Abstract
Background
One particularly promising component of personalized medicine in cancer treatment is targeted therapy, which aims to maximize therapeutic efficacy while minimizing toxicity. However, the number of approved targeted agents remains limited. Expression microarray data for different types of cancer are resources to identify genes that were upregulated. The genes are candidate targets for cancer-targeting agents for future anticancer research and targeted treatments.
Methods and findings
The gene expression profiles of 48 types of cancer from 2,141 microarrays reported in the Gene Expression Omnibus were analyzed. These data were organized into 78 experimental groups, on which we performed comprehensive analyses using two-tailed Student’s t-tests with significance set at P < 0.01 to identify genes that were upregulated compared with normal cells in each cancer type. The resulting list of significantly upregulated genes was cross-referenced with three categories of protein inhibitor targets, categorized by inhibitor type (‘Targets of US Food and Drug Administration (FDA)-approved anticancer drugs’, ‘Targets of FDA-approved nonantineoplastic drugs’, or ‘Targets of non-FDA-approved chemical agents’). Of the 78 experimental groups studied, 57 (73%) represent cancers that are currently treated with FDA-approved targeted treatment agents. However, the target genes for the indicated therapies are upregulated in only 33 of these groups (57%). Nevertheless, the mRNA expression of the genes targeted by FDA-approved treatment agents is increased in every experimental group, including all of the cancers without FDA-approved targeted treatments. Moreover, many targets of protein inhibitors that have been approved by the FDA as therapies for nonneoplastic diseases, such as 3-hydroxy-3-methylglutaryl-CoA reductase and cyclooxygenase-2 and the targets of many non-FDA-approved chemical agents, such as cyclin-dependent kinase 1 and DNA-dependent protein kinase, are also overexpressed in many types of cancer.
Conclusion
This research demonstrates a clinical correlation between bioinformatics data and currently approved treatments and suggests novel uses for known protein inhibitors in future antineoplastic research and targeted therapies.
Keywords: personalized medicine, targeted therapy, protein inhibitor, cancer treatment, upregulated gene expression
Introduction
Cancer is a leading cause of death worldwide, and it is estimated that approximately 15 cancer deaths occur every minute.1 Conventional chemotherapy is still ineffective for many types of cancer, due to heterogeneity among cancer clones and patient genotypes.2 Therefore, the development of personalized medicine will be an important aspect of anticancer treatment in the 21st century.3 One very promising aspect of personalized medicine is its facilitation of targeted therapy, the goal of which is to maximize therapeutic efficacy while minimizing toxicity by specifically antagonizing cancer-related molecules.2
An example of targeted therapy, trastuzumab, has proven very effective in treating HER2-dependent breast cancer in several clinical trials.4–6 The side effects of trastuzumab are much less frequent than those of conventional chemotherapy.4 This drug specifically targets the juxtamembrane portion of the extracellular domain of the HER-2 (ERBB2) receptor, thereby preventing the activation of its intracellular tyrosine kinase.4,7 The US Food and Drug Administration (FDA) has approved trastuzumab for the treatment of metastatic HER2-positive breast cancer.4 Another example of targeted therapy, imatinib mesylate, has been shown to be a very safe and efficacious treatment for chronic myeloid leukemia (CML) and gastrointestinal stromal tumor (GIST).5,8 This drug explicitly inhibits the active fusion product from the Philadelphia (Ph) chromosome of CML and c-kit (CD117), which is overexpressed in GIST, thereby prohibiting the intracellular tyrosine kinase cascade.9 Consequently, the FDA has approved imatinib mesylate for the treatment of CML and GIST.5 However, only a limited number of targeted treatment agents have been developed to date. Only approximately 50 FDA-approved antineoplastic drugs have been introduced, such that many types of cancers still lack targeted therapies.
Currently, many lines of research are focused on identifying novel target/inhibitor pairs for different types of cancers. Several anticancer protein inhibitors, such as perifosine (an AKT inhibitor) and tipifarnib (an RAS and Wnt inhibitor), are currently undergoing clinical trials to investigate their potential as antineoplastic drugs.10,11 Typically, it takes an average of 7 years for these drugs to be approved after they undergo evaluation in clinical trials.12 Therefore, it has been suggested that drugs for incurable diseases should be subjected to an accelerated approval process that allows their use immediately after phase I or II clinical trials.12 Another group of protein inhibitors includes FDA-approved drugs that are used to treat nonneoplastic diseases, such as inhibitors of cyclooxygenase-2 (COX2) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR).13–16 At present, there is limited information about whether drugs from this group have potential uses as anticancer treatments. Finally, there is a large group of chemical agents that are used for in vitro research and which may or may not have antineoplastic properties.
The field of targeted therapy is still in its infancy, and scientists are just beginning to understand more about the cancer genome. Advances in the field of targeted therapy are inevitable because of the improvements in high-throughput technologies, such as whole-genome sequencing and array-based gene expression profiling.2 The Gene Expression Omnibus (GEO) is a repository of high-throughput gene expression data for various types of cancer and thus a potential source of information for identifying genes that are upregulated in different types of cancer.17,18 This information might be a key to discovering novel protein inhibitors for anticancer treatment.
Here, we collected information from the GEO DataSets for different types of cancers and performed comprehensive analyses to identify upregulated genes. These analyses confirmed the correlation between gene expression profiles and currently approved treatments and suggest novel uses for currently available protein inhibitors in future anticancer research and targeted treatments.
Methods
Data collection and template preparation
Expression microarray data for normal cells and cancer cells were obtained from the GEO repository17,18 using different cancer types as search keywords. GEO series (GSE) records were included only if they contained expression array data for both cancer and normal cells. Subsequently, all GSE records were downloaded for template preparation. In this study, we obtained 51 series records (2,141 expression microarrays) for 48 different types of cancer. Two of our records consisted of primary cell cultures and subcutaneous xenograft tumor tissue as the sources of specimens, whereas all of the other records consisted of specimens collected from patients. The primary cell cultures and subcutaneous xenograft tumor tissue have been proven to exhibit typical cancer gene expression of their types, while also decreasing contamination of normal tissues.19,20 These GSE data were organized into 78 experimental groups because some records encompassed several types of cancer. In the template preparation process, the cancer expression microarrays were defined as the ‘experiments’, whereas normal cell samples were defined as the ‘controls’. Supplementary Table 1 shows how we classified the GSE records into 78 experimental groups and includes the GEO accession and platform numbers, the organ and cancer types, and the sources of specimens.
Classification of protein inhibitors
The protein inhibitors were found by searching journal articles,5,21 cancer.gov and ClinicalTrials.gov web sites of US National Institute of Health, and the Sigma-Aldrich product catalog. The inhibitors were then classified in an Excel spreadsheet (Microsoft, Redmond, WA, USA) along with their specific protein targets and chemical or product names. The gene symbols for each protein inhibitor target were obtained from the National Center for Biotechnology Information’s Gene database. The protein inhibitors were categorized as ‘FDA-approved antineoplastic drugs’, ‘FDA-approved nonantineoplastic drugs’, or ‘non-FDA-approved chemical agents’ (Supplementary Tables 2.1–2.3, respectively). The protein inhibitors were classified as ‘FDA-approved antineoplastic drugs’ or ‘FDA-approved nonantineoplastic drugs’ only if the drugs were in the FDA’s databases. For ‘non-FDA-approved chemical agents’, the protein inhibitors were either undergoing clinical trials or tested in vitro. In this study, there were 597 protein inhibitor targets: 47 targets of ‘FDA-approved antineoplastic drugs’, 76 targets of ‘FDA-approved nonantineoplastic drugs’, and 474 targets of ‘non-FDA-approved chemical agents’. Furthermore, Supplementary Table 2.1 shows the diseases corresponding to the FDA-indicated uses for each targeted therapy, and Supplementary Table 2.3 shows the clinical trial phases and identifiers of some ‘non-FDA-approved chemical agents’.
Statistical analysis
Using the prepared templates of expression microarray data and platforms, a two-tailed Student’s t-test was used to compare the mean expression levels in the experiments (cancer cells) and controls (normal cells) for each experimental group. Statistical significance was set at P < 0.01 in every experiment unless otherwise stated. Paired t-tests were used for three of our experimental groups shown in Supplementary Table 1, where the normal and cancer specimens were collected from the same patient. The genes for which the mRNA levels were expressed at significantly higher levels in the experiments compared with the controls were considered ‘upregulated’. The list of genes with upregulated expression patterns was then cross-referenced with the three major categories of protein inhibitors, and the results are reported in Supplementary Table 3. All statistical analysis steps were performed using extensions of the Connection Up- and Down-Regulation Expression Analysis of Microarrays (CU-DREAM) program.22 The P-values of selected GSEs were compared with P-values using one-way analyses of variance (ANOVA).
Results
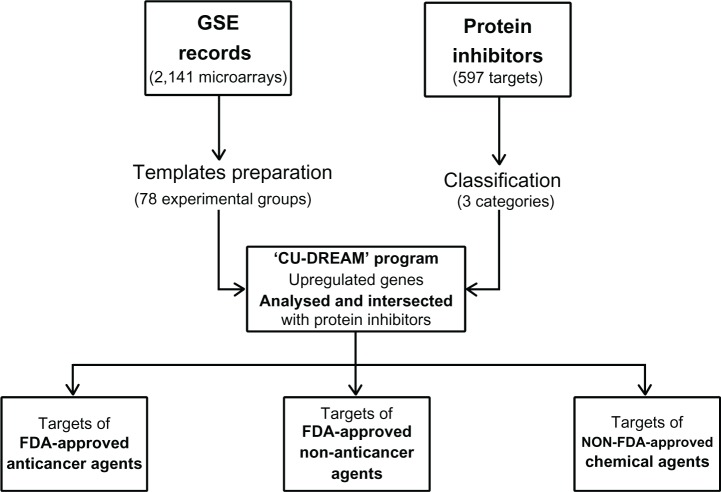
In this study, each experimental group was statistically analyzed, and the resulting list of significantly upregulated genes was cross-referenced with the three categories of protein inhibitor targets presented in Supplementary Tables 2.1–2.3 (Figure 1). The results are shown in Supplementary Table 3, categorized by inhibitor type (‘Targets of FDA-approved anticancer drugs’, ‘Targets of FDA-approved nonantineoplastic drugs’, or ‘Targets of non-FDA-approved chemical agents’). In Supplementary Table 3, the known ‘FDA-indicated targeted agents’ for each type of cancer are also included, referenced from the National Cancer Institute, US National Institutes of Health. The fold changes of genes that were upregulated in cancer are also reported (Supplementary Table 4). We also compared P-values of GSE10927, GSE14922, GSE31671 with P-values using one-way ANOVA. The values of correlation coefficient (r) were 0.973, 0.999, and 0.999, respectively.
Figure 1.
The overall methodological framework of this study.
Notes: First, 51 GSE records (2,141 expression microarrays) were collected for different types of cancer, and the templates for 78 experimental groups were prepared for analysis using the CU-DREAM program. We then obtained 597 protein inhibitors from various databases and classified them into three categories: ‘FDA-approved antineoplastic drugs’, ‘FDA-approved nonantineoplastic drugs’, and ‘non-FDA-approved chemical agents’. The program then analyzed each experimental group to identify genes of which the expression was upregulated in cancer compared with normal cells. Subsequently, the upregulated genes were cross-referenced with the three protein inhibitor lists and were sorted into three categories.
Abbreviations: CU-DREAM, Connection Up- and Down-regulation Expression Analysis of Microarrays; FDA, US Food and Drug Administration; GSE, Gene Expression Omnibus series.
The correlation between bioinformatics data and current treatments
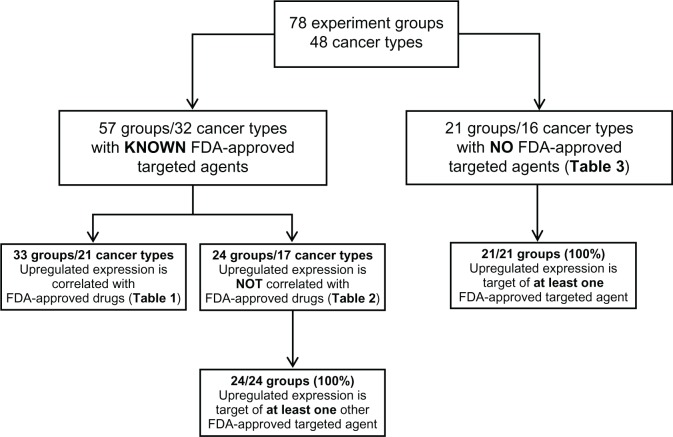
We classified our results based on whether the cancer has a known FDA-approved targeted agent (Figure 2). We found that 57 out of our 78 experimental groups (73%) represent 32 types of cancer with known FDA-approved targeted therapies. Thirty-three out of these 57 groups (58%) have upregulated gene expression that is well correlated with the indicated targeted treatment agent(s). This group of cancers is shown in Table 1, along with their indicated targeted therapies.
Figure 2.
The classification diagram showing the results of this study.
Notes: The classification of the results was based on whether the cancer has a known FDA-approved targeted agent. For the experimental groups with known FDA-approved targeted agents, we evaluated the correlation of the upregulated gene expressions with the indicated targeted treatment agents. For the experimental groups that do not have any FDA-approved targeted agent or do not overexpress the gene that is the target of the FDA-approved agents, we further assessed whether each group overexpresses at least one gene that is associated with an FDA-approved targeted anticancer drug.
Abbreviation: FDA, US Food and Drug Administration.
Table 1.
Cancer types in which an upregulated gene expression is correlated with the FDA-approved drug for that cancer
| Cancer | Targeted protein: current FDA-approved antineoplastic drug(s) |
|---|---|
| Glioblastoma | VEGF: bevacizumab |
| Breast cancer (two groups) | ERBB2: trastuzumab |
| Colorectal cancer (four groups) | VEGF: bevacizumab, EGFR: cetuximab |
| Pediatric AML | CD33: gemtuzumab |
| Chronic myeloid leukemia | ABL1: imatinib |
| Multiple myeloma (three groups) | PSMC (3, 5): bortezomib |
| Soft tissue sarcoma | KIT, PDGFR (A, B): pazopanib |
| Gastrointestinal stromal tumor | KIT, PDGFRB: imatinib |
| Hepatocellular carcinoma (four groups) | PDGFR (A, B), RAF1, BRAF, RET, KIT: sorafenib |
| Lung adenocarcinoma | EGFR: erlotinib, gefitinib, VEGF: bevacizumab |
| Lung non-small-cell cancer | EGFR: erlotinib, gefitinib |
| Renal clear cell cancer (2 groups) | VEGF: bevacizumab, RET, PDGFR (A, B), VEGFR: sorafenib, sunitinib, MTOR: everolimus |
| Renal oncocytoma cell cancer | KIT, VEGFR: sorafenib, VEGF: bevacizumab |
| Basal-like breast cancer | TOP2A: doxorubicin |
| BRCA1-mutated breast cancer (two groups) | TOP2A: doxorubicin |
| Exing sarcoma | TOP2A: doxorubicin |
| Thyroid anaplastic cancer | TOP2A: doxorubicin |
| Thyroid papillary cancer | TOP2A: doxorubicin |
| Squamous cell lung cancer (two groups) | EGFR: cetuximab |
| Fallopian serous carcinoma | TOP2A: liposomal doxorubicin |
| Clear cell ovarian cancer | TOP2A: liposomal doxorubicin |
Note: numbers within parentheses are gene members.
Abbreviations: AML, acute myeloid leukemia; FDA, US Food and Drug Administration; BRCAI, breast cancer I gene.
In addition, 24 of the 57 experimental groups (42%) with known FDA-approved targeted drug treatments do not overexpress the gene that corresponds to the indicated targeted treatment agent. However, all of these experimental groups overexpress at least one gene that is the target of another FDA-approved targeted treatment agent (Table 2).
Table 2.
Cancer types for which the FDA-indicated antineoplastic-targeted agent is not correlated with the upregulated gene(s)
| Cancer | Targeted protein: potential FDA-approved antineoplastic drug(s) |
|---|---|
| Nonbasal breast cancer | BRAF: sorafenib, CTLA4: ipilimumab, CYP19A1: letrozole, DHFR: methotrexate, DNMT1: azacitidine, EGFR: cetuximab, GSR: carmustine, HDAC (2, 4, 7, 11): vorinostat, TOP1: topotecan, TOP2A: doxorubicin, VEGF: bevacizumab |
| Breast invasive ductal cancer | DNMT1: azacitidine, HDAC1: vorinostat, PSMC4: bortezomib |
| Colorectal cancer | PDGFRB: pazopanib, RET: vandetanib |
| Chronic myeloid leukemia (3 groups) | ESR1: tamoxifen, RET: vandetanib, DNMT1: azacitidine, ERBB2: trastuzumab, HDAC3: vorinostat |
| CLL B-cell (3 groups) | CTLA4: ipilimumab, HDAC (1, 2, 6, 7, 9): vorinostat, PSMC (2, 3, 6): bortezomib, NQO1+: carmustine, CD52: alemtuzumab, MTOR: temsirolimus, PDGFRA: pazopanib, TOP1: topotecan |
| Follicular lymphoma | ABL1: imatinib, ESR2: tamoxifen, HDAC7: vorinostat |
| Multiple myeloma | ERBB2: trastuzumab, VEGF: bevacizumab |
| Myelodysplastic syndrome | HDAC2: vorinostat, PSMC2: bortezomib |
| Hepatocellular carcinoma | HDAC2: vorinostat |
| HBV hepatocellular carcinoma | MTOR: temsirolimus, PSMC5: bortezomib |
| HCV hepatocellular carcinoma (4 groups) | HDAC (1, 2, 4, 11): vorinostat, GSR: carmustine, DNMT1: azacitidine, TOP2A: doxorubicin, PSMC2: bortezomib |
| Pancreatic intraductal papillary-mucinous carcinoma (IPMN) | DHFR: methotrexate, GSR, NQO1: carmustine, HDAC1: vorinostat, TOP1: topotecan, TOP2A: doxorubicin |
| Invasive pancreatic cancer originating in the IPMN | TOP2A: doxorubicin, GSR*, NQO1: carmustine, DHFR: methotrexate |
| Chromophobe renal cell carcinoma | BRAF: sorafenib, DHFR: methotrexate, ESR1: tamoxifen, HDAC11: vorinostat, PSMC (2, 3, 6): bortezomib |
| Renal clear cell cancer | CD52: alemtuzumab |
| Renal papillary cell carcinoma | HDAC3: vorinostat, PSMC (1, 2, 3, 4): bortezomib |
Note: numbers within parentheses are gene members.
Abbreviations: CLL, chronic lymphocytic leukemia; HBV, hepatitis B virus; HCV, hepatitis C virus.
Our GSE records also included 21 experimental groups representing 16 types of cancer with no FDA-approved targeted therapies. Surprisingly, every experimental group was found to overexpress genes that are targets of other FDA-approved agents (Table 3).
Table 3.
Cancer without an FDA-indicated antineoplastic-targeted agent, but with suggested FDA-approved antineoplastic drug(s)
| Cancer | Targeted protein: potential FDA-approved antineoplastic drug(s) |
|---|---|
| Adrenocortical carcinoma (two groups) | DHFR: methotrexate, DNMT1: azacitidine, TOP2A: doxorubicin, BRAF, KIT: sorafenib, GSR: carmustine, HDAC6: vorinostat, PSMC2: bortezomib |
| Bladder transitional cell carcinoma (three groups) | ERBB2: trastuzumab, HDAC (1, 2, 3): vorinostat, PSMC (1, 2, 3, 4, 5, 6): bortezomib, RAF1: sorafenib, TOP (2A, 2B): doxorubicin, VEGF: bevacizumab, DHFR: methotrexate, EGFR: cetuximab, GSR, NQO1: carmustine, MTOR: temsirolimus, TOP1: topotecan, DNMT1: azacitidine |
| Brain astrocytoma | ABL1: imatinib, BRAF, PDGFRB, RAF1, VEGFR: sorafenib, DHFR: methotrexate, DNMT1: azacitidine, EGFR: cetuximab, ERBB2: trastuzumab, ESR (1, 2): tamoxifen, HDAC (6, 7, 10): vorinostat, TOP1: topotecan, TOP2A: doxorubicin, VEGF: bevacizumab |
| Brain ependymoma | EGFR: cetuximab, ERBB2: trastuzumab, ESR1: tamoxifen, HDAC3: vorinostat, PDGFRB: pazopanib, TOP2A: doxorubicin |
| Brain pilocytic astrocytoma | ABL1: imatinib, HDAC (1, 6): vorinostat, RAF1: sorafenib, TOP1: topotecan, VEGF: bevacizumab |
| Brain oligodendroglioma | ABL1: imatinib, HDAC6: vorinostat |
| Osteosarcoma (two groups) | HDAC (3, 9): vorinostat, MTOR: temsirolimus, PSMC (2, 5, 6): bortezomib, VEGF: bevacizumab, ABL1: imatinib, GSR: carmustine |
| Cervical squamous cell carcinoma | CYP19A1: letrozole, DHFR: methotrexate, DNMT1: azacitidine, EGFR: cetuximab, HDAC9: vorinostat, NQO1: carmustine, PSMC4: bortezomib, TOP2A: doxorubicin, VEGF: bevacizumab |
| Marginal zone lymphoma | ABL1: imatinib, HDAC (7, 11): vorinostat, PSMC3: bortezomib, RAF1: sorafenib |
| Intestine carcinoid tumor | HDAC5: vorinostat, PSMC2: bortezomib, RET: sorafenib |
| Cholangiocarcinoma | BRAF: sorafenib, DNMT1: azacitidine, GSR, NQO1: carmustine, HDAC (1, 2, 3, 8, 9): vorinostat, PSMC (2, 4): bortezomib, TOP2A: doxorubicin |
| Pleural mesothelioma | DHFR: methotrexate, EGFR: cetuximab, TOP1: topotecan, TOP2A: doxorubicin |
| Nasopharyngeal carcinoma | DHFR: methotrexate, ESR1: tamoxifen, TOP1: topotecan, TOP2As: doxorubicin, VEGF: bevacizumab |
| Prostate cancer (two groups) | ABL1: imatinib, PSMC (2, 3, 6): bortezomib, DNMT1: azacitidine, EGFR: cetuximab, ERBB2: trastuzumab, GSR, NQO1: carmustine, HDAC (3, 6): vorinostat, MTOR: temsirolimus, IL2: denileukin |
| Renal transitional cell carcinoma | HDAC (1, 3, 11): vorinostat, NQO1: carmustine, PSMC (2, 6): bortezomib, TOP (2A, 2B): doxorubicin |
| Vulvar intraepithelial cancer | DHFR: methotrexate, DNMT1: azacitidine, HDAC (1, 2): vorinostat, PSMC (2, 6): bortezomib, TOP1: topotecan, TOP2A: doxorubicin, VEGF: bevacizumab |
Note: Numbers within parentheses are gene members.
Protein inhibitors for novel targeted anticancer therapies
There is significant upregulation of gene expression for protein inhibitor targets other than oncogenes in many of the experimental groups. We had accumulated the frequency of each upregulated protein inhibitor target of ‘FDA-approved nonantineoplastic drugs’ and ‘non-FDA-approved chemical agents’ among the 78 experimental groups.
Table 4 ranks the frequency of the top 30 upregulated genes that are targets of ‘FDA-approved nonantineoplastic drugs’. We further categorized this group into ‘immunomodulatory’ or ‘nonimmunomodulatory’ drug targets. Moreover, Table 5 shows the frequency of the top 20 upregulated genes corresponding to targets of ‘non-FDA-approved chemical agents’.
Table 4.
Upregulation frequency of genes targeted by ‘FDA-approved nonantineoplastic protein inhibitors’
| Targeted protein symbol | Targeted protein | FDA-approved nonantineoplastic drugs | Frequency (n = 78) |
|---|---|---|---|
| Nonimmunomodulatory drugs | |||
| SRD5A1 | Steroid-5-alpha-reductase | Finasteride | 23 |
| COMT | COMT | Tolcapone | 19 |
| ATP1B1 | Adenosine triphosphatase | Digitalis | 16 |
| ALOX5 | 5-lipoxygenase | Zileuton | 15 |
| ITGA2B | GPIIb/IIIa | Abciximab | 15 |
| CA12 | Carbonic anhydrase XII | Acetazolamide | 14 |
| GAA | Alpha-glucosidase | Acarbose | 14 |
| CYP2E1 | Cytochrome P | Disulfram | 13 |
| CYP2C9 | Cytochrome P | Fluconazole | 13 |
| DPP4 | Dipeptidyl-peptidase 4 | Sitagliptin | 12 |
| ACHE | Cholinesterase | Edrophonium | 12 |
| ALDH1B1 | Aldehyde dehydrogenase | Disulfiram | 12 |
| GRIN1 | NMDA receptor | Amantadine | 11 |
| HMGCR | HMG-CoA reductase | Simvastatin | 11 |
| CYP1A2 | Cytochrome P | Ciprofloxacin | 10 |
| ALDH1A3 | Aldehyde dehydrogenase | Disulfiram | 10 |
| CYP2D6 | Cytochrome P | SSRIs, Bupropion | 10 |
| COX2 | Cyclooxygenase II | Celecoxib | 9 |
| CA1 | Carbonic anhydrase I | Acetazolamide | 9 |
| CA5A | Carbonic anhydrase VA | Acetazolamide | 9 |
| CA9 | Carbonic anhydrase IX | Acetazolamide | 9 |
| HRH1 | Histamine receptor H1 | Loratadine | 9 |
| PDE5A | 5-phosphodiesterase | Sildenafil | 8 |
| Immunomodulatory drugs | |||
| PPAT | PPAT | Azathioprine | 29 |
| IMPDH1 | IMPDH1 | Ribavirin | 23 |
| IMPDH2 | IMPDH2 | Ribavirin | 21 |
| PLA2G10 | Phospholipase A2 | Prednisolone | 12 |
| PLA2G12A | Phospholipase A2 | Prednisolone | 12 |
| DHODH | DHODH | Leflunomide | 11 |
| PLA2G15 | Phospholipase A2 | Prednisolone | 9 |
Abbreviation: n, number of tests.
Table 5.
Upregulation frequency for genes targeted by ‘non-FDA-approved chemical agents’
| Targeted protein symbol | Targeted protein | Non-FDA-approved chemical agents | Frequency (n = 78) |
|---|---|---|---|
| CDK1 | Cyclin-dependent kinase I | Alvocidlib, Seliciclib | 33 |
| PRKDC | Protein kinase, DNA-dependent | IC88821, NU7026 | 32 |
| CSNK2A1 | Casein kinase II | Myricetin | 31 |
| PRKCI | Protein kinase C I | HA-100, ML-7 | 31 |
| GAPDH | GAPDH | Iodoacetic acid | 30 |
| CDK2 | Cyclin-dependent kinase 2 | Alvocidlib, Seliciclib | 29 |
| CASP2 | Caspase II | Fluoromethyl ketone | 29 |
| CSNK1A1 | Casein kinase I | IC 261 | 29 |
| MMP11 | Matrix metalloprotease 11 | Bipyridyl reagentPlus | 28 |
| POLR2H | RNA polymerase | Thiolutin | 28 |
| SQLE | Squalene epoxidase | Terbinafine hydrochloride | 28 |
| KRAS | Rat sarcoma viral oncogene | Tipifarnib, Lonafarnib | 27 |
| CLK2 | Cdc2-like kinase | TG003 | 27 |
| HSP90AA1 | Heat shock protein | Tanespimycin | 27 |
| MMP14 | Matrix metalloprotease 14 | Bipyridyl ReagentPlus | 27 |
| CASP3 | Caspase 3 | Fluoromethyl ketone | 27 |
| POLR2G | RNA polymerase | Thiolutin | 27 |
| PARP1 | Poly (ADP-ribose) polymerase | BSI-201, Olaparib | 26 |
| CDK7 | Cyclin-dependent kinase 7 | Alvocidlib, Seliciclib | 26 |
| HSP90AB1 | Heat shock protein | Tanespimycin | 26 |
Abbreviation: n, number of tests.
Discussion
Our comprehensive analysis of expression array data is a useful approach to confirm the connection between bioinformatics data and current treatments. Our results showed that 33 out of 57 experimental groups (58%) overexpress the gene(s) that correspond to the clinical treatments indicated for these cancers. In the current setting, targeted anticancer therapy would be prescribed to these patients.
The data also illustrate the limited availability of targeted therapies for many types of cancer. The majority of experimental groups (45/78, or 58%) do not overexpress the gene that is the target of the FDA-approved drug for the corresponding cancer or do not have any FDA-approved targeted treatment agents. Twenty-four of the 57 experimental groups (42%) with known FDA-approved drugs do not overexpress the gene that is the target of their indicated agents (Figure 2). This result could be hypothesized by several mechanisms. For a gene product to be a therapeutic target, it is not necessary for it to be overexpressed. For example, epidermal growth factor receptor (EGFR), the tyrosine kinase in lung cancer may be expressed at a regular level, but mutation might cause it to over function.23 However, EGFR protein expression was significantly correlated with EGFR mutation and copy number.23 This result also supports the increasing concern of tumor heterogeneity among patient genotypes and within cancer clones (intratumoral heterogeneity).2,24 Thus, patients with these types of cancers cannot currently receive antineoplastic target therapy. However, one or more targets of other FDA-approved anticancer-targeted treatment agents are upregulated in all experimental groups. All of the 24 experimental groups (100%) overexpress at least one gene that is associated with an FDA-approved targeted anticancer drug (Table 2). We recommend that animal studies and in vitro studies with human tissue should be performed to test the possible future use as targeted therapies of these drugs. Synergism is sometimes suggested as a developmental strategy, especially because recent concepts in targeted therapies target multiple ligands.25,26 The analysis of targeted first-line cancer treatments by considering common gene pairs or triplets may be necessary.
In addition, it is interesting that all 21 of the experimental groups (100%) consisting of cancer types for which there is currently no FDA-approved targeted therapy overexpress genes that are targeted in FDA-approved anticancer-targeted therapies (Table 3). The efficacy of the suggested targeted therapies should also be tested in these types of cancer.
Our data also highlight several groups of promising protein inhibitors for future targeted anticancer therapy. The first group consists of targeted inhibitors that have been approved by the FDA for the treatment of nonneoplastic diseases. Some ‘nonimmunomodulatory’ drugs in this group, such as inhibitors of COX2, HMGCR, dipeptidyl peptidase 4 (DPP4), 5-lipoxygenase (ALOX5), and histamine receptor H1 (HRH1), have been widely used to treat nonneoplastic diseases such as allergies, dyslipidemia, diabetes mellitus, and musculoskeletal pain (Table 4). Thus, the potential anticancer mechanisms of these drugs will be very interesting to investigate. This is also known as ‘drug repurposing’. A significant advantage of drug repurposing over conventional drug development is these drugs have already passed a number of toxicity trials where the majority of drugs were disqualified during their development.27–30 If any of these drugs are proven to have anticancer properties, these agents may be very useful as novel targeted treatments.
One good example is HMGCR inhibitors. HMGCR is one of the top 20 most frequently upregulated genes in the experimental groups analyzed herein (Table 4). The prototypical HMGCR inhibitor is simvastatin, which was originally developed as a cholesterol-lowering agent and is among the most therapeutically effective and financially successful pharmaceuticals ever created.15 Our data showed that HMGCR is upregulated in nine types of cancer: acute myeloid leukemia (AML), Ewing sarcoma, soft tissue sarcoma, bladder cancer, breast cancer, hepatocellular carcinoma, nasopharyngeal carcinoma, ovarian cancer, and pancreatic cancer (Supplementary Table 3). Consistent with these data, statins have been reported to have antiproliferative effects on AML, multiple myeloma, breast cancer, prostate cancer, colorectal cancer, lung cancer, pancreatic cancer, ovarian cancer, cervical cancer, and head and neck cancer.31–34 Currently, there are ongoing efforts to demonstrate the significance of the HMGCR or mevalonate pathways in carcinogenesis.35 There have also been several clinical trials combining statins with other conventional chemotherapeutic regimens, the results of which are very promising.15,36–39 It is hoped that statins might be the pioneering example of FDA-approved drugs for nonneoplastic diseases that also have anticancer potential.
Our studies have revealed that other targets of widely used FDA-approved nonantineoplastic therapies, such as COX2, DPP4, ALOX5, and HRH1, are overexpressed in many types of cancer; however, information about the anticancer properties of these agents is still very limited. Several studies have demonstrated the chemopreventive effects of ALOX5 and COX2 inhibitors, but most of these studies have been descriptive and lack a molecular underpinning.40–46 A meta-analysis and multiple randomized controlled trials have shown that selective COX2 inhibitors can reduce the risk of colorectal adenomas and colorectal cancer.13,14,47,48 This is consistent with our data where COX2 is overexpressed in three out of five (60%) experimental groups of colorectal cancer (Supplementary Table 3). It has recently been proposed that because cancer cells experience unique cellular stresses compared with normal cells, the cancer cells must develop tolerance mechanisms through stress support pathways to survive and proliferate in their unusual environments.49 Alterations in cancer cell metabolism may enable cancer cells to achieve their principle distinguishing features.50 Therefore, metabolic reprogramming is considered to be a vulnerability of cancer cells, and targeting this pathway using ‘FDA-approved nonantineoplastic drugs’ may be a potential cancer treatment. Nevertheless, it is possible that some of the overexpressed gene expressions in cancer are the effect of the tumor-driving target and not the principal target itself. We therefore suggest that the roles of these protein inhibitors in oncogenesis be thoroughly studied and determine what signaling pathways, rather than individual genes, are altered in tumors to investigate their potential as antineoplastic treatments. We believe that clinical trials combining these drugs with first-line anticancer treatments in patients diagnosed with cancer are warranted.
In addition, Table 4 also shows that an ‘immunomodulatory’ agent target, phosphoribosylpyrophosphate amidotransferase (PPAT ), results in the highest frequency of upregulated gene expression among all ‘FDA-approved nonantineoplastic drugs’. Surprisingly, many researchers have reported that the prolonged use of PPAT inhibitors, such as azathioprine, can increase the risk of cancer because it chronically suppresses the function of the immune system.51,52 Therefore, a safety assessment prior to the initiation of clinical trials must be required for this category of research to ensure that the potential antineoplastic benefits outweigh the potential side effects.
Furthermore, another group of potential anticancer treatments consisted of the ‘non-FDA-approved chemical agents’. As shown in Table 5, the target with the highest frequency of upregulated gene expression is cyclin-dependent kinase 1 (CDK1). This result is in accordance with research suggesting that CDKs are very promising targets for antineoplastic treatment. Many clinical trials are ongoing to prove the efficacy of CDK inhibitors, such as seliciclib and flavopiridol.53–56 Nevertheless, information regarding the tumorigenicity of many other targets of ‘non-FDA-approved chemical agents’ is still limited. We believe that this group of chemical agents is an excellent candidate for future anticancer research and treatments; however, more mechanistic studies will be required to demonstrate the relationship between the upregulation of these genes and carcinogenesis, and the safety of the inhibitors of these proteins must be assessed.
In this study, we are concerned that some RNA expressions might not positively correlate with the protein expressions as in the central dogma of molecular biology. Various posttranscriptional processes can alter protein expressions. For instance, some mRNAs are strongly preserved within the nucleus, which means that their expression levels will be overestimated relative to protein levels.57 Therefore, it would be useful to apply the knowledge from this study into the next layer of bioinformatics data analysis such as proteomics.
A limitation of this research is that the application of molecular evidence from bioinformatics data may not be sufficient to predict the efficacy of the promising targeted agents or protein inhibitors. Nevertheless, there have been suggestions that appropriately designed clinical trials will provide adequate information to health care providers and patients to make informed clinical decisions.21 However, because large clinical trials are difficult to establish because of the rarity of many types of specific cancer, cancer centers worldwide should collaborate to conduct prospective multicenter proof-of-principle trials. If the outcomes are promising, accelerated approval of these drugs can be highly encouraged.12
Conclusion
There is a strong correlation between the bioinformatics data and current treatments, but many types of cancer still lack FDA-approved targeted therapies. Therefore, it may be reasonable to use targeted treatment agents that are FDA-approved for the treatment of other cancers that overexpress the same target genes. In addition, our study highlights several protein inhibitors that may have potential as targeted anticancer drugs. Further studies should elucidate the comprehensive mechanistic pathways of these novel antineoplastic agents. It will be worth investigating whether in vitro chemical agents and drugs that have been approved for use in the treatment of nonneoplastic diseases have significant effects in oncogenesis.
Author contributions
Conceived and designed the experiments: AM. Performed the experiments: VT, WW, CT, KS, KaC, KL, KhC, OS, PW, WM, CA. Analyzed the data: VT, CV. Contributed reagents/materials/analysis tools: CA, AM. Wrote the manuscript: VT, CV, AM. Agreed with manuscript results and conclusions: All authors. Reviewed data and literature: VT, CV, AM.
Supplementary tables
All supplementary tables are available at http://pioneer.netserv.chula.ac.th/~achatcha/supp/supporting_tables_2013.zip
GEO accession and platform numbers, organs and cancer types, GEO sample numbers (GSMs), and specimen sources for each of the 78 experimental groups.
List of ‘FDA-approved antineoplastic drugs’, of ‘FDA-approved nonantineoplastic drugs’, and ‘non-FDA-approved chemical agents’.
Table S2.1 List of ‘FDA-approved antineoplastic drugs’.
Table S2.2 List of ‘FDA-approved antineoplastic drugs’.
Table S2.3 List of ‘FDA-approved antineoplastic drugs’.
The intersection of significantly upregulated genes identified by two-tailed Student’s t-test analysis of 78 experimental groups with the three categories of protein inhibitors.
The fold change of genes that were upregulated in cancer.
Acknowledgments
The authors would like to thank Professor John J Kavanagh (Chulalongkorn University) and Dr Virote Sriuranpong (Chulalongkorn University) for advice and their critical reviews during the preparation of this manuscript.
Footnotes
Disclosure
This study was supported by Chulalongkorn University, the Four Seasons Hotel Bangkok’s 4th Cancer Care charity fun run in coordination with the Thai Red Cross Society and a 2011 Research Chair Grant from the National Science and Technology Development Agency (NSTDA) of Thailand. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors report no competing or conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Mullard A. Oncology trials gear up for high-throughput sequencing. Nature Rev Drug Discov. 2012;11(5):339–340. doi: 10.1038/nrd3733. [DOI] [PubMed] [Google Scholar]
- 3.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudis CA. Trastuzumab – mechanism of action and use in clinical practice. N Engl J Med. 2007;357(1):39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 5.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353(2):172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 6.Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albanell J, Bellmunt J, Molina R, et al. Node-negative breast cancers with p53 (−)/HER2-neu (−) status may identify women with very good prognosis. Anticancer Res. 1996;16(2):1027–1032. [PubMed] [Google Scholar]
- 8.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 9.Savage DG, Antman KH. Imatinib mesylate–a new oral targeted therapy. N Engl J Med. 2002;346(9):683–693. doi: 10.1056/NEJMra013339. [DOI] [PubMed] [Google Scholar]
- 10.Cho DC, Hutson TE, Samlowski W, et al. Two phase 2 trials of the novel Akt inhibitor perifosine in patients with advanced renal cell carcinoma after progression on vascular endothelial growth factor-targeted therapy. Cancer. 2012;118(8):6055–6062. doi: 10.1002/cncr.27668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margolin KA, Moon J, Flaherty LE, et al. Randomized phase II trial of sorafenib with temsirolimus or tipifarnib in untreated metastatic melanoma (S0438) Clin Cancer Res. 2012;18(4):1129–1137. doi: 10.1158/1078-0432.CCR-11-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabner BA. Early accelerated approval for highly targeted cancer drugs. N Engl J Med. 2011;364(12):1087–1089. doi: 10.1056/NEJMp1100548. [DOI] [PubMed] [Google Scholar]
- 13.Arber N, Eagle CJ, Spicak J, et al. Celecoxib for the prevention of colorectal adenomatous polyps. N Engl J Med. 2006;355(9):885–895. doi: 10.1056/NEJMoa061652. [DOI] [PubMed] [Google Scholar]
- 14.Bertagnolli MM, Eagle CJ, Zauber AG, et al. Celecoxib for the prevention of sporadic colorectal adenomas. N Engl J Med. 2006;355(9):873–884. doi: 10.1056/NEJMoa061355. [DOI] [PubMed] [Google Scholar]
- 15.Clendening JW, Penn LZ. Targeting tumor cell metabolism with statins. Oncogene. 2012;31(48):4967–4978. doi: 10.1038/onc.2012.6. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen SF, Nordestgaard BG, Bojesen SE. Statin use and reduced cancer-related mortality. N Engl J Med. 2012;367(19):1792–1802. doi: 10.1056/NEJMoa1201735. [DOI] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets – 10 years on. Nucleic Acids Res. 2011;39(Database issue):D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgson JG, Yeh RF, Ray A, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11(5):477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanni S, Priolo C, Grasselli A, et al. Epithelial-restricted gene profile of primary cultures from human prostate tumors: a molecular approach to predict clinical behavior of prostate cancer. Mol Cancer Res. 2006;4(2):79–92. doi: 10.1158/1541-7786.MCR-05-0098. [DOI] [PubMed] [Google Scholar]
- 21.Manchana T, Ittiwut C, Mutirangura A, Kavanagh JJ. Targeted therapies for rare gynaecological cancers. Lancet Oncol. 2010;11(7):685–693. doi: 10.1016/S1470-2045(09)70368-7. [DOI] [PubMed] [Google Scholar]
- 22.Aporntewan C, Mutirangura A. Connection up- and down-regulation expression analysis of microarrays (CU-DREAM): a physiogenomic discovery tool. Asian Biomedicine. 2011;5(2):257–262. [Google Scholar]
- 23.Liang Z, Zhang J, Zeng X, Gao J, Wu S, Liu T. Relationship between EGFR expression, copy number and mutation in lung adenocarcinomas. BMC Cancer. 2010;10:376. doi: 10.1186/1471-2407-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid K, Bago-Horvath Z, Berger W, et al. Dual inhibition of EGFR and mTOR pathways in small cell lung cancer. Br J Cancer. 2010;103(5):622–628. doi: 10.1038/sj.bjc.6605761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong S, Zhang XC, Cheng H, et al. Everolimus synergizes with gefitinib in non-small-cell lung cancer cell lines resistant to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Chemother Pharmacol. 2012;70(5):707–716. doi: 10.1007/s00280-012-1946-3. [DOI] [PubMed] [Google Scholar]
- 27.Aronson JK. Old drugs–new uses. Br J Clin Pharmacol. 2007;64(5):563–565. doi: 10.1111/j.1365-2125.2007.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007;448(7154):645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 29.DiMasi JA, Hansen RW, Grabowski HG, Lasagna L. Cost of innovation in the pharmaceutical industry. J Health Econ. 1991;10(2):107–142. doi: 10.1016/0167-6296(91)90001-4. [DOI] [PubMed] [Google Scholar]
- 30.Tartaglia LA. Complementary new approaches enable repositioning of failed drug candidates. Expert Opin Investig Drugs. 2006;15(11):1295–1298. doi: 10.1517/13543784.15.11.1295. [DOI] [PubMed] [Google Scholar]
- 31.Wong WW, Dimitroulakos J, Minden MD, Penn LZ. HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia. 2002;16(4):508–519. doi: 10.1038/sj.leu.2402476. [DOI] [PubMed] [Google Scholar]
- 32.Fritz G. Targeting the mevalonate pathway for improved anticancer therapy. Curr Cancer Drug Targets. 2009;9(5):626–638. doi: 10.2174/156800909789057033. [DOI] [PubMed] [Google Scholar]
- 33.Jakobisiak M, Golab J. Statins can modulate effectiveness of antitumor therapeutic modalities. Med Res Rev. 2010;30(1):102–135. doi: 10.1002/med.20162. [DOI] [PubMed] [Google Scholar]
- 34.Sassano A, Platanias LC. Statins in tumor suppression. Cancer Lett. 2008;260(1–2):11–19. doi: 10.1016/j.canlet.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 35.Clendening JW, Pandyra A, Boutros PC, et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA. 2010;107(34):15051–15056. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawata S, Yamasaki E, Nagase T, et al. Effect of pravastatin on survival in patients with advanced hepatocellular carcinoma. A randomized controlled trial. Br J Cancer. 2001;84(7):886–891. doi: 10.1054/bjoc.2000.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornblau SM, Banker DE, Stirewalt D, et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high-dose Ara-C: a phase 1 study. Blood. 2007;109(7):2999–3006. doi: 10.1182/blood-2006-08-044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidmaier R, Baumann P, Bumeder I, Meinhardt G, Straka C, Emmerich B. First clinical experience with simvastatin to overcome drug resistance in refractory multiple myeloma. Eur J Haematol. 2007;79(3):240–243. doi: 10.1111/j.1600-0609.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 39.Larner J, Jane J, Laws E, Packer R, Myers C, Shaffrey M. A phase I–II trial of lovastatin for anaplastic astrocytoma and glioblastoma multiforme. Am J Clin Oncol. 1998;21(6):579–583. doi: 10.1097/00000421-199812000-00010. [DOI] [PubMed] [Google Scholar]
- 40.Abrahao AC, Castilho RM, Squarize CH, Molinolo AA, dos Santos-Pinto D, Jr, Gutkind JS. A role for COX2-derived PGE2 and PGE2-receptor subtypes in head and neck squamous carcinoma cell proliferation. Oral Oncol. 2010;46(12):880–887. doi: 10.1016/j.oraloncology.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris RE, Beebe-Donk J, Alshafie GA. Reduction in the risk of human breast cancer by selective cyclooxygenase-2 (COX-2) inhibitors. BMC Cancer. 2006;6:27. doi: 10.1186/1471-2407-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris RE, Beebe-Donk J, Alshafie GA. Reduced risk of human lung cancer by selective cyclooxygenase 2 (COX-2) blockade: results of a case control study. Int J Biol Sci. 2007;3(5):328–334. doi: 10.7150/ijbs.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris RE, Beebe-Donk J, Alshafie GA. Similar reductions in the risk of human colon cancer by selective and nonselective cyclooxygenase-2 (COX-2) inhibitors. BMC Cancer. 2008;8:237. doi: 10.1186/1471-2407-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N, Sood S, Wang S, et al. Overexpression of 5-lipoxygenase and cyclooxygenase 2 in hamster and human oral cancer and chemopreventive effects of zileuton and celecoxib. Clin Cancer Res. 2005;11(5):2089–2096. doi: 10.1158/1078-0432.CCR-04-1684. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Wang S, Wu N, et al. Overexpression of 5-lipoxygenase in rat and human esophageal adenocarcinoma and inhibitory effects of zileuton and celecoxib on carcinogenesis. Clin Cancer Res. 2004;10(19):6703–6709. doi: 10.1158/1078-0432.CCR-04-0838. [DOI] [PubMed] [Google Scholar]
- 46.Shi HY, Lv FJ, Zhu ST, Wang QG, Zhang ST. Dual inhibition of 5-LOX and COX-2 suppresses esophageal squamous cell carcinoma. Cancer Lett. 2011;309(1):19–26. doi: 10.1016/j.canlet.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Baron JA, Sandler RS, Bresalier RS, et al. A randomized trial of rofecoxib for the chemoprevention of colorectal adenomas. Gastroenterology. 2006;131(6):1674–1682. doi: 10.1053/j.gastro.2006.08.079. [DOI] [PubMed] [Google Scholar]
- 48.Rostom A, Dube C, Lewin G, et al. Nonsteroidal anti-inflammatory drugs and cyclooxygenase-2 inhibitors for primary prevention of colorectal cancer: a systematic review prepared for the US Preventive Services Task Force. Ann Intern Med. 2007;146(5):376–389. doi: 10.7326/0003-4819-146-5-200703060-00010. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Buell JF, Gross TG, Woodle ES. Malignancy after transplantation. Transplantation. 2005;80(Suppl 2):S254–S264. doi: 10.1097/01.tp.0000186382.81130.ba. [DOI] [PubMed] [Google Scholar]
- 52.Taylor L, Hughes RA, McPherson K. The risk of cancer from azathioprine as a treatment for multiple sclerosis. Eur J Neurol. 2004;11(2):141. doi: 10.1046/j.1351-5101.2003.00721.x. [DOI] [PubMed] [Google Scholar]
- 53.Benson C, White J, De Bono J, et al. A phase I trial of the selective oral cyclin-dependent kinase inhibitor seliciclib (CYC202; R-Roscovitine), administered twice daily for 7 days every 21 days. Br J Cancer. 2007;96(1):29–37. doi: 10.1038/sj.bjc.6603509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bible KC, Peethambaram PP, Oberg AL, et al. A phase 2 trial of flavopiridol (Alvocidib) and cisplatin in platin-resistant ovarian and primary peritoneal carcinoma: MC0261. Gynecol Oncol. 2012;127(1):55–62. doi: 10.1016/j.ygyno.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bose P, Perkins EB, Honeycut C, et al. Phase I trial of the combination of flavopiridol and imatinib mesylate in patients with Bcr-Abl+hematological malignancies. Cancer Chemother Pharmacol. 2012;69(6):1657–1667. doi: 10.1007/s00280-012-1839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Tourneau C, Faivre S, Laurence V, et al. Phase I evaluation of seliciclib (R-roscovitine), a novel oral cyclin-dependent kinase inhibitor, in patients with advanced malignancies. Eur J Cancer. 2010;46(18):3243–3250. doi: 10.1016/j.ejca.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Gry M, Rimini R, Stromberg S, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GEO accession and platform numbers, organs and cancer types, GEO sample numbers (GSMs), and specimen sources for each of the 78 experimental groups.
List of ‘FDA-approved antineoplastic drugs’, of ‘FDA-approved nonantineoplastic drugs’, and ‘non-FDA-approved chemical agents’.
Table S2.1 List of ‘FDA-approved antineoplastic drugs’.
Table S2.2 List of ‘FDA-approved antineoplastic drugs’.
Table S2.3 List of ‘FDA-approved antineoplastic drugs’.
The intersection of significantly upregulated genes identified by two-tailed Student’s t-test analysis of 78 experimental groups with the three categories of protein inhibitors.
The fold change of genes that were upregulated in cancer.