Abstract
Normal mammary gland homeostasis requires the coordinated regulation of protein signaling networks. However, we have little prospective information on whether activation of protein signaling occurs in premalignant mammary epithelial cells, as represented by cells with cytological atypia from women who are at high risk for breast cancer. This information is critical for understanding the role of deregulated signaling pathways in the initiation of breast cancer and for developing targeted prevention and/or treatment strategies for breast cancer in the future. In this pilot and feasibility study, we examined the expression of 52 phosphorylated, total, and cleaved proteins in 31 microdissected Random Periareolar Fine Needle Aspiration (RPFNA) samples by high-throughput Reverse Phase Protein Microarray. Unsupervised hierarchical clustering analysis indicated the presence of four clusters of proteins that represent the following signaling pathways: (1) receptor tyrosine kinase/Akt/mammalian target of rapamycin (RTK/Akt/mTOR), (2) RTK/Akt/extracellular signal-regulated kinase (RTK/Akt/ERK), (3) mitochondrial apoptosis, and (4) indeterminate. Clusters 1 through 3 comprised moderately to highly expressed proteins, while Cluster 4 comprised proteins that are lowly expressed in a majority of RPFNA samples. Our exploratory study showed that the interlinked components of mitochondrial apoptosis pathway are highly expressed in all mammary epithelial cells obtained from high-risk women. In particular, the expression levels of anti-apoptotic Bcl-xL and pro-apoptotic Bad are positively correlated in both non-atypical and atypical samples (unadjusted P < 0.0001), suggesting a delicate balance between the pro-apoptotic and anti-apoptotic regulation of cell proliferation during the early steps of mammary carcinogenesis. Our feasibility study suggests that the activation of key proteins along the RTK/Akt pathway may tip this balance to cell survival. Taken together, our results demonstrate the feasibility of mapping proteomic signaling networks in limited RPFNA samples obtained from high-risk women and the promise of developing rational drug targets or preventative strategies for breast cancer in future proteomic studies with a larger cohort of high-risk women.
Keywords: Breast cancer risk, Atypical mammary cytology, Protein microarray, Biomarker development, Cell survival
Introduction
Mammary gland homeostasis requires a coordinated regulation of cell signaling networks that comprises key regulatory protein kinases, phosphatases, and proteases. Deregulation of signaling pathways that control cell proliferation, differentiation, survival, and death is thought to play an important role in the breast cancer initiation and progression. For example, the superfamily of receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR or related ErbB family) and insulin-like growth factor receptor (IGF-1R), has proliferative and survival roles during mammary ductal outgrowth and branching [1, 2]. However, overexpression of these RTKs can disrupt the development and lead to tumorigenesis [3, 4]. In addition, the superfamily of serine/threonine kinases, such as Protein Kinase B/Akt, plays an important role in mammary carcinogenesis. The three isoforms of Akt (Akt1, Akt2, and Akt3) have distinct roles in this function. For example, the overexpression of Akt2 in ErbB2-over-expressing breast cancer cells resulted in an invasive phenotype [5], while overexpression of Akt1 only accelerated tumor onset [6]. Emerging evidence suggests that overexpression of Akt3 is inversely correlated with estrogen receptor (ER) status [7]. Moreover, cross-talk between signaling pathways may provide alternative mechanisms of cellular deregulation as well as resistance to treatment.
Elucidation of activated signaling pathways in tumor cells has shown promise to identify molecular targets for rational drug design as well as identify patients who may benefit from specific targeted therapeutics [8–13]. In this article, we report the feasibility of profiling mammary epithelial cells obtained from prospective cytological specimens of women who are at a high risk for breast cancer, using Reverse Phase Protein Microarray (RPPM) technology. RPPM is a high-throughput technique for analyzing the expression and activation levels of a large number of signaling proteins in pure cell populations obtained from small clinical samples. The RPPM format immobilizes dozens of protein lysates from different patient samples on one array, followed by incubation with a specific antibody. Thus, a single protein endpoint is measured and directly compared across multiple samples.
We employed Random Periareolar Fine Needle Aspiration (RPFNA) to sample mammary epithelial cells from the whole breast of women who are at a high risk for developing breast cancer. RPFNA has been utilized to assess the short-term breast cancer risk and test for response to chemo-prevention in high-risk women [14, 15]. The presence of cytological atypia in breast RPFNAs of high-risk women confers a 5.6-fold increase of short-term risk for the subsequent development of breast cancer [14]. Recently, we demonstrated that RPFNA is a highly reproducible technique for examining the cytology of replicate samples obtained from the same breasts of high-risk women in a multi-institutional cross-sectional study [16].
In this pilot and feasibility study, we describe the utility of RPPM technology to profile cell signaling pathways in limited RPFNA samples obtained from women at a high risk for breast cancer. Majority of these cytological specimens were obtained from women undergoing lumpectomy, lymph node biopsy, or prophylactic mastectomy in the operating room. Initially, a total of 60 protein endpoints were tested in mammary epithelial cells of 26 high-risk women. We identified three clusters of moderately to highly expressed proteins as well as one cluster of lowly expressed proteins in a majority of aspirates with cytological atypia. Of these clusters, Cluster 3 demonstrated the highest expression of proteins that include specific components of the mitochondrial apoptosis pathway. Our feasibility study demonstrates that the overall levels of key regulators of cytochrome C release determined the susceptibility of cells to programmed cell death or apoptosis. In addition, the majority of RPFNA samples with cytological atypia showed high expression of key cell survival proteins. The preliminary information provided by this pilot study will form the basis of future (a) proteomic studies to test the reproducibility of and changes in protein signaling in cytological samples obtained from both breasts of high-risk women and (b) functional studies with specific targeted agents to identify potential strategies for breast cancer prevention, early detection, and therapeutic intervention.
Materials and methods
Patient population
This study examined the cytological specimens of 26 women who are at a high risk for breast cancer. Specimen collection was conducted from June 2008 to February 2010 under an Institutional Review Board-approved protocol at the Duke University Medical Center.
Eligibility criteria
Women recruited to this study were required to have at least one of the following risk factors for breast cancer: (a) 5-year Gail risk calculation ≥1.7%, (b) a prior excisional biopsy exhibiting atypical hyperplasia, DCIS, or LCIS, (c) a known BRCA1/2 mutation, or (d) a history of invasive breast cancer. In women with history of prior DCIS treated with radiation, RPFNA was performed on the contralateral breasts. However, women with DCIS treated by excision alone (i.e., no radiation) were eligible to undergo bilateral aspiration. In some of the women with prior invasive cancer, the contralateral breasts were aspirated in the operating room (OR).
The primary goal of this pilot study was to profile protein expression of various cell signaling proteins that we hypothesize are deregulated in atypical mammary epithelial cells. We employed RPFNA as a research tool for capturing the molecular changes that may be occurring in these cells, which may or may not progress to become cancer cells. To detect these changes or “field effects” by protein microarray, we have made few exceptions to the RPFNA eligibility criteria on nearly half of the women (with the history of invasive breast cancer) who underwent surgical procedure (Table 1), and allowed random aspirations of the affected breasts.
Table 1.
Patient characteristics
| Total n = 26 | Low Masood n = 7 | High Masood n = 19 | P-value | |
|---|---|---|---|---|
| Age, years | 0.12a | |||
| Mean (SE) | 46.7 (8.7) | 50.4 (1.5) | 45.3 (2.2) | |
| Median | 48 | 50 | 47 | |
| Range | 26–68 | 45–56 | 26–68 | |
| Race, % (n) | 0.37b | |||
| Caucasian | 69 (18) | 86 (6) | 63 (12) | |
| African American | 27 (7) | 14 (1) | 32 (6) | |
| Latina | 4 (1) | 0 (0) | 5 (1) | |
| Menopausal status, % (n) | 0.37b | |||
| Pre | 62 (16) | 43 (3) | 68 (13) | |
| Post/peri | 38 (10) | 57 (4) | 32 (6) | |
| Prior abnormal biopsy, % (n) | 0.64b | |||
| None | 31 (8) | 43 (3) | 26 (5) | |
| DCIS | 23 (6) | 14 (1) | 26 (5) | |
| Cancer | 46 (12) | 43 (3) | 48 (9) | |
| Family history of breast cancer, % (n) | 0.34b | |||
| No | 73 (19) | 57 (4) | 79 (15) | |
| Yes | 27 (7) | 43 (3) | 21 (4) |
Mann–Whitney U-test was employed to compare age between the low and high Masood groups
Fisher’s exact test was used for group comparison with respect to race (Caucasian vs. Non-Caucasian), menopausal status, prior biopsy (absence vs. presence), and family history of breast cancer
Demographic data, family history of breast cancer, menopausal status, and history of abnormal biopsies were collected by patient interview, patient chart reviews, and/or clinical assessment by a physician.
Breast RPFNA and cytology assessment
Aspirates were collected and processed according to the published methods of Carol Fabian and others [14–18]. Cells from the right and left breast were processed separately, yielding one sample per aspirated breast. In this pilot and feasibility study, the majority of the samples were obtained from the operating room due to one or more of the following reasons: prophylactic mastectomy, lumpectomy, or lymph node biopsy. Our RPFNA consent form allows for aspirations in women with suspicious mass in the OR as long as they have been evaluated by a breast surgeon who agrees to have the RPFNA performed on the day of the surgery. These women have core needle biopsies to document the presence or the absence of cancer. Such exception to the RPFNA eligibility criteria helped to increase the accrual of African American women as well collect atypical and/or suspicious cells from women who are potentially at an immediate risk for developing breast cancer. The final decision to aspirate both breasts of women was dependent on the breast surgeon. In some instances, the surgeon elected to aspirate only one breast due to either a time constraint in the operating room or a situation in which the surgical drapes were positioned in such a way that the patient would have to be re-prepped and draped to have a second aspiration (and thus, subjected to additional OR costs).
Cytomorphology assessment and classification of cells by both categorical and semi-quantitative Masood cytology index scores [19, 20] were performed by a blinded, single cytopathologist (C.Z.). Index scores of ≤10 are classified as non-proliferative, 11–13 as hyperplasia (without atypia), 14–17 as atypia (also known as hyperplasia with atypia), and 18–24 as suspicious for malignancy [16–18]. It is noted that the above total scoring system is slightly different from previous studies [14, 15, 19, 20].
Preparation of RPFNA samples for laser capture microdissection
Aspirates were immediately fixed in CytoLyt® (Hologic Inc., Marlborough, MA) containing final concentration of 1% formalin for 24 h at room temperature. RPFNA samples were washed in CytoLyt until the majority of red blood cells were removed and then stored at 4°C. A portion of the cell pellets from each sample were transferred to PreservCyt, followed by cytospin slide preparations via Shandon Cytospin® 4 Cytocentrifuge (Thermo Scientific, Waltham, MA). Individual slides were immediately placed in 75% ethanol for 30 s and hematoxylin stain (Sigma-Aldrich, St. Louis, MO) for 20 s. The slides were dehydrated in a graded series of ethanol (75%, 95% and 100%) for 30 s and xylene for 5 min, followed by air drying. The 75% ethanol and hematoxylin staining solutions were supplemented with Complete™ Protease Inhibitor tablets (Roche Applied Science, Indianapolis, IN). Stained slides were stored in a slide box containing fresh desiccant at room temperature until laser capture microdissection (LCM). Epithelial cells were microdissected with a PixCell II LCM system (MDS Analytical Technologies, Sunnyvale, CA) and stored onto microdis-section caps at −80°C until lysed. Anonymity of each sample was maintained throughout the RPPM analysis at the George Mason University, while data analysis was performed at the Duke University Medical Center.
Reverse phase protein microarrays
As previously described, cellular lysates from microdis-sected mammary epithelial cells were prepared and printed in triplicate onto nitrocellulose-coated slides, using an Aushon 2470 arrayer equipped with 350 μm pins [11–13]. Cellular lysates from A431 ± EGF, HeLa ± pervanadate, or Jurkat ± calyculin served as positive controls for antibody staining. Immunostaining of lysate arrays as well as image analysis of antibody- and SYPRO Ruby-stained slides were performed as previously described [11]. For this study, we initially chose 60 antibodies to examine the broad signaling pathways thought to be involved in breast cancer cell proliferation, survival, apoptosis, and metastasis.
Statistics
The mean relative intensity for each protein, as shown in the supplementary dataset (Online Resource 1), was background subtracted and then normalized to total protein. Unsupervised hierarchical clustering analysis was performed on log 2-transformed values for each protein endpoint, using average linkage of the Pearson correlation coefficient. To determine the associations between the proteomic measurements and patient characteristics, the non-parametric Wilcoxon rank sum test and Spearman correlation coefficient were used for categorical and continuous variables, respectively. Univariate logistic regression was performed on log 2-transformed values to determine whether expression of a particular protein(s) associate with a particular outcome, which in this case is presence of cytological atypia. To determine the associations between the two groups of categorical variables, Fisher’s exact test was employed. Two-sided P-values < 0.05 were reported as significant, and the Benjamini–Hochberg method was employed to estimate the False Discovery Rate from multiple comparisons. The collective association of protein clusters to patient characteristics was evaluated by using Significant Analysis of Function and Expression (SAFE) [21]. SAFE is a resampling-based method for assessing the significance of functional categories while appropriately accounting for co-expression of proteins.
Results
Demographic and clinical characteristics
The present study set included 26 women, five of whom contributed RPFNA samples from both breasts, and 21 women contributed samples from a single breast. RPFNA samples from 65% (17/26) of these women were obtained from the operating room due to (a) removal of suspicious lesion or (b) prophylactic mastectomy given the strong family history of breast cancer or the presence of BRCA1 mutation. In 3 of 17 women who underwent surgery, invasive cancer was diagnosed. Meanwhile, the remaining 35% (3/26) of women in this feasibility study set were aspirated in the clinic. Demographic and clinical data are summarized in Table 1.
Eight of the 26 eligible women did not have history of abnormal biopsy; among these women, two contributed bilateral samples (Table 1). Out of the six women with prior DCIS, two women were treated by excision alone; one woman contributed bilateral samples. In the remaining four women with prior DCIS, only the contralateral breasts were aspirated.
Forty-six percent (12/26) of women with prior invasive breast cancer underwent surgical procedures. The distribution of women among the different surgical procedures is as follows: 33% mastectomy alone, 17% lumpectomy alone, 8% lymph node biopsy alone, 33% mastectomy + lymph node biopsy, and 9% lumpectomy + lymph node biopsy.
Cytology and risk factor associations
Out of the 31 RPFNA samples that were assessed by our modified Masood cytology scoring system, 3% were non-proliferative, 16% were hyperplastic, 75% were atypical, and 6% were suspicious of cancer. Our cytopathologist, Dr. Carola Zalles, assigned both a qualitative assessment (hyperplasia, atypia, or suspicious) and a Masood score. We tested for the categories assigned to the Masood cytology score (hyperplasia: 11–13; atypia: 14–17, and suspicious for malignancy: >17). There was an agreement between the qualitative descriptor and the quantitative Masood score in 97% (30/31) of samples. One sample was not in agreement, which in this case, a Masood score of 14 with a qualitative descriptor of hyperplasia.
Regardless of the technique employed to sample mammary cells, the presence of atypia increases the risk of women to short-term breast cancer [14, 22–25]. We selected a Masood score of 15 and above to indicate the presence of atypia, a surrogate marker of short-term breast cancer, in RPFNA cytology. By stratifying cytological index scores into low and high Masood groups, we are then able to assess the impact of cytological atypia, if any, on risk factors for breast cancer and expression levels of various signaling proteins. Masood scores ≤14 were classified as low Masood group, while Masood scores ≥15 were classified as high Masood group. Approximately, 29% and 71% of the RPFNA samples comprised the low Masood and high Masood groups, respectively. Two of the samples in the high Masood group had suspicious (for malignancy) cytology.
The distribution of women in the two Masood groups is shown on Table 1. Three of the women in the high Masood group were BRCA1 mutation carriers; two of which elected to have bilateral mastectomies. Women in low Masood group are not significantly different from women in high Masood group with respect to age, race, and menopausal status. Moreover, prior abnormal biopsies and family history of breast cancer are not different between these two groups.
A difference of 2 units or more between the Masood scores of bilateral RPFNA samples was considered disparate. Two of the 5 women who contributed bilateral samples had disparate Masood scores. To assign a woman with disparate bilateral index scores to either low or high Masood group, we determined the average of the two scores. For example, an average score of 14.5 was assigned to a high Masood group.
Protein clusters identified by exploratory analysis of RPFNAs
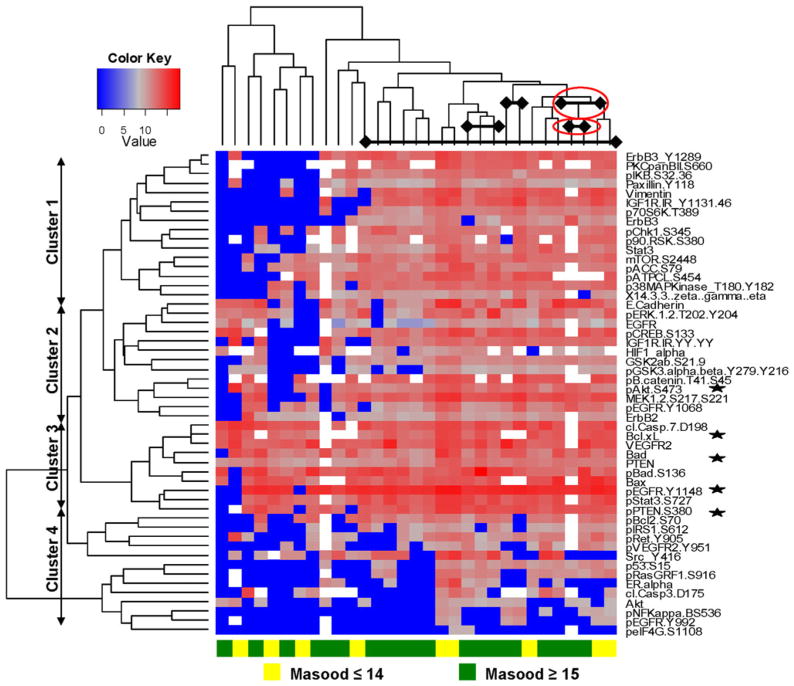
Before analysis, mammary epithelial cells were enriched by LCM to prevent contamination of signal from stroma. 60 proteins were initially examined by RPPM because they are thought to play an important role in deregulation of the broad, pro-survival RTK/PI3K/Akt/mTOR pathway. However, eight proteins were excluded from analysis due to insufficient intensity signals. Unsupervised hierarchical clustering analysis of the 52 protein endpoints revealed three major clusters of proteins (Fig. 1) encompassing the following activated pathways: (1) RTK/Akt/mTOR, (2) RTK/Akt/ERK, and (3) mitochondrial apoptosis. Proteins that regulate or contribute to mitochondrial apoptosis (Cluster 3) showed the highest expression (Fig. 1). There was one cluster of mostly inactivated or low-expressing proteins, such as ER alpha and phosphorylated NFkB S536, which we collectively referred to as indeterminate Cluster 4. In contrast, the unsupervised clustering analysis did not segregate the RPFNA samples into low and high Masood groups (Fig. 1) or patient characteristics, such as race (Caucasian vs. non-Caucasian) or menopausal status (pre vs. post) (data not shown). Moreover, there was no clear similarity in the protein expression profiles of paired samples (i.e., bilateral samples) (Fig. 1).
Fig. 1.
Unsupervised hierarchical clustering analysis of log 2-transformed values of 52 proteins [30]. Four clusters of proteins (rows) were identified in 31 RPFNA cytology samples (columns) with (Masood scores ≥15; green) and without (Masood scores ≤14; yellow) atypia. Shades of blue represent an increase in protein expression, while shades of gray and red represent a decrease in protein expression. White spaces within the heatmap represent missing or not determined values due to insufficient intensity signals from specific samples for a given protein. Bilateral samples from the same individual are indicated by diamonds connected by a solid line; those samples with disparate Masood scores are indicated in red ovals. Representative proteins of the mitochondrial apoptosis and EGFR/PI3K/Akt pathways are also indicated (stars)
Unadjusted Wilcoxon rank sum test indicated differential expression or activation of the following proteins between the samples with low Masood scores and high Masood scores: Bcl-xL (P = 0.003), Bad (P = 0.011), phosphorylated Akt S473 (P = 0.025), phosphorylated EGFR Y992 (P = 0.036), and phosphorylated EGFR Y1068 (P = 0.043). All of these proteins were expressed at a higher level in the low Masood group. However, these apparent associations were not statistically significant after correction with Benjamini–Hochberg method. For the four clusters of proteins identified by unsupervised hierarchical clustering, the distributions of the aforementioned proteins are shown in Table 2. Although no single cluster was statistically significant, components of the RTK/Akt/ERK (Cluster 2), mitochondrial apoptosis (Cluster 3), and indeterminate (Cluster 4) pathways showed negative correlation between the protein expression and high Masood scores (Fig. 1 and Table 3).
Table 2.
Proteins associated with high Masood scores within different protein clusters
| Protein clusters | Number of proteins associated with high Masood scores | Protein names |
|---|---|---|
| Cluster 1 | 0 | N/A |
| Cluster 2 | 2 | Phospho-Akt S473 |
| Phospho-EGFR Y1068 | ||
| Cluster 3 | 2 | Bcl-xL |
| Bad | ||
| Cluster 4 | 1 | Phospo-EGFR Y992 |
Table 3.
Pathway analysis for the four protein clusters with high Masood scores
| Protein clusters | P-value | Mean correlation | Minimal correlation | Maximal correlation |
|---|---|---|---|---|
| Cluster 1 | 0.871 | −0.015 | −0.277 | 0.235 |
| Cluster 2 | 0.187 | −0.106 | −0.386 | 0.177 |
| Cluster 3 | 0.253 | −0.177 | −0.440 | 0.070 |
| Cluster 4 | 0.498 | −0.080 | −0.495 | 0.134 |
Expression of key regulators of mitochondrial apoptosis in breast aspirates of high-risk women
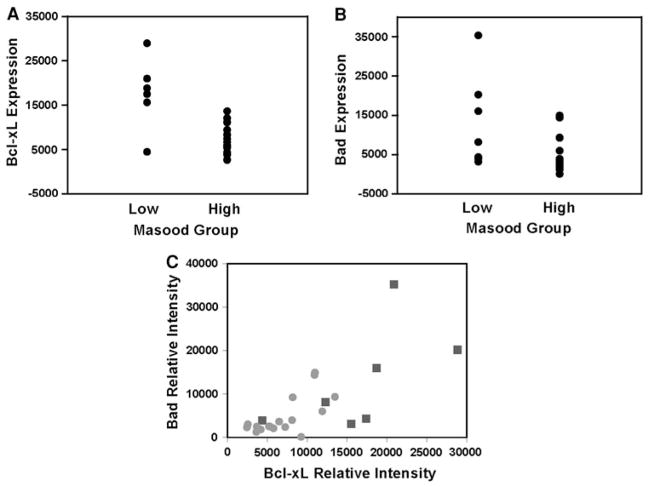
In previous studies, breast aspirates with cytological atypia demonstrated a 5.6-fold increased risk to breast cancer [14]. We next examined whether any of the proteins in the study set associate with atypia, as measured by Masood cytology index scores ≥15, by performing a logistic regression on log 2-transformed values of each protein. Out of the 52 proteins, Bcl-xL (P = 0.021) and Bad (P = 0.026) expression levels seem to associate with the presence of atypia in aspirates of high-risk women. Both proteins belong to a family of B-cell lymphoma-2 (Bcl-2) proteins that play crucial but opposing roles in mitochondrial pathway of the programmed cell death or apoptosis [26, 27]. Upon closer examination, the relative intensity values of each protein (Fig. 2a, b) followed a non-normal distribution (data not shown). Spearman correlation coefficient analysis indicated that the protein levels of Bcl-xL and Bad in all the samples of high-risk women are positively and significantly correlated (r = 0.74, P < 0.0001) (Fig. 2c). When similar correlation analysis was performed by the group classification system, the positive correlation between the Bcl-xL and Bad levels was more statistically significant in the high Masood group (P = 0.009) than in the low Masood group (P = 0.036). However, a comparison of the central tendencies or medians of Bcl-xL/Bad ratios indicated that the low Masood (median = 1.17, range: 0–5.02, SD = 1.74, N = 9) and high Masood (median = 1.47, range: 0–3.27, SD = 1.10, N = 21) groups are not significantly different (P > 0.05). Thus, the relative expression levels of the anti-apoptotic Bcl-xL and the pro-apoptotic Bad appear to be balanced in the breast aspirates of high-risk women.
Fig. 2.
Association between the Bcl-xL and Bad proteins in mammary cytological samples. Plots of a Bcl-xL and b Bad expression levels for samples without atypia (Masood scores ≤14; squares) and with atypia (Masood scores ≥15; circles). c Spearman rank correlation analysis showed a statistically significant positive correlation between the Bcl-xL and Bad levels (P < 0.0001)
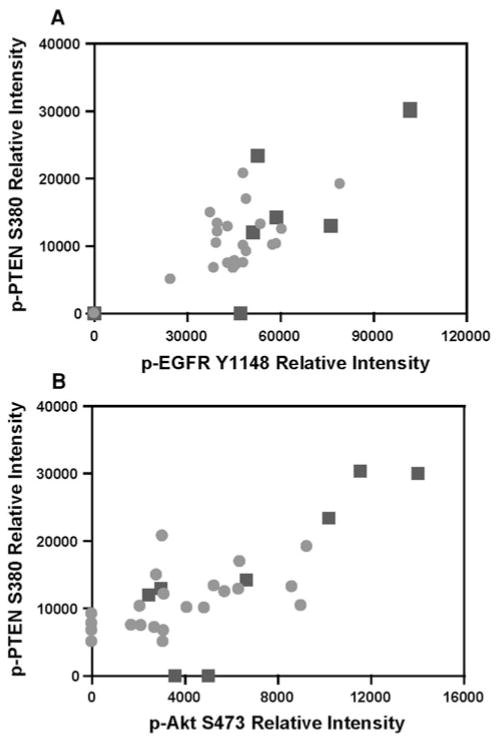
An alternative strategy employed by premalignant cells to evade apoptosis is activation of pro-survival RTK/PI3K/ Akt pathway [24]. To this end, we also examined the correlation between the pairs of key proteins that may play important cell survival roles, specifically proteins from Clusters 2 and 3. As indicated in Fig. 3, we observed moderate but significant correlations in the following pairs of phosphorylated (p) proteins: p-EGFR Y1148 and p-PTEN S380 (r = 0.60, P = 0.0004); p-Akt S473 and p-PTEN S380 (r = 0.63, P = 0.0002).
Fig. 3.
Spearman rank correlation of proteins within pro-survival EGFR/PI3K/Akt pathway. Moderate but significant correlations were observed between the relative protein levels of a phosphorylated (p) phosphatase and tensin homolog S380 (p-PTEN S380) and p-EGFR Y1148 as well as b p-PTEN S380 and p-Akt S473 in RPFNA samples without atypia (squares) and with atypia (circles)
Discussion
Protein microarray technology is an evolving high-throughput technology for examining several proteins simultaneously to determine the roles of cell signaling pathways in normal and diseased states. In combination with the laser capture microdissection, this technology can be employed to examine the signaling pathways of distinct cell populations in limited human specimens. It is becoming clear that the molecular profiling of patients’ samples may contain important information for individualized prognostication and treatment selection. In the present study, we tested the feasibility of using RPPM to interrogate signaling events in mammary epithelial cells of high-risk women obtained by RPFNA. Both survival and apoptotic pathways were examined in this feasibility study set.
The presence of atypia in cytological specimens has been utilized as a surrogate marker of breast cancer risk in high-risk populations [14, 15, 19]. Using a Masood cytology index score of 15 as a cutoff for atypia, we stratified the RPFNA samples into two groups. The samples with Masood index scores ≥15 were collectively considered atypical, while the samples with Masood scores ≤14 were considered non-atypical. Owing to the small sample size of the study set, the two Masood groups were not statistically different in age, race, menopausal status, prior abnormal biopsies, and family history of breast cancer (Table 1).
Initially, we tested whether any protein signature(s) associates with atypia, as measured by Masood scores ≥15. Unsupervised hierarchical clustering analysis indicated no clear separation of samples based on the absence or presence of atypia (Fig. 1, columns). However, there was a clear partitioning of proteins into four clusters, with Cluster 3 showing the highest expression or activation (Fig. 1, rows). Additional exploratory analysis indicated that phosphorylated proteins that promote cell survival (p-EGFR Y1068, p-EGFR Y992, and p-Akt S473) and control cell death (Bcl-xL and Bad) are activated or highly expressed in non-atypical samples, suggesting deregulation of these pathways in the early steps of mammary carcinogenesis.
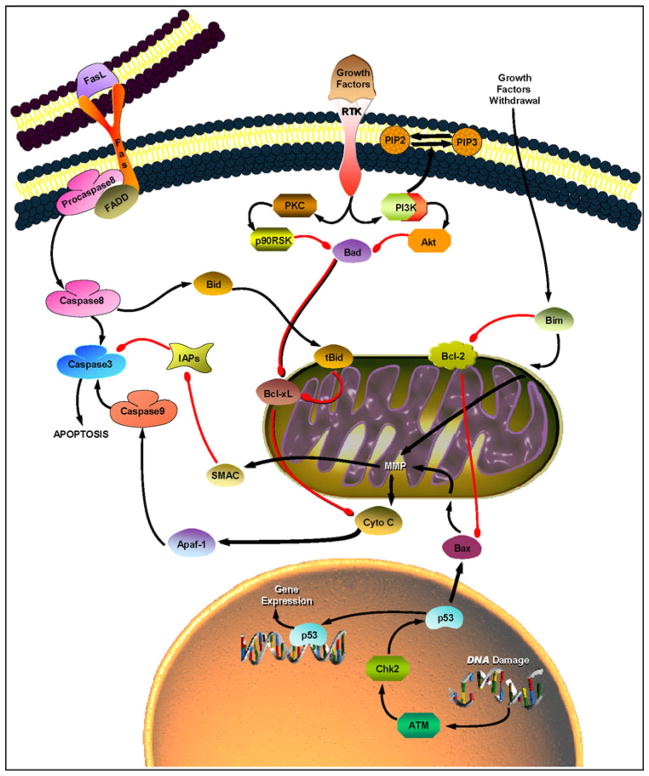
Some of the proteins in Cluster 3 are known regulators of mitochondrial apoptosis (Fig. 4). The intrinsic mitochondrial pathway of apoptosis is induced in response to various cellular stress signals, including direct DNA damage, hypoxia, growth or survival factor withdrawal, and oncogenes [26–29]. In general, these stress signals result in the activation of pro-apoptotic proteins and subsequent interaction with anti-apoptotic proteins, leading to destabilization of the outer mitochondrial membrane and subsequent release of cytochrome C. Consequently, the apoptosome formation (consisting of Apaf-1 and procaspase-9) and the caspase-9/3 activation result in apoptosis (Fig. 4).
Fig. 4.
Two major pathways to apoptosis. The mitochondrial pathway (right) is activated by cellular stress signals, including growth factor withdrawal and DNA damage, and often results in activation of pro-apoptotic Bcl-2 family members (Bad, Bid, Bim, and Bax)1. These pro-apoptotic proteins form dimers with anti-apoptotic Bcl-xL and Bcl-2 at the surface of mitochondria, resulting in mitochondrial membrane permeabilization (MMP) and subsequent release of cytochrome C (Cyto C). Consequently, Apaf-1 and procaspase9 associate with Cyto C to form an apoptosome, resulting in caspase9 activation. Procaspase3 is then cleaved by caspase9 to form caspase3, committing the cell to apoptosis. Inhibition of apoptosis is primarily mediated by components of the RTK/PI3K/Akt signaling pathway. The death receptor-mediated pathway (left) is triggered by members of the Fas (or CD95) receptor superfamily upon stimulation by FasL. This interaction results in the formation of death-inducing complex, which comprises FADD (Fas-associated death domain protein) and procaspase8, and subsequent caspase8/3 activation. The activity of caspase3 is antagonized by IAPs (inhibitors of apoptosis), which in turn are antagonized by SMAC (second mitochondria-derived activator of caspase) [26]
The following Bcl-2 family members are key regulators of mitochondrial apoptosis, specifically in the release of cytochrome C: Bcl-xL, Bcl-2, Bad, Bax, Bim, and Bid. Of these proteins, anti-apoptotic Bcl-xL and Bcl-2 reside in the intracellular membranes of mitochondria and prevent cytochrome C release (Fig. 4), while the remaining pro-apoptotic Bcl-2 family members reside in the cytosol as inactive forms and shuttle to the mitochondria following the activation by proteolysis (i.e., truncated Bid) and dephosphorylation (i.e., non-phosphorylated Bad) [26, 27]. In the present study, significant positive correlation between the Bcl-xL and Bad expression was observed in the breast RPFNAs of high-risk women with and without cytological atypia (Fig. 2c). Overall levels of pro- and anti-apoptotic proteins are thought to dictate a cell’s fate: cells with more pro-death proteins are sensitive to death, while cells with excessive anti-death proteins are usually resistant [26]. Survey of Cluster 3 (Fig. 1) indicated that six of the proteins contribute to cell survival (i.e., Bcl-xL, Bad S136, p-EGFR Y1148, p-Stat3 S727, p-PTEN S380, and VEG-FR2), while four of the proteins promote cell death (cleaved caspase7 D198, Bad, Bax, and PTEN). Given this observation, we predict that a subset of atypical samples in our study set comprised cells that will likely survive and develop to breast cancer. Our future proteomic studies will investigate large number of serial and/or bilateral RPFNAs of high-risk women who develop breast cancer to monitor both the cellular and molecular changes and identify potential targets of therapy. We will also validate our future protein expression profiles with immunohistochemistry (IHC) as previously attempted on a selected protein, using limiting RPFNA samples [30].
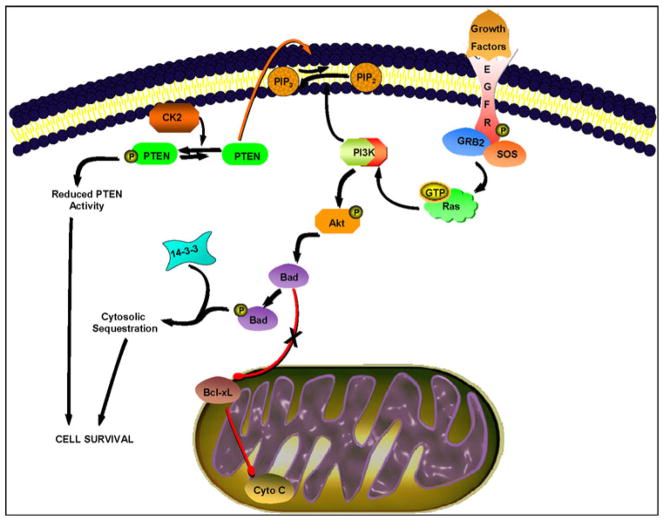
While our study did not examine the expression of Fas, cofactor FADD, or caspase-8, we cannot rule out the activation of death receptor-mediated apoptosis (Fig. 4) in a subset of our breast aspirates. Nonetheless, our results suggest that the activation of RTK family member EGFR at Y1148 leads to subsequent PI3K-mediated activation of downstream effector Akt (Fig. 5), which in turn phos-phorylates Bad at S136 [31–33]. Subsequent interaction of p-Bad S136 with 14-3-3 prevents translocation of Bad to the mitochondria [33, 34]. We propose that the reduction in the pool of unbound or non-phosphorylated Bad results in the decreased formation of Bcl-xL/Bad complex, which is necessary for pore formation to allow for cytochrome C release [35]. Thus, cell survival is favored in samples with activated EGFR/PI3K/Akt pathway (Fig. 5).
Fig. 5.
Proposed pro-survival signaling in RPFNA study set. Activation of p-EGFR Y1148 results in recruitment of adaptors Grb2 and Shc and subsequent activation of Ras and PI3K. PI3K-mediated phosphorylation of membrane-bound phosphatidylinositol-3,4-biphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) recruits Akt to the plasma membrane where it is phosphorylated by phosphoinositide-dependent kinase-1 (PDK-1). Akt inhibits apoptosis by phosphorylating Bad at S136 and preventing its localization to the mitochondria. Phosphorylated Bad S136 is then sequestered by 14-3-3 in the cytosol, resulting in cell survival. Alternatively, casein kinase II (CK2)-mediated phosphorylation of PTEN at S380 may contribute to cell survival
Moreover, our study suggests reduced PTEN activity, as indirectly assessed by p-PTEN S380 levels, in a majority of our samples (Fig. 1, Cluster 3; Fig. 5). Although our study did not examine casein kinase II (CK2) expression levels, PTEN phosphorylation at S380 has been shown to be mediated by CK2, resulting in decreased PTEN stability and subsequent proteosome-mediated degradation [36]. Phosphorylation of PTEN at S380 prevents PTEN from binding to its PDZ domain-containing partners, such as the p85 subunit of PI3K, and localizing to the plasma membrane [37]. Thus, such phosphorylation event will negatively regulate PTEN function as an antagonist of PI3K/ Akt signaling.
Although the small sample size precluded our ability to draw any statistically significant associations between the cytological atypia and the activation of specific proteins or protein clusters, as assessed by Benjamini–Hochberg method, we made several observations that are consistent with other studies in regards to deregulation of cell signaling during mammary carcinogenesis. First, the EGFR/ PI3K/Akt pathway is activated in a subset of atypical mammary epithelial cells from high-risk women (Figs. 1, 3), suggesting the importance of individual components of this pathway in cellular transformation and initiation of breast cancer as previously suggested by others [1–3, 32]. Second, our observed positive and significant correlation between p-PTEN S380 and p-EGFR Y1148 (Fig. 3a) was consistent with previous proteomic study of FNA samples taken from palpable masses of women diagnosed with stage 2 or 3 breast cancer [38]. Third, the balance between the sensitivity and resistance to cell death is regulated by the overall levels of members of the mitochondrial apoptosis pathway (Figs. 2, 4), which in turn is influenced by various stimuli [26–29]. Examination of the mean relative expression levels indicated an increase in p-Bad S136 expression by as much as 2.8-fold compared with positive regulators of cytochrome C release, such as non-phosphorylated Bad and Bax. The increased p-Bad S136 level is presumably due to the activation of the cell survival EGFR/ PI3K/Akt pathway [33, 34]. Fourth, the observation that PTEN stability may be compromised in our study set requires further validation with larger sample size and parallel biochemical experiments. There is an increasing evidence of important functional role of CK2 in modulation of the PTEN activity [36, 37] and global suppression of apoptosis [39].
In addition to small sample size, another limitation of our study is the lack of validation of the proteomic data with immunohistochemistry. The insufficient amount of cells and inadequate preservation of phosphoproteins in these cells precluded our ability to test for the expression of Bcl-xL, Bad, PTEN S380, Akt S473, or EGFR Y1148 by IHC. As mentioned earlier, we will test for correlation of the expression of selected proteins by RPPM and IHC in future studies.
Taken together, our exploratory proteomic analysis of mammary epithelial cells from high-risk women provides future opportunities to (a) identify potential biomarkers of breast cancer risk and progression, (b) develop combination therapies that are aimed at specific molecular targets within pro-survival signaling pathways, such as EGFR, Akt, and 14-3-3, and (c) study in greater detail the role of promising biomarker(s) predictive of response to targeted therapy.
Acknowledgments
The authors thank Susan G. Komen for the Cure foundation for financial support to Drs. Yu and Seewaldt (KG091020) and Dr. Ibarra-Drendall (KG090730). Dr. Seewaldt is also supported by the National Institute of Health/National Cancer Institute (R01CA88799 and R01CA114068). Additional financial support was provided by NCI/AVON Partners in Progress Grant 3 P30 CA014236-32S1 to Drs. Seewaldt and Yu.
Abbreviations
- Apaf-1
Apoptotic protease activating factor-1
- Bad
Bcl2-antagonist of cell death
- Bax
Bcl2 associated X-protein
- Bcl-2
B-cell lymphoma-2
- Bid
Bcl2 interacting protein
- BRCA1/2
Breast cancer-associated gene 1/2
- CK2
Casein kinase II or 2
- DCIS
Ductal carcinoma in situ
- EGFR
Epidermal growth factor receptor
- ER
Estrogen receptor
- ErbB
Erythroblastic leukemia viral oncogene homolog
- ERK
Extracellular signal-regulated kinase
- FADD
Fas-associated via death domain
- IHC
Immunohistochemistry
- IGF-1R
Insulin-like growth factor receptor
- LCIS
Lobular carcinoma in situ
- LCM
Laser capture microdissection
- mTOR
Mammalian target of rapamycin
- OR
Operating room
- PI3K
Phosphatidylinositol 3-kinase
- PTEN
Phosphatase and tensin homolog
- RPFNA
Random Periareolar Fine Needle Aspiration
- RPPM
Reverse phase protein microarray
- RTK
Receptor tyrosine kinase
Footnotes
Original pathway map was obtained from SABiosciences Pathway Central (www.sabiosciences.com).
Contributor Information
Catherine Ibarra-Drendall, Email: ibarr001@mc.duke.edu, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA.
Michelle M. Troch, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
William T. Barry, Department of Biostatistics & Bioinformatics, Duke University Medical Center, Durham, NC 27710, USA
Gloria Broadwater, Department of Biostatistics & Bioinformatics, Duke University Medical Center, Durham, NC 27710, USA.
Emanuel F. Petricoin, III, Center for Applied Proteomics and Molecular Medicine, George Mason University, Manassas, VA 20110, USA.
Julia Wulfkuhle, Center for Applied Proteomics and Molecular Medicine, George Mason University, Manassas, VA 20110, USA.
Lance A. Liotta, Center for Applied Proteomics and Molecular Medicine, George Mason University, Manassas, VA 20110, USA
Siya Lem, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA.
Joseph C. Baker, Jr., Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
Anne C. Ford, Department of Obstetrics and Gynecology, Duke University Medical Center, Durham, NC 27710, USA
Lee G. Wilke, Division of General Surgery, University of Wisconsin, Madison, WI 53792, USA
Carola Zalles, Texas A&M Health Sciences Center, College of Medicine, Round Rock, TX 78665, USA.
Nicole M. Kuderer, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
Abigail W. Hoffman, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
Melanie Shivraj, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA.
Priya Mehta, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA.
Jamila Williams, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA.
Nora Tolbert, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA.
Laurie W. Lee, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
Patrick G. Pilie, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
Dihua Yu, Department of Molecular and Cellular Oncology, MD Anderson Cancer Center, Houston, TX 77030, USA.
Victoria L. Seewaldt, Division of Medical Oncology, Duke University Medical Center, Box 2628, Durham, NC 27710, USA
References
- 1.Wickenden JA, Watson CJ. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res. 2010;12:202. doi: 10.1186/bcr2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes NE, Watson CJ. Mammary gland growth factors: roles in normal development and in cancer. Cold Spring Harb Perspect. 2010;2:a003186. doi: 10.1101/cshperspect.a003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandt R, Eisenbrandt R, Leenders F, Zschiesche W, Binas B, Juergensen C, Theuring F. Mammary gland specific hEGF receptor transgene expression induces neoplasia and inhibits differentiation. Oncogene. 2000;19:2129–2137. doi: 10.1038/sj.onc.1203520. [DOI] [PubMed] [Google Scholar]
- 4.Wood TL, Richert MM, Stull MA, Allar MA. The insulin-like growth factors (IGFs) and IGF binding proteins in postnatal development of murine mammary glands. J Mammary Gland Biol Neoplasia. 2000;5:31–42. doi: 10.1023/a:1009511131541. [DOI] [PubMed] [Google Scholar]
- 5.Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, Slamon DJ. Overex-pression of Akt2/protein kinase Bβ leads to up-regulation of β1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63:196–206. [PubMed] [Google Scholar]
- 6.Hutchinson JN, Jin J, Cardiff RD, Woodgett JR, Muller WJ. Activation of Akt-1 (PKB-alpha) can accelerate ErbB-2-mediated mammary tumorigenesis but suppresses tumor invasion. Cancer Res. 2004;64:3171–3178. doi: 10.1158/0008-5472.can-03-3465. [DOI] [PubMed] [Google Scholar]
- 7.Nakatani K, Thompson DA, Barthel A, Sakaue H, Liu W, Weigel RJ, Roth RA. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J Biol Chem. 1999;274:21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- 8.Gulmann C, Sheehan KM, Kay EW, Liotta LA, Petricoin EF., III Array-based proteomics: mapping of protein circuitries for diagnostics, prognostics, and therapy guidance in cancer. J Pathol. 2006;208:595–606. doi: 10.1002/path.1958. [DOI] [PubMed] [Google Scholar]
- 9.Petricoin EF, III, Bichsel VE, Calvert VS, Espina V, Winters M, Young L, Belluco C, Trock BJ, Lippman M, Fishman DA, Sgroi DC, Munson PJ, Esserman LJ, Liotta LA. Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J Clin Oncol. 2005;23:3614–3621. doi: 10.1200/JCO.2005.02.509. [DOI] [PubMed] [Google Scholar]
- 10.Hait WN. Targeted cancer therapeutics. Cancer Res. 2009;69:1263–1267. doi: 10.1158/0008-5472.CAN-08-3836. [DOI] [PubMed] [Google Scholar]
- 11.Espina V, Liotta LA, Petricoin EF., III Reverse-phase protein microarrays for theranostics and patient tailored therapy. Methods Mol Biol. 2009;520:89–105. doi: 10.1007/978-1-60327-811-9_7. [DOI] [PubMed] [Google Scholar]
- 12.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin EF, III, Liotta LA. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 13.Petricoin EF, III, Espina V, Araujo RP, Midure B, Yeung C, Wan X, Eichler GS, Johann DJ, Jr, Qualman S, Tsokos M, Krishnan K, Helman LJ, Liotta LA. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67:3431–3440. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 14.Fabian CJ, Kimler BF, Zalles CM, Klemp JR, Kamel S, Zeiger S, Mayo MS. Short-term breast cancer prediction by random periareolar fine-needle aspiration cytology and the Gail risk model. J Natl Cancer Inst. 2000;92:1217–1227. doi: 10.1093/jnci/92.15.1217. [DOI] [PubMed] [Google Scholar]
- 15.Fabian CJ, Kimler BF, Brady DA, Mayo MS, Chang CH, Ferraro JA, Zalles CM, Stanton AL, Masood S, Grizzle WE, Boyd NF, Arneson DW, Johnson KA. A phase II breast cancer chemoprevention trial of oral alpha-difluoromethylornithine: breast tissue, imaging, and serum and urine biomarkers. Clin Cancer Res. 2002;8:3105–3117. [PubMed] [Google Scholar]
- 16.Ibarra-Drendall C, Wilke LG, Zalles C, Scott V, Archer LE, Lem S, Yee LD, Lester J, Kulkarni S, Murekeyisoni C, Wood M, Wilson K, Garber J, Gentry C, Stouder A, Broadwater G, Baker JC, Jr, Vasilatos SN, Owens E, Rabiner S, Barron AC, Seewaldt VL. Reproducibility of random periareolar fine needle aspiration in a multi-institutional Cancer and Leukemia Group B (CALGB) cross-sectional study. Cancer Epidemiol Biomarkers Prev. 2009;18:1379–1385. doi: 10.1158/1055-9965.EPI-08-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bean GR, Scott V, Yee L, Ratliff-Daniel B, Troch MM, Seo P, Bowie ML, Marcom PK, Slade J, Kimler BF, Fabian CJ, Zalles CM, Broadwater G, Baker JC, Jr, Wilke LG, Seewaldt VL. Retinoic acid receptor-beta2 promoter methylation in random periareolar fine needle aspiration. Cancer Epidemiol Biomarkers Prev. 2005;14:790–798. doi: 10.1158/1055-9965.EPI-04-0580. [DOI] [PubMed] [Google Scholar]
- 18.Bean GR, Ibarra Drendall C, Goldenberg VK, Baker JC, Jr, Troch MM, Paisie C, Wilke LG, Yee L, Marcom PK, Kimler BF, Fabian CJ, Zalles CM, Broadwater G, Scott V, Seewaldt VL. Hypermethylation of the breast cancer-associated gene 1 promoter does not predict cytologic atypia or correlate with surrogate end points of breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:50–56. doi: 10.1158/1055-9965.EPI-06-0598. [DOI] [PubMed] [Google Scholar]
- 19.Zalles C, Kimler BF, Kamel S, McKittrick R, Fabian CJ. Cytology patterns in random aspirates from women at high and low risk for breast cancer. Breast J. 1995;1:343–349. [Google Scholar]
- 20.Masood S, Frykberg ER, McLellan GL, Scalapino MC, Mitchum DG, Bullard JB. Prospective evaluation of radiologically directed fine-needle aspiration biopsy of nonpalpable breast lesions. Cancer. 1990;66:1480–1487. doi: 10.1002/1097-0142(19901001)66:7<1480::aid-cncr2820660708>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 21.Barry WT, Nobel AB, Wright FA. A statistical framework for testing functional categories in microarray data. Ann Appl Stat. 2008;2:286–315. [Google Scholar]
- 22.Skolnick MH, Cannon-Albright LA, Goldgar DE, Ward JH, Marshall CJ, Schumann GB, Hogle H, McWhorter WP, Wright EC, Tran TD, et al. Inheritance of proliferative breast disease in breast cancer kindreds. Science. 1990;250:1715–1720. doi: 10.1126/science.2270486. [DOI] [PubMed] [Google Scholar]
- 23.Wrensch MR, Petrakis NL, Miike R, King EB, Chew K, Neuhaus J, Lee MM, Rhys M. Breast cancer risk in women with abnormal cytology in nipple aspirates of breast fluid. J Natl Cancer Inst. 2001;93:1791–1798. doi: 10.1093/jnci/93.23.1791. [DOI] [PubMed] [Google Scholar]
- 24.Page DL, Dupont WD. Anatomic markers of human premalignancy and risk of breast cancer. Cancer. 1990;66:1326–1335. doi: 10.1002/1097-0142(19900915)66:14+<1326::aid-cncr2820661405>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 25.Marshall LM, Hunter DJ, Connolly JL, Schnitt SJ, Byrne C, London SJ, Colditz GA. Risk of breast cancer associated with atypical hyperplasia of lobular and ductual types. Cancer Epidemiol Biomarkers Prev. 1997;6:297–301. [PubMed] [Google Scholar]
- 26.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 27.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 29.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 30.Pilie PG, Ibarra-Drendall C, Troch MM, Broadwater G, Barry WT, Petricoin EF, III, Wulfkuhle JD, Liotta LA, Lem S, Baker JC, Jr, Stouder A, Ford AC, Wilke LG, Zalles CM, Mehta P, Williams J, Shivraj M, Su Z, Geradts J, Yu D, Seewaldt VL. Protein microarray analysis of mammary epithelial cells from obese and nonobese women at high risk for breast cancer: feasibility data. Cancer Epidemiol Biomarkers Prev. 2011;20:476–482. doi: 10.1158/1055-9965.EPI-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foley J, Nickerson NK, Nam S, et al. EGFR signaling in breast cancer: bad to the bone. Semin Cell Dev Biol. 2010;21:951–960. doi: 10.1016/j.semcdb.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 34.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 35.Zha J, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer SJ. BH3 domain of BAD is required for heterodimerization with BCL-XL and pro-apoptotic activity. J Biol Chem. 1997;272:24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 36.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J Biol Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 38.Rapkiewicz A, Espina V, Zujewski Ja, Lebowitz PF, Filie A, Wulfkuhle J, Camphausen K, Petricoin EF, III, Liotta LA, Abati A. The needle in the haystack: application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer. 2007;111:173–184. doi: 10.1002/cncr.22686. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad KA, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2—a key suppressor of apoptosis. Adv Enzymol Regul. 2008;48:179–187. doi: 10.1016/j.advenzreg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]