Abstract
Nitric oxide and its derivative peroxynitrites are generated by host defense system to control bacterial infection. However certain Gram positive bacteria including Staphylococcus aureus possess a gene encoding nitric oxide synthase (SaNOS) in their chromosome. In this study it was determined that under normal growth conditions, expression of SaNOS was highest during early exponential phase of the bacterial growth. In oxidative stress studies, deletion of SaNOS led to increased susceptibility of the mutant cells compared to wild-type S. aureus. While inhibition of SaNOS activity by the addition of L-NAME increased sensitivity of the wild-type S. aureus to oxidative stress, the addition of a nitric oxide donor, sodium nitroprusside, restored oxidative stress tolerance of the SaNOS mutant. The SaNOS mutant also showed reduced survival after phagocytosis by PMN cells with respect to wild-type S. aureus.
1. Introduction
Staphylococcus aureus is a Gram-positive bacterial pathogen that colonizes anterior nares and mucosal surfaces in humans and is responsible for causing a wide array of diseases from mild skin infections to life-threatening conditions such as bacteremia, pneumonia, and endocarditis [1–4]. The emerging resistant strains of S. aureus exacerbate efforts to control or properly treat staphylococcal infections [5].
The host immune system responds to bacterial infections in a concerted manner to eliminate this pathogen. This involves recruitment of polymorphonuclear leukocytes and macrophages to the site of infection and ingestion of invading bacteria. Uptake of bacteria triggers oxygen-dependent and oxygen-independent microbicidal pathways in the phagocytic cells. The oxygen-dependent pathway generates superoxide anion (O2 −) that serves as a precursor for additional reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), hydroxyl radical, singlet oxygen, hypochlorous acid (HOCl), and peroxynitrite [6–9].
S. aureus utilizes various strategies to defend itself against host immune attack. It produces antioxidant enzymes such as superoxide dismutase that converts superoxide anion to H2O2, catalase that converts H2O2 to water and oxygen, and alkyl hydroperoxide reductases that detoxify H2O2, peroxynitrites and hydroperoxides [10, 11]. In addition to their ability to protect from host's oxidants, S. aureus infections impose oxidative stress in a host [12]. During infection with a methicillin resistant S. aureus strain, host neutrophils respond by an increase in nitric oxide production [12]. Nitric oxide (NO) is a free radical synthesized by nitric oxide synthase.
Certain Gram-positive bacteria express homologs of nitric oxide synthases (NOS) that have been extensively studied in eukaryotic species. In these species, NOS-derived nitric oxide (NO) is involved in vasodilation, neurotransmission, and host defense [7, 13, 14], but the functions of bacterial NOS are still being defined. Recent genome sequencing has revealed that NOS-like protein exists in many bacteria including Streptomyces (StNOS), Deinococcus (DrNOS), Staphylococcus (SaNOS), and Bacillus (BsNOS) species [15]. Bacterial NOS enzymes are homologous with the mammalian NOS, but lack an associated NOS reductase and N-terminal β-hairpin hook that binds Zn2+, the dihydroxypropyl side chain of H4B, and the adjacent subunit of the oxygenase dimer [15–18].
It has also been reported that in Bacillus subtilis, NO protects bacterial cells from reactive oxygen species [19]. In addition, the in vivo survival of Bacillus anthracis was dependent on its own NOS activity [20]. NOS activity was also shown to protect from oxidative stress, and deletion of the gene encoding NOS reduced the virulence of a methicillin resistant S. aureus [21]. In this study, SaNOS-derived NO was seen to be protective in a methicillin sensitive S. aureus from lethal oxidative stress conditions, suggesting its moderate role in stress tolerance.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
All experiments were carried out using the methicillin sensitive S. aureus strain SH1000 (wild-type) [22], its isogenic SaNOS deletion mutant, and the mutant complemented with SaNOS in trans. Bacterial cultures were grown in tryptic soy broth/agar (TSB/TSA; Becton Dickinson) at 37°C in a shaking (220 rpm) or static incubator. When needed, tetracycline (10 μg mL−1) and chloramphenicol (10 μg mL−1) were added to the growth medium.
2.2. DNA Manipulations and Analysis
Plasmid DNA was isolated using the Qiaprep kit (Qiagen Inc.); chromosomal DNA was isolated using a DNAzol kit (Molecular Research Center) from lysostaphin-treated S. aureus cells as per the manufacturer's instructions. All restriction and modification enzymes were purchased from Promega. DNA manipulations were carried out using standard procedures. PCR was performed with the PTC-200 Peltier Thermal Cycler (MJ Research). Oligonucleotide primers were obtained from Sigma Genosys.
2.3. Construction of SaNOS Mutant
To construct a mutation in the SaNOS gene, primers P1 (5′-ACGAATTCTGCTAGCCTTTGTTG-3′) and P2 (5′-GGATCCCAAAATAAACGACCAATGC-3′) were used to amplify an 831 bp DNA fragment using genomic DNA from S. aureus strain SH1000 as the template. This amplicon represents SaNOS left flanking fragment (starting 207 nt downstream of the SaNOS start codon and going upstream). Another set of primers, P3 (5′-GGATCCATTATCTCCAACATTG-3′) and P4 (5′-TCTAGAATCAGCCTGAACGAAAAATCG-3′), was used to amplify an 850 bp DNA fragment representing SaNOS right flanking fragment (starting 120 nt upstream of the SaNOS stop codon and going downstream). These two fragments were ligated together into vector pTZ18R [23] and a unique BamHI site was engineered between the ligated fragments. To the BamHI site of this fragment (lacking most of the SaNOS gene; 750 nt out of a total of 1074 nt of the SaNOS gene), a 2.2 kb tetracycline resistance cassette was cloned. The resulting construct was used as a suicidal plasmid to transform S. aureus RN4220 cells by electroporation. Transformants were selected on TSA plates containing 10 μg mL−1 tetracycline that led to a single crossover event where the mutated SaNOS from the plasmid was integrated into the bacterial genome leaving the wild-type SaNOS intact. These merodiploids were used to resolve the mutation in the SaNOS gene using a phage 80α transduction procedure as described previously [24, 25]. Mutation in the SaNOS was verified by PCR. For genetic complementation of the SaNOS mutant, a 2.4 kb DNA fragment was PCR amplified using primers P1 and P4 and S. aureus SH1000 genomic DNA as template. The amplicon represents a fragment starting from 624 nt upstream and spanning 730 nt downstream of the SaNOS gene that was cloned into the shuttle plasmid pCU1 [26] and subsequently transferred to the SaNOS mutant of S. aureus strain SH1000.
2.4. Quantitative Real-Time RT-PCR (qRT-PCR) Assays
qRT-PCR assays were carried out as described [27] using primers P5 (ATGGTGCTAAAATGGCTTGGC) and P6 (GCTTCGTCAGTAACATCTCTTG) to determine optimum expression of SaNOS during different stages of S. aureus growth in TSB. Bacterial cells were harvested from early- (OD600 = 0.6), mid- (OD600 = 1.8), late-exponential (OD600 = 3.0), and stationary (OD600 = 4.2) phase cultures. Total RNA extracted from these cells was used in qRT-PCR assays as described [27].
2.5. Determination of Nitric Oxide Synthase Activity
Total protein was extracted from lysostaphin treated S. aureus cells grown to OD600 = 0.6 as described previously [28]. The NOS activity was determined using NOS activity assay kit (Cayman Chemical Company) and radioactive 3H arginine monohydrochloride as substrate (Amersham Biosciences).
2.6. Determination of H2O2 Susceptibility
For these studies, S. aureus cells from early exponential phase cultures OD600 = 0.6 were treated with 350 mM H2O2 for 30 min. The surviving bacteria were enumerated by serial dilution and plating on TSA agar plates. L-arginine serves as a substrate for the nitric oxide synthase in the production of NO. Wild-type S. aureus cultures in TSB were added with L-arginine (1 mM final concentration) at OD600 = 0.5 and subsequently at OD600 = 0.6 were stressed with 350 mM H2O2 to determine if the addition of L-arginine affected NO production and the oxidative stress tolerance. Additionally, the wild-type S. aureus cells were collected from cultures grown to OD600 = 0.3 and were resuspended in similar volume of TSB containing 5 mM L-NAME (Tocris Bioscience), an inhibitor of NOS activity. At an OD600 = 0.6, these NOS-inhibited cells were stressed with 350 mM H2O2 for 30 min and the surviving bacteria were counted. To further ascertain the role of nitric oxide in the protection of S. aureus cells, the SaNOS mutant cells at OD600 = 0.5 were treated with 2.5 mM concentration of an NO donor, sodium nitroprusside (SNP) (Sigma). At OD600 = 0.6, these SNP-treated cells were stressed with 350 mM H2O2 for 30 min, and the surviving bacteria were counted.
2.7. Phagocytic Killing of S. aureus SaNOS Mutant
The promyelocytic HL-60 cells (ATCC) were grown in Iscove's Modified Dulbecco's Medium (IMDM) (ATCC) with 20% fetal bovine serum (Fisher) and were treated with 1.3% DMSO (Fisher) for 5 days to induce their differentiation into neutrophil-like cells [29, 30]. Morphology of differentiated cells was confirmed by Giemsa staining under inverted microscope. The oxidative burst inside neutrophil cells was determined by the reduction of nitroblue tetrazolium. The differentiated neutrophils were used for phagocytic killing using a method described previously [9] with slight modification. In brief, the neutrophils (1 × 106) were added with S. aureus cells (2.5 × 106) (MOI 1 : 2.5) in a 24-well plate. The plate was centrifuged at 4000 rpm for 10 min and incubated in a CO2 incubator at 37°C for 1 h. The supernatant was gently aspirated and the neutrophils were lysed by the addition of IMDM containing 0.025% Triton X-100. The number of surviving bacteria was enumerated by making serial dilutions and plating of this lysate on TSA plate.
2.8. Statistical Analysis
All results are reported as the mean ± SD of at least three independent experiments. Data were analyzed with Dunnett's Method in one-way analysis of variance or with Student-Newman-Keuls Method in two-way analysis of variance using statistical analysis computer programs (SigmaPlot for Windows, version 12.0, Systat Software, Inc.). Statistical significance was set at P < 0.05.
3. Results and Discussion
3.1. Construction of SaNOS Deletion Mutant in S. aureus
To investigate the role of the S. aureus nitric oxide synthase and NO produced by this enzyme, the SaNOS gene was deleted and replaced with a tetracycline cassette by site-directed mutagenesis. The deletion of SaNOS gene was confirmed by PCR (Figure 1).
Figure 1.
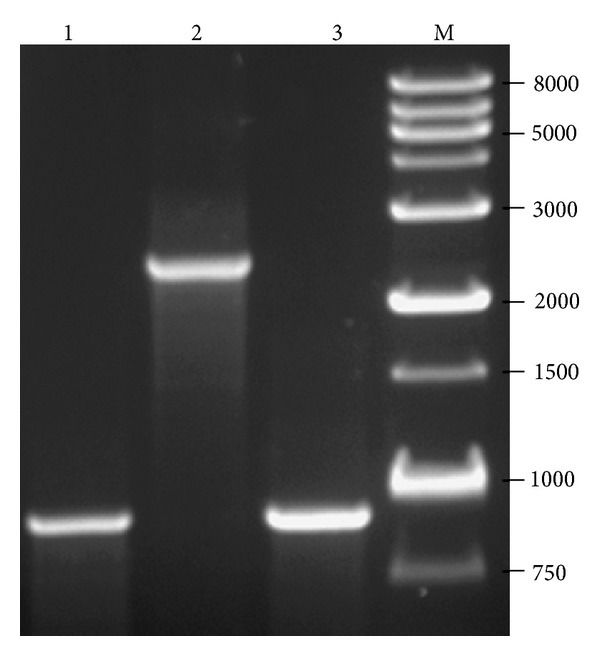
PCR verification of a mutation in the SaNOS gene in S. aureus. Primers P7 (5′-ATACAGAAGAAGAACTTATTTATGG-3′) and P8 (5′- CACCTCTACTAACTTAATGATGG-3′) were used in the PCR that allowed amplification of a 963 bp product (lane 1) when genomic DNA from wild-type S. aureus strain SH1000 was used. These primers amplified a ~2.4 kb fragment when genomic DNA from the SaNOS mutant of S. aureus strains SH1000 was used as template (lanes 2). Lane 3: PCR product when genomic DNA from the SaNOS mutants of S. aureus strains SH1000 complemented in trans with SaNOS was used as template. The larger PCR product is not seen because of complementation with wild-type SaNOS gene on a high copy plasmid pCU1. Lane M: DNA ladder.
3.2. Expression of SaNOS and NOS Enzymatic Activity in S. aureus
In qRT-PCR assays, maximum expression of SaNOS in strain SH1000 was determined during the early stage of the bacterial growth (Table 1). The expression of SaNOS declined dramatically during the late stages of the bacterial growth and was least during the stationary phase (Table 1). A higher bacterial NO production was also noted during the early stages of macrophage infection by B. anthracis [19]. The determination of NOS activity, based on the conversion of L-arginine to citrulline, indicated that SaNOS was functional and was able to use L-arginine as the substrate (Table 2). The level of citrulline in the SaNOS mutant was similar or below the background level; a reaction mixture that contained only the L-arginine substrate and no protein extract was added to this reaction mixture (Table 2). The complementation of the SaNOS mutant with SaNOS gene on a high copy plasmid led to a significant increase in the NOS activity in this complemented strain (Table 2). Similar NOS activities in these strains were also verified by measuring the nitrite and nitrate levels using Griess reagent (data not shown).
Table 1.
Expression of SaNOS in S. aureus during different phases of growth.
| Growth stage | SaNOS expression* |
|---|---|
| Early-exponential | 100% |
| Mid-exponential | 19.48% |
| Late-exponential | 10.73% |
| Stationary | 4.90% |
*Expression of SaNOS is shown relative to its transcript level during early-exponential phase of growth.
Table 2.
Nitric oxide synthase activity in different S. aureus strains.
| Strain | NOS activity (%)* |
|---|---|
| SH1000 | 3.95 ± 1.61 |
| SH1000ΔSaNOS | 0 |
| Complemented strain | 26.47 ± 3.95 |
*%Citrulline formed in relation to total L-arginine used in the assay. Citrulline conversion in the mutant strain was below the background level (control reaction with no protein extract). Values represent average of three independent experiments ± standard deviation.
3.3. Lack of SaNOS in S. aureus Reduces Its Survival under Oxidative Stress
The impact of the deletion of SaNOS was investigated for its growth in TSB. There was no change in the growth of the mutant strain and it was comparable to the growth of the wild-type S. aureus (data not shown). Under stress conditions such as salt (1.5 mM NaCl) and pH (6.0 or 8.5), the growth rate of the SaNOS was comparable to the growth rate of the wild-type S. aureus (data not shown). Also, in the presence of 1.1 mM H2O2, the growth of the SaNOS mutant of S. aureus SH1000 was comparable to the wild-type strain (data not shown). However, it has been shown that the priming of the B. subtilis cells with nitric oxide for 5 sec leads to a significant increase in their resistance to the exposure of a much higher H2O2 concentration (370 mM) [19].
In qRT-PCR assays, maximum expression of SaNOS was determined in the cells from the early exponential phase (OD600 = 0.6). Thus, cultures at this density were used in H2O2 susceptibility assays. When wild-type and the SaNOS mutant cells were treated with a lethal dose of 350 mM H2O2, there were significantly more surviving wild-type bacteria (>1000-fold) compared to the SaNOS mutant bacteria under identical experimental conditions (Figure 2(a)). Addition of L-arginine is expected to increase the production of nitric oxide and thus is expected to also increase the resistance of S. aureus cells grown in the presence of L-arginine. Addition of L-arginine indeed increased the resistance of the wild-type S. aureus cells but caused no increase in the survival of the SaNOS mutant (Figure 2(a)). Complementation of SaNOS mutant with the SaNOS gene on a plasmid partially restored the ability of these bacteria to survive H2O2 stress when it was grown with or without L-arginine (Figure 2(a)). When the NOS activity was inhibited in the wild-type S. aureus by the addition of L-NAME, a competitive inhibitor of the NOS enzymatic activity, it dramatically reduced the bacterial survival (Figure 2(b)) under oxidative stress. In addition, when sodium nitroprusside (an NO donor) was added to the SaNOS mutant cells, there was significant increase (>300-fold) in the survival of the mutant bacteria when they were exposed to H2O2 (Figure 2(c)). These results, collectively, suggest the role of a functional nitric oxide synthase in the protection of S. aureus cells from oxidative stress conditions.
Figure 2.
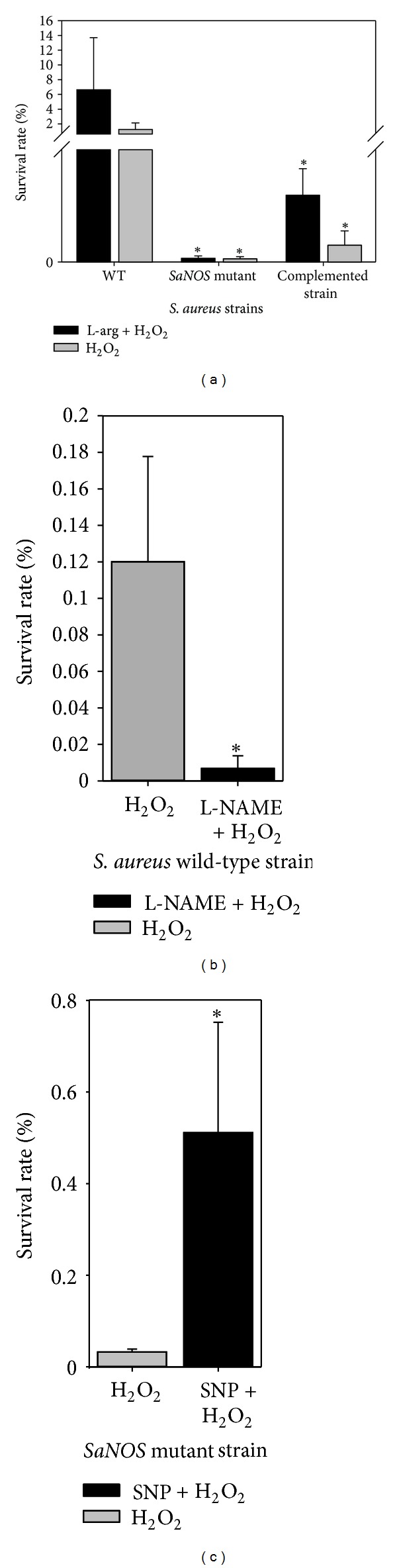
(a) Survival of S. aureus SH1000, its isogenic SaNOS mutant, and the mutant complemented with SaNOS gene in trans from a lethal dose (350 mM) of H2O2 with and without supplementation with 1 mM L-arginine. (b) Survival of wild-type S. aureus SH1000 pretreated with 5 mM L-NAME from 350 mM H2O2. (c) Survival of SaNOS mutant of S. aureus SH1000 pre-treated with 2.5 mM sodium nitroprusside from 350 mM H2O2. *Significant at P < 0.05.
3.4. Phagocytic Killing of the SaNOS Mutant
Neutrophils are a critical component of innate immunity and are essential in controlling bacterial infections in a host. Experiments were carried out to determine if the lack of a functional NOS decreased the survival of the S. aureus bacteria when it was allowed to interact with neutrophils. In these experiments, the SaNOS mutant showed significantly reduced survival compared to the wild-type S. aureus (Figure 3). These SaNOS mutant bacteria were also used to determine their survival compared to wild-type S. aureus in a murine intraperitoneal model as described previously [24, 25]. However, there was no decrease in the survival of the SaNOS mutant when compared to the wild-type S. aureus bacteria (data not shown). The ability of the SaNOS mutant cells to make biofilms was also comparable to the wild-type S. aureus cells (data not shown).
Figure 3.
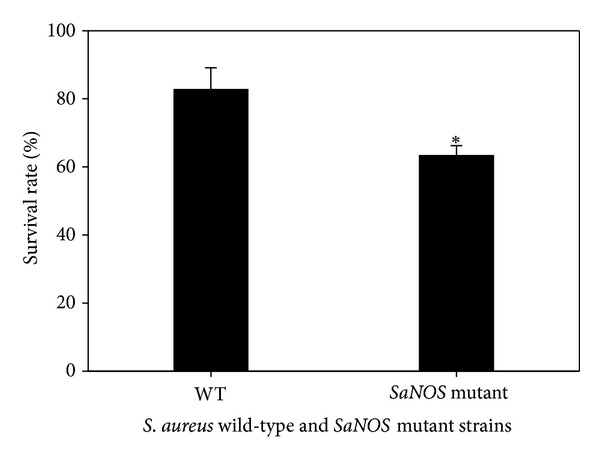
S. aureus survival in neutrophil cells. Neutrophil cells were infected (MOI 1 : 2.5) with wild-type S. aureus SH1000 and its isogenic SaNOS mutant for 1 h at 37°C and then plated on TSA plate. *Significant at P < 0.05.
In recent years, the presence of NOS has been viewed with great interest for its role in bacterial physiology and virulence. Presence of NOS was determined to be a key factor in the defense of B. subtilis and B. anthracis from reactive oxygen species generated by the neutrophils and macrophages [19, 20]. It was shown that exposure to nitric oxide enhanced catalase activity in B. subtilis [19]. We observed a slight reduction in catalase activity in the SaNOS mutant relative to its level in the wild-type S. aureus (data not shown). S. aureus bacteria are known to produce a very high level of catalase activity. A lower level of superoxide dismutase activity was determined in the SaNOS mutant of a methicillin resistant S. aureus [21]. The reduced catalase and superoxide dismutase activity levels might be the reasons of the reduced survival of the SaNOS mutant under oxidative stress. Lack of the ability of the S. aureus cells to produce NO increased the susceptibility to reactive oxygen species and host antimicrobial peptides [21]. The level of the expression of the staphylococcal NOS was induced by exposure to cell wall-active antibiotics and it was also determined to be a factor in conferring resistance to these antibiotics in a methicillin resistant S. aureus [21]. Surprisingly, in that study, the lack of a functional NOS increased the resistance of S. aureus to aminoglycosides [21].
Studies utilizing a methicillin resistant S. aureus showed reduced virulence subsequent to NOS deletion [21]. Infection with the mutant cells resulted in smaller abscess formation compared to the S. aureus cell with a functional NOS suggesting its role in staphylococcal virulence [21]. In our studies that utilized a methicillin sensitive S. aureus, there was no difference in the survival of the SaNOS mutant in a mouse. There was also no appreciable difference in the survival or growth of the SaNOS mutant of S. aureus SH1000 under mild stress conditions. The difference in the survival was only detected when the SaNOS mutant and the wild-type bacteria were exposed to a lethal dose of H2O2. The reduction in virulence of S. aureus subsequent to SaNOS deletion in the recent report [21] can be attributed to strain differences (methicillin-resistant versus methicillin-sensitive S. aureus) and to a difference in the type of animal model used to study the virulence. These strain differences are significant as host neutrophils respond differently when they are exposed to methicillin-resistant S. aureus compared to during infection with methicillin-sensitive S. aureus [12]. NO production decreased in neutrophils in mice infected with vancomycin sensitive S. aureus and exposed to vancomycin but the decrease in neutrophilic NO production was insignificant when the mice were infected with vancomycin resistant S. aureus and exposed to vancomycin [12].
During the phagocytic process to control bacterial infections, the respiratory burst generates two very potent toxic substances, H2O2 and superoxide anions (O2 −). A model has been proposed describing how bacterial NO might be protective from the toxic action of these reactive oxygen species [19, 20]. It is suggested that the O2 − fails to cross the bacterial cell wall and membrane and limits the production of peroxynitrites inside the bacterial cell from a reaction between bacterial NO and phagocytic O2 −. Although H2O2 can diffuse inside the bacterial cell, a higher bacterial catalase should degrade it to protect the bacterial cells from any damage.
Considering the fact that the SaNOS was seen to be significant only during extreme conditions of stress and has a varied role in antibiotic stress tolerance and virulence, more studies need to be carried out to determine the significance of this enzyme in S. aureus.
Conflict of Interests
The authors do not have any conflict of interests with the content of the paper.
Acknowledgments
The authors thank Deborah Hudman for her valuable assistance with statistical analysis. This work was supported by Grant 1R15AI090680-01 from the National Institutes of Health to V. K. Singh.
References
- 1.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) Current Opinion in Microbiology. 2012;15(5):588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Stenehjem E, Rimland D. MRSA nasal colonization burden and risk of MRSA infection. American Journal of Infection Control. 2012 doi: 10.1016/j.ajic.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins RR, David MZ, Salata RA. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus . Journal of Medical Microbiology. 2012;61, part 9:1179–1193. doi: 10.1099/jmm.0.043513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowy FD. Medical progress: Staphylococcus aureus infections. The New England Journal of Medicine. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 5.Rivera AM, Boucher HW. Current concepts in antimicrobial therapy against select gram-positive organisms: methicillin-resistant Staphylococcus aureus, penicillin-resistant pneumococci, and vancomycin-resistant enterococci. Mayo Clinic Proceedings. 2011;86(12):1230–1243. doi: 10.4065/mcp.2011.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nature Reviews Microbiology. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari CK, Souto PC, França EL, Honorio-França AC. Oxidative and nitrosative stress on phagocytes' function: from effective defense to immunity evasion mechanisms. Archivum Immunologiae et Therapia Experimentalis. 2011;59(6):441–448. doi: 10.1007/s00005-011-0144-z. [DOI] [PubMed] [Google Scholar]
- 8.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annual Review of Immunology. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 9.Voyich JM, Braughton KR, Sturdevant DE, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. Journal of Immunology. 2005;175(6):3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 10.DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infectious Disease Clinics of North America. 2009;23(1):17–34. doi: 10.1016/j.idc.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu GY. Molecular pathogenesis of Staphylococcus aureus infection. Pediatric Research. 2009;65(5, part 2):71R–77R. doi: 10.1203/PDR.0b013e31819dc44d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty SP, Pramanik P, Roy S. Staphylococcus aureus infection induced oxidative imbalance in neutrophils: possible protective role of nanoconjugated vancomycin. ISRN Pharmacol. 2012;2012 doi: 10.5402/2012/435214.435214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopincova J, Puzserova A, Bernatova I. Biochemical aspects of nitric oxide synthase feedback regulation by nitric oxide. Interdisciplinary Toxicology. 2011;4(2):63–68. doi: 10.2478/v10102-011-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariotto S, Menegazzi M, Suzuki H. Biochemical aspects of nitric oxide. Current Pharmaceutical Design. 2004;10(14):1627–1645. doi: 10.2174/1381612043384637. [DOI] [PubMed] [Google Scholar]
- 15.Crane BR. The enzymology of nitric oxide in bacterial pathogenesis and resistance. Biochemical Society Transactions. 2008;36, part 6:1149–1154. doi: 10.1042/BST0361149. [DOI] [PubMed] [Google Scholar]
- 16.Bird LE, Ren J, Zhang J, et al. Crystal structure of SANOS, a bacterial nitric oxide synthase oxygenase protein from Staphylococcus aureus . Structure. 2002;10(12):1687–1696. doi: 10.1016/s0969-2126(02)00911-5. [DOI] [PubMed] [Google Scholar]
- 17.Brunel A, Santolini J, Dorlet P. Electron paramagnetic resonance characterization of tetrahydrobiopterin radical formation in bacterial nitric oxide synthase compared to mammalian nitric oxide synthase. Biophysical Journal. 2012;103(1):109–117. doi: 10.1016/j.bpj.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pant K, Bilwes AM, Adak S, Stuehr DJ, Crane BR. Structure of a nitric oxide synthase heme protein from Bacillus subtilis . Biochemistry. 2002;41(37):11071–11079. doi: 10.1021/bi0263715. [DOI] [PubMed] [Google Scholar]
- 19.Gusarov I, Nudler E. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shatalin K, Gusarov I, Avetissova E, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(3):1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Sorge NM, Beasley FC, Gusarov I, et al. Methicillin-resistant Staphylococcus aureus bacterial nitric oxide synthase affects antibiotic sensitivity and skin abscess development. The Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.M112.448738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. δb modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. Journal of Bacteriology. 2002;184(19):5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mead DA, Szczesna-Skorupa E, Kemper B. Single-stranded DNA “blue” t7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Engineering, Design and Selection. 1986;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- 24.Singh VK, Hattangady DS, Giotis ES, et al. Insertional inactivation of branched-chain α-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Applied and Environmental Microbiology. 2008;74(19):5882–5890. doi: 10.1128/AEM.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus . Microbiology. 2007;153(9):3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 26.Augustin J, Rosenstein R, Wieland B, et al. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. European Journal of Biochemistry. 1992;204(3):1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 27.Singh VK, Syring M, Singh A, Singhal K, Dalecki A, Johansson T. An insight into the significance of the DnaK heat shock system in Staphylococcus aureus . International Journal of Medical Microbiology. 2012;302(6):242–252. doi: 10.1016/j.ijmm.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Singh VK, Jayaswal RK, Wilkinson BJ. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiology Letters. 2001;199(1):79–84. doi: 10.1111/j.1574-6968.2001.tb10654.x. [DOI] [PubMed] [Google Scholar]
- 29.Collins SJ. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood. 1987;70(5):1233–1244. [PubMed] [Google Scholar]
- 30.Tarella C, Ferrero D, Gallo E. Induction of differentiation of HL-60 cells by dimethyl sulfoxide: evidence for a stochastic model not linked to the cell division cycle. Cancer Research. 1982;42(2):445–449. [PubMed] [Google Scholar]


