Abstract
Copy number variations (CNVs), either DNA gains or losses, have been found at common regions throughout the human genome. Most CNVs neither have a pathogenic significance nor result in disease-related phenotypes but, instead, reflect the normal population variance. However, larger CNVs, which often arise de novo, are frequently associated with human disease. A genetic contribution has long been suspected in VACTERL (Vertebral, Anal, Cardiac, TracheoEsophageal fistula, Renal and Limb anomalies) association. The anomalies observed in this association overlap with several monogenetic conditions associated with mutations in specific genes, e.g. Townes Brocks (SALL1), Feingold syndrome (MYCN) or Fanconi anemia. So far VACTERL association has typically been considered a diagnosis of exclusion. Identifying recurrent or de novo genomic variations in individuals with VACTERL association could make it easier to distinguish VACTERL association from other syndromes and could provide insight into disease mechanisms. Sporadically, de novo CNVs associated with VACTERL are described in literature. In addition to this literature review of genomic variation in published VACTERL association patients, we describe CNVs present in 68 VACTERL association patients collected in our institution. De novo variations (>30 kb) are absent in our VACTERL association cohort. However, we identified recurrent rare CNVs which, although inherited, could point to mechanisms or biological processes contributing to this constellation of developmental defects.
Key Words: Chromosomal anomalies, Copy number variation, Esophageal atresia, Tracheoesophageal fistula, VACTERL, VATER
Copy number analysis has proven to be a powerful tool for identifying genes and genomic regions that contribute to the occurrence of congenital malformations. Common copy number variations (CNVs), regions of variable DNA gains or losses, account for a significant proportion of the healthy human genome [Iafrate et al., 2004; Sebat et al., 2004]. Most CNVs are inherited polymorphisms that have no appreciable effect on health. However, there are many examples of de novo or rare CNVs that have clearly been associated with human diseases, e.g. Wolf-Hirschhorn syndrome (OMIM 194190) and 22q11.2 deletion syndrome (OMIM 192430) [Girirajan et al., 2011]. Pathologic CNVs are often larger (>500 kb) in size and are usually not inherited from an unaffected parent [Miller et al., 2010].
Implementation of new molecular cytogenetic techniques, such as microarray-based comparative genomic hybridization and single nucleotide polymorphism arrays, has revealed previously unidentified genotypic aberrations which can now be correlated with phenotypic anomalies. As a result, numerous publications have implicated specific pathogenic CNVs in intellectual disability, congenital anomalies such as cleft lip, microcephaly, renal malformations [Southard et al., 2012], and neurological conditions including autism and schizophrenia [Girirajan et al., 2011]. It may very well be that, as in other congenital anomalies, there is a role for pathogenic CNVs in VACTERL association (OMIM 192350) etiology.
VACTERL association is a heterogeneous condition defined by 6 core structural defects (Vertebral, Anal, Cardiac, TracheaEsophageal fistula, Renal and Limb anomalies) which occur together more commonly than would be expected by chance alone.
These defects are also observed in several other monogenetic conditions caused by intragenic mutations, e.g. Townes-Brocks syndrome (OMIM 107480; SALL1) – whose features include imperforate anus, cardiac defects, renal anomalies, and hand defects, most often affecting the thumb – and Feingold syndrome (OMIM 164280; MYCN) – whose features can include esophageal atresia, cardiac anomalies, renal anomalies, and abnormalities of the hand and fingers [Solomon, 2011]. In addition to gene mutations, CNVs have been described as causal factor in several VACTERL-like syndromes. These include Goldenhar/OAVS (OMIM 141400) [Huang et al., 2010], Townes-Brocks syndrome [Bardakjian et al., 2009], X-linked VACTERL-H (OMIM 314390) [Chung et al., 2011], MURCS association (OMIM 601076) [Uliana et al., 2008], OEIS complex (OMIM 258040) [Kosaki et al., 2005; El-Hattab et al., 2010], TAR syndrome (OMIM 274000) [Klopocki et al., 2007], 13q32 deletion syndrome [Walsh et al., 2001], and 22q11.2 deletion syndrome [Ryan et al., 1997].
Due to the abundance of overlapping defects in various organs, the scarcity of known causal factors and its heterogeneous phenotype, VACTERL association is typically considered a diagnosis of exclusion. In general, the diagnosis is made when at least 3 of the 6 associated core defects are present and all other phenotypical overlapping syndromes have been excluded [Solomon et al., 2012].
The role of genetics in VACTERL association has long been suspected. VACTERL is usually a sporadic finding, but familial cases do exist [Hilger et al., 2012]. Moreover, in about 9% of VACTERL patients one of the relatives has 1 of the 6 core VACTERL features [Solomon et al., 2010b]. In some rare cases, genetic defects have been described, such as a polyalanine expansion [Wessels et al., 2010], nuclear [Damian et al., 1996; Garcia-Barceló et al., 2008; Szumska et al., 2008] or mtDNA [Thauvin-Robinet et al., 2006; Solomon et al., 2011] mutations, and numerical or structural chromosome aberrations [Shaw-Smith, 2006; Felix et al., 2007; Solomon et al., 2010a]. The resolution to detect these chromosomal anomalies has increased significantly with the introduction of micro-array technology, as current technologies allow detection of genomic imbalances down to only a few kb in size. Although the role of CNV and chromosomal aberrations in congenital anomalies is well established, little is known of their role in VACTERL association etiology. It is possible that recurrent or de novo genomic variations contribute to the development of some cases of VACTERL association. Identifying such changes could make it easier to distinguish VACTERL association from other syndromes and other potentially related conditions and could provide insight into disease mechanisms.
Materials and Methods
Literature Review
We reviewed the literature to identify both numerical or structural chromosomal anomalies and copy number variations described in individuals with VACTERL association. We followed the inclusion criteria for VACTERL association (3 or more of the core VACTERL elements, fig. 1) and excluded the patients with a confirmed genetic syndrome.
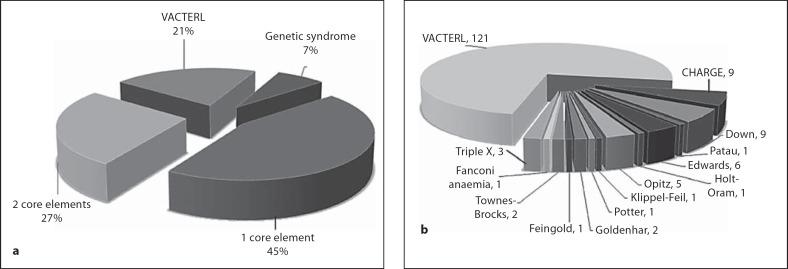
Fig. 1.
Inclusion criteria. a In total 45% of the EA/TEF cohort had 1 core component (TEF) and 27% had one additional core element. 163 patients out of 567 (21%) entries in the Rotterdam EA/TEF cohort met the criteria for VACTERL association. b 42 Patients had a confirmed genetic syndrome (7%) and were excluded from the VACTERL cohort.
Study Population
Since 1988, the Erasmus MC – Sophia Children's Hospital Department of Pediatric Surgery unit has been collecting clinical data and, when possible, DNA, from VACTERL patients. This cohort is part of a larger esophageal atresia and/or tracheoesophageal fistula (EA/TEF) cohort (n = 567) in which patient sampling and registration are based on the existence of either EA/TEF regardless of additional anomalies. Patients were selected using the same criteria as for the literature review. In total, DNA from 68 out of 121 VACTERL patients was analyzed for copy number variations.
Analysis of Copy Number Variation
CNV analysis was performed using Illumine 12-HumanCytoSNP, Human 610-Quad or Omni Express Bead Chips (Illumina Inc., San Diego, Calif., USA) according to manufacturer's instructions. Arrays are processed according to their manufacturer's standard protocol. Normalized output was generated with Illumina's Genome Studio program (Illumina) and CNV was visualized in Nexus CN6.1. (Biodiscovery Inc, El Segundo, Calif., USA). Inheritance of CNVs was determined only if they were larger than 30 kb, contained genes and were either unique or had a low frequency in the general population.
Results and Discussion
Published CNVs Identified in Patients with VACTERL Association
No large studies looking for CNVs in individuals with VACTERL association have been published. However, several case reports have been published that describe chromosomal anomalies and recurrent and de novo CNVs in VACTERL patients (table 1). Most of the published de novo genetic anomalies that have been identified in individuals with VACTERL association are unique. However, some changes are recurrent and have been identified in more than one VACTERL patient.
Table 1.
Chromosomal anomalies, recurring and de novo CNVs seen in VACTERL patients
| Chromosome | Type | Remarks (hg18) | Inheritance | Reference |
|---|---|---|---|---|
| 1q41 | gain | ** | de novo | – |
| 2q37.3 | gain | ** | de novo | – |
| 2q22–q24.2 | deletion | del(2)(q22q24.2) | de novo | Woods et al., 1993 |
| 3q28 | loss | chr3:189,395,885–189,951,376 | de novo | Arrington et al., 2010 |
| 5q11 | loss | chr5:51,185,650–55,001,348 | ICSI; father NA | de Jong et al., 2010 |
| 6q25.3–q27 | loss | * | NA | – |
| 6q13–q15 | deletion | del(6)(q13q15) | de novo | McNeal et al., 1977 |
| 7 | duplication | trisomy 7 | de novo | Schinzel, 2001 |
| 8q24.22–q24.3 | gain | * | NA | – |
| 8q24.3 | gain | ** | de novo | – |
| 10q22–qter | duplication | dup(10)(q22) | de novo | Field et al., 1983 |
| 10q25.3 | gain | chr10:116,250,268–116,546,953 | inherited-mat | |
| 10q25.3 | gain | chr10:116,261,258–116,515,586 | inherited-mat | |
| 11q23–qter | duplication | 47,XY+der(22)t(11;22)(q23;q11.2) | de novo | Prieto et al., 2007 |
| 12 | duplication | r(12) | de novo | Cinti et al., 2001 |
| 13,r(13) | duplication | trisomy 13 | de novo | Felix et al., 2007; Lorentz et al., 2002 |
| 16p12.1 | loss | chr16:21,854,140–22,342,140 | inherited-mat | de Jong et al., 2010 |
| 16q24.1 | loss | chr16:82,908,199–86,405,076 | de novo | Stankiewicz et al., 2009 |
| 17q23.2–q24.3 | deletion | del(17)(q23.2q24.3) | de novo | Levin et al., 1995 |
| 17q22–q23.3 | deletion | del(17)(q22q23.3) | de novo | Marsh et al., 2000 |
| 18q12.1 | loss | * | NA | – |
| 18q22.2–qter | deletion | del(18)(q22.2) | de novo | Dowton et al., 1997 |
| 18,r(18) | duplication | trisomy 18 | de novo | Felix et al., 2007; van der Veken et al., 2010 |
| 21 | duplication | trisomy 21 | de novo | Felix et al., 2007 |
| 22q11.2 | gain | chr22:17,281,004–19,792,353 | de novo | Schramm et al., 2011 |
| 22q11.2 | gain | chr22:17,017,139–18,665,833 | inherited-mat | |
| 22pter–q11.2 | duplication | 47,XY+der(22)t(11;22)(q23;q11.2) | de novo | Prieto et al., 2007 |
| X | duplication | triple X | de novo | Brosens et al., 2012 |
| Xp22.3 | gain | chrX:242,432–1,318,727 | inherited-pat | Brosens et al., 2012 |
| Xp22.3 | gain | chrX:327,015–1,889,115 | inherited-mat | Brosens et al., 2012 |
Abstract Shin et al. [2011], American Society of Human Genetics;
abstract Hilger et al. [2012], European Society of Human Genetics; NA = data not available; mat = maternal; pat = paternal; ICSI = intracytoplasmic sperm injection; recurrent CNV in bold.
The first is on chromosome 17 where 2 overlapping deletions have been reported affecting chromosome band 17q23 in both patients. This region contains many genes, but includes 2 candidate genes, TBX2 and TBX4, which encode T-box transcription factors. Heterozygous loss of function mutations in TBX4 have been shown to cause small patella syndrome (OMIM 147891), an autosomal dominant skeletal dysplasia characterized by patellar aplasia or hypoplasia and by anomalies of the pelvis and feet [Bongers et al., 2004]. TBX2 has not been implicated in human disease, but homozygous Tbx2 knockout mice are embryonic lethal and have cardiac anomalies and polydactyly [Harrelson et al., 2004].
The second shared locus is chromosomal band 8q24.3, which is duplicated in 2 individuals with VACTERL. This locus harbours many genes including GLI4 that encodes a member of the krueppel C2H2-type zinc-finger protein family. Although the exact function of GLI4 has not been determined, we note that Gli2–/– and Gli3–/– mice have a VACTERL phenotype [Kim et al., 2001]. Therefore, we consider GLI4 to be an excellent candidate gene.
CNVs Identified in the Rotterdam VACTERL Cohort
In the Rotterdam VACTERL cohort, we did not observe any clearly de novo CNVs. However, one VACTERL patient with trachea agenesis had a maternally inherited 488-kb 16p12.1 deletion and a 3.7-Mb deletion on chromosome 5q11.2 [de Jong et al., 2010]. The 5q11.2 deletion was not inherited from the mother. There was no DNA available from the unknown sperm donor. Several genes are located in this large deletion. Among the top ranked genes by the Endeavour gene prioritization tool are ITGA1, which regulates mesenchymal stem cell proliferation [Ekholm et al., 2002] and FST, an activin-binding protein.
Although de novo CNVs were not identified, all of the patients in this cohort had one or more large (>100 kb) CNVs (fig. 2). Most of these CNVs were known polymorphisms whose frequencies in normal controls make them unlikely to contribute to VACTERL association. However, we observed 3 regions with CNVs which are rarely seen in the general population but were shared by more than one of our VACTERL patients (table 1). These recurring variations can point to pathways or mechanisms involved in disease etiology, especially when they have an extremely low frequency in the general population.
Fig. 2.
CNVs in the Rotterdam cohort and recurring published CNV and structural chromosomal anomalies. In this karyogram, our institution's unique and rare (underlined) gain (blue) and loss (red) are depicted alongside the chromosomal bands in which they are located. Common polymorphisms are not visualized. At 3 loci, recurrent gains either from literature (8q24.3), in our cohort (10q25.3) or both (22q11) (blue regions on ideogram). Two published recurrent chromosomal anomalies lead to a deletion of band 17q23 (red region on ideogram). With arrows, de novo CNVs are depicted.
The first of these rare recurrent CNV in our cohort consisted of maternally inherited 300-kb duplications in band 10q25.3. This region contains the actin-binding LIM protein family member 1 gene (ABLIM1) which encodes a protein that may play a role in binding cytoplasmatic proteins to the actin cytoskeleton [Roof et al., 1997]. Interestingly, Arrington et al. [2010] found a 451-kb interstitial deletion on chromosome 3q28 involving only the LIM domain containing preferred translocation partner in lipoma (LPP) gene in an individual with esophageal atresia with tracheoesophageal fistula, hypospadia, cardiac, renal, and rib anomalies. This change was not found in the individual's mother, but a paternal sample was not available, making it impossible to determine if this was a de novo change or was inherited from an unaffected family member. In our cohort, no CNVs affecting LPP were identified [Hernández-García et al., 2012], but the presence of deletions affecting both ABLIM1 and LPP in some VACTERL association patients (e.g. esophageal atresia with tracheoesophageal fistula, hypospadia, horseshoe kidneys, hemivertebrae, and urinary reflux) suggests that disturbances of the cytoskeleton may contribute to VACTERL phenotypes.
The second recurring CNV was a duplication affecting chromosome 22q11.2. The patient affected with all of the 6 core VACTERL features and her mother had a 22q11.2 micro duplication overlapping 1.4 Mb of the de novo duplication in a VACTERL patient described by Schramm et al. [2011]. This patient had vertebral fusion, anal atresia, right-sided duplicated kidney, and additional non-VACTERL deformations.
The third recurring CNV involves a gain of the short stature homeobox-containing gene (SHOX), which plays an important role in limb development [Vickerman et al., 2011]. Duplications involving SHOX were identified in 2 VACTERL patients; both had limb anomalies and esophageal atresia with tracheoesophageal fistula [unpubl. observations]. Moreover, one of the 2 patients had horseshoe kidneys, hypospadia and dysmorphic features and the second patient an atrial septum defect. The duplications were inherited, one from the patient's mother and the other from the patient's father.
CNVs inherited from an unaffected parent are often considered to be nonpathogenic [Shaw-Smith et al., 2004]. However, the absence of a phenotype in the parent does not exclude a causal relationship. Differences in phenotypes seen in individuals carrying the same CNV can be due to several mechanisms, i.e. differences in environmental exposures, the combination of 2 recessive alleles in the affected individual, variable expressivity, incomplete penetrance, skewed X-inactivation, or a 2-hit CNV model. In this last model, it is postulated that second hits, or alterations in another genomic region, can affect the same biological process and that the additive effects of these changes result in a particular phenotype [Girirajan et al., 2010; Veltman and Brunner, 2010]. Large CNVs with low population frequencies are excellent candidates for a 2-hit model.
Due to the scarcity of published de novo CNVs and absence of such changes in our cohort, we believe that relatively large de novo CNVs are not a common cause of VACTERL association. Furthermore, we cannot exclude the possibility that smaller, inherited CNVs can contribute to the development of VACTERL phenotypes. We did observe several small unique and rare inherited CNVs in our cohort, which on their own are likely benign but could contribute to VACTERL association in a 2-hit model. The latest high-resolution arrays contain millions of probes and can detect CNVs as small as 2 kb. If we want to progress our knowledge about the role of small CNV in VACTERL association etiology, we must know more about frequency and distribution of these small variants in large normal control populations.
To summarize, we believe that CNVs can play a role in VACTERL association by shifting the balance from normal to abnormal development in combination with other genetic, environmental and/or stochastic factors. These changes may also focus attention on genes, pathways or processes that are frequently affected, by mutations or CNVs, in individuals with VACTERL association. By studying larger cohorts, it may be possible to identify additional recurrent CNVs that contribute to VACTERL phenotypes which could, in turn, provide insight into the etiology of VACTERL association.
References
- Arrington CB, Patel A, Bacino CA, Bowles NE. Haploinsufficiency of the LIM domain containing preferred translocation partner in lipoma (LPP) gene in patients with tetralogy of Fallot and VACTERL association. Am J Med Genet A. 2010;152A:2919–2923. doi: 10.1002/ajmg.a.33718. [DOI] [PubMed] [Google Scholar]
- Bardakjian TM, Schneider AS, Ng D, Johnston JJ, Biesecker LG. Association of a de novo 16q copy number variant with a phenotype that overlaps with Lenz microphthalmia and Townes-Brocks syndromes. BMC Med Genet. 2009;10:137. doi: 10.1186/1471-2350-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers EM, Duijf PH, van Beersum SE, Schoots J, Van Kampen A, et al. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosens E, de Jong EM, Barakat TS, Eussen H, D'haene B, Baere E, Poddighe PJ, Galjaard R, Gribnau J, Brooks AS, Tibboel D, de Klein A. Congenital malformations of the gastro-intestinal tract, triple X syndrome and the role of the short stature homeobox-containing gene. 2012 submitted. [Google Scholar]
- Chung B, Shaffer LG, Keating S, Johnson J, Casey B, Chitayat D. From VACTERL-H to heterotaxy: variable expressivity of ZIC3-related disorders. Am J Med Genet A. 2011;155A:1123–1128. doi: 10.1002/ajmg.a.33859. [DOI] [PubMed] [Google Scholar]
- Cinti R, Priolo M, Lerone M, Gimelli G, Seri M, et al. Molecular characterisation of a supernumerary ring chromosome in a patient with VATER association. J Med Genet. 2001;38:E6. doi: 10.1136/jmg.38.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damian MS, Seibel P, Schachenmayr W, Reichmann H, Dorndorf W. VACTERL with the mitochondrial np 3243 point mutation. Am J Med Genet. 1996;62:398–403. doi: 10.1002/(SICI)1096-8628(19960424)62:4<398::AID-AJMG13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- de Jong EM, Douben H, Eussen BH, Felix JF, Wessels MW, et al. 5q11.2 deletion in a patient with tracheal agenesis. Eur J Hum Genet. 2010;18:1265–1268. doi: 10.1038/ejhg.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton SB, Hing AV, Sheen-Kaniecki V, Watson MS. Chromosome 18q22.2→qter deletion and a congenital anomaly syndrome with multiple vertebral segmentation defects. J Med Genet. 1997;34:414–417. doi: 10.1136/jmg.34.5.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm E, Hankenson KD, Uusitalo H, Hiltunen A, Gardner H, et al. Diminished callus size and cartilage synthesis in alpha 1 beta 1 integrin-deficient mice during bone fracture healing. Am J Pathol. 2002;160:1779–1785. doi: 10.1016/s0002-9440(10)61124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab AW, Skorupski JC, Hsieh MH, Breman AM, Patel A, et al. OEIS complex associated with chromosome 1p36 deletion: a case report and review. Am J Med Genet A. 2010;152A:504–511. doi: 10.1002/ajmg.a.33226. [DOI] [PubMed] [Google Scholar]
- Felix JF, Tibboel D, de Klein A. Chromosomal anomalies in the aetiology of oesophageal atresia and tracheo-oesophageal fistula. Eur J Med Genet. 2007;50:163–175. doi: 10.1016/j.ejmg.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Field B, Smith A, Sillence D. Malformation syndrome of chromosome 10q duplication and the VATER association. Ann Genet (Paris) 1983;26:31–33. [Google Scholar]
- Garcia-Barceló MM, Wong KK, Lui VC, Yuan ZW, So MT, et al. Identification of a HOXD13 mutation in a VACTERL patient. Am J Med Genet A. 2008;146A:3181–3185. doi: 10.1002/ajmg.a.32426. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, et al. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- Hernández-García A, Brosens E, Zaveri HP, de Jong EM, Yu Z, et al. Contribution of LPP copy number and sequence changes to esophageal atresia, tracheoesophageal fistula, and VACTERL association. Am J Med Genet A. 2012;158A:1785–1787. doi: 10.1002/ajmg.a.35391. [DOI] [PubMed] [Google Scholar]
- Hilger A, Schramm C, Draaken M, Mughal SS, Dworschak G, et al. Familial occurrence of the VATER/VACTERL association. Pediatr Surg Int. 2012;28:725–729. doi: 10.1007/s00383-012-3073-y. [DOI] [PubMed] [Google Scholar]
- Huang XS, Xiao L, Li X, Xie Y, Jiang HO, et al. Two neighboring microdeletions of 5q13.2 in a child with oculo-auriculo-vertebral spectrum. Eur J Med Genet. 2010;53:153–158. doi: 10.1016/j.ejmg.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kim PC, Mo R, Hui Cc C. Murine models of VACTERL syndrome: role of sonic hedgehog signaling pathway. J Pediatr Surg. 2001;36:381–384. doi: 10.1053/jpsu.2001.20722. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Schulze H, Strauss G, Ott CE, Hall J, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki R, Fukuhara Y, Kosuga M, Okuyama T, Kawashima N, et al. OEIS complex with del(3)(q12.2q13.2) Am J Med Genet A. 2005;135:224–226. doi: 10.1002/ajmg.a.30733. [DOI] [PubMed] [Google Scholar]
- Levin ML, Shaffer LG, Lewis RA, Gresik MV, Lupski JR. Unique de novo interstitial deletion of chromosome 17, del(17)(q23.2q24.3) in a female newborn with multiple congenital anomalies. Am J Med Genet. 1995;55:30–32. doi: 10.1002/ajmg.1320550110. [DOI] [PubMed] [Google Scholar]
- Lorentz CP, Jalal SM, Thompson DM, Babovic-Vuksanovic D. Mosaic r(13) resulting in large deletion of chromosome 13q in a newborn female with multiple congenital anomalies. Am J Med Genet. 2002;111:61–67. doi: 10.1002/ajmg.10457. [DOI] [PubMed] [Google Scholar]
- Marsh AJ, Wellesley D, Burge D, Ashton M, Browne C, et al. Interstitial deletion of chromosome 17 (del(17)(q22q23.3)) confirms a link with oesophageal atresia. J Med Genet. 2000;37:701–704. doi: 10.1136/jmg.37.9.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal RM, Skoglund RR, Francke U. Congenital anomalies including the VATER association in a patient with del(6)q deletion. J Pediatr. 1977;91:957–960. doi: 10.1016/s0022-3476(77)80903-7. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JC, Garcia NM, Elder FF, Zinn AR, Baker LA. Phenotypic expansion of the supernumerary derivative (22) chromosome syndrome: VACTERL and Hirschsprung's disease. J Pediatr Surg. 2007;42:1928–1932. doi: 10.1016/j.jpedsurg.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Roof DJ, Hayes A, Adamian M, Chishti AH, Li T. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol. 1997;138:575–588. doi: 10.1083/jcb.138.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel A. ed 2. Berlin: Walter de Gruyter GmbH & Co; 2001. Catalogue of Unbalanced Chromosome Aberrations in Man. [Google Scholar]
- Schramm C, Draaken M, Bartels E, Boemers TM, Aretz S, et al. De novo microduplication at 22q11.21 in a patient with VACTERL association. Eur J Med Genet. 2011;54:9–13. doi: 10.1016/j.ejmg.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C. Oesophageal atresia, tracheo-oesophageal fistula, and the VACTERL association: review of genetics and epidemiology. J Med Genet. 2006;43:545–554. doi: 10.1136/jmg.2005.038158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD. VACTERL/VATER association. Orphanet J Rare Dis. 2011;6:56. doi: 10.1186/1750-1172-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Bous SM, Bianconi S, Pineda-Alvarez DE. Consideration of VACTERL association in patients with trisomy 21. Clin Dysmorphol. 2010a;19:209–211. doi: 10.1097/MCD.0b013e32833b2b9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Pineda-Alvarez DE, Raam MS, Cummings DA. Evidence for inheritance in patients with VACTERL association. Hum Genet. 2010b;127:731–733. doi: 10.1007/s00439-010-0814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Patel A, Cheung SW, Pineda-Alvarez DE. VACTERL association and mitochondrial dysfunction. Birth Defects Res A Clin Mol Teratol. 2011;91:192–194. doi: 10.1002/bdra.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon B, Bear K, Kimonis V, de Klein A, Scott D, et al. Clinical geneticists’ views of VACTERL/VATER association. Am J Med Genet. 2012 doi: 10.1002/ajmg.a.35638. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southard AE, Edelmann LJ, Gelb BD. Role of copy number variants in structural birth defects. Pediatrics. 2012;129:755–763. doi: 10.1542/peds.2011-2337. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumska D, Pieles G, Essalmani R, Bilski M, Mesnard D, et al. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 2008;22:1465–1477. doi: 10.1101/gad.479408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauvin-Robinet C, Faivre L, Huet F, Journeau P, Glorion C, et al. Another observation with VATER association and a complex IV respiratory chain deficiency. Eur J Med Genet. 2006;49:71–77. doi: 10.1016/j.ejmg.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Uliana V, Giordano N, Caselli R, Papa FT, Ariani F, et al. Expanding the phenotype of 22q11 deletion syndrome: the MURCS association. Clin Dysmorphol. 2008;17:13–17. doi: 10.1097/MCD.0b013e3282ef97ee. [DOI] [PubMed] [Google Scholar]
- van der Veken LT, Dieleman MM, Douben H, van de Brug JC, van de Graaf R, et al. Low grade mosaic for a complex supernumerary ring chromosome 18 in an adult patient with multiple congenital anomalies. Mol Cytogenet. 2010;3:13. doi: 10.1186/1755-8166-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. Understanding variable expressivity in microdeletion syndromes. Nat Genet. 2010;42:192–193. doi: 10.1038/ng0310-192. [DOI] [PubMed] [Google Scholar]
- Vickerman L, Neufeld S, Cobb J. Shox2 function couples neural, muscular and skeletal development in the proximal forelimb. Dev Biol. 2011;350:323–336. doi: 10.1016/j.ydbio.2010.11.031. [DOI] [PubMed] [Google Scholar]
- Walsh LE, Vance GH, Weaver DD. Distal 13q deletion syndrome and the VACTERL association: case report, literature review, and possible implications. Am J Med Genet. 2001;98:137–144. doi: 10.1002/1096-8628(20010115)98:2<137::aid-ajmg1022>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Wessels MW, Kuchinka B, Heydanus R, Smit BJ, Dooijes D, et al. Polyalanine expansion in the ZIC3 gene leading to X-linked heterotaxy with VACTERL association: a new polyalanine disorder? J Med Genet. 2010;47:351–355. doi: 10.1136/jmg.2008.060913. [DOI] [PubMed] [Google Scholar]
- Woods CG, Koehn DC, McFadden DE, Evans JA, Van Allen MI. A case report of a child with an interstitial deletion of chromosome 2 (q22;q24.2) and the VATER association phenotype. Proc Greenwood Genet Ctr. 1993;12:119–120. [Google Scholar]