Abstract
Background
Acinetobacter baumannii is an opportunistic microorganism with an increasing role in nosocomial outbreaks. For the last 2 decades, a growing number of carbapenem-resistant A. baumannii strains have been identified, including the metallo-beta-lactamases (MBLs) producers. The study aimed to investigate the genetic relatedness of, and MBLs production among, a collection of A. baumannii isolates from Poland.
Material/Methods
This study involved 78 clinical isolates of carbapenem-resistant A. baumannii. Strain typing of the isolates was performed using PCR-RAPD. The presence of MBLs was phenotypically determined using different double disc synergy tests (DDST), the imipenem/EDTA combination disk test (CDT) and Etest MBL. blaIMP and blaVIM genes were detected using a duplex PCR assay.
Results
The isolates were divided into 18 PCR-RAPD patterns. Among 18 examined isolates, 94.4% were MBL-positive by the phenotypic method relying on comparing the bacteria growth inhibition zones diameters between imipenem/EDTA and imipenem discs, 88.9% using Etest MBL, 66.7% using the double disc synergy test with ceftazidime, imipenem, meropenem and EDTA, and 88.9% using a corresponding method with 2-MPA. The existence of blaIMP was identified in 8 (10.3%) strains.
Conclusions
MBLs production was an important mechanism of carbapenem resistance among A. baumannii isolates in Poland. Laboratories should routinely screen for MBLs among A. baumannii isolates.
Keywords: Acinetobacter baumannii, carbapenem-resistant Gram-negative rods, carbapenem resistance detection, metallo-beta-lactamases (MBLs)
Background
Acinetobacter baumannii significantly contributes to nosocomial infections [1]. The increasing rate of occurrence is mainly due to the ability of A. baumannii to survive in different environments, including nutritionally poor ones, and ability to acquire different antimicrobial resistance genes [2,3]. For the last 2 decades, increasing numbers of carbapenem-resistant A. baumannii strains have been reported worldwide [4]. Resistance to carbapenems may result from metallo-beta-lactamases (MBLs) synthesis, hydrolyzing almost all beta-lactam antibiotics. MBLs are not inhibited by classic beta-lactams inhibitors (clavulanic acid, tazobactam, sulbactam). However, MBLs use Zn2+ ions as cofactors and can therefore be inhibited in vitro by EDTA [5]. A. baumannii mainly produces the IMP-, VIM- and SIM-like MBLs enzymes [4]. NDM-1 has been also recently been described in A. baumannii[6]. MBLs-positive A. baumannii strains are a serious therapeutic problem due to co-resistance to numerous antibiotic groups and difficulties in treating the infections caused by them.
The aim of this study was to evaluate the antimicrobial susceptibility patterns of A. baumannii isolates obtained from Poland and to estimate their genetic relatedness and ability to synthesize MBLs.
Material and Methods
Bacterial strains
The study included 78 A. baumannii clinical isolates resistant to at least 1 carbapenem, collected between 2008 and 2009 from patients hospitalized at Dr. Antoni Jurasz University Hospital No. 1 in Bydgoszcz, Poland. One environmental isolate derived from the same hospital was also included. Identification of the isolates to the species level was done using the ID 32 E and API 20 NE (bioMérieux) tests, according to the manufacturer’s recommendations.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing for meropenem, imipenem, gentamicin, amikacin, tobramycin, ciprofloxacin, levofloxacin, and trimethoprim/sulfamethoxazole was performed by the agar disc diffusion method according to the recommendations of the National Reference Centre for Antimicrobial Susceptibility Testing. Resistance to imipenem, meropenem and doripenem was also determined using Etest (bioMérieux) according to the manufacturer’s instructions. Results were interpreted using clinical breakpoints as defined by the Clinical and Laboratory Standards Institute [7,8].
PCR-RAPD typing
DNA for PCR-RAPD typing purpose was isolated using GeneMATRIX Bacterial & Yeast Genomic DNA Purification Kit (EURx). PCR-RAPD reaction was conducted using the method previously described by Carr et al. with GCTTGTGAAC primer [9]. Final volume of the reaction (25 μl) consisted of 0.4 μl of Taq DNA polymerase (5 U/μl, Solis BioDyne), 2.5 μl of buffer B (0.8 M Tris-HCl, 0.2 M (NH4)2SO4, 0.2% w/v Tween-20, Solis BioDyne), 3.5 μl of MgCl2 (25mM, Solis BioDyne), 0.25 μl of dNTP mixture (20 mM, Solis BioDyne), 1.8 μl of 10 × primer (100 pmol/μl, Genomed), 15.55 μl of water (Molecular Biology Grade Water, Eppendorf) and 1 μl of bacterial DNA. Amplification was done in a GeneAmp PCR System 2700 thermocycler (Applied Biosystems) according to the following temperature profile: first denaturation step at 92°C for 30 seconds, 34 cycles, each consisting of 3 steps at 92°C denaturation for 30 seconds, annealing at 40°C for 1 minute and primer extension at 72°C for 3 minutes. The final step included 30 seconds at 92°C, 1 minute at 40°C, and the final extension of the primer for 10 minutes at 72°C. PCR-RAPD reaction products were separated by electrophoresis in 2% agarose gel (1xTBE) at 13.5 V/cm for 2.5 hours in the SUB-CELL® GT (BioRad) apparatus. After the electrophoresis step, gel was stained with ethidium bromide solution for 20 minutes, washed for 20 minutes in deionized water and observed in UV light. Molecular sizes of the obtained PCR-RAPD reaction products were compared to molecular size marker 100–3000 bp (Solis BioDyne). Visualizations of the stained gel were collected on Gel Doc 2000 system and Quantity One program (BioRad).
Detection of MBLs
MBLs activity was phenotypically investigated using: (i) different double disk synergy tests for ceftazidime, imipenem, and meropenem, along with EDTA or 2-mercaptopropionic acid (2-MPA) discs (DDST); (ii) imipenem/EDTA combination disc test (CDT); and (iii) Etest MBL (bioMérieux). Bacterial suspension of 0.5 MacFarland was placed on Mueller-Hinton agar in all MBLs detection methods. In double disc synergy tests with EDTA and 2-MPA, ceftazidime (30 μg), imipenem (10 μg) and meropenem (10 μg) discs were placed 2 cm apart from the sterile disc with EDTA and 2-MPA addition, respectively. Bacteria inhibition zone deformation or enhancement for any used antibiotic disc from the side of MBLs inhibitor was interpreted separately as the A. baumannii strain’s ability to MBLs production in disc diffusion methods. At least 5 mm bacteria inhibition zone size enhancement for imipenem impregnated with EDTA was also interpreted as MBLs-positive strain in the method of comparison inhibition zones diameters. Using Etest MBL imipenem and imipenem/EDTA MIC values ratio higher or equal to 8, or deformation of the inhibition zone, was interpreted as a positive result. P. aeruginosa strains producing IMP- or VIM-like MBLs (carrying blaIMP or blaVIM genes) and ATCC 27853 served as positive and negative controls of the reaction, respectively. A duplex PCR assay was performed to detect the occurrence of IMP- and VIM-like MBLs genes as previously described [10]. PCR reaction products were separated by electrophoresis on 1% agarose gel in 1xTBE at 9 V/cm for 1 hour in MINI SUB™ DNA CELL (BioRad) apparatus. Visualizations were recorded and documented in Gel Doc 2000 system using Quantity One (BioRad) program.
Results
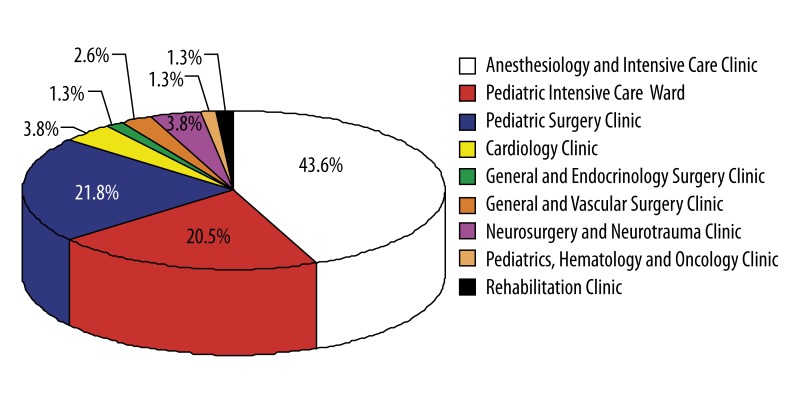
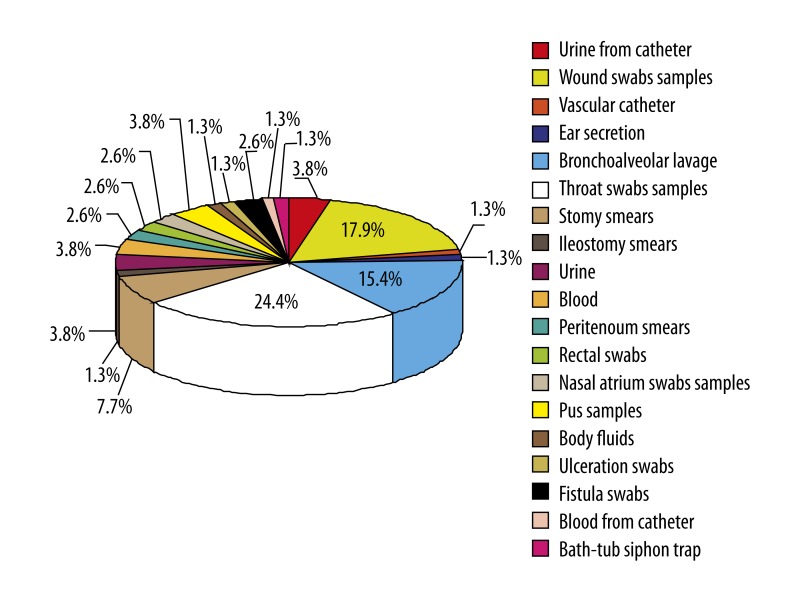
About 85% of the isolates were obtained from patients from the Anesthesiology and Intensive Care Clinic, Pediatric Intensive Care Ward, and Pediatric Surgery Clinic. Most of the isolates were derived from throat and wound swabs and bronchoalveolar lavage (Figures 1, 2). Seventy-seven (~99%) of the isolates were multi-drug resistant (MDR), showing reduced susceptibility to antimicrobial agents from ≥3 classes regularly used for treatment. Resistance to at least 1 carbapenem was confirmed in all the isolates. Among the examined 78 carbapenem-resistant A. baumannii strains, 94.99% were resistant to meropenem, 89.7% to imipenem, and 88.5% to doripenem. All of the examined strains were resistant to amikacin and ciprofloxacin, 98.7% to trimethoprim/sulfamethoxazole, 61.5% to levofloxacin, 53.4% to gentamicin, and 15.4% to netilmicin. Only 1 isolate (~1%) was resistant to tobramycin and colistin.
Figure 1.
Origin of the examined carbapenem-resistant A. baumannii (n=78) strains.
Figure 2.
Clinical material that examined carbapenem-resistant A. baumannii (n=78) strains was isolated from.
The genetic similarity analysis revealed 18 PCR-RAPD patterns (Figure 3). Five of the PCR-RAPD patterns were represented by 1 isolate/pattern, while 13 included ≥2 isolates/pattern.
Figure 3.
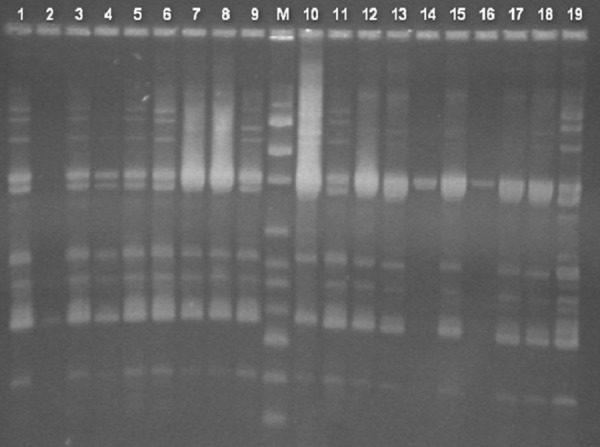
Gel electrophoresis of PCR-RAPD amplification products for the examined A. baumannii strains. M – DNA size marker of 100–3000 bp, lanes 1–19 numbers of examined strains.
Each double disc diffusion method with ceftazidime, imipenem, meropenem and EDTA revealed 66.7% of MBLs-positive strains. The enhancement of bacteria growth inhibition zones was strongest for imipenem and ceftazidime. Using the same method but with 2-MPA, 88.9% of the strains were classified as MBLs-positive and the deformation of the bacteria growth inhibition zones were observed more frequently for imipenem and meropenem. In the comparison method of bacterial growth inhibition zones diameters between imipenem/EDTA and imipenem discs, MBLs synthesis was revealed in 94.4% of the strains. Using Etest MBL, positive results were obtained for 88.9% of strains (Table 1).
Table 1.
Detection of MBLs in 18 A. baumannii isolates from Poland.
| Method | Number/percentage of positive isolates)* | Number/percentage of positive isolates | ||
|---|---|---|---|---|
| IPM | MEM | CAZ | ||
| DDST using IPM, MEM, CAZ and EDTA disks | 10/55.6% | 1/5.6% | 7/38.9% | 12/66.7% |
| DDST using IPM, MEM, CAZ and 2-MPA disks | 12/66.7% | 8/44.4% | 7/38.9% | 16/88.9% |
| CDT | 17/94.4% | |||
| Etest MBL | 16/88.9% | |||
IPM – imipenem; MEM – meropenem; CAZ – ceftazidime.
The presence of IMP-like MBLs coding genes was identified in 8 isolates, while none of the isolates contained the blaVIM-4 gene. The blaIMP-like-positive isolates were mainly derived from patients of the Anesthesiology and Intensive Care Clinic.
Discussion
A. baumannii is an important opportunistic bacterial pathogen responsible for serious infections. The most common are: bacteremias, pneumonias, meningitis, urinary tract and wound infections [1]. Carbapenem-resistant A. baumannii has increasingly become a serious therapeutic problem worldwide [11–13]. In this regard, limited data has been published on carbapenem-resistant A. baumannii in Poland [14].
Genetic similarity analysis by PCR-RAPD revealed 18 different patterns among the isolates. The existence of isolates with undistinguished molecular patterns indicated the occurrence of bacterial spread between patients. The spread between different hospital wards and clinics might be due to transfer of the patients or mediated by health-care personnel and/or medical devices [12]. This idea is supported by the isolation, from a bath-tub siphon trap, of an A. baumannii strain in which PCR-RAPD pattern was observed, as well as strains derived from patients cured in 4 different wards.
Among the strains examined with phenotypic methods, the highest percentage of MBLs producers was obtained using the comparison method of bacterial growth inhibition zones between imipenem/EDTA and imipenem discs. The obtained results indicate high significance of MBLs in occurrence of A. baumannii resistance to carbapenems.
The existence of 18 MBLs-positive A. baumannii isolates was observed using phenotypic methods; 8 (44.4%) of them were confirmed as IMP-like enzymes producers. A study from Korea demonstrated that 48.4% of the A. baumannii isolates resistant to carbapenems produced blaIMP-1, while blaVIM was produced by only 3.2% of the isolates [15]. Another study reported the presence of blaIMP and blaVIM in 61% and 29% of A. baumannii isolates, respectively [16]. A. baumannii isolates with blaIMP-1 gene were also described in Korea and Brasil [17,18]. The lack of blaVIM was similarly observed among isolates from India [19]. Lack of MBLs coding genes blaIMP-1 and blaVIM-4 in the isolates showing positive results in phenotypic methods probably indicated the existence of other genes, like blaSIM[19]. The carbapenem-resistant A. baumannii strains with no phenotypic or genotypic sign of MBLs production may possess other enzymes mediating carbapenem resistance, such as OXA-type lactamases [19]. OXA-type carbapenemases belong to class D according to the classification of Ambler. OXA carbapenemases inactivate penicillins and cephalosporins, and most of them also hydrolyze carbapenems [20]. Naturally occurring OXA carbapenemases are OXA-51-like enzymes. To date, up to 45 variants of OXA-51 have been identified in A. baumannii isolates from medical centers worldwide [21]. Acquired OXA carbapenemases can be divided into 3 clusters, based upon the variant sequence homology: OXA-23, OXA-40, and OXA-58. These groups of enzymes are chromosomally and plasmid encoded [3].
Phenotypic methods have been previously reported to be sensitive for the detection of MBLs in A. baumannii. The technique is very easy and economical and can be incorporated into the routine testing of any busy microbiology laboratory. The genotypic methods are based on analysis of genetic material that is unique and invariable for every organism, in contrast to phenotypic features. PCR method is highly accurate and reliable. MBLs detection using the E-test and other phenotypic methods is not reliable because there can be false-positive results [22].
Conclusions
A. baumannii isolates resistant to carbapenems are mainly obtained from throat and wound swab samples derived from patients of the Anesthesiology and Intensive Care Clinic and Pediatric Surgery Clinic.
The majority of the isolates are susceptible to colistin and tobramycin.
The CDT is the most sensitive of the phenotypic methods used for MBLs detection.
PCR-RAPD demonstrates a high level of genetic diversity among the isolates, although intra-hospital spread of some strains is noted.
MBL-coding genes represent an important mechanism of carbapenem resistance among A. baumannii isolates from Poland.
Footnotes
Source of support: This research was financially supported by the Nicolaus Copernicus University with funds from the maintenance of the research potential of the Department of Microbiology
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez F, Hujer AM, Hujer KM, et al. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garnacho-Montero J, Amaya-Villar R. Multiresistant Acinetobacter baumannii infections: epidemiology and management. Curr Opin Infect Dis. 2010;23:332–39. doi: 10.1097/QCO.0b013e32833ae38b. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826–36. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 5.Laudy AE. Karbapenemazy-enzymy mogące hydrolizować szerokie spektrum beta-laktamów. Zakażenia. 2003;4:32–38. [in Polish] [Google Scholar]
- 6.Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011;66:1255–59. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 7.Gniadkowski M, Żabicka D, Hryniewicz W. Krajowy Ośrodek Referencyjny ds. Lekowrażliwości Drobnoustrojów. Warszawa: 2009. Rekomendacje doboru testów do oznaczania wrażliwości bakterii na antybiotyki i chemioterapeutyki 2009. [in Polish] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Eighteenth informational supplement, M100-S18. CLSI Wayne; Pa: 2008. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 9.Carr E, Eason H, Feng S, et al. RAPD-PCR typing of Acinetobacter isolates from activated sludge systems designed to remove phosphorus microbiologically. J Appl Microbiol. 2001;90:309–19. doi: 10.1046/j.1365-2672.2001.01245.x. [DOI] [PubMed] [Google Scholar]
- 10.Pitout JD, Gregson DB, Poirel L, et al. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–35. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarrilli R, Giannouli M, Tomasone F, et al. Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J Infect Dev Ctries. 2009;3:335–41. doi: 10.3855/jidc.240. [DOI] [PubMed] [Google Scholar]
- 12.Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–62. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 13.Gordon NC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents. 2010;35:219–26. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Wróblewska MM, Towner KJ, Marchel H, Luczak M. Emergence and spread of carbapenem-resistant strains of Acinetobacter baumannii in a tertiary-care hospital in Poland. Clin Microbiol Infect. 2007;13:490–96. doi: 10.1111/j.1469-0691.2007.01694.x. [DOI] [PubMed] [Google Scholar]
- 15.Sung JY, Kwon KC, Park JW, et al. Dissemination of IMP-1 and OXA type beta-lactamase in carbapenem-resistant Acinetobacter baumannii. Korean J Lab Med. 2008;28:16–23. doi: 10.3343/kjlm.2008.28.1.16. [DOI] [PubMed] [Google Scholar]
- 16.Peymani A, Nahaei MR, Farajnia S, et al. High prevalence of metallo-beta-lactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn J Infect Dis. 2011;64:69–71. [PubMed] [Google Scholar]
- 17.Gales AC, Tognim MC, Reis AO, et al. Emergence of an IMP-like metallo-enzyme in an Acinetobacter baumannii clinical strain from a Brazilian teaching hospital. Diagn Microbiol Infect Dis. 2003;45:77–79. doi: 10.1016/s0732-8893(02)00500-x. [DOI] [PubMed] [Google Scholar]
- 18.Jeong SH, Bae IK, Park KO, et al. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J Microbiol. 2006;44:423–31. [PubMed] [Google Scholar]
- 19.Uma Karthika R, Srinivasa Rao R, Sahoo S, et al. Phenotypic and genotypic assays for detecting the prevalence of metallo-beta-lactamases in clinical isolates of Acinetobacter baumannii from a South Indian tertiary care hospital. J Med Microbiol. 2009;58:430–35. doi: 10.1099/jmm.0.002105-0. [DOI] [PubMed] [Google Scholar]
- 20.Perez F, Hujer AM, Hujer KM, et al. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012;35:317–25. [PubMed] [Google Scholar]
- 22.Omair M, Usman J, Kaleem F, et al. Evaluation of combined disc method for the detection of metallo-β-lactamase producing Gram negative bacilli. Malaysian Journal of Microbiology. 2012;8:21–25. [Google Scholar]