Abstract
Epimorphic regeneration in humans of complex multitissue structures is primarily limited to the digit tip. In a comparable mouse model, the response is level-specific in that regeneration occurs after amputation at the distal end of the terminal phalanx, but not more proximally. Recent isolation of stromal cells from CD1 murine phalangeal elements two and three (P2 and P3) allow for comparative studies of cells prevalent at the amputation plane of a more proximal region (considered nonregenerative) and a more distal region (considered regenerative), respectively. This study used adherent, suspension, and collagen gel cultures to investigate cellular processes relevant to the initial response to injury. Overall, P2 cells were both more migratory and able to compact collagen gels to a greater extent compared to P3 cells. This observed increased capacity of P2 cells to generate traction forces was likely related to the higher expression of key cytoskeletal proteins (e.g., microfilament, nonkeratin intermediate filaments, and microtubules) compared to P3 cells. In contrast, P3 cells were found to be more proliferative than P2 cells under all three culture conditions and to have higher expression of keratin proteins. In addition, when cultured in suspension rather than on adherent surfaces, P3 cells were both more proliferative and had greater gene expression for matrix proteins. Together these results add to the known inherent differences in these stromal cells by characterizing responses to the physical microenvironment. Further, while compaction by P2 cells confirm that collagen gels is a useful model to study wound healing, the response of P3 cells indicate that suspension culture, in which cell–cell interactions dominate like in the blastema, may be better suited to study regeneration. Therefore, this study can help develop clinical strategies for promoting regeneration through increased understanding in the properties of cells involved in endogenous repair as well as informed selection of useful in vitro models.
Introduction
Species such as salamanders and newts can undergo epimorphic regeneration, which includes the replacement of whole limbs.1 In mice2–4 and humans,5,6 however, regeneration of complex multitissue structures is primarily limited to regeneration of the distal digit tip. Animal models have been pivotal in determining key signaling pathways7,8 and cell sources9,10 involved in regeneration. In addition, recent tissue engineering studies have begun to test treatment modalities to help promote whole digit and limb regeneration.11,12 Use of in vitro techniques with mammalian cells, however, is also essential to increase understanding of the cellular processes involved in injury responses to amputation.
It is unclear the relative contribution of the different endogenous cells to the regenerative process. It was originally thought that the blastema was a homogenous population of dedifferentiated cells that form the base of tissue regrowth.13 More recent studies have found that multiple lineage-restricted tissue stem/progenitor cells contribute to the blastema in the urodele limb and mouse digit tip.9,10,14 Regardless of cell source, complete repair of the digit tip ultimately involves multiple specialized phenotypes, including endothelial cells, mesenchymal stem cells, fibroblasts, and skeletal cells. Comparison of the native cells from regenerating and nonregenerating regions of the digit can be useful to help identify cellular attributes necessary for the restoration of lost tissue.
Regenerative processes in mammalian digit tips is level-specific in that amputation at the distal end leads to regeneration while a more proximal injury leads to wound healing.3 These outcome differences occur despite fairly similar cellular and tissue components at the original site of injury. Recent isolation of skeletal cells from mouse phalangeal element three (regenerating region; P3) and phalangeal element two (nonregenerating region; P2)15 allow for studies with a major phenotype prevalent at the amputation plane. Comparative in vitro studies using these cells will improve understanding of the processes that limit or drive regeneration.
Complex aspects of the in vivo microenvironment are known to mediate cell processes. Use of adherent, suspension and scaffold-based cultures in vitro can help establish the effects of physical configuration on cell proliferation, migration, and function. The objective of these early studies with P2 and P3 cells was to determine phenotypic differences in response to culture environment.
Materials and Methods
Phalangeal element (P2 and P3) cells
Cells (a generous gift from Dr. Ken Muneoka of Tulane University) were previously isolated from week 8 adult CD1 mice through digestion of the skeletal connective tissue of phalangeal elements (separated from the adjacent skin, fur, fat pad, nail, and ligament tissue) of digits II–IV.15 The adherent cells from mouse phalangeal element 2 (P2: from middle phalanx) and 3 (P3: from terminal phalanx) were then expanded using fibronectin-coated (Fn; 3.5 μg/cm2) dishes in culture medium, which consisted of Dulbecco's modified Eagle's medium/molecular cellular developmental biology (MCDB 201) medium supplemented with insulin-transferrin-sodium selenite+1 (Sigma), 5% embryonic stem cell-qualified fetal bovine serum (Invitrogen), 10−9 M dexamethasone (Sigma), 10−4 ascorbic acid 2-phosphate (Sigma), 50 μg/mL platelet-derived growth factor-ββ, 50 μg/mL epidermal growth factor (R&D Systems), 1000 U/mL leukemia inhibitory factor (EMD Millipore), and antibiotics.16
Culture conditions
Cells were cultured under both two- and three-dimensional (2D and 3D, respectively) conditions. In adherent 2D culture, cells were seeded at 8000 per cm2 on Fn-coated tissue culture plastic (Fn-TCP). To provide a 3D culture environment, cells were either put into suspension (SUS) or collagen gels (GEL).17 For SUS cultures, expanded cells were placed into bacteriological Petri dishes (0.5 E6 cells/100-mm dish) and continuously agitated on an orbital shaker (40 rpm). Cells were maintained for up to 12 days, with culture medium and dishes changed every other day. For GEL cultures, expanded cells were encapsulated into type I collagen gels (2 mg/mL) with an initial seeding density of 0.2 E6 cells/mL. Each gel (0.75 mL) was polymerized overnight in 12-well plates and then released to allow for unconstrained compaction. GEL samples were maintained for up to 8 days with medium changed every 2 days (25 mL/gel).
Cell proliferation
Cell proliferation was evaluated by quantification of number and cell cycle phase. Number was determined for cells recovered from trypsinized Fn-TCP samples and collagenase-digested (600 U/mL, type 2; Worthington Biochemical) GEL samples using the Z1 Coulter Counter® (counts >6 μm). Cell cycle phases were assessed by recovering single-cell solutions either by trypsinization of Fn-TCP samples or dissociation of SUS samples, staining with DRAQ5 (Biostatus), analyzing fluorescence using an FACS Canto (BD Biosciences), and fitting for phase distribution using FCS Express Software v4.
Scratch test
Cell migration was evaluated for confluent monolayers of P2 and P3 cells on Fn-coated glass slides. A 1000 μL pipet tip was used to create a scratch ∼240 μm wide that was then imaged at 0, 3, and 6 h. For better visualization of outstretched cellular processes, samples were fixed at 6 h, stained for F-actin using phalloidin (Invitrogen), and imaged using standard fluorescent microscopy.
Gene expression
At the end of culture, cells from Fn-TCP samples and cell clusters from SUS samples were lysed, homogenized using Qiagen QIAshredders, and stored at −80°C until further processing. RNA was isolated from the frozen cell lysates and the GEL samples using the Qiagen RNeasy and RNeasy Lipid Tissue kits, respectively, and each sample was quantified using a Nanodrop® spectrophotometer. Standard analysis of mRNA levels for each sample was done on cDNA converted from 1 μg RNA (Invitrogen Superscript® III First-strand synthesis) and analyzed using SYBR® Green on a StepOnePlus™ polymerase chain reaction (PCR) System (Applied Biosystems). Primers were custom designed (Primer Express® Software v3) for microfilaments (ACTA1, ACTA2), intermediate filaments (KRT6A, KRT8, KRT13, LMNA, and VIM), microtubule (TUBA1B), and extracellular matrix (ECM) proteins (FN1, COL1A1, and COL4A1) (primer sequences are listed in Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/tea). Gene expression levels were quantified using standard curves and are all reported as normalized to GAPDH expression.
Analysis of genes associated with signal transduction pathways was performed using an RT2 Profiler™ PCR array (SA Biosciences). RNA from three independent samples per group were converted to cDNA and analyzed using the vendor's specified reagents and instructions. For each sample, 84 signal transduction genes were normalized to housekeeping genes. Fold regulation between groups was used to evaluate differences in expression due to either cell type or culture condition (Complete listing of genes, fold regulation, and p-values are found in Supplementary Table S2.).
Microscopy
Images of Fn-TCP, SUS, and GEL samples were taken periodically during culture, as well as after histological processing at terminal timepoints. Phase images were taken of Fn-TCP and SUS samples, while macroscopic pictures were taken of GEL samples. To quantify changes in cell cluster and collagen gel size over time, cross-sectional areas of individual SUS and GEL samples were calculated using analysis (ImageJ software) of calibrated images. Spatial distribution of cells in 3D culture conditions was also visualized, where fixed SUS and GEL samples were paraffin processed, cut into 7 μm sections, stained with hematoxylin and eosin, and imaged under brightfield conditions.
Statistical analysis
Data are presented as mean±SEM. A Student's t-test was used to compare P2 versus P3 cells, as well as FN-TCP versus SUS culture conditions. These separate comparisons between cell types or between culture conditions were considered significant with p-values of <0.05.
Results
Culture on tissue culture plastic (Fn-TCP)
P2 and P3 cells attached and proliferated on Fn-TCP, although images of P3 cultures showed a greater presence of light-refractive edges (Fig. 1A, B). Quantification of cell number after 2½ days resulted in a 5.7- and 8.6-fold increase over the initial seeding density for P2 and P3 cells, respectively (Fig. 1C). The P3 value, however, was 50% higher (p<0.05), indicating a greater proliferative rate for P3 cells. Cell cycle analysis was consistent with this finding, in that the fitted percentage for the DNA replicating S-phase was approximately two-fold higher for P3 cells (31% for P2 vs. 56% for P3 in Fig. 1D and E, respectively). These observed differences in proliferation between P2 and P3 cells may be due to cell cycle inhibitors, as both CDKN2A and CDKN2B were found to be upregulated (each >300×with p<0.001) in P2 cells using PCR arrays (Fig. 2A).
FIG. 1.
Morphology and proliferation on fibronectin-coated tissue culture plastic (Fn-TCP). Phase images at lower (left) and higher (right) magnification of P2 (A) and P3 (B) cells grown on Fn-TCP. (C) Cell number for P2 and P3 cultures were counted after 2.5 days, with the dotted line representing the initial cell number (mean±SEM, n=5, *p<0.05). Histograms show fluorescence due to DRAQ5 staining for P2 (D) and P3 (E) cultures, with fitted curves for cell cycle analysis overlaid to determine percentages in the G0/G1, S, and G2 phases (indicated by separate colors). Color images available online at www.liebertpub.com/tea
FIG. 2.
Gene expression of signaling pathways. Relative expression of 84 signaling-related genes is displayed on scatter plots of P2 Fn-TCP versus P3 Fn-TCP (A) and P3 SUS versus P3 Fn-TCP (B). Genes with changes ≤2-fold are indicated in black (and lie within the region marked by the lines). Changes in expression levels ≥2-fold are indicated in red (upregulation) or green (downregulation). Relative expression of CDKN2A and CDKN2B (single arrowheads), as well as matrix metalloprotease 10 (double arrowheads) are shown. Fn-TCP, fibronectin-coated tissue culture plastic; SUS, suspension. Color images available online at www.liebertpub.com/tea
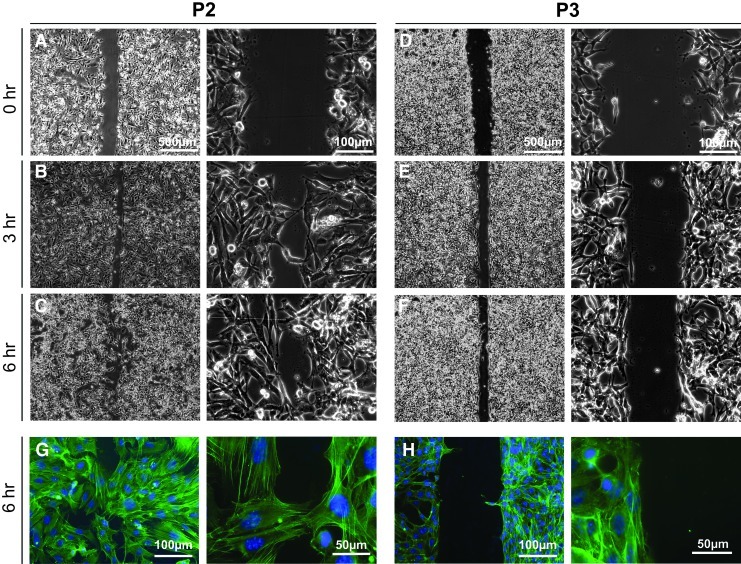
Response of P2 and P3 cells to a generated scratch indicated differences in migratory potential. The initial scratch was similar in size (240 μm) for samples of both groups (Fig. 3A, D). Within 3 h, P2 cells had begun to span the distance with a small number of cells from opposing sides in direct contact (Fig. 3B). Larger sections of the edges came into contact by 6 h (Fig. 3C), with staining for F-actin highlighting individual P2 cells reaching across the gap (Fig. 3G). In contrast, for P3 cultures the gap between the edges of the original scratch narrowed (Fig. 3D–F), but the margins remained largely defined (Fig. 3F) with few cells reaching across (Fig. 3H). These results therefore indicate that P2 cells may have an increased migratory capacity compared to P3 cells.
FIG. 3.
Scratch test on Fn-coated glass slides. Lower (left) and higher (right) magnification phase images were taken of P2 (A–C, G) and P3 (D–F, H) cultures immediately (A, D), 3 h (B, E), and 6 h (C, F, G, H) after creating a scratch of confluent monolayers. Samples fixed at 6 h and stained with phalloidin were imaged fluorescently to visualize outstretched cellular processes. Color images available online at www.liebertpub.com/tea
Cytoskeletal gene expression is markedly different for P2 and P3 cells on Fn-TCP. To span the variety of cytoskeletal proteins that are present in cells, we assessed genes for microtubules, intermediate filaments, and microfilaments. P2 cultures had a 1.5-fold increase for TUBA1B (Fig. 4A), a 2-fold increase for both LMNA and VIM (Fig. 4B), and a 4-fold increase for ACTA-1 and -2 (Fig. 4C). Keratin intermediate filaments, however, were more highly expressed in P3 samples with a four-fold increase for both KRT6A and KRT13 and an increase of more than an order of magnitude for KRT8 (Fig. 4D). Thus, P3 cells are more proliferative while P2 cells are more migratory on adherent culture, possibly due to cytoskeletal differences.
FIG. 4.
Cytoskeletal gene expression on Fn-TCP. P2 and P3 cultures were analyzed for microtubules (A: TUBA1B), intermediate filaments (B: LMNA, VIM; D: KRT6A, KRT8, KRT13), and microfilaments (C: ACTA1, ACTA2). Values presented are mean±SEM (n=5), with asterisks indicating p-values (*p<0.05, **p<0.01).
Culture in suspension (SUS)
Single-cell solutions of P3 cells placed into continuously agitated suspension culture form cell clusters that increased in size with time. P3 cells through aggregation and/or proliferation formed clusters comprised of multiple cells within 4 days (Fig. 5D). At day 8, clusters were still loosely connected and transparent (Fig. 5E), though most seemed tightly packed with dense central regions by day 12 (Fig. 5F). Overall cluster size also changed with time (Fig. 5J–L), with the average cross-sectional area increasing from 2350 μm2 at day 4 to 7720 μm2 at day 12.
FIG. 5.
Morphology and proliferation in SUS. Lower (left) and higher (right) magnification phase images were taken of P2 (A–C) and P3 (D–F) cell clusters after 4 (A, D), 8 (B, E), and 12 (C, F) days. Histograms show fluorescence due to DRAQ5 staining for P2 (G) and P3 (H) SUS samples, with fitted curves for cell cycle overlaid to determine percentages in the G0/G1, S, and G2 phases (indicated by separate colors). (I) Phase image of P2 cells attached to Petri dishes at day 5. Histograms of size analysis for P2 (blue) and P3 (red) cell clusters after 4 (J), 8 (K), and 12 (L) days. Color images available online at www.liebertpub.com/tea
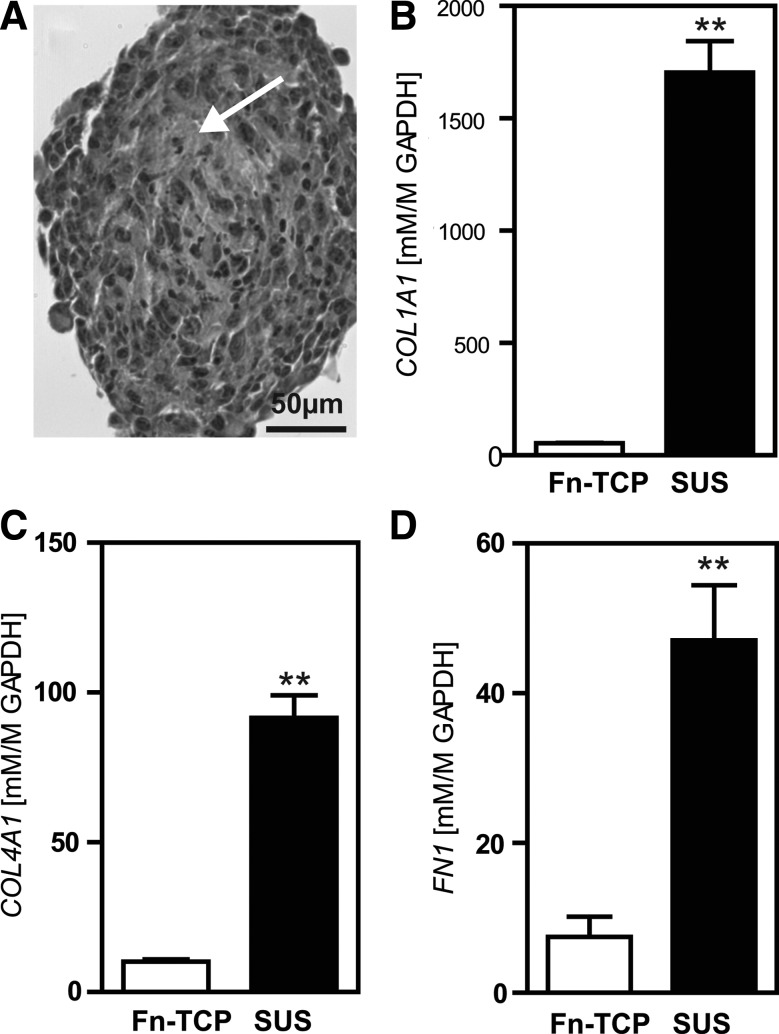
Histological sections of P3 SUS samples indicated granular matrix between individual cells within the clusters (Fig. 6A, arrow). Quantification of mRNA did show a significant (p<0.01) upregulation of gene expression of the ECM proteins collagen type I, collagen type IV, and fibronectin for P3 cells in SUS compared to Fn-TCP (Fig. 6B–D). Further, overall greater matrix dynamics for SUS cultures was also due to a notable increase compared to Fn-TCP controls (>1500×; p<0.001) for matrix metalloprotease 10 (Fig. 2B and Supplementary Table S2). Therefore in suspension, P3 cells seem to aggregate and proliferate, with the capacity to synthesize and remodel secreted matrix proteins.
FIG. 6.
Extracellular matrix of P3 cells in SUS. (A) Phase image of a D8 P3 cell cluster, where arrow indicates granular matrix between cells. Relative gene expression of collagen type I (B), collagen type IV (C), and fibronectin (D) of P3 cells at D8 grown in SUS compared to Fn-TCP. Values presented are mean±SEM (n=3–5, **p<0.01).
P2 cells may have a preference for culture on adherent surfaces rather than in suspension. As early as day 4, differences between cell types were detected where P2 clusters in SUS were markedly fewer (Fig. 5A) and averaged only 820 μm2 in size (33% of time matched P3 samples). From day 4 to day 8, P2 clusters increased in size but remained smaller than in comparable P3 samples (Fig. 5B, J, K). The persisting P2 clusters at day 12, however, were similar in size to those in P3 samples (Fig. 5L) but extremely few in number (Fig. 5C). This lack of P2 cells in SUS may be partly due to the negligible percentage of proliferating cells (<1% in the S-phase at day 4; Fig. 5G), unlike P3 cells which maintained proliferative rates similar to that in Fn-TCP (59%; Fig. 5H). Further, P2 cells were observed attached to the non-TC-treated surfaces despite frequent Petri dish changes (Fig. 5I), indicating an aversion to suspension conditions.
Culture in type I collagen gels (GEL)
Encapsulation in collagen gels was found to support the in vitro culture of both P2 and P3 cells. Proliferation rates were quite low for both cell types, as there were less than two population doublings over 4 days (Fig. 7A), though P3 cell number was slightly higher (p<0.05). Histological sections revealed that cells were still homogenously and sparsely distributed after 4 days with an initial seeding density of 1.5 E5 cells per 0.75 mL GEL. By day 8, however, P2 samples had an accumulation of cells at the free GEL boundary (Fig. 7, arrows) that was absent in P3 samples. This observed accumulation, despite a low proliferation rate in P2 GEL samples, and the previous results of the scratch test in adherent culture, may be due to a more migratory nature of P2 cells compared to P3 cells.
FIG. 7.
Cell number and distribution in GEL samples. (A) Cell number was determined for P2 and P3 GEL samples after 4 days of culture, with the dotted line representing the initial cell number (mean±SEM, n=4, *p<0.05). Brightfield images of P2 (B, D) and P3 (C, E) samples at 4 (B, C) and 8 (D, E) days are shown, with arrows indicating the edge of the collagen gels in cross section.
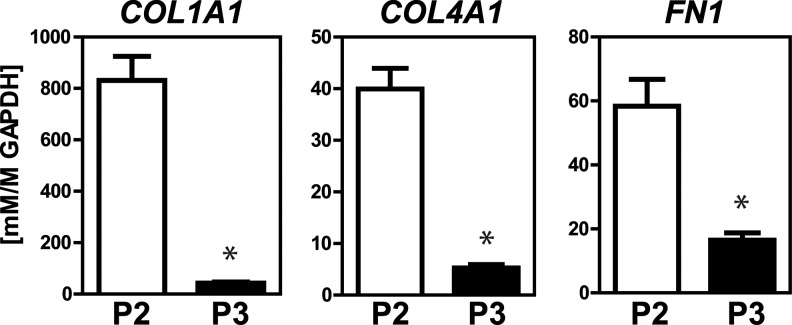
P2 and P3 cells are able to respond to the matrix environment in GEL culture. Qualitative images show that over 8 days the gels compacted (Fig. 8A), which is known to be due to active cellular processes, including force generation.18 Quantification of the cross-sectional area of multiple independent samples over time (Fig. 8B) revealed that P2 cells compacted the GEL not only more quickly (by 66% vs. 22% at day 4) but also to greater levels (by 84% vs. 67% at day 8). Considering the role of the cytoskeleton with force generation, it was not unexpected that P2 cells had significantly higher gene expression levels of microtubules, nonkeratin intermediate filaments, and microfilaments (Fig. 8D). As was found on Fn-TCP, P3 cells did have higher levels of keratin expression (Fig. 8C); however, these proteins are not usually implicated in traction force. In addition, P2 cells had >4-fold expression levels (p<0.05 for each) compared to P3 cells of extracellular matrix proteins collagen type I, collagen type IV, and fibronectin (Fig. 9). Thus, in an exogenously matrix-rich environment, such as collagen type I gels, P2 cells compared to P3 cells have a greater ability to remodel the surrounding microenvironment.
FIG. 8.
Compaction and cytoskeletal expression in GEL samples. Macroscopic images are shown (A) for P2 (left) and P3 (right) GEL samples at days 1 and 8, with cross-sectional area plotted for each day to quantitate compaction (B; mean±SEM, n=6–15). Cytoskeletal gene expression was analyzed in day 8 samples for keratin filaments (C: KRT6A, KRT8, and KRT13), as well as other cytoskeletal proteins (D: TUBA1B, VIM, LMNA, ACTA1, and ACTA2). Values presented are mean±SEM (n=3), with asterisks indicating p-values (*p<0.05, **p<0.01).
FIG. 9.
Extracellular matrix in GEL samples. P2 and P3 GEL samples at day 8 were analyzed for relative gene expression of collagen type I, collagen type IV, and fibronectin. Values presented are mean±SEM (n=3, *p<0.05).
Discussion
These in vitro studies explored cellular processes of P2 and P3 cells in 2D and 3D culture. P3 cells were found to be more proliferative when compared to P2 cells for adherent, suspension, and collagen gel cultures. Moreover, P2 cells were almost completely nonproliferative when presented with no exogenous matrix proteins, as in suspension culture. Cell mobility, conversely, was primarily observed in P2 samples that were able to span gaps on 2D glass slides and accumulate at the free edges of 3D gels. In addition, P2 cells contracted collagen gels to a greater degree than P3 cells. These observed changes in cell functionality may be partly due to the stark differences in expression of cytoskeletal proteins, which were all greater in P2 cells except for the keratins.
The use of cells isolated from mouse phalangeal elements two and three, which are considered regions of nonregenerative and regenerative potential, respectively, allow for in vitro comparative studies using mammalian cells. Here we used simple culture paradigms that controlled the microenvironmental cues and focused on a single phenotype, skeletal cells. There is the possibility of population heterogeneity, as evidenced by a small distinct subpopulation in the P2-SUS cultures at later timepoints. These studies, however, were still able to identify discernible population differences based on original tissue location in terms of proliferation, migration, force generation, and cytoskeleton expression. The higher expression of keratin proteins in the P3 cells compared to the P2 cells, expected due to the close proximity and potential regenerative involvement of the nail bed at the terminal phalanx,19 substantiate that the differences observed in these in vitro cultures are likely reflective of inherent in situ cell differences. Ultimate regeneration with tissue outgrowth in vivo, however, not only depends on factors intrinsic to the cells, but also extrinsic cues.20 Thus, subsequent in vitro studies need to provide a more complex microenvironment, allowing spatiotemporal regulation of cytokines, heterotypic cell–cell interactions, and cell–matrix binding, to properly investigate the different phases of regeneration.
Wound healing and regeneration have distinct goals, that is, survival versus restoration of function, that lead to different tissue outcomes. Wound healing commences with an inflammatory response, followed by re-epithelization and matrix synthesis, and is completed by matrix remodeling.21 Regeneration in the mouse digit tip undergoes similar initial processes, but then progresses to blastema formation, and tissue outgrowth.22 Though regeneration ultimately forms complex multitissue structures, it has been suggested that differences in the early cellular responses,13 including proliferation, migration, and contraction, may prescribe the end result.23 Thus, the model systems used here were selected to investigate events relevant to the initial response after amputation.
Cell proliferation is a necessary component not only for tissue outgrowth, but also for blastema formation. Cells in the blastema are not just those resident at the time of injury but also their progeny, with certain phenotypes over-represented compared to original availability.9,10,24 In our studies, we found that cells from the regenerative P3 element had markedly higher proliferative rates in vitro compared to similar cells from the nonregenerative P2 element, with the greatest differences seen in suspension cultures. Such differences may be due to regulation of cell cycle inhibitors, which were more highly expressed for P2 cells. In conjunction with the propensity of P2 cells to migrate when presented with proteins (either as a basement layer in 2D or a collagenous tissue in 3D), the inability to initially amass skeletal cells as part of a stable blastema cell cluster may be one hurdle for regeneration.
The standard in vitro wound healing models have not been optimized to study regeneration. Contraction of granulation tissue is a key event during wound healing25 and is mimicked in collagen gels where stromal cells induce observable levels of compaction.26 Consistent with the notion that P2 cells participate in wound healing as opposed to regeneration, it was found that P2 cells in gels were able to migrate and induce compaction to a greater extent than P3 cells. Further, the markedly lower levels of ECM expression in P3-GEL samples suggest that collagen gels may not be suitable to study cells associated with regeneration. Instead, suspension culture, in which cell–cell interactions dominate over cell–matrix interactions,27,28 seems to be a more appropriate in vitro model to study early blastema events. As would be expected from cells native to a regenerative region, P3 cells proliferated and expressed high levels of ECM-relevant proteins when cultured in suspension. Thus, while collagen gels are frequently used to study wound healing,29 suspension cultures may be a useful model to study regenerative processes.
One of the more notable distinctions between P2 and P3 cells was cytoskeletal gene expression. As the cytoskeleton is associated with force generation,30 the higher levels of microfilament and microtubule expression in P2 cells are consistent with observations of greater migration and gel compaction. In contrast, the higher levels of keratin expression in P3 cells may not only be due to the proximity of the source tissue to the nail bed, but may be associated with the recent implication of keratins in stem cell self-renewal.10 Thus, the cytoskeleton may play a major role in the early cellular responses to injury, including proliferation, migration, and wound closure. Collectively, these studies suggest that attempts to steer injury responses from wound healing toward regeneration may involve cytoskeletal modulation.
The default response to amputation in humans is one of survival through wound healing, including an immune response to ward off infection31 and scar tissue formation to seal the injury.32 The evolved efficacy of this response may in fact hinder the more sophisticated process of regeneration.32,33 While documented cases of regeneration at the distal digit tip5,6 signals potential for promoting a robust regenerative response in humans, there is yet no obvious biological approach to leverage during treatment. For example, it is unclear whether to focus on redevelopment; endogenous repair of the mouse digit tip relies on secondarily evolved processes,3 but exogenously promoted repair recapitulates developmental pathways.7 In addition, the importance of blastema formation in mammalian regeneration has yet to be established, as recent studies indicate that multiple fate-restricted tissue stem cells contribute to tissue restoration.9,10 Further, while the P2 and P3 phenotypes are sourced from regions with inherently different regenerative potentials, it cannot be currently excluded that primary causal differences may be due to other factors, such as proximity to the nail bed or different scarring responses. Yet comparative studies such as ours help advance the biological understanding of endogenous cell sources that will translate into treatment modalities for digit and limb amputation.
In conclusion, our studies elucidated the distinct responses of P2 and P3 cells to different culture environments, implicated the cytoskeleton in these responses, and evaluated the relative value of each culture model for the study of regenerative processes. Specifically, we found that P3 cells were overall more proliferative while P2 cells were more migratory. In addition, P2 cells had higher expression of microfilaments, microtubules, and intermediate filaments, with the clear exception of the keratin proteins that were more highly expressed in P3 cells. Finally, we propose that suspension culture may be a better in vitro system to study regenerative processes than collagen gel culture, which is classically used for wound healing studies. While these types of studies lack the complete spectrum of environmental cues present during in vivo regeneration, the limited-factor approach of in vitro studies allows for confirmation of mechanisms thought to be critical in vivo, as well as exploration into new mechanisms and metrics of regeneration.
Determining the keys to regeneration would radically alter medical treatment, yet is still among the greatest biological challenges. Study in nonmammalian species, such as salamanders and frogs, and the few available mammalian models have provided insight into some of the pathways that govern regeneration. It is not possible, however, to perform controlled mechanistic studies during the complicated regenerative process in vivo. Thus, the use of in vitro models that can spatially and temporally control environmental factors will be critical to help translate basic science studies into therapies for regeneration.
Supplementary Material
Acknowledgments
The authors thank Tulane University, the Louisiana Board of Regents (KML), and the National Institutes of Health (No. P20 GM103629) for supporting this work. In addition, the authors thank Dr. Ken Muneoka, Jaime Castillo, and members of the Muneoka laboratory at Tulane University for the generous gift of the P2 and P3 cells.
Disclosure Statement
No competing financial interests exist.
References
- 1.Masaki H. Ide H. Regeneration potency of mouse limbs. Dev Growth Differ. 2007;49:89. doi: 10.1111/j.1440-169X.2007.00909.x. [DOI] [PubMed] [Google Scholar]
- 2.Borgens R.B. Mice regrow the tips of their foretoes. Science. 1982;217:747. doi: 10.1126/science.7100922. [DOI] [PubMed] [Google Scholar]
- 3.Han M. Yang X. Lee J. Allan C.H. Muneoka K. Development and regeneration of the neonatal digit tip in mice. Dev Biol. 2008;315:125. doi: 10.1016/j.ydbio.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernando W.A. Leininger E. Simkin J. Li N. Malcom C.A. Sathyamoorthi S., et al. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev Biol. 2011;350:301. doi: 10.1016/j.ydbio.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas B.S. Conservative management of guillotine amputation of the finger in children. Aust Paediatr J. 1972;8:86. doi: 10.1111/j.1440-1754.1972.tb01793.x. [DOI] [PubMed] [Google Scholar]
- 6.Lee L.P. Lau P.Y. Chan C.W. A simple and efficient treatment for fingertip injuries. J Hand Surg [Br] 1995;20:63. doi: 10.1016/s0266-7681(05)80019-1. [DOI] [PubMed] [Google Scholar]
- 7.Yu L. Han M. Yan M. Lee E.C. Lee J. Muneoka K. BMP signaling induces digit regeneration in neonatal mice. Development. 2010;137:551. doi: 10.1242/dev.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wills A.A. Kidd A.R., 3rd Lepilina A. Poss K.D. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehoczky J.A. Robert B. Tabin C.J. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc Natl Acad Sci U S A. 2011;108:20609. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinkevich Y. Lindau P. Ueno H. Longaker M.T. Weissman I.L. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hechavarria D. Dewilde A. Braunhut S. Levin M. Kaplan D.L. BioDome regenerative sleeve for biochemical and biophysical stimulation of tissue regeneration. Med Eng Phys. 2010;32:1065. doi: 10.1016/j.medengphy.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal V. Johnson S.A. Reing J. Zhang L. Tottey S. Wang G., et al. Epimorphic regeneration approach to tissue replacement in adult mammals. Proc Natl Acad Sci U S A. 2010;107:3351. doi: 10.1073/pnas.0905851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardiner D.M. Ontogenetic decline of regenerative ability and the stimulation of human regeneration. Rejuvenation Res. 2005;8:141. doi: 10.1089/rej.2005.8.141. [DOI] [PubMed] [Google Scholar]
- 14.Kragl M. Knapp D. Nacu E. Khattak S. Maden M. Epperlein H.H., et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y. Wang K. Karapetyan A. Fernando W.A. Simkin J. Han M. Rugg E.L. Muneoka K. Connective tissue fibroblast properties are position-dependent during mouse digit tip regeneration. PLOS ONE. 2013;8:547. doi: 10.1371/journal.pone.0054764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y. Jahagirdar B.N. Reinhardt R.L. Schwartz R.E. Keene C.D. Ortiz-Gonzalez X.R., et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 17.Pineda E. Nerem R. Ahsan T. Differentiation patterns of pluripotent stem cells in two and three dimensional culture. Cells Tissues Organs. 2012 doi: 10.1159/000346166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez P. Bausch A.R. The compaction of gels by cells: a case of collective mechanical activity. Integr Biol (Camb) 2009;1:252. doi: 10.1039/b822897c. [DOI] [PubMed] [Google Scholar]
- 19.Han M. Yang X. Farrington J.E. Muneoka K. Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development. 2003;130:5123. doi: 10.1242/dev.00710. [DOI] [PubMed] [Google Scholar]
- 20.Bryant S.V. Gardiner D.M. Muneoka K. Limb development and regeneration. Am Zool. 1987;27:675. [Google Scholar]
- 21.Broughton G., 2nd Janis J.E. Attinger C.E. The basic science of wound healing. Plast Reconstr Surg. 2006;117:12S. doi: 10.1097/01.prs.0000225430.42531.c2. [DOI] [PubMed] [Google Scholar]
- 22.Bryant S.V. Endo T. Gardiner D.M. Vertebrate limb regeneration and the origin of limb stem cells. Int J Dev Biol. 2002;46:887. [PubMed] [Google Scholar]
- 23.Yokoyama H. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev Growth Differ. 2008;50:13. doi: 10.1111/j.1440-169X.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 24.Han M. Yang X. Taylor G. Burdsal C.A. Anderson R.A. Muneoka K. Limb regeneration in higher vertebrates: developing a roadmap. Anat Rec B New Anat. 2005;287:14. doi: 10.1002/ar.b.20082. [DOI] [PubMed] [Google Scholar]
- 25.Tomasek J.J. Gabbiani G. Hinz B. Chaponnier C. Brown R.A. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 26.Wallace D.G. Rosenblatt J. Collagen gel systems for sustained delivery and tissue engineering. Adv Drug Deliv Rev. 2003;55:1631. doi: 10.1016/j.addr.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Huang H. Kamm R.D. Lee R.T. Cell mechanics and mechanotransduction: pathways, probes, and physiology. Am J Physiol Cell Physiol. 2004;287:C1. doi: 10.1152/ajpcell.00559.2003. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz M.A. DeSimone D.W. Cell adhesion receptors in mechanotransduction. Curr Opin Cell Biol. 2008;20:551. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grinnell F. Fibroblasts, myofibroblasts, and wound contraction. J Cell Biol. 1994;124:401. doi: 10.1083/jcb.124.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alenghat F.J. Ingber D.E. Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002;2002:pe6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- 31.Singer A.J. Clark R.A. Cutaneous wound healing. N Engl J Med. 1999;341:738. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 32.Harty M. Neff A.W. King M.W. Mescher A.L. Regeneration or scarring: an immunologic perspective. Dev Dyn. 2003;226:268. doi: 10.1002/dvdy.10239. [DOI] [PubMed] [Google Scholar]
- 33.Gurtner G.C. Werner S. Barrandon Y. Longaker M.T. Wound repair and regeneration. Nature. 2008;453:314. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.