Abstract
While the virtual physiological human (VPH) project has made great advances in human modelling, many of the tools and insights developed as part of this initiative are also applicable for facilitating mechanistic understanding of the physiology of a range of other species. This process, in turn, has the potential to provide human relevant insights via a different scientific path. Specifically, the increasing use of mice in experimental research, not yet fully complemented by a similar increase in computational modelling, is currently missing an important opportunity for using and interpreting this growing body of experimental data to improve our understanding of cardiac function. This overview describes our work to address this issue by creating a virtual physiological mouse model of the heart. We describe the similarities between human- and mouse-focused modelling, including the reuse of VPH tools, and the development of methods for investigating parameter sensitivity that are applicable across species. We show how previous results using this approach have already provided important biological insights, and how these can also be used to advance VPH heart models. Finally, we show an example application of this approach to test competing multi-scale hypotheses by investigating variations in length-dependent properties of cardiac muscle.
Keywords: mouse modelling, cardiac multi-scale model, parameter sensitivity, genetic knockout studies, heterogeneous length-dependent activation
1. Introduction
The virtual physiological human (VPH) project aims to create a framework for collaborative investigation of the human body as a single complex system [1,2]. Its many sub-projects are focused on creating tools to share resources and observations, as well as developing integrated models of the mechanical, physical and biochemical functions of a human body. Although human models are most directly relevant for clinical purposes, animal models represent a target with often significantly more available experimental data, from both healthy animals and those manipulated in a variety of ways. Of particular relevance, over the past 20 years, there has been an increasing use of mice in experimental research. The number of procedures on mice has steadily risen over the past 20 years, to the extent that in the past year in the UK 71 per cent of all experimental procedures were carried out on mice [3], while the use of other species such as rats and guinea pigs has steadily decreased.
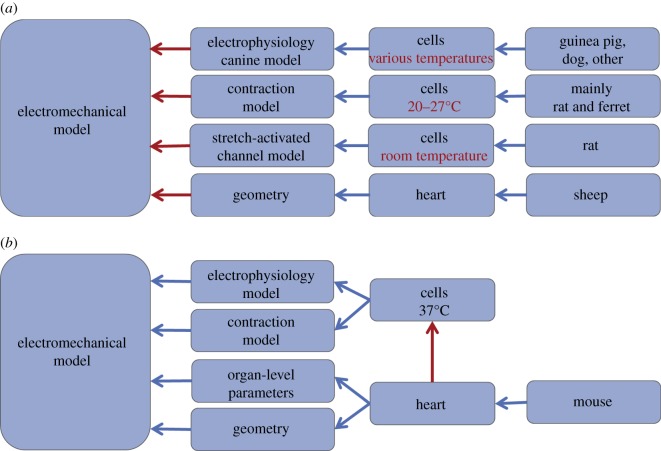
However, while the experimental literature has revealed many important scientific insights, these murine experimental programmes have not been complemented by a similar focus in computational modelling. This lack of mouse cardiac models is a significant limitation in interpreting the growing body of experimental data. Furthermore, the majority of cardiac frameworks are parametrized using data, or couple electrophysiology, contraction, tissue properties and cardiac geometry from different species, resulting in models whose results cannot be quantitatively compared with experimental data. It is within this context that we assert that the mouse represents the ideal animal for careful species-specific modelling, with significant potential to develop a framework in which simulation results can be quantitatively compared with in vivo function. Such a temperature- and species-consistent model can be seen as a framework for hypothesis testing, representing various assumptions about the physiology, experimental protocols, and their associated uncertainties. Thus, remaining differences between model and data are more likely to lead to improved understanding of the physiology and experimental methods, rather than being explained by model limitations and inconsistencies. Figure 1 schematically illustrates these challenges and the resulting predictive power in examples of both a species-consistent and a species-inconsistent electromechanical model.
Figure 1.
Limitations in consistent and inconsistent electromechanical models. The major potential causes for a mismatch between in vivo cardiac function and the electromechanical model are indicated in red. (a) An example inspired by a recent inconsistent electromechanical model. This model integrates data from several species, and from cellular experiments performed at low temperatures. This creates significant potential for the resulting model not to match experimental data. Furthermore, the different species makes it difficult to state what hypothesis the final model represents and what data the model should be able to reproduce. (b) Our approach to mouse modelling. By creating models based on consistent data, the final model represents a virtual mouse heart at body temperature. However, there remains a potential cause for an incorrect prediction in factors by which the heart and the rest of the body affect individual cardiac myocytes, which can be missing in measurements of in vitro cellular data.
Cardiac models are arguably one of the most developed types of multi-scale organ models, both in general and in the context of the VPH project [2,4,5]. This has led to many sophisticated tools for integrating data within cardiac models that are also applicable across species. Among these cardiac frameworks, in this study, we focus on electromechanical models, which integrate mechanical and electrical function across both the cellular and whole organ scale. The motion of cardiac tissue significantly affects the tension generation of muscle cells in the heart, and these feedback systems are of key importance in regulating the heart's pump function. First, length dependence of tension ensures that the heart pump function is increased with blood volume (the Frank–Starling law), through increasing the tension generated by cells as they are stretched. Second, velocity-dependent mechanisms generate complex deformation-dependent effects on tension generation: as cells shorten, the tension generated will decrease first, and then recover above baseline levels on a longer time scale. Integrating these feedback mechanisms and creating an electromechanical model that is consistent with respect to both species and temperature using electrophysiology and contraction data across spatial scales represents a significant undertaking.
This study summarizes the progress on creating this virtual physiological mouse (VPM) model of the heart at physiological temperature and heart rate, in parallel with clinically focused VPH activities. Section 2 reviews some important concepts that are shared between human and mouse cardiac modelling, including the ways in which previous tools developed as part of the VPH initiative have been used to develop the mouse heart framework. Section 3 summarizes several essential ways in which mouse models differ from human cardiac models, and how we have used these differences to our advantage in improving our understanding of murine cardiac physiology. Specifically, we show how the remaining potential for major differences between model predictions and experimental data in a species-consistent virtual mouse heart model can be limited through careful parametrization with knowledge of uncertainty. We then discuss how these advances in modelling the mouse heart can help improve electromechanical models across species. Finally, we present an example application by extending the VPM framework to test a novel hypothesis about cardiac tension generation, and discuss how the framework can be further developed and applied in future work.
2. Similarities between cardiac modelling in the virtual physiological human and virtual physiological mouse
This section reviews some important concepts underpinning the development of our mouse heart model which are applicable across species. Section 2.1 describes the use of VPH tools in developing our model, demonstrating the potential of these tools for greater applicability to cardiac modelling beyond the human context. In §2.2, we describe the use of our research in computational methods for human electromechanics, and their suitability in mouse modelling. Next, we describe our work on model parametrization, which is of increasing importance in computational modelling in general. In this section, we describe our techniques for improving model parametrization through linking experimental constraints to model parameters while visualizing uncertainty.
2.1. Virtual physiological human tools
The creation of a new species-specific electromechanical model represented a significant challenge. Fortunately, several VPH tools are readily applicable to assist with this process. First, the electrophysiology models we use are available as a CellML implementation [6,7]. CellML is an open standard based on XML, and used to store mathematical models across programming languages and modelling tools [8]. Availability of previous work in such a portable language allowed for model reuse free of the errors often associated with the process of re-implementing models, by both our group as well as other researchers [9]. Second, these tools have also allowed us to publish our novel mouse contraction model [10], allowing other researchers in the field to apply and extend the model efficiently. Particularly, the cellular open resource tool [11] was instrumental in streamlining this process, allowing model equations to be entered in a human-readable format, and the resulting model to be tested for correctness and consistency.
Furthermore, we made use of data processing and meshing tools [12] developed as part of the VPH project euHeart [13]. These tools, which were developed for personalization of human cardiac geometry from a patient's medical MRI data, were able to be applied for the processing of mouse data as well, generating a geometry that more accurately captures the mouse ventricle from MRI data. This required only a minor modification, the re-scaling of the template mesh to be compatible with the smaller size of a mouse heart.
Thus, even though the focus of the VPH project is on human modelling, the tools developed are useful across species, and have already helped in developing our model of the mouse heart.
2.2. Computational efficiency
Key to integrating experimental data in electromechanics is managing the available time needed to run simulations. Efficient simulations allow for a wider range of modelling investigations, such as parameter sensitivity investigations and running models for several dozen beats. We previously created a set of efficient computational methods for solving cardiac electromechanical simulations in the context of human geometry and cell models [14]. This work included a re-parametrized human contraction model, an efficient cell model solver for a specific family of electrophysiology models, as well as optimizations to the mechanics solution procedure. In converting this human modelling framework to be used for modelling a mouse heart, we found that there are both differences and similarities in solving such electromechanical models. The effort in optimizing the cell model solver was largely non-portable to mouse models, owing to differences in the specific electrophysiology models used. However, as the mouse heart has a similar conduction velocity to human tissue, the required mesh resolution for accurate solutions to continuum models of electrophysiology is similar. As a result of the small size of a mouse heart, this leads to a greatly reduced computational cost for the electrical part of the simulation compared with human-scale simulations. This allows for a computationally efficient calculation of a converged solution of the electrical activation of the heart, and results in the mechanics part of the solution process dominating simulation run-time using standard methods. Alternatively, the small size of the mouse heart could also allow more detailed discrete models to be used to represent whole organ electrophysiology. Such models may be better at representing sharp transmural gradients in repolarization that are important for studying arrhythmias [15].
By optimizing the mechanics solution process in our human framework, we found that the computational cost for this part of the computation can be reduced by approximately a factor of 50 compared with a standard Newton iteration process. As the mechanics solution process is largely independent of scale, this speed-up is similarly applicable to mouse modelling. Combined with the smaller computational requirements for mouse electrophysiology, multiple models can now quickly be solved on dense electrophysiology meshes on a desktop computer. These advances have enabled extensive studies in sensitivity analyses and model fitting in mice without the need for supercomputing facilities. Thus, our computational framework initially developed for human electromechanical models is potentially even more suitable for mouse models, as the optimizations developed are more effective in this context.
2.3. Model parametrization
As outlined in §1, there are a number of important issues relating to model parametrization which are becoming increasingly important in model development. These include the consistency of data sources, knowledge of uncertainty in the model and variability [16–18]. Our VPM framework seeks to balance the requirements of transparently integrating the best available experimental data from mice for our particular focus, while both being simple enough to be constrained by these data and still representing the underlying biophysical processes involved in contraction.
With this goal, we included the constraining effect of experimental data on the choice of model parameters using a novel visualization [19]. Figure 2a shows an idealized example of this visualization technique. We start with a computational model of a biological system that has several parameters not directly constrained by a single experimental measurement. Instead, several high-level outputs of the model are constrained by experimental data, each of which limits the parameter space of the model. The full visualization of the parameter space would require vast computational resources, and the result would be difficult to interpret. However, a standard one-dimensional parameter variation can also miss important effects related to the coupling between parameters, and can likewise result in measures that are difficult to interpret. To overcome these issues, we chose to visualize pairs of parameters in the model, together with the experimental constraints. Investigating a single pair of parameters at a time, we can visualize each of these constraints as a region in the two-dimensional parameter space. The intersection of all of these constraints results in a ‘viable region’ for the model parameters. This region shows how well constrained the parameters are by experimental data. Furthermore, the boundaries for this region indicate which experimental data were most useful in constraining it, which can help prioritize future experiments carried out for model development.
Figure 2.
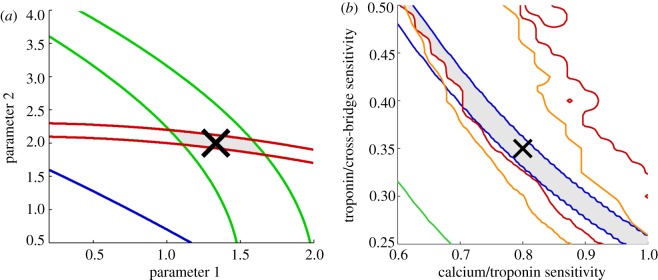
Visualization of model constraints. (a) A synthetic example. The two model parameters are constrained using three experimental constraints, visualized in green, red and blue. Each of the coloured lines represents the boundary of a region in which a specific experimental constraint is met, given variation in two parameters. The grey region represents the ‘viable region’ for the model parameters, which is the intersection of all the regions. This example visualization shows the green constraint as being most useful to refine experimentally, which would especially lead to a better estimate of parameter 1. The blue experimental constraint is too weak to be useful, being met in the vast majority of models, and would not be useful to refine, or include in fitting these model parameters. (b) Previous model results, for the sensitivity of troponin C binding to calcium (‘calcium/troponin sensitivity’) and the sensitivity of cross-bridge binding and tension generation to the fraction of bound troponin C (‘troponin/cross-bridge sensitivity’). In this particular case, the ‘blue’ constraint, related to the amount of tension generated, determines most of the constraint for both model parameters. The red and orange constraints also limit the region for parameter 2, although both parameters are still highly coupled. To indicate the relation of the final choice of parameters in the model to these constraints, this choice can be indicated in the visualization, such as by the ‘X’ in (b).
In our cellular contraction model, six parameters influence isometric tension development. The full exploration of all pairs of parameters would also be difficult to interpret, owing to the large number of such pairs, many of which would be uninformative. Instead of including all of these cases, we show only those that are most informative, clearly highlighting areas of uncertainties instead of attempting to hide this information. In previous work, we included three of these visualizations, representing pairs of highly coupled parameters. By showing pairs of highly coupled parameters with a clear interpretation in all cases, we were able to identify areas of uncertainty in parameters and suggest where new experimental results could lead to improvements in the model by introducing new or improved constraints into the parametrization process. We have provided a specific example of this in figure 2b. This plot shows our sensitivity analysis for the two parameters that describe the sensitivity of troponin C to calcium and the sensitivity of cross-bridge binding to troponin C. As most experimental measurements are related to crossbridge dynamics and the amount of force, the dynamics of the intermediate biological process are not as constrained, and these two parameters are highly coupled as a result. However, magnitudes and rates of tension development limit both parameters to a narrow band, as shown in the viable region in the sensitivity analysis. This shows that decreasing the uncertainty in one of these parameters would help constrain both of them better.
A suggested refinement to this technique is the inclusion of probability densities of the experimental data, which would lead to a most probable point in the parameter plot. However, for the purposes of our model, and probably many other biological models, the continuous variation of model outputs as a function of their parameters as well as the fairly symmetric uncertainty profile in the correct model outputs means that much of this information is already implicitly present in the distance from the boundaries. Thus, this would lead to a more complex visualization, making the information presented more difficult to understand, without adding significant value.
3. Advantages of the virtual physiological mouse heart model
As mentioned in §1, a model of a common experimental animal presents an important opportunity for using rich datasets with temperature- and species-specific data. Healthy human cardiac myocytes suitable for experimentation are rare, and this is unlikely to change without significant technological advances. Sections 3.1 and 3.2 discuss these essential differences, and demonstrate how richer datasets can enable different kinds of research. These differences are presented in the context of recent results obtained in both previous and current work that exemplify the advances that are possible in a mouse model.
3.1. Results in studying multi-scale deformation-dependent effects
Among the advantages of a species-consistent electromechanical model, as shown in figure 1b, are the limited possibilities for a significant model prediction mismatch. Combining this knowledge with information about experimental variability, the limitations of experimental preparation and the parameter sensitivity visualization discussed in §2.3, we can also correct for these remaining uncertainties in the context of a multi-scale model.
As mentioned in §1, among the most important multi-scale effects included in a detailed electromechanical model are feedback effects by which the deformation of the heart influences tension generation of individual cells. These include the length dependence of tension giving rise to the Frank–Starling law of the heart, and the velocity dependence of tension. Including these effects is important for predicting the mechanical behaviour of the heart. However, their underlying mechanisms remain controversial, and in vitro experiments can be limited in providing data for such inherently multi-scale effects. In our previous work, the development of an integrated mouse model enabled the correction of limitations in cellular experimental data, and revealed the important role that velocity-dependent coupling mechanisms play in generating ventricular pressure [19]. While model results initially predicted that the shape of ventricular pressure would closely follow the shape seen in isometric tension generation, measurements of ventricular pressure show a significantly different shape. These measurements show a rapid increase in pressure, followed by a slowing of pressure development, leading to a plateau phase in which pressure is nearly constant while blood is being ejected. Only at the end of this phase does pressure rapidly decrease. Through knowledge of uncertainties in the cellular experimental data, along with a knowledge of the effects of different parameters through extensive parameter sensitivity analyses, we were able to identify velocity-dependent coupling mechanisms as a key factor in generating a ‘plateau’ of nearly constant pressure throughout systole. We also identified a limited number of areas in cellular data, where experimental data were known to be potentially less reliable owing to the limitations of cellular preparations, and thus were able to customize our hypothesis testing to complement reliability of data. In this particular case, the unreliability was related to the fact that cells in vitro generate less tension than they do in vivo. In the case of a mixed-species model such as figure 1a, there would have been many more potential explanations for these differences, as well as a lack of availability of data in correcting these limitations.
Paralleling experimental efforts in the life sciences to relate animal data to human function, these frameworks also have significant potential for improving electromechanical models in other species, in which there are similar differences between pressure and tension generation. From our results, we expect that, in electromechanical models of other species, including velocity-dependent coupling mechanisms is likely also to be of key importance in accurately predicting ventricular pressure. Furthermore, these results also allow us to identify how the reliability of experimental data on the velocity dependence can be improved, by indicating the average tension generated during the experiment, which gives a clearer indication of the quality of the experimental preparation.
3.2. Results in studying genetically modified mice
Another major difference between human and mouse modelling is the consistency of data for hearts in health and disease. The majority of invasive cardiac measurements on humans are in diseased patients, and these can have widely varying disease profiles and underlying causes of heart failure. Furthermore, there is a general lack of healthy control data, owing to practical and ethical problems of performing invasive measurements on healthy people.
By contrast, in animal models, there are nearly always healthy controls available as a point of comparison, as this is standard experimental protocol. Furthermore, studies of heart failure in animals tend to involve a specific defect that has been bred into a line, or introduced through genetic manipulation. This process, in turn, allows for detailed studies of a particular type of failure, and testing several hypotheses or intervention without risking human life.
Indeed, the majority of animals now have either been bred with a genetic defect or have been genetically manipulated [3]. Other than their use in studying heart failure, these genetically modified animals are also useful for improving our knowledge of many of the processes involved in cardiac contraction [20–23]. The mouse is uniquely suitable for genetic manipulation of mammals, with many genetic variants now being investigated using high-throughput techniques [24], generating a vast amount of data. However, the results of a genetic knockout can often be complex, resulting in many changes in cellular and tissue function. In these cases, a consistent modelling framework provides the ability to integrate data collected in these animals from cellular preparations and in vivo, and make quantitative connections. This allows us to learn more about the physiological mechanisms that are activated in a compensatory response in disease.
We have recently applied our mouse modelling framework to investigate the progression of heart failure in a specific genetic knockout in mice. In these experimental studies, the Serca2 gene, which is an important part of calcium regulation in the cell, was knocked out in adult mice [21]. This led to a significant decrease in the calcium transient, but only a moderate degree of heart failure [25]. Such moderate impairment of pump function was surprising considering the fact that an increase in cellular calcium concentration is the main underlying cause for cardiac contraction. Previous experimental and modelling studies have revealed compensatory mechanisms that limit the decrease in the calcium transient [6,7,26,27]. Most recently, our framework has been applied to look at the temporal progression of heart failure in these genetically modified animals [28]. Results from this study again confirm the importance of a consistent computational model for making quantitative connections between cellular and whole organ data. While the previous experimental results measuring calcium transients were thought to accurately represent cardiac function at the cellular and whole organ scale, investigation of these data using our mouse modelling framework revealed that they are quantitatively incompatible. Specifically, modelling results showed that these calcium transients observed in knockout mice do not result in viable ventricular function, even when taking into account potential compensatory mechanisms by which cells could generate more tension from the observed small change in calcium concentration.
These results have inspired new experiments with the purpose of uncovering factors missing in the in vitro experiments compared with the in vivo environment. After considering potential factors by which calcium dynamics are affected differently in vivo, the experimental protocols on isolated cells were expanded to quantify such effects. Specifically, in the β-adrenergic system, a response by the sympathetic nervous system was shown to be of key importance, allowing the mice to survive the significant decrease in sarco/endoplasmic reticulum Ca2+-ATPase (SERCA). This biological system is normally activated in the classic fight-or-flight response, and can be chronically active in disease. When this response was triggered in isolated cells, and the resulting calcium transients used as inputs to the VPM model, the resulting predicted ventricular function accurately matched that of previous pressure–volume measurements in these knockout mice. Thus, in these genetic knockout mice, there appear to be several advantages to increased β-adrenergic stimulation, showing that this kind of stimulation may not be as universally deleterious as sometimes thought. Other studies of heart failure in genetically modified mice by Xiao et al. [29] have also found beneficial effects of β-adrenergic stimulation, and suggest that deleterious side-effects can be prevented by selectively blocking β1 receptors whose activation can lead to cell death, while maintaining β2-based stimulation.
4. Hypothesis testing in the mouse heart model
Another advantage of a consistent murine heart model is the ability to test competing hypotheses in silico, in areas where there are less available experimental data. The ability to inspect results in great detail can both point to the plausibility of competing hypotheses, as well as inspire new experimental work that can distinguish between competing mechanisms. To further demonstrate this potential in this study, we now provide a further example of the value of this cardiac model by using it to investigate potential hypotheses for the interaction between the heterogeneity in length dependence and generated tension.
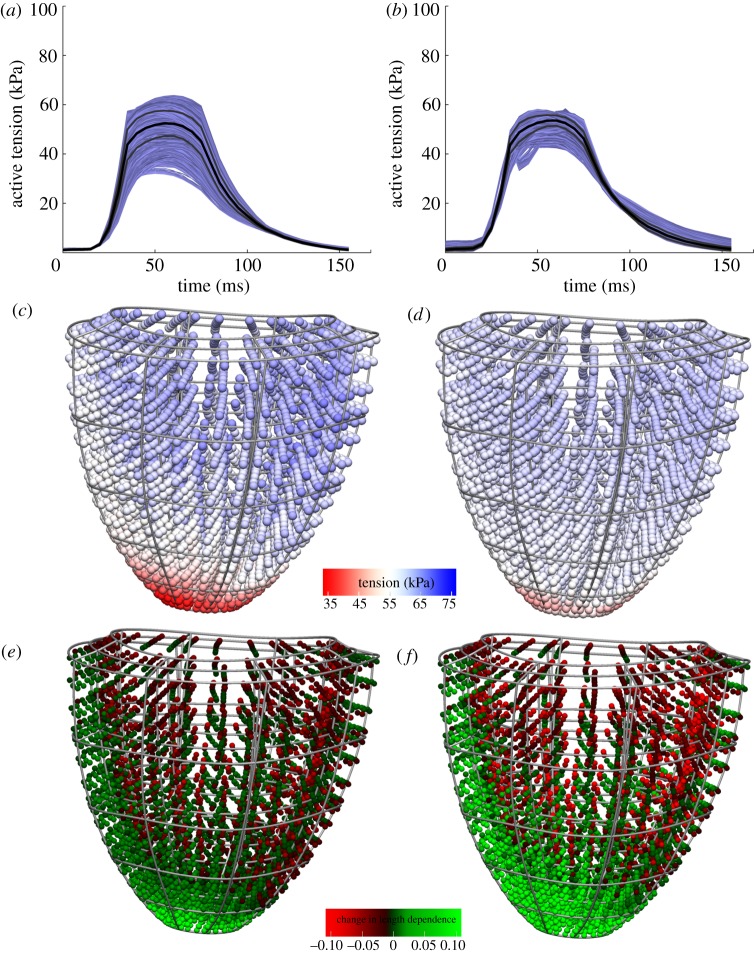
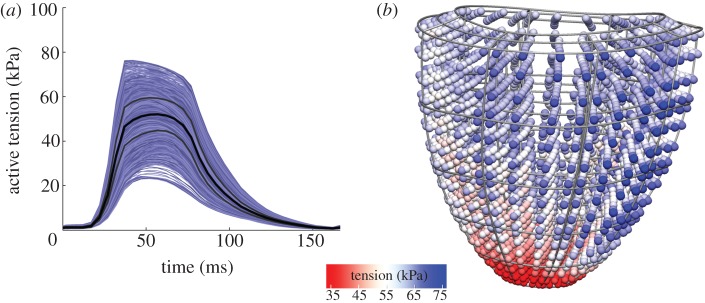
Cardiac cells are roughly cylindrical, defining a ‘fibre direction’ along their long axis. Owing to the sensitivity of stretch along this direction (as already discussed earlier), as well as the variation in this fibre direction throughout the heart, activation of cells throughout the heart can be highly heterogeneous in a simulation. Figure 3 shows an example of this variation in tension throughout the heart beat in the VPM heart model.
Figure 3.
Heterogeneity of tension development. (a) Default simulation results showing a large heterogeneity in tension generation in the myocardium. The graph shows tension development in all Gauss points of the elements not adjacent to the apex or base, to reduce the effects of boundary conditions. The black line indicates mean tension, and the grey lines indicate mean ±s.d. (b) Three-dimensional visualization of the complex apex-base and transmural heterogeneity in this distribution. This visualization shows the distribution of active tension at the time point of maximal mean active tension.
Some heterogeneity in stretch is to be expected in the heart, owing to the aforementioned complex fibre structure. However, there are several assumed drawbacks to a large heterogeneity in tension from a biological perspective. As tension is highly correlated with mechanical work, a heterogeneous distribution of tension is mechanically less efficient, and leads to variation in demand for oxygen and nutrients across the heart. When most of the pump function of the heart is dependent on a relatively small region of tissue, this region has the highest demand for oxygen, making it more vulnerable to failure. Such failure then would be likely to lead to severely impaired pump function, as the regions most likely to fail are those responsible for the majority of the heart's pump function.
Furthermore, research in the past decade has uncovered complex heterogeneity in length-dependent regulation [30,31]. The majority of length dependence of tension in cardiac myocytes is attributed to the regulation of contractile proteins by titin [32]. Titin is a giant protein, which functions as a molecular spring, plays an important structural role in cardiac cells, and is responsible for most passive tension in cardiac cells [33]. Different forms of titin can have missing segments of the protein, leading to shorter, stiffer, variants. Changes in such ‘isoform’ proportions over time have been observed across species in response to chronic cardiac stress [30], and the protein has also been implicated in ischaemic human heart disease [34]. Studies in recent years have uncovered more data on this important protein in experiments on both wild-type and genetically modified mice [22,31,35], although its functional role in the beating heart remains debated.
Section 4.1 provides an example of an application of the VPM model to investigate the heterogeneity of tension generation in light of the previous experimental results and modelling predictions described in this section. Specifically, by investigating several simplified models of different hypotheses and considering their plausibility and effect on tension generation, our goal is to learn more about the potential role for length-dependent mechanisms in homogenizing tension generation in the heart.
4.1. Transmural heterogeneity
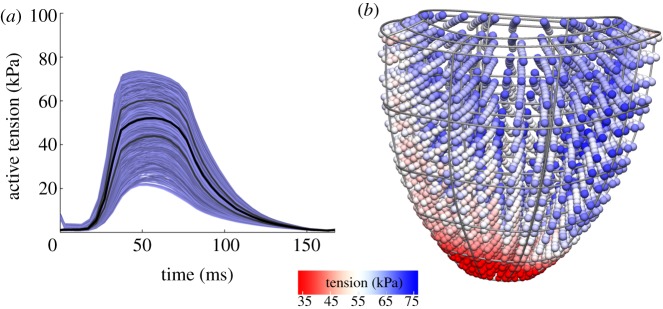
Recent experimental data have shown heterogeneity in length dependence [31]. However, the implications of this effect on whole organ pump function are still unclear. Our modelling approach is to investigate several potential underlying mechanisms that could lead to such heterogeneity, with the assumption that their main function is to homogenize tension. These models are developed to be a simple and abstract representation of a biological mechanism, with the goal of studying these phenomena in general, and not based on the underlying regulation of cardiac proteins. The electronic supplementary material describes the mathematical and computational techniques behind these models in detail. Our first potential mechanism is one of transmural heterogeneity, i.e. we model cells as more sensitive to being stretched depending on whether they are on the outside (epicardium) or inside (endocardium) of the heart. Such variations are already well known from electrophysiology, showing that there exist genetic and developmental markers that can lead to different spatial patterns of gene expression.
Figure 4 shows the results for optimal transmural variation, that is, the choice of transmural variation that minimized mean heterogeneity in tension over the cardiac cycle. These show only a mild decrease in heterogeneity, whereas a complex pattern of tension generation remains. First, tension generation near the apex (bottom of the heart) is low, and transmural variation cannot correct for this. Even ignoring the low apical tension generation, depending on the geometry of the heart, different regions may have more or less transmural variation, which makes a single choice for the strength of this variation throughout the heart less effective. Furthermore, the complex fibre structure can result in the mid-myocardial region being significantly different while epi- and endocardium are similar, which is also not well corrected for by increasing tension generation in either side while decreasing the other.
Figure 4.
(a) Heterogeneity of tension development with transmural variation in length-dependent activation. (b) Simulation results showing the heterogeneity after a transmural adjustment in length dependence. Details of the model equations are shown in the electronic supplementary material, §S2.1.
4.2. Length- and tension-dependent feedback
Studies showing co-expression of different titin variants in single cells [36] and changes in these different forms of titin over time [30] suggest that there are likely to be more complex mechanisms present in the heart that regulate its expression than the fixed transmural variation. To investigate the effects of such mechanisms, we have created several phenomenological models of potential feedback mechanisms and used them to investigate differences in tension generation after a period of feedback. The first of these models changes the length-dependent properties of tension generation in the cells depending on the average stretch cells experience over a longer time scale. Specifically, we shift these cells towards activation at higher lengths in regions where cells are consistently longer than average, and vice versa. Similarly, we have created a model that shifts length-dependent activation towards higher lengths in cells that generate above average tension over a cardiac cycle, and towards lower lengths in cells that generate below average tension.
Figure 5 shows the results of these simulations, from which we can see that both these mechanisms are more likely to result in more homogeneous tension than a simple transmural variation. These simulations also show that tension-dependent feedback is more effective in adjusting the low apical tension generation, but in both cases transmural variation is significantly reduced. In the case of tension-dependent feedback, we can see that there is a clear increase in heterogeneity during the later phases in which the heart is more relaxed. This may point to an inherent conflict between minimizing variability in tension across the entire cardiac cycle.
Figure 5.
Heterogeneity of tension development after length- and tension-dependent feedback on length-dependent activation properties. Simulation results showing (a) decreased heterogeneity after length-dependent feedback; (b) a larger decrease in heterogeneity after tension-dependent feedback. Both results are shown 55 ms into the 30th heart beat, near peak tension levels. (a,b) Show tension development throughout the heart beat; (c,d) show the heterogeneity near peak tension; and (e,f) show the adjustment in length-dependent activation Δλ. Details of the model equations are shown in the electronic supplementary material, §S2.2.
5. Discussion and conclusion
This study summarizes the work on an integrative electromechanical framework for mouse modelling, the use of VPH tools in its development, the advantages of this model and its potential for interpreting experimental data. The framework has been used to infer data about coupling mechanisms from ventricular data provided from healthy mice, via integration within species-specific models with carefully quantified uncertainty embedded within the parametrization process. Furthermore, the VPM framework has been used to provide insights into the progression of heart function in genetically modified mice, temporally tracking increasing heart failure and its compensatory mechanisms. This approach has already inspired physiologists to perform new experiments, where model results show that previously measured cellular and ventricular function is quantitatively incompatible without incorporating the effects of additional systemic physiological mechanisms that influence the heart. These physiological insights were made possible by careful species- and temperature-specific modelling, resulting in clear model-based hypotheses. However, such model development should be seen in the context of previously obtained knowledge of the dependencies of various physiological processes on species and temperature. For example, as measurements such as action potential morphology [37] and calcium to troponin binding [38] are known to be not very temperature dependent, they can be taken from lower temperature data. By contrast, the function of ion pumps is increased by 1.5–2.5 times for a 10°C increase in temperature [39], and the dynamics of cross bridges can be even more sensitive to such changes [38]. Thus, the development of a species-specific model requires careful attention to both the experimental conditions and the influence of these conditions on the experimental measurements used for model development.
Finally, in this study, we have used the framework to test several potential mechanisms for homogenizing tension generation in the heart, identifying further areas for experimental studies. In this particular investigation, we found that heterogeneity of tension generation is complex, and does not consist of simple transmural or apex-to-base gradient. While a simple transmural heterogeneity does little to improve heterogeneous tension development, dynamic mechanisms such as length- and tension-dependent feedback were shown to be more effective. Furthermore, our results show that a tension-dependent mechanism is likely to have consequences for diastolic filling of the ventricle. This is caused by an increased heterogeneity in the usually very low diastolic tension giving rise to a higher mean diastolic tension that can impair filling. Mechanisms related to passive tension of titin are already known to exist in cardiac cells, making them more likely to be co-opted for other pathways. Thus, if there are titin-based mechanisms present that work to homogenize tension, given these model results we hypothesize that such regulation is likely to be length or passive tension dependent. Novel experiments that determine in vivo variation in titin isoforms could lend further experimental support to these findings. As our study was carried out with the assumption of homogeneous calcium dynamics throughout the ventricle, we cannot exclude mechanisms in calcium dynamics and excitation–contraction coupling which homogenize tension. Indeed, such variation is known to exist from experiments, with both transmural and apical/septal variations in action potential and excitation–contraction coupling having been characterized by experiments and mathematical models [15,40,41], and has been represented by other mouse models [15]. We can conclude from our results that, if such a mechanism is dominant, including apex-to-base heterogeneity is likely to be important, as we showed a linear transmural gradient in excitation–contraction coupling is less effective at homogenizing tension. Given sufficient data to characterize the heterogeneity in calcium dynamics across the heart, this information could be included in future work to investigate the effects of this mechanism on tension heterogeneity. Alternatively, calcium dynamics could be set to result in optimal homogeneity, similar to the feedback-type models in this study, and results compared with experimental data where available. Finally, we have used homogeneity in tension as a substitute for work performed by the cell. The suitability of this approximation should be considered as well in any future work.
In this study, we have discussed the similarities and advantages that cardiac models of mice have compared with a human model. Although mice have many advantages as an experimental animal, including their small size, low maintenance costs, rapid gestation, availability of inbred lines and ease of genetic manipulation, they also have a number of limitations [20]. Their small heart size limits some experiments and devices such as catheters are more likely to influence measurements. Although mice, rats and other rodents are evolutionarily closer to humans than the larger mammals used for cardiac experiments [42], they are significantly different in several ways, which creates limitations for using them to understand human physiology [43]. When compared with humans, mice have higher heart rates and a significantly different action potential morphology, which has implications for studying electrophysiological diseases in particular, such as long QT syndrome [44]. Second, calcium cycling is more dependent on intracellular cycling, which increases the effects of impaired SERCA pump function. Thus, the genetic knockout of the Serca2 gene potentially overestimates the effect of decreased levels of SERCA protein. Nevertheless, the mouse still relies on very similar underlying physiological mechanisms for electrophysiology and tension generation, which makes many of the results obtained in these animals qualitatively valid in other mammalian species.
In conclusion, a virtual physiological mouse heart model has the potential to be the basis for a wide variety of research in mice, and has already led to several new physiological insights. Through providing tools and biological insights, the VPM framework can advance cardiac modelling across species.
Acknowledgements
The authors acknowledge support from the Virtual Physiological Rat Project (NIH 1 P50 GM094503-01), the Medical Research Council under grant no. G0800980 and the Engineering and Physical Sciences Research Council under grant nos. EP/F043929/1 and EP/G007527/2.
References
- 1.Viceconti M, Clapworthy G, Van Sint Jan S. 2008. The virtual physiological human: a European initiative for in silico human modelling. J. Physiol. Sci. 58, 441–446 10.2170/physiolsci.RP009908 (doi:10.2170/physiolsci.RP009908) [DOI] [PubMed] [Google Scholar]
- 2.Kohl P, Noble D. 2009. Systems biology and the virtual physiological human. Mol. Syst. Biol. 5, 292. 10.1038/msb.2009.51 (doi:10.1038/msb.2009.51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UK Home Office 2011. Statistics of scientific procedures on living animals: Great Britain 2011. See http://www.homeoffice.gov.uk/publications/science-research-statistics/research-statistics/other-science-research/spanimals11/. [Google Scholar]
- 4.Fink M, et al. 2011. Cardiac cell modelling: observations from the heart of the cardiac physiome project. Prog. Biophys. Mol. Biol. 104, 2–21 10.1016/j.pbiomolbio.2010.03.002 (doi:10.1016/j.pbiomolbio.2010.03.002) [DOI] [PubMed] [Google Scholar]
- 5.Smith NP, Crampin EJ, Niederer SA, Bassingthwaighte JB, Beard DA. 2007. Computational biology of cardiac myocytes: proposed standards for the physiome. J. Exp. Biol. 210, 1576–1583 10.1242/jeb.000133 (doi:10.1242/jeb.000133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Louch WE, Niederer SA, Andersson KB, Christensen G, Sejersted OM, Smith NP. 2011. Calcium dynamics in the ventricular myocytes of SERCA2 knockout mice: a modeling study. Biophys. J. 100, 322–331 10.1016/j.bpj.2010.11.048 (doi:10.1016/j.bpj.2010.11.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Louch WE, Niederer SA, Aronsen JM, Christensen G, Sejersted OM, Smith NP. 2012. Sodium accumulation in SERCA knockout-induced heart failure. Biophys. J. 102, 2039–2048 10.1016/j.bpj.2012.03.045 (doi:10.1016/j.bpj.2012.03.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd CM, Lawson JR, Hunter PJ, Nielsen PF. 2008. The CellML model repository. Bioinformatics 24, 2122–2123 10.1093/bioinformatics/btn390 (doi:10.1093/bioinformatics/btn390) [DOI] [PubMed] [Google Scholar]
- 9.Magyar J, et al. 2012. Rem-GTPase regulates cardiac myocyte L-type calcium current. Channels 6, 166–173 10.4161/chan.20192 (doi:10.4161/chan.20192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.cellML website 2012. Model exposures: an analysis of deformation-dependent electromechanical coupling in the mouse heart. See http://models.cellml.org/e/e4/view [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garny A, Noble D, Hunter PJ, Kohl P. 2009. Cellular open resource (COR): current status and future directions. Phil. Trans. R. Soc. A 367, 1885–1905 10.1098/rsta.2008.0289 (doi:10.1098/rsta.2008.0289) [DOI] [PubMed] [Google Scholar]
- 12.Lamata P, Niederer S, Nordsletten D, Barber DC, Roy I, Hose DR, Smith N. 2011. An accurate, fast and robust method to generate patient-specific cubic hermite meshes. Med. Image Analysis 15, 801–813 10.1016/j.media.2011.06.010 (doi:10.1016/j.media.2011.06.010) [DOI] [PubMed] [Google Scholar]
- 13.Smith N, et al. 2011. euHeart: personalized and integrated cardiac care using patient-specific cardiovascular modelling. Interface Focus 1, 349–364 10.1098/rsfs.2010.0048 (doi:10.1098/rsfs.2010.0048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Land S, Niederer SA, Smith NP. 2012. Efficient computational methods for strongly coupled cardiac electromechanics. IEEE Trans. Biomed. Eng. 59, 1219–1228 10.1109/TBME.2011.2112359 (doi:10.1109/TBME.2011.2112359) [DOI] [PubMed] [Google Scholar]
- 15.Bondarenko VE, Rasmusson RL. 2010. Transmural heterogeneity of repolarization and Ca2+ handling in a model of mouse ventricular tissue. Am. J. Physiol. 299, H454–H469 10.1152/ajpheart.00907.2009 (doi:10.1152/ajpheart.00907.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkar AX, Christini DJ, Sobie EA. 2012. Exploiting mathematical models to illuminate electrophysiological variability between individuals. J. Physiol. 590, 2555–2567 10.1113/jphysiol.2011.223313 (doi:10.1113/jphysiol.2011.223313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niederer SA, Fink M, Noble D, Smith NP. 2009. A meta-analysis of cardiac electrophysiology computational models. Exp. Physiol. 94, 486–495 10.1113/expphysiol.2008.044610 (doi:10.1113/expphysiol.2008.044610) [DOI] [PubMed] [Google Scholar]
- 18.Committee on Mathematical Foundations of Verification, Validation, and Uncertainty Quantification; Board on Mathematical Sciences and Their Applications, Division on Engineering and Physical Sciences, National Research Council 2012. Assessing the reliability of complex models: mathematical and statistical foundations of verification, validation, and uncertainty quantification. Washington, DC: The National Academies Press [Google Scholar]
- 19.Land S, Niederer SA, Aronsen JM, Espe EKS, Zhang L, Louch WE, Sjaastad I, Sejersted OM, Smith NP. 2012. An analysis of deformation-dependent electromechanical coupling in the mouse heart. J. Physiol. 590, 4553–4569 10.1113/jphysiol.2012.231928 (doi:10.1113/jphysiol.2012.231928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeishi Y, Walsh RA. Cardiac hypertrophy and failure: lessons learned from genetically engineered mice. Acta Physiol. Scand. 173, 103–111 10.1046/j.1365-201X.2001.00890.x (doi:10.1046/j.1365-201X.2001.00890.x) [DOI] [PubMed] [Google Scholar]
- 21.Andersson KB, et al. 2009. Mice carrying a conditional Serca2(flox) allele for the generation of Ca2+ handling-deficient mouse models. Cell Calcium 46, 219–225 10.1016/j.ceca.2009.07.004 (doi:10.1016/j.ceca.2009.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee EJ, Peng J, Radke M, Gotthardt M, Granzier HL. 2010. Calcium sensitivity and the Frank–Starling mechanism of the heart are increased in titin N2B region-deficient mice. J. Mol. Cell. Cardiol. 49, 449–458 10.1016/j.yjmcc.2010.05.006 (doi:10.1016/j.yjmcc.2010.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel JR, Pleitner JM, Moss RL, Greaser ML. 2012. Magnitude of length-dependent changes in contractile properties varies with titin isoform in rat ventricles. Am. J. Physiol. 302, H697–H708 10.1152/ajpheart.00800.2011 (doi:10.1152/ajpheart.00800.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skarnes WC, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 10.1038/nature10163 (doi:10.1038/nature10163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson KB, et al. 2009. Moderate heart dysfunction in mice with inducible cardiomyocyte-specific excision of the Serca2 gene. J. Mol. Cell. Cardiol. 47, 180–187 10.1016/j.yjmcc.2009.03.013 (doi:10.1016/j.yjmcc.2009.03.013) [DOI] [PubMed] [Google Scholar]
- 26.Louch WE, et al. 2010. Sodium accumulation promotes diastolic dysfunction in end-stage heart failure following Serca2 knockout. J. Physiol. 588, 465–478. 10.1113/jphysiol.2009.183517 (doi:10.1113/jphysiol.2009.183517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swift F, Franzini-Armstrong C, Øyehaug L, Enger UH, Andersson KB, Christensen G, Sejersted OM, Louch WE. 2012. Extreme sarcoplasmic reticulum volume loss and compensatory T-tubule remodeling after Serca2 knockout. Proc. Natl Acad. Sci. USA 109, 3997–4001. 10.1073/pnas.1120172109 (doi:10.1073/pnas.1120172109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Land S, Louch WE, Niederer SA, Aronsen JM, Christensen GA, Sjaastad I, Sejersted OM, Smith NP. In press. Beta-adrenergic stimulation maintains cardiac function in Serca2 knockout mice. Biophys. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao RP, Zhu W, Zheng M, Chakir K, Bond R, Lakatta EG. 2004. Heping Cheng subtype-specific β-adrenoceptor signaling pathways in the heart and their potential clinical implications. Trends Pharmacol. Sci. 25, 358–365 10.1016/j.tips.2004.05.007 (doi:10.1016/j.tips.2004.05.007) [DOI] [PubMed] [Google Scholar]
- 30.Hein S, Gaasch WH, Schaper J. 2002. Giant molecule titin and myocardial stiffness. Circulation 106, 1302–1304 10.1161/01.CIR.0000031760.65615.3B (doi:10.1161/01.CIR.0000031760.65615.3B) [DOI] [PubMed] [Google Scholar]
- 31.Cazorla O, Lacampagne A. 2011. Regional variation in myofilament length-dependent activation. Pflugers Archiv. Eur. J. Physiol. 462, 15–28 10.1007/s00424-011-0933-6 (doi:10.1007/s00424-011-0933-6) [DOI] [PubMed] [Google Scholar]
- 32.Fukuda N, Terui T, Ohtsuki I, Ishiwata S, Kurihara S. 2009. Titin and troponin: central players in the Frank–Starling mechanism of the heart. Curr. Cardiol. Rev. 5, 119–124 10.2174/157340309788166714 (doi:10.2174/157340309788166714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granzier HL, Labeit S. 2004. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 94, 284–295 10.1161/01.RES.0000117769.88862.F8 (doi:10.1161/01.RES.0000117769.88862.F8) [DOI] [PubMed] [Google Scholar]
- 34.Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, Linke WA. 2002. Titin isoform switch in ischemic human heart disease. Circulation 106, 1333–1341 10.1161/01.CIR.0000029803.93022.93 (doi:10.1161/01.CIR.0000029803.93022.93) [DOI] [PubMed] [Google Scholar]
- 35.Cazorla O, Wu Y, Irving TC, Granzier H. 2001. Titin-based modulation of calcium sensitivity of active tension in mouse skinned cardiac myocytes. Circ. Res. 88, 1028–1035 10.1161/hh1001.090876 (doi:10.1161/hh1001.090876) [DOI] [PubMed] [Google Scholar]
- 36.Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitás K, Labeit S, Granzier H. 2000. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ. Res. 86, 59–67 10.1161/01.RES.86.1.59 (doi:10.1161/01.RES.86.1.59) [DOI] [PubMed] [Google Scholar]
- 37.Brouillette J, Clark RB, Giles WR, Fiset C. 2004. Functional properties of K+ currents in adult mouse ventricular myocytes. J. Physiol. 559, 777–798 10.1113/jphysiol.2004.063446 (doi:10.1113/jphysiol.2004.063446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Little SC, Biesiadecki BJ, Kilic A, Higgins RSD, Janssen PML, Davis JP. 2012. The rates of Ca2+ dissociation and cross-bridge detachment from ventricular myofibrils as reported by a fluorescent cardiac troponin C. J. Biol. Chem. 287, 27 930–27 940 10.1074/jbc.M111.337295 (doi:10.1074/jbc.M111.337295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon TR, Wang F, Puglisi J, Weber C, Bers DM. 2004. A mathematical treatment of integrated Ca dynamics within the ventricular myocyte. Biophys. J. 87, 3351–3371 10.1529/biophysj.104.047449 (doi:10.1529/biophysj.104.047449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dilly KW, Rossow CF, Scott Votaw V, Meabon JS, Cabarrus JL, Santana LF. Mechanisms underlying variations in excitation-contraction coupling across the mouse left ventricular free wall. J. Physiol. 572, 227–241 10.1113/jphysiol.2005.102020 (doi:10.1113/jphysiol.2005.102020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bondarenko VE, Szigeti GP, Bett GCL, Kim SJ, Rasmusson RL. 2004. Computer model of action potential of mouse ventricular myocytes. Am. J. Physiol. 287, H1378–H1403 10.1152/ajpheart.00185.2003 (doi:10.1152/ajpheart.00185.2003) [DOI] [PubMed] [Google Scholar]
- 42.Springer MS, Meredith RW, Janecka JE, Murphy WJ. 2011. The historical biogeography of mammalia. Phil. Trans. R. Soc. B 366, 2478–2502 10.1098/rstb.2011.0023 (doi:10.1098/rstb.2011.0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niederer SA, Land S, Omholt SW, Smith NP. 2012. Interpreting genetic effects through models of cardiac electromechanics. Am. J. Physiol. 303, H1294–H1303 10.1152/ajpheart.00121.2012 (doi:10.1152/ajpheart.00121.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salama G, London B. 2007. Mouse models of long QT syndrome. J. Physiol. 578, 43–53 10.1113/jphysiol.2006.118745 (doi:10.1113/jphysiol.2006.118745) [DOI] [PMC free article] [PubMed] [Google Scholar]