mechanical behaviour of the different airway tissues enabling CFD validation as well as investigation of airway dynamics [62]. The cell–tissue-level models are being developed in parallel with ex vivo tissue models within the realm of AirPROM.
One of the many challenges that will be faced within this project will be bringing together disparate science from several partners across Europe, and in particular bringing together models and data across different spatial and temporal scales. One way in which we are tackling this issue is by forming a truly collaborative working approach, whereby a detailed understanding of the models and the data is first obtained. The AirPROM knowledge management platform also provides a tool to aid in the linking across these scales. To break this down into more manageable segments, the model verification, clinical validation and development will occur in multiple iterative cycles each with increasing throughput and automation working towards a ‘turn-key’ platform. AirPROM will thus develop a validated patient-specific multi-scale predictive computational airway model underpinned by extensive clinical data. Validation will be undertaken cross-sectionally, following interventions and after longitudinal follow-up to incorporate both spatial and temporal dimensions. Clinical interventions that we are aiming to study, via both clinical measurement and modelling techniques, include pharmaceutical interventions with highly specific molecular therapies and surgical intervention procedures (i.e. bronchial thermoplasty and lung volume reduction surgery), both targeting stratified patient populations.
One of the many strengths of AirPROM is the close interaction between clinicians, patients and pharmaceutical companies, so that the project is aware of the needs of the patients, as well as the practicality of the application of models in the clinic for drug discovery and development. We have already engaged the prospective ‘users’ and ‘providers’ and started to translate the platforms into usable applications: collaborations with pharmaceutical companies are already developing into joint projects. AirPROM will bridge the critical gaps in our clinical management of airway diseases, by providing validated models to predict disease progression and response to treatment and the platform to translate these patient-specific tools, so as to pave the way to improved, personalized management of airway diseases.
Acknowledgements
This work is funded by the EU Seventh Framework Programme FP7/2007–2013 under grant agreement no. 270194 and is presented on behalf of the whole AirPROM consortium (www.airprom.eu).
Footnotes
FP6 BioBridge; FP7 AirPROM, nanoMILE, MeDALL, STATegra, SYNERGY-COPD, SysCLAD; BMBF MedSYS CancerMotiSys, m4 smart; FFG COMET OncoTyrol.
References
- 1.Decramer M, Janssens W, Miravitlles M. 2012. Chronic obstructive lung disease. Lancet 379, 1341–1351 10.1016/S0140-6736(11)60968-9 (doi:10.1016/S0140-6736(11)60968-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization 2008. World health statistics. Geneva, Switzerland: WHO [Google Scholar]
- 3.Hogg JC. 2006. State of the art. Bronchiolitis in chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 3, 489–493 10.1513/pats.200603-065MS (doi:10.1513/pats.200603-065MS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez C, Chen Y, Westgate P, Liu L, Murray S, Curtis J. 2012. Relationship between quantitative CT metrics and health status and BODE in chronic obstructive pulmonary disease. Thorax 67, 399–406 10.1136/thoraxjnl-2011-201185 (doi:10.1136/thoraxjnl-2011-201185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agusti A, Barnes P. 2012. Update in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 185, 1171–1176 10.1164/rccm.201203-0505UP (doi:10.1164/rccm.201203-0505UP) [DOI] [PubMed] [Google Scholar]
- 6.Repapi E, Sayers I, Wain L, Burton P, Johnson T, Obeidat M. 2010. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44 10.1038/ng.501 (doi:10.1038/ng.501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinbami L, Moorman J, Bailey C, Zahran H, King M, Johnson C. 2012. Trends in asthma prevalence, health care use, and mortality in the United States, 2001–2010. NCHS Data Brief 94, 1–8 [PubMed] [Google Scholar]
- 8.Partridge M, van der Molen T, Myrseth S, Busse WW. 2006. Attitudes and actions of asthma patients on regular maintenance therapy: the INSPIRE study. BMC Pulm. Med. 13, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brightling CE, Gupta S, Gonem S, Siddiqui S. 2012. Lung damage and airway remodelling in severe asthma. Clin. Exp. Allergy 42, 638–649 10.1111/j.1365-2222.2011.03917.x (doi:10.1111/j.1365-2222.2011.03917.x) [DOI] [PubMed] [Google Scholar]
- 10.Haldar P, Pavord ID, Shaw DE, Berry DE, Thomas M, Brightling CE, Wardlaw AJ, Green RH. 2008. Cluster analysis and clinical asthma phenotypes. Am. J. Respir. Crit. Care Med. 178, 218–224 10.1164/rccm.200711-1754OC (doi:10.1164/rccm.200711-1754OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavord ID, Korn S, Howarth P, Bleecker E, Buhl R, Keene O. 2012. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet 380, 651–659 10.1016/S0140-6736(12)60988-X (doi:10.1016/S0140-6736(12)60988-X) [DOI] [PubMed] [Google Scholar]
- 12.Holgate S. 2012. Trials and tribulations in identifying new biologic treatments for asthma. Trends Immunol. 33, 238–246 10.1016/j.it.2012.02.003 (doi:10.1016/j.it.2012.02.003) [DOI] [PubMed] [Google Scholar]
- 13.Castro M, Rubin A, Laviolette M, Fiterman J, De Andrade Lime M, Shah P. 2010. Effectiveness and safety of bronchial thermoplasty in the treatment of severe asthma: a multicenter, randomized, double-blind, sham-controlled clinical trial. Am. J. Respir. Crit. Care Med. 181, 116–124 10.1164/rccm.200903-0354OC (doi:10.1164/rccm.200903-0354OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bel E, et al. 2011. Diagnosis and definition of severe refractory asthma: an international consensus statement from the Innovative Medicine Initiative (IMI). Thorax 66, 910–917 10.1136/thx.2010.153643 (doi:10.1136/thx.2010.153643) [DOI] [PubMed] [Google Scholar]
- 15.Ziegler-Heitbrock L, et al. 2012. The EvA study: aims and strategy. Eur. Respir. J. 40, 823–829 10.1183/09031936.00142811 (doi:10.1183/09031936.00142811) [DOI] [PubMed] [Google Scholar]
- 16.Heaney L, Brightling C, Menzies-Gow A, Stevenson M, Niven R. 2010. British Thoracic Society Difficult Asthma Network. Refractory asthma in the UK: cross-sectional findings from a UK multicentre registry. Thorax 65, 787–794 10.1136/thx.2010.137414 (doi:10.1136/thx.2010.137414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antezana E, Kulper M, Mironov V. 2009. Biological knowledge management: the emerging role of the semantic web technologies. Brief Bioinform. 10, 392–407 10.1093/bib/bbp024 (doi:10.1093/bib/bbp024) [DOI] [PubMed] [Google Scholar]
- 18.Stein LD. 2003. Integrating biological databases. Nat. Rev. Genet. 4, 337–345 10.1038/nrg1065 (doi:10.1038/nrg1065) [DOI] [PubMed] [Google Scholar]
- 19.Hu H, Murai R, Liebman M. 2008. Biomedical informatics in translational research. Boston, MA: Artech House Publishers [Google Scholar]
- 20.Losko S, Heumann K. 2009. Semantic data integration and knowledge management to represent biological network associations. Methods Mol. Biol. 563, 241–258 10.1007/978-1-60761-175-2_13 (doi:10.1007/978-1-60761-175-2_13) [DOI] [PubMed] [Google Scholar]
- 21.Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 10.1038/75556 (doi:10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang A, Barrett J, Bentley T, Markwell D, Price C, Spackman K, Stearns MQ. 2001. Mapping between SNOMED RT and clinical terms version 3: a key component of the SNOMED CT development process. Proc. AMIA Symp. 2001, 741–745 [PMC free article] [PubMed] [Google Scholar]
- 23.Hucka M, et al. 2003. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531 10.1093/bioinformatics/btg015 (doi:10.1093/bioinformatics/btg015) [DOI] [PubMed] [Google Scholar]
- 24.Lloyd CM, Halstead M, Nielsen P. 2004. CellML: its future, present and past. Prog. Biophys. Mol. Biol. 85, 433–450 10.1016/j.pbiomolbio.2004.01.004 (doi:10.1016/j.pbiomolbio.2004.01.004) [DOI] [PubMed] [Google Scholar]
- 25.Le Novère N, et al. 2005. Minimum information requested in the annotation of biochemical models (MIRIAM). Nat. Biotechnol. 23, 1509–1515 10.1038/nbt1156 (doi:10.1038/nbt1156) [DOI] [PubMed] [Google Scholar]
- 26.Wimalaratne S, Grenon P, Hoehndorf R, Gkoutos G, de Bono B. 2012. An infrastructure for ontology-based information systems in biomedicine: RICORDO case study. Bioinformatics 28, 448–450 10.1093/bioinformatics/btr662 (doi:10.1093/bioinformatics/btr662) [DOI] [PubMed] [Google Scholar]
- 27.Schulz M, Krause F, Le Novère N, Klipp E, Liebermeister W. 2011. Retrieval, alignment, and clustering of computational models based on semantic annotations. Mol. Syst. Biol. 7, 512. 10.1038/msb.2011.41 (doi:10.1038/msb.2011.41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Celli B. 2006. COPD, inflammation and its modulation by phosphodiesterase 4 inhibitors: time to look beyond the FEV1. Chest 129, 5–6 10.1378/chest.129.1.5 (doi:10.1378/chest.129.1.5) [DOI] [PubMed] [Google Scholar]
- 29.Jones PW. 2001. Health status measurement in chronic obstructive pulmonary disease. Thorax 56, 880–887 10.1136/thorax.56.11.880 (doi:10.1136/thorax.56.11.880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Backer LA, Vos W, De Backer J, Van Holsbeke C, Vinchurkar S, De Backer W. 2012. The acute effect of budesonide/formoterol in COPD: a multi-slice computed tomography and lung function study. Eur. Respir. J. 40, 298–305 10.1183/09031936.00072511 (doi:10.1183/09031936.00072511) [DOI] [PubMed] [Google Scholar]
- 31.De Backer L, Vos WG, Salgado R, De Backer JW, Devolder A, Verhulst SL, Claes R, Germonpre PR, De Backer WA. 2011. Functional imaging using computer methods to compare the effect of salbutamol and ipratropium bromide in patient-specific airway models of COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 6, 637–646 10.2147/COPD.S21917 (doi:10.2147/COPD.S21917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Backer JW, Vos WG, Devolder A, Verhulst SL, Germonpré P, Wuyts FL, Parizel PM, De Backer W. 2008. Computational fluid dynamics can detect changes in airway resistance in asthmatics after acute bronchodilation. J. Biomech. 41, 106–13 10.1016/j.jbiomech.2007.07.009 (doi:10.1016/j.jbiomech.2007.07.009) [DOI] [PubMed] [Google Scholar]
- 33.De Backer L, et al. 2011. The effects of long-term noninvasive ventilation in hypercapnic COPD patients: a randomized controlled pilot study. Int. J. Chron. Obstruct. Pulmon. Dis. 6, 615–624 10.2147/COPD.S22823 (doi:10.2147/COPD.S22823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Backer JW, Vos WG, Gorlé CD, Germonpré P, Partoens B, Wuyts FL, Parizel PM, De Backer W. 2008. Flow analyses in the lower airways: patient-specific model and boundary conditions. Med. Eng. Phys. 30, 872–879 10.1016/j.medengphy.2007.11.002 (doi:10.1016/j.medengphy.2007.11.002) [DOI] [PubMed] [Google Scholar]
- 35.De Backer JW, Vos WG, Vinchurkar S, Claes R, Drollmann A, Wulfrank D, Parizel PM, Germonpre P, De Backer W. 2010. Validation of computational fluid dynamics in CT-based airway models with SPECT/CT. Radiology 257, 854–862 10.1148/radiol.10100322 (doi:10.1148/radiol.10100322) [DOI] [PubMed] [Google Scholar]
- 36.de Rochefort L, et al. 2007. In vitro validation of computational fluid dynamic simulation in human proximal airways with hyperpolarized 3He magnetic resonance phase-contrast velocimetry. J. Appl. Physiol. 102, 2012–2023 10.1152/japplphysiol.01610.2005 (doi:10.1152/japplphysiol.01610.2005) [DOI] [PubMed] [Google Scholar]
- 37.Parra-Robles J, Ajraoui S, Deppe M, Parnell S, Wild J. 2010. Experimental investigation and numerical simultion of 3He gas diffusion in simple geometries: implications for analytical models of 3He MR lung morphometry. J. Magn. Reson. 204, 228–238 10.1016/j.jmr.2010.02.023 (doi:10.1016/j.jmr.2010.02.023) [DOI] [PubMed] [Google Scholar]
- 38.Fichele S, Woodhouse N, Swift AJ, Said Z, Paley MNJ, Kasuboski L, Mills GH, van Beek EJR, Wild JM. 2004. MRI of helium-3 gas in healthy lungs: posture related variations of alveolar size. J. Magn. Reson. Imaging 20, 331–335 10.1002/jmri.20104 (doi:10.1002/jmri.20104) [DOI] [PubMed] [Google Scholar]
- 39.Tawhai MH, Bates JHT. 2011. Multi-scale lung modeling. J. Appl. Physiol. 110, 1466–1472 10.1152/japplphysiol.01289.2010 (doi:10.1152/japplphysiol.01289.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aikawa T, Shimura S, Sasaki H, Ebina M, Takishima T. 1992. Marked goblet cell hyperplasia with mucus accumulation in the airways of patients who died of severe acute asthma attack. Chest 101, 916–921 10.1378/chest.101.4.916 (doi:10.1378/chest.101.4.916) [DOI] [PubMed] [Google Scholar]
- 41.Carroll NG, Mutavdzic S, James AL. 2002. Increased mast cells and neutrophils in submucosal mucous glands and mucus plugging in patients with asthma. Thorax 57, 677–682 10.1136/thorax.57.8.677 (doi:10.1136/thorax.57.8.677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elias J, Zhu Z, Chupp G, Homer R. 1999. Airway remodeling in asthma. J. Clin. Invest. 104, 1001–1006 10.1172/JCI8124 (doi:10.1172/JCI8124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brewster CE, Howarth P, Djukanovic R, Wilson J, Holgate S, Roche W. 1990. Myofibroblasts and subepithelial fibrosis in bronchial asthma. Am. J. Respir. Cell Mol. Biol. 3, 507–511 [DOI] [PubMed] [Google Scholar]
- 44.McParland B, Macklem P, Pare P. 2003. Airway wall remodeling: friend or foe? J. Appl. Physiol. 95, 426–434 [DOI] [PubMed] [Google Scholar]
- 45.Niimi A, Matsumoto H, Takemura M, Ueda T, Chin K, Mishima M. 2003. Relationship of airway wall thickness to airway sensitivity and airway reactivity in asthma. Am. J. Respir. Crit. Care Med. 168, 983–988 10.1164/rccm.200211-1268OC (doi:10.1164/rccm.200211-1268OC) [DOI] [PubMed] [Google Scholar]
- 46.Grimm V. 1999. Ten years of individual-based modelling in ecology: what have we learned, and what could we learn in the future? Ecol. Model. 115, 129–148 10.1016/S0304-3800(98)00188-4 (doi:10.1016/S0304-3800(98)00188-4) [DOI] [Google Scholar]
- 47.Holcombe M, et al. 2012. Modelling complex biological systems using an agent-based approach. Integr. Biol. 4, 53–64 10.1039/c1ib00042j (doi:10.1039/c1ib00042j) [DOI] [PubMed] [Google Scholar]
- 48.Adra S, Sun T, MacNeil S, Holcombe M, Smallwood R. 2010. Development of a three dimensional multiscale computational model of the human epidermis. PLoS ONE 5, e8511. 10.1371/journal.pone.0008511 (doi:10.1371/journal.pone.0008511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brook BS, Peel SE, Hall IP, Politi AZ, Sneyd J, Bai Y, Sanderson MJ, Jensen OE. 2010. A biomechanical model of agonist-initiated contraction in the asthmatic airway. Respir. Physiol. Neurobiol. 170, 44–58 10.1016/j.resp.2009.11.006 (doi:10.1016/j.resp.2009.11.006) [DOI] [PubMed] [Google Scholar]
- 50.Venegas JG, et al. 2005. Self-organized patchiness in asthma as a prelude to catastrophic shifts. Nature 434, 777–782 10.1038/nature03490 (doi:10.1038/nature03490) [DOI] [PubMed] [Google Scholar]
- 51.Donovan G. 2011. Multiscale mathematical models of airway constriction and disease. Pulm. Pharmacol. Ther. 24, 533–539 10.1016/j.pupt.2011.01.003 (doi:10.1016/j.pupt.2011.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amin SD, Mujamdar A, Frey U, Suki B. 2010. Modeling the dynamics of airway constriction: effects of agonist transport and binding. J. Appl. Physiol. 109, 553–563 10.1152/japplphysiol.01111.2009 (doi:10.1152/japplphysiol.01111.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark AR, Tawhai MH, Burrowes KS. 2011. The interdependent contributions of gravitational and structural features of the lung to the distribution of pulmonary perfusion in a multi-scale model of the pulmonary circulation. J. Appl. Physiol. 110, 943–955 10.1152/japplphysiol.00775.2010 (doi:10.1152/japplphysiol.00775.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burrowes KS, Clark AR, Marcinkowski A, Wilsher ML, Milne DG, Tawhai MH. 2011. Pulmonary embolism: predicting disease severity. Phil. Trans. R. Soc. A 13, 4255–4277 10.1098/rsta.2011.0129 (doi:10.1098/rsta.2011.0129) [DOI] [PubMed] [Google Scholar]
- 55.Burrowes KS, Buxton RB, Prisk GK. 2012. Assessing the potential errors of MRI-based measurements of pulmonary blood flow using a detailed network flow model. J. Appl. Physiol. 113, 130–141 10.1152/japplphysiol.00894.2011 (doi:10.1152/japplphysiol.00894.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tawhai MH, Clark AR, Donovan GM, Burrowes KS. 2011. Computational modeling of airway and pulmonary vascular structure and function: development of a ‘lung physiome’. CRC Crit. Rev. Biomed. Eng. 39, 319–336 10.1615/CritRevBiomedEng.v39.i4.50 (doi:10.1615/CritRevBiomedEng.v39.i4.50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tawhai MH, Hunter PJ, Tschirren J, Reinhardt JM, McLennan G, Hoffman EA. 2004. CT-based geometry analysis and finite element models of the human and ovine bronchial tree. J. Appl. Physiol. 97, 2310–2321 10.1152/japplphysiol.00520.2004 (doi:10.1152/japplphysiol.00520.2004) [DOI] [PubMed] [Google Scholar]
- 58.Swan AJ, Clark AR, Tawhai MH. 2012. A computational model of the topographic distribution of ventilation in healthy human lungs. J. Theor. Biol. 300, 222–231 10.1016/j.jtbi.2012.01.042 (doi:10.1016/j.jtbi.2012.01.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tawhai M, Nash N, Lin C, Hoffman E. 2009. Supine and prone differences in regional lung density and pleural pressure gradients in the human lung with constant shape. J. Appl. Physiol. 107, 912–920 10.1152/japplphysiol.00324.2009 (doi:10.1152/japplphysiol.00324.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy K, Smith JR, Kolsum U, Borrill Z, Vestbo J, Singh D. 2009. COPD phenotype description using principal components analysis. Respir. Res. 10, 41. 10.1186/1465-9921-10-41 (doi:10.1186/1465-9921-10-41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fingleton J, Weatherall M, Beasley R. 2011. Towards individualised treatment in COPD. Thorax 66, 363–364 10.1136/thx.2010.155564 (doi:10.1136/thx.2010.155564) [DOI] [PubMed] [Google Scholar]
- 62.Timmerman B, Gibbons G, Bryanston-Cross P. 2012. Flow measurements in patient-specific conducting airways models. In 16th Int. Symp. on Applications of Laser Techniques to Fluid Mechanics, Lisbon, Portugal, 9–12 July 2012 [Google Scholar]