Abstract
MicroRNAs (miRNAs) play important roles in plant development through regulation of gene expression by mRNA degradation or translational inhibition. Despite the fact that tomato (Solanum lycopersicum) is the model system for studying fleshy fruit development and ripening, only a few experimentally proven miRNA targets are known, and the role of miRNA action in these processes remains largely unknown. Here, by using parallel analysis of RNA ends (PARE) for global identification of miRNA targets and comparing four different stages of tomato fruit development, a total of 119 target genes of miRNAs were identified. Of these, 106 appeared to be new targets. A large part of the identified targets (56) coded for transcription factors. Auxin response factors, as well as two known ripening regulators, COLORLESS NON-RIPENING (CNR) and APETALA2a (SlAP2a), with developmentally regulated degradation patterns were identified. The levels of the intact messenger of both CNR and AP2a are actively modulated during ripening, by miR156/157 and miR172, respectively. Additionally, two TAS3-mRNA loci were identified as targets of miR390. Other targets such as ARGONAUTE 1 (AGO1), shown to be involved in miRNA biogenesis in other plant species, were identified, which suggests a feedback loop regulation of this process. In this study, it is shown that miRNA-guided cleavage of mRNAs is likely to play an important role in tomato fruit development and ripening.
Key words: Degradome, fruit development, microRNA, post-transcriptional regulation, tomato, transcription factors.
Introduction
MicroRNAs (miRNAs) are small, 21–22 nucleotide RNA molecules encoded in the genomes of plants, animals, and even viruses. They are derived from single-stranded RNA precursors that can form stem–loop structures (Bartel, 2004; Chen, 2009; Song et al., 2011). MiRNAs regulate the expression of genes by binding to complementary sequences in specific mRNAs. This binding leads either to cleavage-induced degradation of the mRNA or suppression of its translation. Both processes eventually result in down-regulation of the product of the target gene (Meyers et al., 2006). In plants, the complementarity between miRNA and its targets is very high. The cleavage of the target mRNA occurs in the middle of the mRNA–miRNA duplex (between the 10th and 11th nucleotide from the 5’ end of the miRNA) (Rhoades et al., 2002; German et al., 2008). A miRNA may regulate the expression of multiple genes, while it is also possible that multiple miRNAs control a single gene. Therefore, the identification of their target mRNAs is essential for the functional analysis of miRNAs.
Many plant miRNAs are conserved among species and have been implicated in processes such as development, abiotic stress tolerance, signal transduction, and resistance to pathogens (Jones-Rhoades et al., 2006; Ori et al., 2007; Sun, 2011). Recently, using next-generation sequencing technology, many new, non-conserved miRNAs with low abundance could be identified. For example, high-throughput sequencing was first used to identify tomato small RNAs, including miRNAs from young green fruits and, more recently, from several fruit developmental stages (Moxon et al., 2008; Mohorianu et al., 2011; Zuo et al., 2012). In these studies, 103 conserved miRNAs belonging to 24 families and 10 non-conserved miRNAs comprising nine families were identified. The targets of many of these miRNAs were predicted to be transcription factors (TFs). These include, for example, COLORLESS NON-RIPENING (CNR), a gene encoding a SQUAMOSA promoter-binding protein (SBP) (Manning et al., 2006), a positive regulator of tomato fruit ripening, and a tomato APETALA2 (SlAP2a) gene that was shown to be a negative regulator of ripening (Chung et al., 2010; Karlova et al., 2011). These two examples are members of TF families, of which some members may be targets of miR156/157 or miR172, respectively. Indeed, CNR was shown to be an in vivo target of miR156 in tomato (Moxon et al., 2008), but the importance, or even the occurrence during ripening, of miRNA-directed cleavage for both genes is unknown. Thus, identifying miRNAs that regulate the expression of TFs controlling tomato fruit development and ripening, or of genes otherwise involved in these processes, will contribute to a better understanding of the regulation of fleshy fruit development. Moreover, the recent annotation of the complete tomato genome sequence (Tomato Genome Consortium, 2012) allows for miRNA–mRNA pair identification on a genome-wide scale.
High-throughput degradome library sequencing, or parallel analysis of RNA ends (PARE), has been developed for genome-wide-scale identification of miRNA target genes. This method was successfully used to discover many new miRNA–mRNA target pairs in Arabidopsis, rice, soybean, a moss, and grapevine (Addo-Quaye et al., 2008; German et al., 2008; Li et al., 2010; Pantaleo et al., 2010; Song et al., 2011).
Here the high-throughput degradome library sequencing for global identification of targets of miRNAs from four different stages of tomato fruit development is described. New target genes of miRNAs were detected, many of which encoded TFs, which hints at the importance of miRNAs in regulating tomato fruit development. Known ripening regulators such as the CNR and AP2a TFs were also identified as targets of miR156 and miR172, respectively, with developmentally regulated degradation patterns in tomato fruit. The findings suggest the important role of miRNAs in tomato fruit development starting from the initiation of the fruit until the final ripe stage.
Materials and methods
Plant material
Tomato (Solanum lycopersicum) variety ‘Ailsa Craig’ plants were grown under standard greenhouse conditions. At anthesis, flowers were labelled, and developing fruits were harvested at 5 DPA (days post-anthesis), mature green (MG), breaker (Br), and red ripe (RR) stage (7 d post-breaker).
Library preparation and sequencing
Fruits were frozen in liquid nitrogen, ground, and RNA was extracted using an InviTrap Spin Plant RNA mini kit (Stratec, Berlin, Germany). Purified poly(A) RNA was used to produce PARE libraries by Vertis Biotechologie AG (Freising, Germany) essentially following the protocol of German et al. (2008, 2009) with slight modifications for high-throughput sequencing. Following MmeI digestion, the resulting 42bp fragments were gel purified and ligated to Illumina 3’ TruSeq adaptors (Illumina, San Diego, CA, USA). Subsequently, the ligation products were PCR-amplified with a common 5’ primer and library-specific index primers for multiplex sequencing. Pooled libraries were sequenced on an Illumina HiSeq 2000 by ServiceXS BV (Leiden, The Netherlands) and upon deconvolution yielded ~26–36 million raw reads per library. The sequence data from this study were submitted to the Gene Expression Omnibus repository under accession number GSE42661.
Sequence analysis
Raw reads were processed for removal of 5’ and 3’ adaptor sequences and filtered for quality, oligo(A) reads up to 15As, for tomato chloroplast DNA, and for small non-coding RNAs as present in the Rfam 10.1 database (Gardner et al., 2011). This was performed using parts of the FASTX Toolkit, version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/), and Vmatch version 2.0 (http://www.vmatch.de/), wrapped in Perl and Bash scripts. After filtering, the remaining reads were mapped on the sense strand of the ITAG Release 2.3 cDNA models (SL2.40 genome build) using SHRiMP v2.0 (David et al., 2011) allowing one mismatch and retaining only the best scoring hit or hits in the event of equally scoring hits. Mapped reads were matched to potential miRNA target sites in tomato cDNAs, identified using the TargetFinder v1.6 script (Allen et al., 2005; Fahlgren et al., 2007), using an updated version of the CleaveLand pipeline (Addo-Quaye et al., 2009a ), CleaveLand v3.0.1 (kindly provided by Dr M. Axtell), according to the instructions of the author. In order to allow for inaccurate target cleavage or variations in miRNA 5’ ends, the pipeline was modified to recognize targets cleaved at the 9th or 11th position of the miRNA, as well as at the 10th position. In CleaveLand 3, per single miRNA the likelihood for one or more random matches, at a given position, to the degradome data is calculated, using the binomial distribution. Here, the binomial probability is the fraction of eligible bases occupied by the degradome data, and the number of ‘trials’ for the binomial distribution is the cumulative number of alignments at a given score. Because of the large number of likelihoods that are calculated, a false discovery rate (FDR) correction was applied, for each category, using the FDR procedure of Benjamini and Hochberg (1995), to give q-values that are adjusted for multiple testing and that can be interpreted in terms of the expected FDR at a chosen threshold. Tag numbers for any position were normalized for library size by using the formula TP10M (tags per 10 million)= (raw abundance/total filtered tags–cDNA matches)×10 000 000.
Overlap between tags and coordinates of gene models, untranslated regions (UTRs), coding sequences (CDSs), exons, and introns (kindly provided by Stephane Rombauts), extracted with BedTools (Quinlan and Hall, 2010), was determined using SAMtools (Li et al., 2009).
Gene expression analysis
For comparison of degradome tag frequency and gene expression analysed by microarray hybridization, PARE-derived sequences from each library were treated as RNAseq data in the CLC Genomics Workbench (CLC bio, Aarhus, Denmark), giving a normalized expression value [reads per kilobase transcript per million reads (RKPM)] for all four libraries. Expression data were independently derived from MG, Br/turning, and RR stage cv. Ailsa Craig fruit RNA hybridization to the Affymetrix EU-SOL TOM3 microarray. Hybridizations were performed in triplicate with independent biological samples, background-corrected, and normalized. The methods and gene expression data from this study were submitted to the Gene Expression Omnibus repository under accession number GSE42783.
Gene expression analysis by quantitative PCR
RNA extraction, cDNA synthesis, and real-time quantitative PCR (qPCR) were performed as described before (Karlova et al., 2011). The following primer pairs were used: AP2a spanning the miRNA target site (AGACTCCGAGCAGCAACAATGTAGG and TCCTTATTATCTGCTGGGGGAATCC), AP2a 3’ UTR (CC CCAGCAGATAATAAGGACTCAA and GAAATTAATGAAT GGTCAAGGTCTC), CNR spanning the miRNA target site (GCA TCAATGGCAGCCAAATAACC and CGGCTTATTCTTTTTGT CCTTCCC) and CNR 3’ UTR (TTCCCGGATTTCTAAGCAA ATTGT and GTTGGAATGTCAACATGGATATGCA).
RLM-5’RACE
Total RNA (15 µg) from MG stage fruits was purified using TriPure isolation reagent (Roche, The Netherlands). mRNA was purified using the Dynabeads mRNA purification kit (Life Technologies, CA, USA). RNA ligase-mediated rapid amplification of 5’ cDNA ends (RLM-5’RACE) was performed with the FirstChoice RLM-RACE kit according to the manufacturer’s instructions (Ambion), with gene-specific primers for the SlDRM5 gene: 5’-AGACCGTCCCTGCTCACCCT-3’ and 5’-GTTGTTCTAAGCGGTCTCCA-3’. The final PCR product was extracted and purified from a 2% agarose gel, cloned into pGEMT-easy (Promega, Madison, WI, USA), and plasmid DNA from 10 different colonies was sequenced.
Results
Degradome library generation, sequencing, and sequence analysis
In order to identify targets of known and putative miRNAs from tomato, PARE libraries (Addo-Quaye et al., 2008; German et al., 2008) of tomato cv. Ailsa Craig fruits of four developmental stages [5 days after pollination (5 DAP), MG, Br, and RR] were prepared. Between 26 and 36 million short sequence reads were obtained from each library, representing the 5’ ends of uncapped, polyadenylated RNAs. The reads were subsequently trimmed and filtered for contaminating small ribosomal and nuclear RNAs from the Rfam database as well as for organellar RNAs. The filtered tag reads were mapped on the ITAG v2.3 tomato predicted cDNA set, as well as on exons, introns, CDSs, and 5’- or 3’-UTRs separately (results shown in Supplementary Table S1 available at JXB online). Approximately 80–85% of all filtered tags could be mapped to the (predicted) S. lycopersicum transcriptome, suggesting that part of the tags map to as yet unannotated genes. A relatively large fraction (10%) mapped on regions annotated as introns, suggesting either erroneous annotation or the frequent retention of introns by alternative splicing. Similar findings have been reported previously for the degradome analysis in Arabidopsis and rice (German et al., 2008; Li et al., 2010). In the present analysis, at least one specific tag could be detected for an average of 25 000 out of the ~35 000 predicted cDNAs in each of the four libraries, and, after RNAseq analysis of the data, 20 976 cDNAs showing at least 1 RKPM and 12 239 genes showing ≥10 RKPM, demonstrating the great depth of coverage of the tomato transcriptome in these experiments.
Previous studies revealed that the miRNA-guided cleavage is quite precisely between the 10th and 11th nucleotide from the 5’ end of the miRNA in the complementary region of the target transcript. Therefore, the 5’ position of tags that were mapped to tomato cDNAs were matched to predicted miRNA target sites (position 10 of the miRNA) using the CleaveLand pipeline version 3.0.1, a modified version of the original pipeline (Addo-Quaye et al., 2009a ). In this pipeline, tags are first mapped on cDNAs and the number of tags with the 5’ nucleotide corresponding to each position in the mRNA is counted, which can be depicted in a t (target)-plot. t-plot peaks corresponding to position 10 of an aligned miRNA are reported (Fig. 1). Additionally, to be able to capture products of less accurate cleavage, as well as to account for naturally occurring variation in miRNA length at its 5’-processing site, the procedure was repeated for cleavage positions 9 and 11. As noted before (Addo-Quaye et al., 2008), processes other than miRNA-directed cleavage (e.g. decapping followed by 5’–3’ breakdown of mRNA) may give rise to uncapped mRNA fragments that were also detected by the procedure, and indeed many tags and t-plot peaks not corresponding to a known miRNA target site were observed. For these reasons, the total number of tags detected per transcript is generally also representative of its expression level in a certain fruit developmental stage. This is demonstrated by the correlation between normalized tag numbers per kilobase transcript per million reads (presented as RKPM) from the libraries in this study and independently derived microarray expression data for comparable samples (Supplementary Fig. S1 at JXB online).
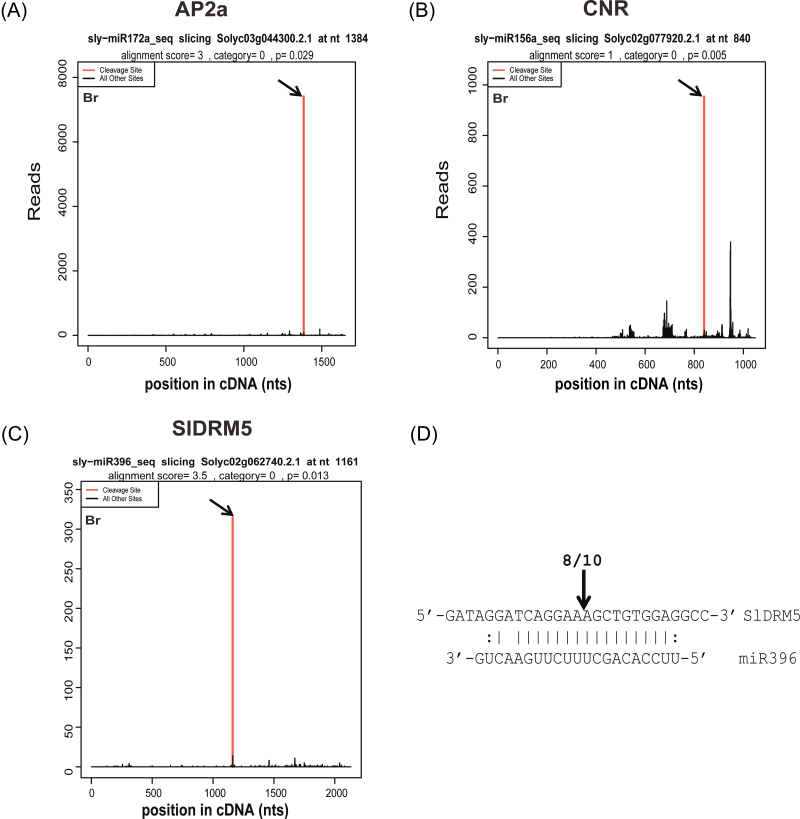
Fig. 1.
Target plots of validated target mRNAs. The abundance of each sequenced read is plotted as a function of the position of its 5’ end in the transcript. In red are the peaks of signatures at the predicted cleavage site of the corresponding miRNAs (arrows). (A) Cleavage features in SlAP2a mRNA by miR172a from the breaker (Br) fruit stage degradome library. (B) Cleavage features in CNR mRNA by miR156a at the Br fruit stage. (C) Cleavage features in SlDRM5 mRNA by miR396 at the Br fruit stage. (D) RLM-RACE confirmation of the SlDRM5 cleavage site by miR396. The numbers above the vertical arrow indicate the number of sequences found at the exact miR396 cleavage site.
CleaveLand classifies t-plot peaks in four categories (0–4), with peaks of categories 0–3 having >1 read per peak. Category 0 are peaks that represent a single maximum in a particular transcript, category 1 are peaks that are equal to the maximum, with more than one maximum per transcript, category 2 are peaks lower than the maximum but higher than the median of a transcript, and category 3 are peaks with an equal or less than median number of reads (for examples, see Supplementary Fig. S2 at JXB online). Category 4 peaks have only 1 read. The statistical significance of an observed peak–miRNA match is represented by the P-value, the likelihood of a peak in a particular position occurring by chance, which is determined by both the total number of nucleotides/positions for each category and the miRNA–target alignment score. This score was determined by the Targetfinder script (Allen et al., 2005; Fahlgren et al., 2007), where all alignments with a penalty score up to 7 were included (listed in Supplementary Table S2). For this procedure, all miRNAs reported to be present or expressed in tomato, collected from miRbase (www.mirbase.org) and from the literature, were used. The entire output can be viewed in the Gene Expression Omnibus repository under accession number GSE42661. The protocol was repeated with the tomato Unigene collection from the Solanaceae Genomics Network (SGN) database, in order to find targets in transcripts that have not been mapped on the tomato genome sequence and hence have no corresponding cDNA sequence. This resulted in identification of nine additional targets with category 0 or 1 peaks (Table 1).
Table 1.
miRNA–target mRNA pairs with a category 0 or 1 peak identified in at least two libraries or in one library with q-value ≤0.05 and cleavage peak height >1 TP10M.A comprehensive list of all category 0 and 1 peaks, with P- and q-values and normalized read numbers can be found in Supplementary Table S3 at JXB online.
| Target mRNA | ITAG2.3 annotation | Common name | Cleavage position | Peak categories | Reference |
|---|---|---|---|---|---|
| miR156/157 | |||||
| Solyc02g077920.2.1 | Squamosa promoter binding-like protein | CNR | 10 | 0 | Moxon et al. (2008) |
| Solyc04g045560.2.1 | Squamosa promoter binding-like protein | SlySBP2/SlSPL2 | 10 | 0 | Moxon et al. (2008) |
| Solyc05g012040.2.1 | Squamosa promoter binding-like protein | SlySBP6b/SlSPL6b | 10 | 0 | |
| Solyc05g015510.2.1 | Squamosa promoter binding-like protein | SlySBP10 | 10 | 0 | |
| Solyc05g015840.2.1 | Squamosa promoter binding-like protein | SlySBP13 | 10 | 1 | |
| Solyc07g062980.2.1 | Squamosa promoter binding-like protein | 11 | 0 | ||
| Solyc10g078700.1.1 | Squamosa promoter binding-like protein | SlySBP15/SlSPL15 | 10 | 0 | |
| miR158 | |||||
| No category 0 or 1 peaks with P ≤0.05 | |||||
| miR159 | |||||
| Solyc01g009070.2.1 | MYB transcription factor | GAMyb-like1 | 10 | 0 | |
| Solyc01g090530.1.1 | MYB transcription factor | 10 | 0 | ||
| Solyc06g073640.2.1 | MYB transcription factor | GAMYB-like2 | 10 | 0 | |
| Solyc01g098770.1.1a | Xylanase inhibitor | 10 | 0 | ||
| miR160 | |||||
| Solyc06g075150.2.1 | Auxin response factor 16 | ARF16 | 10 | 0 | |
| Solyc11g013470.1.1 | Auxin response factor 17 (Fragment) | ARF17 | 10 | 0 | Moxon et al. (2008) |
| Solyc11g069500.1.1 | Auxin response factor 16 | ARF10 | 10 | 0 | Hendelman et al. (2012) |
| miR161 | |||||
| No category 0 or 1 peaks | |||||
| miR162 | |||||
| Solyc10g008310.1.1 | Unknown Protein (AHRD V1) | 10 | 0 | ||
| miR164 | |||||
| Solyc03g115850.2.1 | NAC domain protein | 10 | 0 | Moxon et al. (2008) | |
| Solyc06g069710.2.1 | NAC domain protein | 10 | 1 | ||
| Solyc07g062840.2.1 | NAC domain protein | 10 | 0 | ||
| Solyc07g066330.2.1 | NAC domain protein | 10 | 0 | ||
| miR165/166 | |||||
| Solyc08g066500.2.1 | class III homeodomain-leucine zipper | 10 | 0 | ||
| Solyc12g044410.1.1 | class III homeodomain-leucine zipper | 10 | 0 | Moxon et al. (2008) | |
| miR167 | |||||
| No category 0 or 1 peaks | |||||
| miR168 | |||||
| Solyc06g072300.2.1 | ARGONAUTE 1 | 10 | 0 | ||
| SGN-U563432 | AGO1-1 (Nicotiana tabacum) | 10 | 0 | ||
| miR169 | |||||
| Solyc02g068500.2.1 | Thioredoxin o | – | 10 | 0 | |
| Solyc06g068930.1.1 | Jasmonate ZIM-domain protein 2 | – | 10 | 0 | |
| Solyc01g006930.2.1 | Nuclear transcription factor Y subunit A-3 | – | 10 | 0 | |
| Solyc01g087240.2.1 | Nuclear transcription factor Y subunit A-3 | – | 10 | 0 | |
| Solyc03g121940.2.1 | Nuclear transcription factor Y subunit A-3 | – | 10 | 0 | |
| Solyc08g062210.2.1 | Nuclear transcription factor Y subunit A-3 | – | 10 | 0 | |
| miR170/171 | |||||
| Solyc01g090950.2.1 | GRAS family transcription factor | – | 10 | 0 | Moxon et al. (2008) |
| Solyc08g078800.1.1 | GRAS family transcription factor | – | 10 | 0 | |
| Solyc11g013150.1.1 | GRAS family transcription factor | – | 10 | 0 | |
| miR172 | |||||
| Solyc02g064960.2.1 | AP2-like ethylene-responsive transcription factor | SlAP2b | 10 | 0 | |
| Solyc02g093150.2.1 | AP2-like ethylene-responsive transcription factor | SlAP2c | 10 | 0 | Itaya et al. (2008); Moxon et al. (2008) |
| Solyc03g044300.2.1 | AP2-like ethylene-responsive transcription factor | SlAP2a | 10 | 0 | |
| Solyc04g049800.2.1 | AP2-like ethylene-responsive transcription factor | 10 | 0 | ||
| Solyc06g075510.2.1 | AP2-like ethylene-responsive transcription factor | SlAP2e | 10 | 0 | |
| Solyc09g007260.2.1 | AP2-like ethylene-responsive transcription factor | 10 | 0 | ||
| Solyc10g084340.1.1 | AP2-like ethylene-responsive transcription factor | 10 | 0 | ||
| Solyc11g072600.1.1 | AP2-like ethylene-responsive transcription factor | SlAP2d | 10 | 0 | |
| SGN-U584830 | Glutamate permease | 10 | 0 | ||
| miR173 | |||||
| Solyc10g005490.2.1 | Protein strawberry notch homolog 1 | 10 | 0 | ||
| miR319 | |||||
| Solyc07g062680.1.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | Lanceolate | 11 | 0 | Ori et al. (2007) |
| Solyc12g014140.1.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | 11 | 0 | ||
| Solyc02g077250.2.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | 11 | 0 | ||
| Solyc05g012840.1.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | 11 | 0 | ||
| Solyc07g053410.2.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | 11 | 0 | ||
| Solyc08g048370.2.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | 11 | 0 | ||
| Solyc08g048390.1.1 | Transcription factor CYCLOIDEA (TCP transcription factor) | 11 | 0 | ||
| miR390 | |||||
| Solyc01g058100.2.1 | Unknown Protein (AHRD V1) | Sly-TASI3a | 10 | 0 | Li et al. (2012) |
| SGN-U587265 (2×) | Sly-TASI3b | 10 | 0 | ||
| Solyc12g098430.1.1 | Actin-related protein 2/3 complex subunit 4 | 10 | 0 | ||
| miR393 | |||||
| Solyc02g079190.2.1 | Auxin F-box protein 5 | 10 | 0 | ||
| Solyc06g008780.1.1 | Auxin F-box protein 5 | TIR1-like protein | 10 | 0 | |
| Solyc09g074520.2.1 | Auxin F-box protein 5 | TIR1 | 10 | 0 | |
| miR394 | |||||
| Solyc05g015520.2.1 | F-box family protein | 10 | 0 | ||
| miR395 | |||||
| Solyc04g054730.2.1 | High affinity sulphate transporter 2 | 10 | 0 | ||
| miR396 | |||||
| Solyc12g096070.1.1 | Growth-regulating factor 5 | 10 | 0 | ||
| Solyc08g005430.2.1 | Growth-regulating factor 4 | 10 | 0 | ||
| Solyc03g082430.1.1 | Growth-regulating factor 4 | 10 | 0 | ||
| Solyc08g075950.1.1 | Growth-regulating factor 3 | 10 | 0 | ||
| Solyc08g083230.1.1 | Growth-regulating factor 3 | 10 | 0 | ||
| Solyc10g083510.1.1 | Growth-regulating factor 2 | 10 | 0 | ||
| Solyc07g041640.2.1 | Growth-regulating factor 1 | 10 | 0 | ||
| Solyc04g077510.2.1 | Growth regulating factor 1 (Fragment) | 10 | 0 | ||
| Solyc01g066730.2.1 | MADS-box protein | 10 | 0 | ||
| Solyc02g062740.2.1 | DNA (Cytosine-5-)-methyltransferase 3 | SlDRM5 | 10 | 0 | |
| Solyc12g088670.1.1 | Cathepsin B-like cysteine proteinase | TDI-65 | 10 | 0 | |
| Solyc02g032870.2.1 | 5’-Tyrosyl-DNA phosphodiesterase | 10 | 0 | ||
| Solyc05g054050.2.1a | Glutamate decarboxylase | 10 | 0 | ||
| miR397 | |||||
| No category 0 or 1 peaks with P < 0.05 | |||||
| miR398 | |||||
| Solyc06g082890.2.1 | S-like RNase (Fragment) | 10 | 1 | ||
| Solyc11g066390.1.1 | Superoxide dismutase | 10 | 0 | ||
| miR399 | |||||
| Solyc03g096770.1.1 | Unknown Protein (AHRD V1) | 10 | 0 | ||
| Solyc12g015630.1.1 | Genomic DNA chromosome 5 P1 clone MDF20 | 10 | 0 | ||
| miR403 | |||||
| Solyc02g069260.2.1 | ARGONAUTE 1c | 10 | 0 | Moxon et al. (2008) | |
| miR408 | |||||
| Solyc01g104400.2.1 | Blue copper protein | 11 | 1 | ||
| Solyc04g074740.2.1 | Blue copper-like protein | 11 | 0 | ||
| miR414 | |||||
| No category 0 or 1 peaks with P < 0.05 | |||||
| miR482/miR-Y | |||||
| No category 0 or 1 peaks | |||||
| miR828 | |||||
| No category 0 or 1 peaks | |||||
| miR858 | |||||
| Solyc05g053330.2.1 | MYB transcription factor | 10 | 0 | ||
| Solyc08g065910.1.1 | Myb-related transcription factor | 11 | 0 | ||
| Solyc12g049300.1.1 | MYB transcription factor | 11 | 0 | ||
| Solyc06g034000.1.1 | Myb-related transcription factor | 11 | 0 | ||
| Solyc03g083260.1.1a | Unknown Protein (AHRD V1) | 11 | 0 | ||
| miR894 | |||||
| Solyc01g095920.2.1 | O-Methyltransferase family | 10 | 1 | ||
| miR1916 | |||||
| Solyc02g014150.2.1a | Photosystem II stability/assembly factor Ycf48-like protein | 10 | 1 | ||
| miR1917 | |||||
| Solyc06g059920.1.1a | Sesquiterpene synthase 2 | 10 | 1 | ||
| miR1918 | |||||
| Solyc05g005330.2.1 | Cc-nbs-lrr, resistance protein | 10 | 1 | ||
| miR1919 | |||||
| No category 0 or 1 peaks | |||||
| miR5301*/472/482 b | |||||
| Solyc11g065780.1.1 | Cc-nbs, resistance protein fragment | 10 | 0 | ||
| SGN-U571260 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| SGN-U598669 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc02g036270.2.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc07g005770.2.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc07g009180.1.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc08g076000.2.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc04g009070.1.1 | Nbs, resistance protein fragment | 10 | 0 | ||
| SGN-U585460 | Glutamate permease | 10 | 0 | Mohorianu et al. (2011) | |
| Solyc04g025160.2.1 | Arsenite ATPase transporter (Eurofung) | 10 | 0 | Mohorianu et al. (2011) | |
| miR4376 | |||||
| No category 0 or 1 peaks | |||||
| miR5301 | |||||
| Solyc09g020000.2.1 | MYB-like TF | 10 | 0 | ||
| miR5302 | |||||
| Solyc08g078760.1.1 | AT5g47580/MNJ7_17 | 10 | 0 | ||
| miR5303 | |||||
| Solyc08g079800.2.1 | Growth-regulating factor 12 | 10 | 0 | ||
| SGN-U564554 | S-locus F-box-like protein b | 10 | 0 | ||
| mir6022 | |||||
| Solyc01g005720.2.1 | LRR receptor-like serine/threonine- protein kinase, RLP | 10 | 0 | ||
| Solyc01g005760.2.1 | LRR receptor-like serine/threonine- protein kinase, RLP | 10 | 0 | ||
| Solyc01g005870.1.1 | LRR receptor-like serine/threonine- protein kinase, RLP | 10 | 0 | ||
| Solyc01g006550.2.1 | LRR receptor-like serine/threonine- protein kinase, RLP | 10 | 0 | ||
| Solyc07g052920.2.1a | Unknown Protein (AHRD V1) | 10 | 0 | ||
| miR6023 | |||||
| Solyc08g075710.2.1 | Amino acid permease-like protein | 10 | 1 | ||
| Solyc09g055760.2.1 | T-snare (AHRD V1***- Q2HS23_ MEDTR) | 10 | 0 | ||
| miR6024 | |||||
| Solyc05g005330.2.1 | Cc-nbs-lrr, resistance protein with an R1-specific domain | 10 | 1 | ||
| Solyc05g008070.2.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc10g008230.1.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc11g070000.1.1 | Nbs, resistance protein fragment | 10 | 0 | ||
| SGN-U569340 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| SGN-U583156 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| miR6026 | |||||
| No category 0 or 1 peaks | |||||
| miR6027 | |||||
| Solyc04g009120.1.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc04g009150.1.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc04g009660.2.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc05g012890.1.1 | Cc-nbs-lrr, resistance protein with an R1-specific domain | 10 | 0 | ||
| Solyc05g012910.2.1 | Cc-nbs-lrr, resistance protein with an R1-specific domain | 10 | 0 | ||
| Solyc09g098130.1.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
| Solyc10g047320.1.1 | Cc-nbs-lrr, resistance protein | 10 | 0 | ||
a Targets with a P-value ≥0.05
b miR5301*, miR472, and miR482 have overlapping target sites and similar predicted targets. Sly-miR482 (miR-Y in Moxon et al.) is different from the canonical miR482.
c Although the iTAG annotation lists this gene as an AGO1 homologue, it is in fact more homologous to the Arabidopsis AGO2 gene (AT1G31280).
Based on the signature abundance at the putative miRNA target sites and as reported before (Addo-Quaye et al., 2008, 2009b ; Li et al., 2010; Song et al., 2011), the cleavage products have been categorized into three classes. class I miRNA-guided cleavage products are the most significant (t-plots with category 0 and 1 peak and P-value ≤0.05, Fig. 1). The targets belonging to this class are shown in Table 1 and discussed below. A comprehensive list of category 0 or 1 targets, their position within the transcript, peak height, and P-value for each library can be found in Supplementary Table S3 at JXB online. Additionally, a Benjamini–Hochberg correction was performed for multiple testing of the P-values, and the resulting q-values (FDRs) for categories 0 and 1 are reported. In the class II targets category (t-plots with category 2 peaks), the signature abundance was higher than the median on the transcript, but below the maximum. In total, for all four libraries 3956 putative cDNA targets belonging to class II were found. The rest of the cleavage signatures with low abundance were grouped in class III (category 3 and 4 peaks); 4444 targets for the four libraries together were found.
Identification of conserved and non-conserved targets of conserved miRNAs
Two categories of miRNAs, conserved and non-conserved, are present in land plants (for a review, see Sun, 2011). Conserved miRNAs are present in different plant lineages, often have high expression, and are encoded by multiple loci in the genomes. Non-conserved miRNAs are usually species specific and expressed at low levels, encoded by a single gene. From the 24 conserved miRNA families in tomato (Mohorianu et al., 2011; Zhai et al., 2011; Zuo et al., 2012), targets classified as category I were identified for 21 of them (Table 1). For miR167, miR397, and miR827, no targets grouped in class I were found. In most cases in plants, targets of an miRNA family belong mainly to the same gene family (Table 1). However, some miRNAs can additionally cleave targets not belonging to the conserved target gene families (Sun, 2011). For most of the conserved miRNAs in the present study, members of the conserved target gene families were found as targets in tomato as well. Many appear also to target one or more non-conserved target genes. Of the latter, a large fraction had low tag counts at the peak position and a P-value >0.05, and therefore are considered not significant in this study. However, some are significant and warrant further study of their relevance in tomato. The miRNA that was found to target the highest number of different genes from unrelated families and all belonging to class I targets, is miR396, which was shown previously to be equally expressed in the tomato fruit from the MG stage onwards (Mohorianu et al., 2011). For instance, miR396 has been found to target a 50S ribosomal protein only in rice, and growth-related factor (GRF) TF genes in diverse species (Sun, 2011). From the 10 predicted tomato GRF TF targets of miR396, strong evidence (category 0 peaks) was found for eight of them (Table 1). Besides the GRFs, it was found that in tomato fruits miR396 also targets homologues of a MADS box gene, a DNA (cytosine-5-)-methyltransferase (Fig. 1C) gene, a cathepsin B-like cysteine proteinase gene, and a tyrosyl-DNA phosphodiesterase gene (Table 1), in addition to several genes with lower significance (P > 0.05). The miR396-directed processing of the DNA (cytosine-5-)-methyltransferase cDNA was independently confirmed by RLM-RACE analysis of the mRNA degradation products (Fig. 1D). This gene was identified as one of four tomato domains rearranged methyltransferases (DRMs), SlDRM5, putatively involved in DNA methylation (Teyssier et al., 2008).
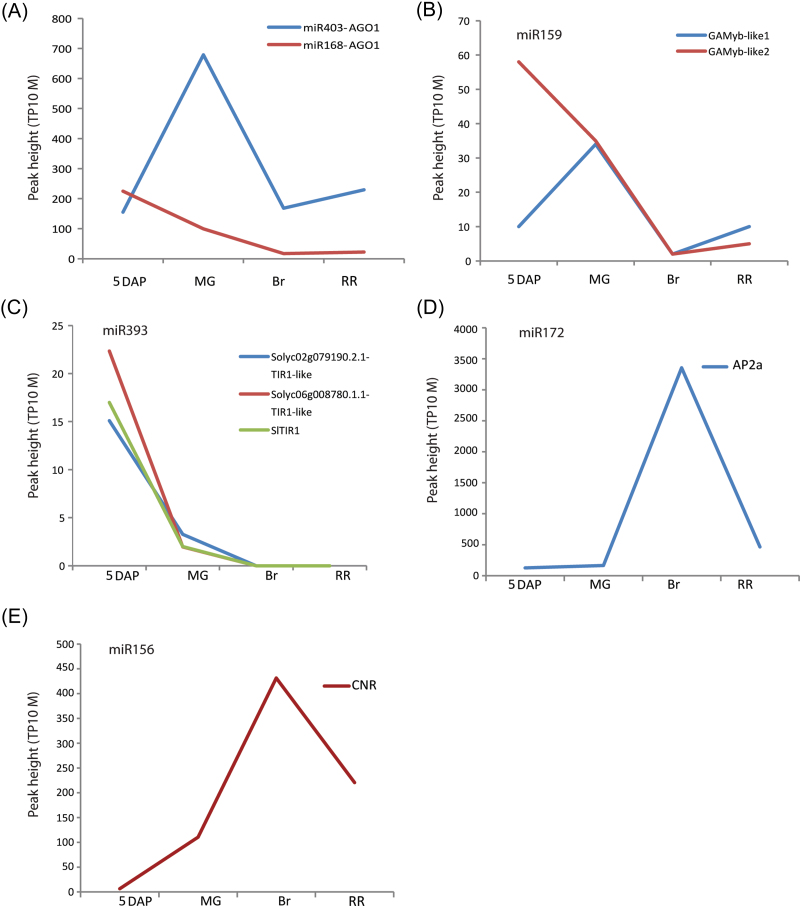
For some conserved miRNAs, such as miR162, only a few targets have been validated in any other species, but it is generally accepted that this miRNA regulates small RNA (including miRNA) production by targeting Dicer-like (DCL) RNase genes (Xie et al., 2003). Of the two RNase-encoding mRNAs predicted to be targets of miR162 in tomato, evidence for targeting of one (Solyc10g005130.2.1) was found but above the P-value cut-off (Supplementary Table S3 at JXB online). For miR403, a member of the ARGONAUTE family different from that targeted by miR168 in tomato (Moxon et al., 2008) was identified as a target (Table 1). Interestingly, the degradation pattern of the AGO2-like target of miR403 was increased at the MG stage and decreasing at the Br stage, while the AGO1-like target of miR168 degradation is lower and already decreases at the MG stage (Fig. 2A). This degradation pattern correlates with miR403 expression. MiR403 was shown to be highly expressed at the MG stage and decreases at the Br stage (Zuo et al., 2012).
Fig. 2.
miRNA-directed cleavage product abundance at different fruit stages. Normalized sequenced read abundance [peak height=tags per 10 million (TP10M)] on the y-axis. (A) Cleavage product abundances of AGO1-like (Solyc02g069260.2.1) by miR403 and of AGO1-like (Solyc06g072300.2.1) by miR168. (B) Cleavage product abundances of GAMYB-like 1 and of GAMYB-like 2 by miR159. (C) Cleavage product abundances of SlTIR1, and of two other TIR1-like mRNAs by miR393. (D) Cleavage product abundance of AP2a by miR172. (E) Cleavage product abundance of CNR by miR156.
For the rest of the conserved miRNAs, the predicted conserved targets were confirmed. For example, out of 13 MYB TF genes predicted to be a target of miR159, three with degradome tags at any developmental stage were found, and all these had at least one category 0 peak. All three are also predicted miR319 targets, with slightly higher TargetFinder penalty values (Supplementary Table S2 at JXB online). Two of the identified MYB TF genes are GAMYB-like 1 and GAMYB-like 2 genes (Gong and Bewley, 2008), which were shown to play a role in seed development in rice and Arabidopsis (Kaneko et al., 2004; Reyes and Chua, 2007). The present data show that while GAMYB-like 2 degradation is higher at the 5 DAP fruit stage, the cleavage of GAMYB-like 1 is higher at the MG stage (Fig. 2B). In conclusion, from the above-described target genes of miR159, different degradation patterns were observed, which is probably due to the difference in the transcript levels of these targets and not due to the level of miR159, which was shown to be equally expressed in different stages of tomato fruit development (Buxdorf et al., 2010).
MiR156 is generally known to target TF genes of the SBP family, which are involved in phase changes during plant development (Wu and Poethig, 2006). Out of 10 tomato SBP genes predicted to be targets of miR156, seven with a category 0 peak in at least one stage were detected. Among these are the two previously validated targets, one of which is CNR (Solyc02g077920.2.1), a regulator of tomato fruit ripening (Thompson et al., 1999; Manning et al., 2006). Members of another TF gene family, AP2 homologues, were shown to be targeted by miR172, both through mRNA degradation and through translational inhibition (Chen, 2004), and as such it affects flowering time and phase transition as well as floral organ identity (Aukerman and Sakai, 2003). All eight tomato AP2 genes predicted to be targets of miR172 were found to have category 0 tag peaks in almost all developmental stages tested. Among these are SlAP2a, b, c, d, and e, which were all predicted to be targets of miR172 (Karlova et al., 2011), but of which only AP2c is a previously validated target (Itaya et al., 2008; Moxon et al., 2008). SlAP2b and SlAP2c cannot be distinguished based on the peak tag sequence as their homology extends 20 nucleotides 3’ of the miR172 processing site. As they are both expressed in the green fruit stages (Karlova et al., 2011), they are probably both targets. The three other, newly identified miR172 targets are members of an AP2 subclade, containing SlAP2d and SlAP2e, as well as Arabidopsis flowering time regulators SCHNARCHZAPFEN (SNZ) and SCHLAFMÜTZE (SMZ), which is distinct from the subclade that contains SlAP2a, SlAP2b, SlAP2c, and Arabidopsis AP2 (Karlova et al., 2011). For the conserved miR319, the TCP family TF genes were identified as targets. From seven predicted tomato TCP TF targets of miR319, evidence was found for cleavage of all mRNAs, but interestingly the cleavage in all cases was found to be at position 11 relative to the 5’ end of Sly-miR319 as submitted to miRbase. As the submitted sequence is one nucleotide longer at the 5’ end than any of the published miR319 sequences from other species, it is assumed that the predominant miR319 species in tomato is similar to that of other species, which would bring the cleavage position to position 10. For miR164, four tomato No Apical Meristem/Cup-shaped Cotyledon (NAC) TF genes were identified as targets. Previously, miR164, through its NAC targets, were shown to be involved in regulation of organ boundaries and lateral root formation (Laufs et al., 2004; Mallory et al., 2004; Guo et al., 2005). As was shown before, miR166 in other species (Sun, 2011) targets TF genes of the class III Homeodomain Leucine Zipper (HD-ZIP) family. Evidence of targeting for two tomato HD-ZIP targets was found, including one that had been validated as a target before (Moxon et al., 2008). As expected for miR169 and miR170/171, the A-subunit genes of the Nuclear Factor Y (NF-Y) or HAP transcription factors (Combier et al., 2006) and the GRAS/SCR (Scarecrow-related) TF family (Llave et al., 2002), respectively, were identified as targets. The results show that during tomato fruit development some conserved miRNAs are regulating non-conserved targets in addition to the conserved targets. In total, 16 non-conserved tomato-specific targets were identified for nine conserved miRNAs (Table 1).
Identification of targets of non-conserved miRNAs
The non-conserved miRNAs are found only in specific species. These miRNAs are often expressed at lower levels than the conserved miRNAs and therefore are more difficult to discover (Sun, 2011; Zuo et al., 2012). Of the 22 non-conserved tomato miRNAs identified previously (Sun, 2011; Zuo et al., 2012), targets in class I could be identified for 15 in tomato fruits (Table 1). Only for miR158, miR161 (Zuo et al., 2012), miR828, miR1919 (Moxon et al., 2008), miR5300, miR5304 (Mohorianu et al., 2011), and miR6026 (Li et al., 2012) significant target cleavage could not be detected. This may be explained by the low expression of these miRNAs in the tomato fruit, or by the absence of targets. A total of 36 target genes for the non-conserved tomato miRNAs were found in this study. This includes new targets not shown before in tomato. These new targets all belonged to class I (Table 1). As shown previously for soybean (Song et al., 2011), the targets of non-conserved miRNAs were not enriched in TFs. The present data confirmed these results. In tomato fruits, targets of the non-conserved miRNAs miR158, miR894, and miR1917 encode enzymes. Other targets of miR393, miR1918, miR5301*, miR6022, miR6023, miR6024, and miR6027 are likely to be involved in signal transduction and/or disease resistance (see below, and Table 1). Exceptions were miR858 and miR5301, for which targets belonging to the MYB class TF family were identified, as well as for miR5303 with a GRF TF as target. The status of miR5301 as a non-conserved mature miRNA unique in tomato is somewhat in doubt as it derives from the complementary strand of miR5301*, which in turn is the tomato homologue of miR482, a conserved miRNA (see below). Members of the MYB TF families were shown before to be targets of miR858 in Arabidopsis (Fahlgren et al., 2007) and tomato (Moxon et al., 2008). As for miR319, the conserved targets for miR858 were found to be cleaved at a site corresponding to position 11 of the published miR858 sequence, as was also reported by Moxon et al. (2008) for another target (Solyc08g082890). In Arabidopsis, MYB12 was shown to function as an activator of flavonoid biosynthesis. Flavonoids in the tomato fruit are found particularly in the peel, where their synthesis is regulated by a MYB12 homologue, which is a predicted target of miR858 (Supplementary Table S2 at JXB online) (Ballester et al., 2010). The targets identified here are other tomato homologues of MYB12, which suggests that also in tomato miR858 together with its targets could be involved in regulation of the biosynthesis of flavonoids or other metabolites (Ballester et al., 2010).
Tomato immune receptor mRNAs targeted by microRNAs
Among the miRNAs that are conserved in different plant species, 22 nucleotide miR472 and miR482, which have similar sequences, target R (Resistance)-like genes, resulting in the formation of secondary, trans-acting small interfering RNAs (tasi-RNAs) (Cuperus et al., 2010). In tomato fruits, both miRNAs have been shown to be expressed (Pilcher et al., 2007; Itaya et al., 2008; Moxon et al., 2008). Sly-miR482 is shifted two nucleotides relative to miR472 and (non-tomato) miR482. MiR5301* (miR-W* in Mohorianu et al., 2011), derived from the complementary strand of the miR5301 precursor, has a sequence intermediary between miR472 and (non-tomato) miR482 (Supplementary Fig. S3 at JXB online). Not surprisingly, miR472, miR482, and miR5301* are predicted to have largely similar targets among the CC-NBS-LRR (coiled coil–nucleotide binding–leucine-rich repeat domain) genes. The present degradome analysis only identified category 0 peaks in these targets for miR472 and miR5301* (Table 1). Additionally, a previously identified in vivo target of mir5301* (Solyc04g025160.2.1, encoding an Arsenite ATPase transporter homologue) was found (Table 1). Thus miR472 or miR5301* activity in developing tomato fruits has been shown, but no clear evidence of miR482 activity was found.
Recently a set of nine new microRNAs in Solanaceae (tobacco, potato, tomato), which target R genes, were described (Li et al., 2012). Of these, five (miR6022–miR6024, miR6026, and miR6027) were shown to occur in tomato. The present data shows that during tomato fruit development miRNAs involved in the regulation of immune response-related genes are active.
Signatures associated with the TAS3 precursors in tomato
In Arabidopsis, miR390 targets a single TAS3-precursor mRNA, and induces the phased production of tasiRNAs from that precursor, starting from the cleaved 3’ miR390 target site. Two dominant siRNA products from this process were shown to target two auxin response factor genes, ARF3 and ARF4, in Arabidopsis (Allen et al., 2005). In tomato, the miR390-derived product of a TASI3 homologue, cDNA Solyc01g058100.2.1 (Table 1), was detected, as well as an as yet unannotated TASI3 homologue, SGN-U587265, which has two detected miR390 cleavage sites. Additionally, evidence was found for miR390-directed processing of one other tomato transcript (Table 1, Solyc12g098430.1.1), which shows 97% identity with the putative arp2/3 complex 20kDa subunit from castor bean (GenBank accession no. EEF44192.1).
Cleavage of miRNA targets involved in tomato fruit development and ripening
Previous studies in tomato have shown that a large part of the predicted miRNA targets are TFs (Mohorianu et al., 2011; Zuo et al., 2012). The present results prove those predictions; that is, a large fraction of the miRNA targets that were identified as class I targets at four different fruit developmental stages code for TFs. Several TF genes and genes that control fruit development and ripening have been found. Among those identified targets are ARF genes, a putative auxin receptor, SlTIR1, as well as fruit ripening regulators such as CNR and AP2a. The plant hormone auxin was shown to be a key regulator for fruit development. Recently, Ren et al. (2011) demonstrated that the tomato SlTIR1 gene plays an important role at the flower-to-fruit transition. SlTIR1 was found to be expressed in floral buds and flowers at the anthesis stage. From anthesis to post-anthesis when fruit set is expected to occur, the expression of SlTIR1 declines in the ovary and sepal. Here it was found that SlTIR1 as well as two other TIR1 homologues are targets of miR393, with a peak of degradation in 5 DAP fruits (Fig. 2C). These data suggest that miR393 is involved in controlling the expression of TIRs during tomato fruit set, which may be essential for the process. Overexpression of SLTIR1 in tomato resulted in parthenocarpy and also in altered gene expression of ARF genes, such as SLARF6 and ARF7 (Ren et al., 2011). Another ARF gene, SlARF8 was shown to be involved in tomato fruit set (Goetz et al., 2007). Several members of the ARF family predicted to be targets of miR160 (Mallory et al., 2005) were found in the degradome libraries, including the two previously validated as targets (Moxon et al., 2008; Hendelman et al., 2012). Taken together, the data suggest that miRNAs in tomato are involved in the initiation of fruit growth by controlling the expression of ARF genes.
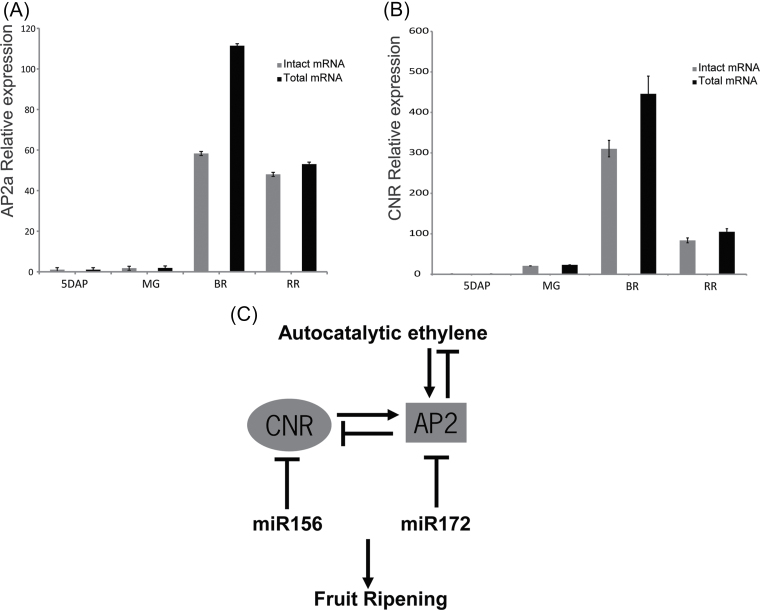
Fruit formation and fruit yield were affected in tomato by overexpression of miR156 (Zhang et al., 2011). MiR156 is known to target mRNA of the TF gene CNR (Solyc02g077920.2.1), a regulator of tomato fruit ripening (Thompson et al., 1999; Manning et al., 2006). Another ripening regulator, AP2a, was identified in the libraries as a target of miR172. AP2a was shown to regulate fruit ripening through negative regulation of ethylene biosynthesis and signalling (Chung et al., 2010; Karlova et al., 2011). For both ripening regulators CNR and AP2a, an increased amount of cleavage product was observed at the Br stage as compared with the other three fruit stages, which coincides with the highest expression level of the genes as shown by qPCR (Figs 2D, E, 3A, B). Both miR156 and miR172 are expressed at different fruit stages and their expression decreases at the RR (Br+7) stage (Mohorianu et al., 2011). To determine exactly how much of the CNR and AP2a transcripts are present as cleavage products, expression analysis by qPCR was used. For this purpose, two sets of primers were designed for each mRNA. One primer pair fits the 3’ UTR of the transcript (and thus detects both intact messenger and the 3’ product of cleavage). The other primer pair spans the miRNA target site and thus detects only the non-cleaved mRNA. As shown in Fig. 3 and in agreement with the degradome data (Fig. 2D, E), increased cleavage of the AP2a (48%) and CNR (30%) transcripts was observed at the Br stage as compared with the other fruit stages tested. Since AP2 and CNR are important regulators of fruit ripening, their expression increases at the Br stage, which is concurrent with an increase in ethylene production. Here it is shown that not only the expression, but also the miRNA-dependent cleavage of AP2a and CNR increases at the same Br stage. Apparently both miR156 and miR172 significantly modulate the intact mRNA levels of their targets during ripening, without completely abolishing them.
Fig. 3.
Relative level of degradation of AP2a and of CNR mRNA by miR172 and miR156, respectively. Relative expression profiles of AP2a and CNR obtained by quantitative reverse transcription PCR with two different primers sets: in grey, primers spanning the miRNA cleavage site (detecting the level of the non-cleaved mRNA); in black, primers for the 3’ UTR, downstream of the cleavage sites (detecting both the intact and the cleaved mRNA) in tomato wild-type fruits at the 5 DAP, MG, Br, and RR stage. Error bars represent the SE. The expression levels are normalized relative to 5 DAP, which was set at 1, for each gene separately. (A) Relative expression profile of AP2a. (B) Relative expression profile of CNR. (C) Model of tomato fruit ripening control by CNR, AP2a, miR156, and miR172. The transcription factor genes AP2a and CNR are required for the normal ripening process. AP2a has a negative regulatory function in ethylene synthesis besides its positive ripening regulatory functions. CNR and AP2a regulate each other’s expression in opposite ways and their expression is also negatively regulated by miR156 and miR172, respectively.
Discussion
Conserved and non-conserved targets of the conserved miRNAs in tomato
Control of gene expression involves a variety of transcriptional and post-transcriptional processes. The abundance of a given mRNA depends on the equilibrium of its expression and degradation rates. In the last decade, a new class of small RNAs, miRNAs were shown to be involved in the regulation of gene expression by mRNA degradation, and many of their targets encoded members of TF families. To identify the function of tomato miRNAs, the relatively new method PARE was used for global analysis of mRNA cleavage products of miRNA action, comparing the mRNA degradomes from different stages of tomato fruit development. The data confirm, as was shown for other species, that miRNAs and their targeted mRNAs are clearly conserved in flowering plants (Li et al., 2010; Pantaleo et al., 2010; Song et al., 2011). For the 24 conserved and 22 non-conserved miRNAs identified previously for tomato, targets from class I could be detected for 19 and 15 miRNAs, respectively. Homologues of the targets of the conserved miRNAs were also detected in the degradome studies of other species. For example, miRNAs 156, 172, 160, 164, and 319 were shown to target mRNAs of primarily one family of (different) TFs each in Arabidopsis, rice, grapevine, and soybean (for a review, see Sun, 2011). Because some of these miRNAs and their targets were found to exist in mosses (Addo-Quaye et al., 2009b ), it is assumed that regulation of gene expression by miRNAs arose early in plant evolution, showing again the importance of the miRNA-dependent RNA degradation process. In this study new targets of conserved as well as of non-conserved miRNAs in tomato could be identified. For example, for the conserved miR396, apart from members of the expected GRF TF family, five different targets not shown in other plant species before were identified. One of these, SlDRM5, is a homologue of DNA (cytosine-5-)-methyltransferase 3, and is probably involved in DNA methylation in tomato. The function of these newly identified targets, and their regulation by miRNA in tomato fruit development remains to be determined.
Targets of non-conserved miRNAs
For the non-conserved tomato miRNAs, 36 targets were identified, of which 35 were new targets not shown before in tomato. As shown previously in degradome studies in soybean (Song et al., 2011), the targets of the non-conserved miRNAs were not enriched in TFs. Interestingly, among the non-conserved miRNA targets were miRNAs that regulate innate immune responses. The data show that although some of the non-conserved miRNAs and their targets are hard to detect, because of their low expression, using degradome libraries from different fruit developmental stages is a successful strategy.
The role of miRNAs in tomato fruit development
Most fruit crops require pollination and fertilization to ensure fruit set. The phytohormone auxin is a key regulator of tomato fruit set and development (Woodward and Bartel, 2005). Recently, it was shown that the tomato SlTIR1 (a putative auxin receptor) plays an important role in fruit set (Ren et al., 2011). Using the PARE method, it could be shown that miRNAs may regulate initiation of fruit development through miR393 and miR167 targeting of SlTIR1 and ARFs, respectively.
Ripening affects several fruit quality traits, such as colour, flavour, and soluble solid content. In the cultivated tomato, it comprises a series of biochemical and physiological events, including softening, pigment change, development of flavour components, autocatalytic ethylene production, and climacteric respiratory behaviour (for a review, see Klee and Giovannoni, 2011). Two TFs that play a major role in regulation of tomato fruit ripening, CNR and AP2a, were known to be a target, or a putative target, of miR156/157 and miR172, respectively (Karlova et al., 2011; Moxon et al., 2008). In this study, it was confirmed that SlAP2a is indeed a target of miR172 in tomato and, moreover, it was shown that both TF mRNAs are actively targeted during fruit development and ripening. The appearance of the t-plots of both mRNAs would suggest that most mRNA is present only in a cleaved state as there is one dominant peak at the cleavage position. In fact, qPCR experiments that distinguish uncleaved mRNA from total transcript show that for both genes intact and cleaved mRNAs are present simultaneously. The miRNA-directed cleavage of both CNR and AP2a mRNAs increases at the Br stage of the fruit, to at least 30% and 48%, respectively, which coincides with the peak of ethylene production. These numbers may be higher if the respective 3’ cleavage products have a faster turnover. In the RR stage, the contribution of 3’ cleavage products declines for both mRNAs, although more so for SlAP2a, while its intact mRNA level remains almost equal. This fits the earlier observed expression profile of miR172 during fruit development determined by northern blotting, which showed miR172 levels peaking at Br to Br+5 d and declining at Br+7 d (Mohorianu et al., 2011). For CNR, the level of its intact mRNA as well as of its 3’ cleavage product decreases during that stage. miR156/157 was shown to be expressed more equally from immature green stages onward, but also declined at Br+7 d (Mohorianu et al., 2011). Although for both miRNAs fruit is not the organ with the highest expression in tomato, these levels are apparently well suited for achieving an attenuating effect on the intact mRNA levels of two important ripening regulators. AP2a expression was shown to be induced by ethylene, while AP2a is a negative regulator of ethylene biosynthesis (Karlova et al., 2011). Interestingly, AP2a was shown to be a repressor of CNR expression, while CNR positively regulates the expression of AP2a. The results presented here identify miR156/157 and miR172 as additional components of a regulatory network in tomato fruit ripening (Fig. 3C). The results suggest that miR156 and miR172 function in fruit ripening by modulating the activity of AP2a (which inhibits ethylene biosynthesis) and of CNR, particularly at and around the Br stage. As AP2a and CNR are considered as an inhibitor and an activator of ripening, respectively, this adds another layer of complexity to the regulatory network model (Fig. 3C). In this model, the function of miR156/157 and miR172 in fruit ripening is to fine-tune the expression of CNR and AP2a to appropriate levels in the different stages of tomato fruit ripening. All these events are contributing in a balanced manner to the process of normal tomato fruit development and ripening. This also implies that natural or induced variation in miRNA expression could be used in breeding programmes aimed at modifying ripening parameters in tomato.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Relationship between independent microarray hybridization-derived transcript abundance and transcript abundance derived from treatment of degradome data as RNAseq.
Figure S2. Examples of t-plots depicting categories.
Figure S3. Alignment of mir472/482-related sequences.
Table S1. Summary of the degradome tag mapping to the tomato genome.
Table S2. miRNA target prediction results for tomato cDNAs (ITAG2.3) and SGN-Unigenes.
Table S3. Normalized CleaveLand v3 output matching degradome tags to predicted miRNA cleavage sites for targets with at least one category 0 or 1 peak in the four libraries.
Acknowledgements
We would like to thank Dr Michael Axtell (Pennsylvania State University) for providing us with the CleaveLand v3 scripts ahead of publication and for his advice on using the scripts, and Dr Stephane Rombauts (Flanders Institute of Biotechnology, Ghent University) for providing us with the tomato gene model details. This work was supported by the Netherlands Genomics Initiative (NGI) ‘Horizon Breakthrough’ ZonMw grant no. 40-41009-98-9054 to RK, and by the Centre for BioSystems Genomics to RdeM.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. 2008. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Current Biology 18, 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Miller W, Axtell MJ. 2009a. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. 2009b. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15, 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, et al. 2010. Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SLMYB12 leads to pink tomato fruit color. Plant Physiology 152, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statististical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Buxdorf K, Hendelman A, Stav R, Lapidot M, Ori N, Arazi T. 2010. Identification and characterization of a novel miR159 target not related to MYB in tomato. Planta 232, 1009–1022. [DOI] [PubMed] [Google Scholar]
- Chen X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology 25, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. 2010. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. The Plant Journal 64, 936–947. [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, De Billy F, et al. 2006. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes and Development 20, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Structural and Molecular Biology 17, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics 27, 1011–1012. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, et al. 2007. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2, e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, et al. 2011. Rfam: Wikipedia, clans and the ‘decimal’ release. Nucleic Acids Research 39, D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Luo S, Schroth G, Meyers BC, Green PJ. 2009. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nature Protocols 4, 356–362. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, et al. 2008. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nature Biotechnology 26, 941–946. [DOI] [PubMed] [Google Scholar]
- Goetz M, Hooper LC, Johnson SD, Rodrigues JC, Vivian-Smith A, Koltunow AM. 2007. Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiology 145, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Bewley DJ. 2008. A GAMYB-like gene in tomato and its expression during seed germination. Planta 228, 563–572. [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. The Plant Cell 17, 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendelman A, Buxdorf K, Stav R, Kravchik M, Arazi T. 2012. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Molecular Biology 78, 561–576. [DOI] [PubMed] [Google Scholar]
- Itaya A, Bundschuh R, Archual AJ, Joung J-G, Fei Z, Dai X, Zhao PX, Tang Y, Nelson RS, Ding B. 2008. Small RNAs in tomato fruit and leaf development. Biochimica et Biophysica Acta 1779, 99–107. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57, 19–53. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, et al. 2004. Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. The Plant Cell 16, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. 2011. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. The Plant Cell 23, 923–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. 2011. Genetics and control of tomato fruit ripening and quality attributes. Annual Review of Genetics 45, 41–59. [DOI] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. 2004. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. 2012. MicroRNA regulation of plant innate immune receptors. Proceedings of the National Academy of Sciences, USA 109, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. 2010. Transcriptome-wide identification of microRNA targets in rice. The Plant Journal 62, 742–759. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell 17, 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. 2004. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Current Biology 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics 38, 948–952. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Souret FF, Lu C, Green PJ. 2006. Sweating the small stuff: microRNA discovery in plants. Current Opinion in Biotechnology 17, 139–146. [DOI] [PubMed] [Google Scholar]
- Mohorianu I, Schwach F, Jing R, Lopez-Gomollon S, Moxon S, Szittya G, Sorefan K, Moulton V, Dalmay T. 2011. Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. The Plant Journal 67, 232–246. [DOI] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme, Pilcher RL, Moulton V, Dalmay T. 2008. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Research 18, 1602–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, et al. 2007. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nature Genetics 39, 787–791. [DOI] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. 2010. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. The Plant Journal 62, 960–976. [DOI] [PubMed] [Google Scholar]
- Pilcher RLR, Moxon S, Pakseresht N, Moulton V, Manning K, Seymour G, Dalmay T. 2007. Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226, 709–717. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y. 2011. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. Journal of Experimental Botany 62, 2815–2826. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. 2007. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal 49, 592–606. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS. 2011. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biology 11, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. 2011. MicroRNAs and their diverse functions in plants. Plant Molecular Biology 18, 17–36. [DOI] [PubMed] [Google Scholar]
- Teyssier E, Bernacchia G, Maury S, How, Kit A, Stammitti-Bert L, Rolin D, Gallusci P. 2008. Tissue dependent variations of DNA methylation and endoreduplication levels during tomato fruit development and ripening. Planta 228, 391–399. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Tor M, Barry CS, Vrebalov J, Orfila C, Jarvis MC, Giovannoni JJ, Grierson D, Seymour GB. 1999. Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiology 120, 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Annals of Botany 95, 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3 . Development 133, 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. 2003. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Current Biology 13, 784–789. [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, de Paoli E, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes and Development 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z. 2011. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Letters 585, 435–439. [DOI] [PubMed] [Google Scholar]
- Zuo J, Zhu B, Fu D, Zhu Y, Ma Y, Chi L, Ju Z, Wang Y, Zhai B, Luo Y. 2012. Sculpting the maturation, softening and ethylene pathway: the influences of microRNAs on tomato fruits. BMC Genomics 13, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.