Abstract
Strigolactones (SLs) are newly identified hormones that regulate multiple aspects of plant development, infection by parasitic weeds, and mutualistic symbiosis in the roots. In this study, the role of SLs was studied for the first time in the model plant Lotus japonicus using transgenic lines silenced for CAROTENOID CLEAVAGE DIOXYGENASE 7 (LjCCD7), the orthologue of Arabidopsis More Axillary Growth 3. Transgenic LjCCD7-silenced plants displayed reduced height due to shorter internodes, and more branched shoots and roots than the controls, and an increase in total plant biomass, while their root:shoot ratio remained unchanged. Moreover, these lines had longer primary roots, delayed senescence, and reduced flower/pod numbers from the third round of flower and pod setting onwards. Only a mild reduction in determinate nodule numbers and hardly any impact on the colonization by arbuscular mycorrhizal fungi were observed. The results show that the impairment of CCD7 activity in L. japonicus leads to a phenotype linked to SL functions, but with specific features possibly due to the peculiar developmental pattern of this plant species. It is believed that the data also link determinate nodulation, plant reproduction, and senescence to CCD7 function for the first time.
Key words: Arbuscular mycorrhizal fungi (AMF), carotenoid cleavage dioxygenase, determinate nodulation, leaf senescence, Lotus japonicus, reproduction, shoot and root branching, strigolactone.
Introduction
Plant development is plastic and environmentally sensitive, and is regulated by hormones acting as long-range signals to integrate developmental, genetic, and environmental inputs. In the control of shoot and root branching, for example, large variations in plant architecture can be generated in a single genotype in response to a number of cues (Domagalska and Leyser, 2011; Müller and Leyser, 2011). The role of classic plant hormones, such as auxin and cytokinins, in the regulation of shoot and root branching and in the maintenance of coordinated growth between root and shoot has been extensively studied (Sachs, 2005; Hwang et al., 2012). Recently, strigolactones (SLs) have been identified as a new class of branch-inhibiting hormones that seem to fine-tune the regulation of shoot branching further (Gomez-Roldan et al., 2008; Umehara et al., 2008; Xie et al., 2010). In the past decade, genotypes affected in the SL pathway were identified, and include ramosus (rms) mutants in pea, decreased apical dominance (dad) in petunia, more axillary branch (max) in Arabidopsis, and high-tillering dwarf (htd or d) in rice (Beveridge and Kyozuka, 2009).
Besides regulating shoot architecture, SLs contribute to shaping the root system, by affecting primary root length, adventitious root formation, lateral root initiation and subsequent outgrowth, and root hair elongation (Koltai, 2011, 2012; Ruyter-Spira et al., 2011; Rasmussen et al., 2012). Moreover, a broad range of developmental roles was postulated for SLs in the alleviation of secondary seed dormancy induced by high temperature (Toh et al., 2012), hypocotyl elongation (Hu et al., 2010; Tsuchiya et al., 2010), secondary growth (Agusti et al., 2011), light harvesting (Mayzlish-Gati et al., 2010; Tsuchiya et al., 2010), reproductive development (Snowden et al., 2005; Kohlen et al., 2012), and leaf senescence (Snowden et al., 2005; Ledger et al., 2010). SLs seem to be specifically involved in the establishment of the symbiosis with nitrogen-fixing bacteria in legumes, namely in the formation of indeterminate nodules (Soto et al., 2010; Foo and Davies, 2011; Foo et al., 2013). Finally, besides their hormonal role, SLs exuded into the rhizosphere stimulate the germination of parasitic plants (Yoneyama et al., 2010; Lechat et al., 2012) and the branching of arbuscular mycorrhizal fungi (AMF) (Akiyama et al., 2005; Bouwmeester et al., 2007; Yoshida et al., 2012). A recent report suggests that the hormonal effect on the elongation of rhizoids (the equivalent of root hairs in higher plants) in ancestral species of the green lineage pre-dates the exogenous function as signalling molecules in the rhizosphere (Delaux et al., 2012).
SLs are a family of carotenoid-derived terpenoid lactones mostly produced in roots (Matusova et al., 2005). The precursor all-trans-β-carotene is converted by the recently characterized β-carotene isomerase D27 into 9-cis-β-carotene (Alder et al., 2012). The isomerized substrate is then cleaved by two double bond-specific cleavage enzymes (carotenoid cleavage dioxygenases, CCDs), CCD7 and CCD8. The 9’, 10’ bond of 9-cis-β-carotene is cleaved by CCD7, yielding β-ionone (C13) and 10’-apo-β-carotenal (C27). The latter compound is subsequently cleaved and cyclized by CCD8 into a bioactive SL precursor named carlactone (Booker et al., 2004; Schwartz et al., 2004; Alder et al., 2012). Several orthologues of these two CCDs have been characterized in Arabidopsis, pea, petunia, rice, and tomato (Morris et al., 2001; Sorefan et al., 2003; Booker et al., 2004; Snowden et al., 2005; Zou et al., 2006; Arite et al., 2007; Drummond et al., 2009; Vogel et al., 2010; Kohlen et al., 2012). CCD8 orthologues are also characterized in kiwifruit, chrysanthemum, maize, and the moss Physcomitrella patens (Ledger et al., 2010; Liang et al., 2010; Proust et al., 2011; Guan et al., 2012). MAX1, a class-III cytochrome P450 protein of Arabidopsis, has also been proposed to act in the SL biosynthetic pathway, namely converting carlactone into 5-deoxystrigol, but its biochemical action still needs to be resolved experimentally (Booker et al., 2005; Alder et al., 2012). Other genes putatively involved in SL biosynthesis have been identified in several species (SlORT1 and AtPPD5), but their exact function is so far unknown (Koltai et al., 2010b ; Roose et al., 2011). No orthologues of any of the above biosynthetic genes have been characterized in Lotus japonicus yet.
Lotus japonicus is a perennial legume of temperate climates, and a model plant for several developmental processes and interactions with soil (micro)organisms (Handberg and Stougaard, 1992; Lohar and Bird, 2003); the first SL molecule described as a branching factor for AMF, 5-deoxystrigol, was isolated from this plant (Akiyama et al., 2005). Additionally, L. japonicus has a phyllotaxis distinct from that of other model plants such as Arabidopsis, pea, petunia, tomato, and rice. In fact, all of its cotyledonary axillary buds develop immediately into lateral shoots even in very young seedlings. This, and the regular, proliferative accessory (axillary) meristem initiation and immediate development of axillary shoots, makes L. japonicus an attractive experimental model to study the regulation of accessory meristem initiation and development (Alvarez et al., 2006). Namely, how SLs specifically affect the architecture of a plant with this kind of phyllotaxis is not known at present. Also, L. japonicus develops determinate nodules, differently from pea and Medicago sativa, the two species investigated so far for the role of SLs in nodulation.
In this study, the cloning of the Lotus orthologue of CCD7 (LjCCD7), and an overall characterization of its role in the regulation of plant architecture, reproductive development, senescence, and root symbiosis are reported. The results also link CCD7 expression with the regulation of determinate nodulation, reproduction, and senescence in L. japonicus. These latter phenotypes have not been reported so far for any CCD7 orthologue.
Materials and methods
RNA isolation, cDNA synthesis, and quantitative reverse transcription–PCR (RT–qPCR)
For transcript quantification and rapid amplification of cDNA ends (RACE) cloning purposes, RNA was isolated from freshly harvested tissues of wild-type L. japonicus ecotype Gifu B-129 and transgenic lines in the same background with Tripure reagent (Roche). On-column DNase digestion was performed with an RNase-free DNase kit, and total RNA was further purified by an RNeasy Plant Mini Kit (both Qiagen). RNA quality and integrity were checked by NanoDrop ND-2000 and standard gel electrophoresis. A 1 μg aliquot of total RNA was reverse transcribed to cDNA with an iScript cDNA synthesis kit (Bio-Rad). RT–qPCRs were set up in 20 μl using the iQ SYBR Green Supermix on the iQ5 Real-Time PCR system (Bio-Rad). LjCCD7-specific primers are listed in Supplementary Table S1 available at JXB online. Ubiquitin (LjUBI) transcript was used as a normalizer (Yokota et al., 2009). Quantification followed the 2-Δ ΔCt method.
5’- and 3’-RACE
The putative LjCCD7 gene was identified based on the EST (expressed sequence tag) in the Kazusa database (http://www.kazusa.or.jp/lotus/) aligning best under BlastP default settings with query sequences from Arabidopsis (GI: 330255400) and pea (GI: 90019042). The SMART RACE cDNA amplification kit (Clontech) was used to amplify the unknown ends of the coding sequence in a cDNA pool from Lotus roots, and cloning was performed using the Advantage 2 PCR enzyme system (Clontech) and primers UPM with GSP1 or GSP2 (Supplementary Table S1 at JXB online). PCR conditions for both cDNA ends were five cycles at 94 °C for 30 s, 72 °C for 3min; five cycles at 94 °C for 30 s, 70 °C for 30 s, 72 °C for 3min; and 25 cycles at 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3min. RACE products were electrophoresed, tested with primers NestF and NestR (Supplementary Table S1 at JXB online), cloned in the pGEM-T easy vector (Promega), and sequenced (BMR Genomics, Padova). The full-length sequence of LjCCD7 (1866bp) was assembled virtually with the Vector NTI Advance 11.0 and amplified from the aforementioned cDNA batch with primers LDF and LDR (Supplementary Table S1 at JXB online). The PCR product was cloned into pGEM-T to generate pGEM-T_LjCCD7, and sequenced. The resulting coding sequence is deposited in GenBank (ID: GU441766).
Plasmid constructs
For expression in Escherichia coli of glutathione S-transferase (GST)–LjCCD7, an EcoRI fragment was subcloned from pGEM-T_LjCCD7 into pGEX-5X-3. To generate the LjCCD7 silencing vector, a 250bp region (from 1451bp downstream of the transcription start site to the stop codon, within the last exon) was amplified with Pfu polymerase (Promega) and primers RNAiF and RNAiR (Supplementary Table S1 at JXB online). The PCR product was directly sequenced and cloned in pDONR221 to generate the entry clone pDONR_LjCCD7. Finally, a single-step Gateway-based reaction was performed with pDONR_LjCCD7 and the destination vector pTKO2 (Snowden et al., 2005). In parallel, the negative control (NC) construct pTKO2_pENTR was generated with the empty entry vector pENTR4 and the destination vector pTKO2. Both pTKO2_LjCCD7 and pTKO2_pENTR constructs were sequenced with primer pairs 572F/1133R and 1388F/1847R (Supplementary Table S1 at JXB online).
Protein expression and in vitro enzyme assays
Cultures of E. coli BL21 (600ml) harbouring pGEX_LjCCD7 or the empty vector in 2× YT medium (per litre: 16g of tryptone, 10g of yeast extract, and 5g of NaCl) were grown at 28 °C until A600=0.5. Recombinant protein expression was induced by 0.2mM isopropyl-β-d-1-thiogalactopyranoside at 18 °C for 24h. Escherichia coli cells were harvested by centrifugation at 4 °C, resuspended in cold STE buffer (100mM NaCl, 10mM TRIS, 1mM EDTA, pH 8.0) containing 100 μg ml–1 lysozyme, incubated on ice for 15min, and then lysed by sonication. The recombinant protein was purified with glutathione–Sepharose 4B (GE Healthcare), visualized by 10% SDS–PAGE, and electrotransferred to a blotting membrane (Fluorotrans, Fluka). Western blot analysis was performed by blocking with 5% non-fat dehydrated milk in TBST buffer (10mM TRIS-HCl, 150mM NaCl, and 0.05% Tween-20), followed by incubation with polyclonal antibodies against GST and then against alkaline phosphatase (Sigma) diluted respectively at 1:5000 and 1:10 000 in TBST buffer with 0.5% milk. Extensive washing in TBST buffer was followed by nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate staining (Sigma).
For in vitro enzymatic assays, reactions were performed as previously reported (Schwartz et al., 2004; Marasco et al., 2006) with minor modifications. Soluble proteins were quantified with the Bradford reagent (Bio-Rad). Aliquots of the affinity-purified GST–LjCCD7 protein (50–100 μg in 100 μl of glutathione elution buffer) were brought to 400 μl with 100mM TRIS buffer, pH 7.0 containing 300mM NaCl, 0.5mM FeSO4, 10mM dithiothreitol (DTT), 5mM ascorbate, and 0.05% Triton X-100. After 20min of equilibration, 80 μl of substrate (0.5mM Type II β-carotene in acetonitrile; presumably all-trans, Sigma C4582) were added. Note that even if 9-cis-β-carotene is the real substrate of CCD7 (Alder et al., 2012), trans-β-carotene can also be cleaved (though not as efficiently), and is the commercially available isomer. Reaction tubes were gently shaken in the dark at 28 °C for 5h before being quenched with 50 μl of 33% formaldehyde for 10min at 37 °C. A 600 μl aliquot of acetonitrile was added to each tube and the organic layer was saved for HPLC analysis. A Perkin Elmer series 200 HPLC system equipped with a diode array was used for detection. Separation was performed using a C18 reversed-phase column (250 mm×4.6mm internal diameter, 5 μm particles; YMC Europe) with the solvent system methanol:water (70:30, v/v) containing 0.1% ammonium acetate (B) and methanol (A) as described (Marasco et al., 2006).
Plant transformation and regeneration
The transformation of L. japonicus ecotype Gifu B-129 was performed via Agrobacterium tumefaciens strain EHA105 harbouring pTKO2_LjCCD7 or pTKO2_pENTR (NC) as detailed previously (Barbulova et al., 2005) with slight changes. Briefly, roots, shoots, and leaves were excised from 3-week-old seedlings grown in Petri dishes on Gamborg B5 (Sigma) and separately conditioned onto callus-inducing medium (CIM) (Lombari et al., 2003) for an additional week. Segments were dipped for 25min into A. tumefaciens cultures grown to A600=0.4–0.6 at 28 °C in YEB medium (per litre: 5g of peptone, 5g of beef extract, 1g of yeast extract, 5g of sucrose, and 0.5g of MgCl2). The explants were then dried quickly on sterile filter paper and transferred onto fresh CIM plates. After 3 d of co-cultivation in the dark, bacterial slime was rinsed away in sterile water. Explants were dried on sterile filter paper and transferred on CIM plus 500mg l–1 cefotaxime and 200mg l–1 carbenicillin. After 2 d, transformed explants were selected on CIM plus 500mg l–1 cefotaxime, 200mg l–1 carbenicillin, and 100mg l–1 kanamycin. After 3–4 weeks, the kanamycin-resistant sectors were transferred on shoot-inducing medium (SIM) (Barbulova et al., 2005) containing 500mg l–1 cefotaxime, 100mg l–1 kanamycin, and 0.5mg l–1 thidiazuron. After 2 weeks, calli from which rudimentary shoots were emerging were transferred on SIM plus 500mg l–1 cefotaxime, 100mg l–1 kanamycin, and 0.05mg l–1 thidiazuron for shoot elongation. Regenerated T0 plants with 1.5–2.0cm roots were transferred into pots filled with 2:1 perlite:Arabidopsis special soil (Horticoop) for seed setting. Their seeds (T1 generation) were surface-sterilized (70% ethanol for 1min and 2.5% NaClO for 25min), stratified at 4 °C for 2–3 d on jellified Gamborg B5, then germinated at 24 °C in a growth chamber (16h light/8h dark). Then they were transferred in perlite-filled pots in a growth chamber (20–21 °C, 16h light/8h dark); 3-week-old seedlings from >40 independent T0 transgenic lines were screened by PCR with primer pairs KanF/KanR and RNAiR1/pTKO2-1847R (Supplementary Table S1 at JXB online) for the integration of the transgenic cassette. Among those with an SL-related phenotype, lines PG, P9, and P16 were selected for further studies because of their obvious shoot phenotype and homogeneous degree of target gene silencing and SL depletion. At the T0 generation, these were hemizygous (as ascertained by PCR screening of the T1 progeny obtained from self-pollination). Homozygous T1 individuals were retained on the basis of progeny segregation analysis and propagated to T2 and T3. All plants included in all experiments were tested for cassette integration and LjCCD7 transcript levels in roots (if possible), and showed an obvious shoot phenotype.
Plant phenotyping
Seedlings of the wild type and RNA interference (RNAi) line P16 (unless otherwise stated) were used for metabolic (T1 generation) and phenotypic analysis (T0, T1, T2, and/or T3 generations, depending on the phenotypic character; for details, see the figure legends). Wild-type and/or transgenic NC plants were used interchangeably as controls, since no differences in LjCCD7 expression, SL content, or general morphology were recorded. Surface-sterilized seeds were pre-conditioned on wet filter paper for 2 d, and then moved into a growth chamber (16h light/8h dark, 24 °C). Two-week-old seedlings were grown in 18cm diameter pots filled with Arabidopsis special soil and perlite (1:2) in a greenhouse (16h light/8h dark, 20 °C), watered daily, and given ‘Hornum’ nutrients (Handberg and Stougaard, 1992) twice per week. Different traits were evaluated at different plant ages, as indicated in the Results.
For all statistical analyses on shoot branching, only branches longer than 0.5cm were included. Root parameters (total length, area, and volume) were calculated by the WinRhizo software on digital images of whole apparatuses. For primary root length and stem width measurements, organs were photographed and analysed by ImageJ software. For chlorophyll quantification, two compound leaves, located at the same position on the main stem, were collected from three individual plants for each of the RNAi lines (PG, P9, and P16) and analysed as reported (Ni et al., 2009). From the third round of seed setting until plants were 11 months old, flowers were counted on three plants for each of the three RNAi lines and NCs, and pods were harvested. The definitions used to describe Lotus phyllotaxis are as in Alvarez et al. (2006).
SL extraction and LC-MS/MS analysis
Two-week-old seedlings of transgenic line P16 and transgenic NC (15 each) were transplanted into an X-stream 20 aeroponic system (Nutriculture) and supplied with 5 litres of modified half-strength Hoagland solution (Hoagland and Arnon, 1950) refreshed twice a week. Experiments were performed 3–4 weeks later, when roots were fully developed but before the emergence of flowers. Root exudates were collected, purified, and concentrated as described previously (Liu et al., 2011). One day before exudate collection, roots were thoroughly rinsed with distilled water and the nutrient solution was refreshed to eliminate accumulated SLs. After exudate collection, roots from five plants for each sample were pooled and stored at –80 °C for later use. The experiment was repeated three times.
SL extraction from root exudates and tissues was performed as previously reported (López-Ráez et al., 2010) with minor modifications. Exudates were firstly condensed into 50ml 60% acetone fractions eluted from a C18 column (GracePure 5000mg 20ml–1), of which 2ml fractions were used, and each mixed with 200 μl of 0.1 nmol ml–1 [2H]6-5-deoxystrigol in acetone as internal standard for further purification. Once samples were well evaporated under a speed vacuum, the residues were dissolved in 50 μl of ethyl acetate and diluted with 4ml of hexane for silica column purification (GracePure 200mg 3ml–1). Elutions of 40% or 60% ethyl acetate in hexane were combined and dried. The residues were dissolved in 200 μl of acetonitrile:water (25:75), filtered, and used for ultraperformance liquid chromatography tandem spectrometry (UPLC-MS/MS) analysis. Fresh frozen root samples (0.5g each) were ground and extracted with 2ml of 0.05 nmol ml–1 [2H]6-5-deoxystrigol, used as internal standard, in ethyl acetate. The samples were sonicated for 15min in a Branson 3510 ultrasonic bath (Branson Ultrasonics, USA). Samples were centrifuged for 15min at 2500 g; the supernatant was gently transferred to 4ml glass vials, and the pellet was re-extracted with 2ml of ethyl acetate without internal standard. Supernatants were combined, and ethyl acetate evaporated under vacuum. The following steps were performed as described above in root exudate purification. A Xevo tandem mass spectrometer (Waters) equipped with an electrospray ionization (ESI) source and coupled to an Acquity UPLC system (Waters) was used for SL detection and identification as described (Kohlen et al., 2012). Data were analysed with MassLynx 4.1 (Waters).
Rhizobia and AMF infection assays
Mesorhizobium loti strain R7A was used for nodulation assays. Slightly lignified shoots were cut off 4-month-old plants (line P16), and set for root induction in rockwool. When roots were ~0.5–1.0cm long (1.5–2.0 weeks), plants were transferred to perlite for another week, and irrigated with B&D nutrient solution (Broughton and Dilworth, 1971). Each plant was inoculated with 30ml of bacterial culture grown at 28 °C in YMB medium (per litre: 0.4g of yeast extract, 10.0g of mannitol, 5.0g of K2HPO4, 0.2g of MgSO4, 0.1g of NaCl, 0.5g of sucrose) and diluted to a final A600 of ~0.05. Nodules and lateral roots were counted 2 weeks post-inoculation.
AMF colonization assessment was carried out on plants infected with Gigaspora margarita spores in ‘Millipore sandwiches’ (Novero et al., 2002). Briefly, seeds sterilized with sulphuric acid were pre-germinated on 0.6% water agar. Roots were placed between two nitrocellulose membranes (5cm diameter, 0.45 μm pores; Millipore) with 20 fungal spores sterilized by chloramine T (3%) and streptomycin sulphate (0.3%). After 4 weeks of co-culture, roots were sampled and stained with cotton blue (0.1% in lactic acid) to visualize fungal structures. For each genotype, intraradical colonization was evaluated in three plants and at least 160cm of roots under an optical microscope (Trouvelot et al., 1986).
Statistical analysis
One-way analysis of variance (ANOVA) was performed on all data sets by using GenStat for Windows. If needed, data were also subjected to Student’s t-test.
Results
Cloning of LjCCD7 and in vitro confirmation of its enzymatic activity
MAX3 from Arabidopsis and RMS5 from pea were used to interrogate the EST and genomic database at the Kazusa DNA Research Institute. The EST aligning best to the queries (LjSGA_131670.1, 873bp) was assumed to derive from their bona fide Lotus orthologue, designated LjCCD7. Based on the partial coding sequence available, the missing cDNA ends of LjCCD7 were obtained by 5’- and 3’-RACE followed by direct amplification of the full-length coding sequence. The predicted polypeptide was 74% identical to RMS5 and 58% identical to MAX3. A phylogenetic tree (Supplementary Fig. S1 at JXB online) was produced from an alignment of several CCD7 homologues from land plants and moss, as well as of some putative homologues identified in cyanobacteria (Cui et al., 2012). The CCD7 proteins clustered into several clades, with LjCCD7 in the subclade of predicted leguminous homologues within the dicot clade, as expected (Supplementary Fig. S1 at JXB online).
To verify whether the phylogenetic prediction corresponded to a conserved enzymatic function, the carotenoid cleavage activity of LjCCD7 was tested in vitro. To do so, recombinant GST–LjCCD7 was affinity purified from E. coli cells, and its apparent size (~98kDa) checked by SDS–PAGE and western blotting (Supplementary Fig. S2A, B at JXB online). After 5h incubation in the presence of β-carotene (C40) as substrate, two compounds were detected in the presence of GST–LjCCD7 and not with GST alone. One of them had a visible spectrum peak and retention time similar to β-ionone (C13; Supplementary Fig. S2C). The second was deduced to be 10’-apo-β-carotenal (Supplementary Fig. S2D); that is, the complementary C27 that would result from the cleavage of the 9’, 10’-bond of β-carotene by CCD7. These results agreed with previous work in Arabidopsis and tomato (Booker et al., 2004; Schwartz et al., 2004; Vogel et al., 2010) and proved that the cloned cDNA corresponds to a protein endowed with carotenoid cleavage activity in vitro. With the outcome of phylogenetic analysis shown in Supplementary Fig. S1, they support the notion that the identified gene encodes the L. japonicus CCD7 orthologue.
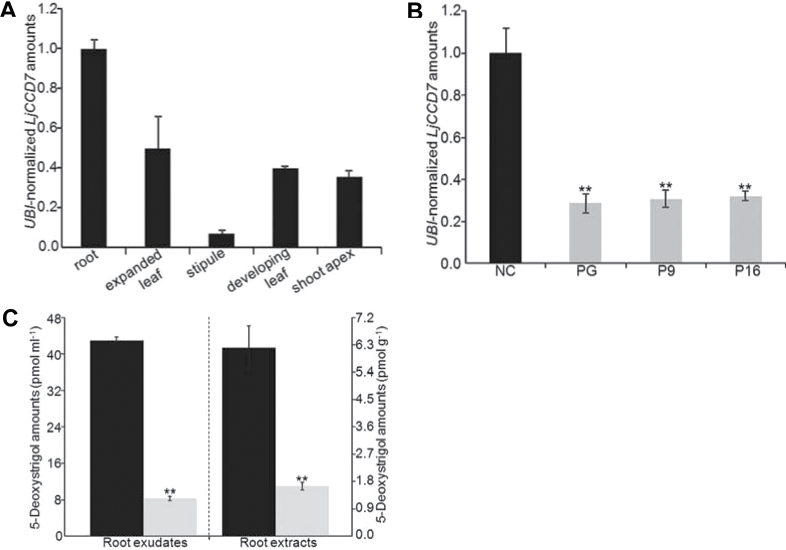
To study the expression pattern of LjCCD7, RT–qPCR was performed on RNA samples from a range of vegetative tissues from 8-week-old wild-type plants. As in Arabidopsis (Booker et al., 2004), pea (Johnson et al., 2006), petunia (Drummond et al., 2009), and tomato (Vogel et al., 2010), LjCCD7 was predominantly expressed in roots, ~2-fold more than in leaves and the shoot apex. LjCCD7 transcript was also detected in stipules, although 13 times less abundantly than in roots (Fig. 1A).
Fig. 1.
LjCCD7 transcription pattern, and molecular and metabolic characterization of CCD7-silenced lines. (A) LjCCD7 transcript abundance in various vegetative tissues relative to root expression level in 8-week-old wild-type L. japonicus. Values are the average of n biological and three technical replicates ±SE, normalized to LjUBI transcript levels. n=6 for root and shoot apex, n=5 for expanded or developing leaf, n=4 for stipule. (B) Relative LjCCD7 transcript amounts in roots of RNAi lines PG, P9, and P16 (grey bars), compared with a line transformed with the empty vector (negative control, NC; black bar). Values are normalized to LjUBI transcript amounts, and displayed as means of n biological and three technical replicates ±SE (n=3 for each RNAi line and n=5 for controls, all at the T1 generation). The asterisks indicate statistically significant differences for P < 0.01. (C) LC-MS/MS analysis of 5-deoxystrigol content in root exudates and extracts of 6/7-week-old, aeroponically grown plants of the wild type (black bars) and of the CCD7-knockdown line P16 at generation T1 (grey bars). Twenty-four hours before collection of root exudates, the nutrient solution was refreshed for 15 plants. In root extracts, three independent samples were used and each consisted of five plants. Error bars represent the SE for three analytical replicates. The asterisks indicate a statistically significant difference for P < 0.01.
Generation and molecular characterization of transgenic lines impaired in LjCCD7 expression
To elucidate the biological function of LjCCD7 in planta, >40 independent RNAi lines were generated. Three of them (PG, P9, and P16) displayed ~70% less LjCCD7 transcript in roots relative to corresponding transgenic control plants (Fig. 1B), and were selected for further analysis. CCD7 is a key biosynthetic gene for SLs, so the presence of all known SLs in the root exudates and extracts of wild-type Lotus was initially checked (not shown); 5-deoxystrigol was the only one detected, in accordance with previous work (Sugimoto and Ueyama, 2008). 5-Deoxystrigol was therefore quantified in both root exudates and extracts of wild-type L. japonicus and transgenic seedlings (line P16). In all samples, one distinct peak was detected that had the retention time and transitions corresponding to 5-deoxystrigol. 5-Deoxystrigol levels were higher in root exudates than in root extracts, both in wild-type seedlings and in the CCD7-silenced line. Metabolite abundance in RNAi plants was decreased by 81% and 73% in root exudates and extracts, respectively, relative to the non-silenced control seedlings (Fig. 1C).
Decreased LjCCD7 transcript correlates with changes in shoot and root morphology
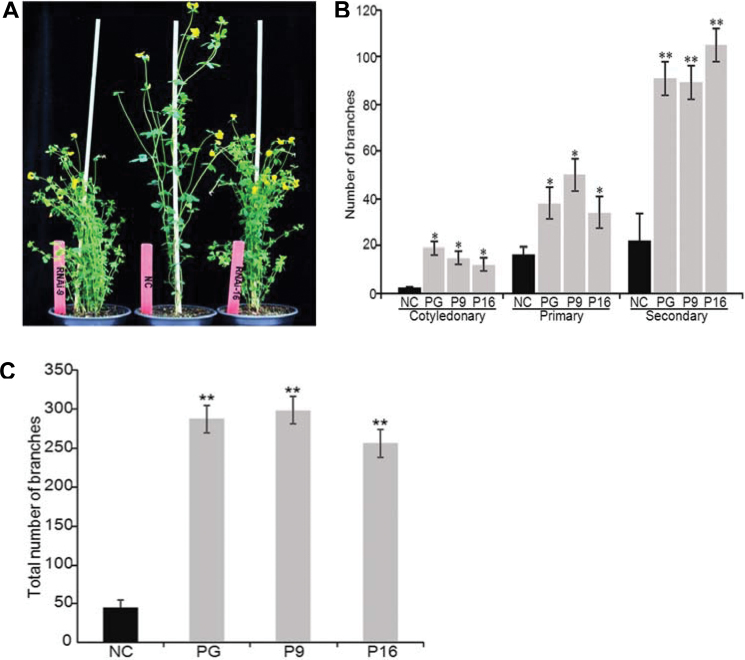
Since SLs regulate plant architecture, experiments were conducted to assess whether the reduction in LjCCD7 transcript and SL content was associated with altered morphology in RNAi line P16; lines P9 and PG, in which LjCCD7 was silenced comparably, were also evaluated. From 3 weeks after germination, the RNAi lines exhibited a clearly stunted and bushy phenotype (Fig. 2A). Shoot branches were counted at two time points. Eight-week-old transgenic plants were significantly more branched than controls transformed with the empty vector: RNAi lines displayed 5.3, 2.1, and 4.0 times more cotyledonary, primary, and secondary aerial branches, respectively (Fig. 2B). When 6 months old, plants of the PG, P9, and P16 lines displayed 6.4, 6.6, and 5.7 times more total shoot branches relative to the controls, respectively (Fig. 2C). No branches of secondary or higher order occurred in the transgenic control plants at this or later stages, in contrast to RNAi lines (Supplementary Fig. S3A, B at JXB online). RNAi plants also had a significantly reduced main stem height (39, 40, and 29% shorter than the transgenic NC line, respectively; Fig. 3A). Reduced plant height was not due to a reduced number of nodes in the RNAi lines (not shown). Rather, internodes were significantly shorter in silenced plants compared with the corresponding control, particularly in high-order nodes. In fact, internodes IV–VII of the main stems were ~30–40% shorter in line P16 than in the wild type (Fig. 3B). The same was observed for most internodes above node III in secondary shoot branches (Fig. 3C). SLs have been shown to control the thickening of stems and roots (Agusti et al., 2011). As expected, RNAi line P16 displayed an ~39% reduction of secondary shoot diameter compared with the control (Fig. 3D, E).
Fig. 2.
Shoot architecture analysis of RNAi lines. (A) Representative comparison of shoot branching appearance of RNAi line P16 and negative controls (NCs) transformed with the empty vector, both 10 weeks old and at the T1 generation. (B) Cotyledonary, primary, and secondary aerial branches of 8-week-old RNAi plants from lines PG, P9, and P16 (grey bars) and the NC (black bars). Each value represents the mean ±SD (n=4 and n=3 T1 plants for control and each RNAi genotype, respectively). The asterisks indicate statistically significant differences for *P < 0.05 and **P < 0.01. (C) Total shoot branches of 6-month-old T1 RNAi lines PG, P9, and P16 (grey bars) and the NC (black bar). Each value represents the mean ±SD (n=14 for control versus n=3 for each RNAi line). The asterisks indicate statistically significant differences for P < 0.01. (This figure is available in colour at JXB online.)
Fig. 3.
Shoot morphology of SL-depleted versus control plants. (A) Plant height of 6-month-old T1 transgenic negative control (NC, black bar) and RNAi lines PG, P9, and P16 (grey bars). Error bars represent the SD of the means (n = 10 and n=3 for NC and each RNAi line, respectively). Asterisks indicate statistically significant differences between each RNAi line and the NC, for P < 0.01. (B and C) Comparison of internode length between 2.5-month-old wild-type plants (black bars) and RNAi line P16, T1 generation (grey bars). The internode above the cotyledons was set as the first node in the acropetal direction, so internode I lies between collar and cotyledonary node I, and so forth. Both the internodes from the main stem (B) and basal secondary shoots (C) were used for analysis. Data are means ±SD (n ≥ 6). Asterisks indicate statistically significant differences for *P < 0.05 and **P < 0.01. (D) Representative display of secondary stem width in SL-depleted (RNAi) and NC plants. The picture was taken on 3-month-old T1 plants of RNAi line P16 and transgenic plants transformed with the empty vector. Bar scale=1cm. (E) Quantification of secondary stem width. Data were obtained on 3-month-old T1 plants and are presented as means ±SD (n=22 for NC, black bar; and n=45 for RNAi line P16, grey bar). The asterisks indicate significantly different values for P < 0.01. (This figure is available in colour at JXB online.)
SLs affect root development, including lateral root formation, and primary root and root hair development (Koltai, 2011; Ruyter-Spira et al., 2011). To assess the function of SLs in Lotus root development, plants of RNAi line P16 were grown either in aeroponic tanks or in pots. Under all conditions, the RNAi line consistently showed more lateral roots and longer primary roots (Fig. 4A). Five-week-old plants grown in aeroponic tanks were used for quantitative analysis: RNAi plants had a 1.6–2.0 times higher total root length, area, and volume than controls (Fig. 4B, C). Notably, the primary root of the RNAi line was significantly longer than in the corresponding controls grown either in the aeroponic system or in pots.
Fig. 4.
Phenotypic characterization of roots in SL-depleted and control plants. (A) Root morphology of 4-week-old plants representative of the wild type and RNAi line P16, T1 generation (left panel). The plants in the picture were grown for 1 week in Petri dishes in a growth chamber, 1 week in perlite, and another 2 weeks in aeroponic tanks. Root shape of 3.5-month-old transgenic negative control (NC) and RNAi line P16, T2 generation (right panel), cultivated in pots filled with Arabidopsis soil (Horticoop) and perlite (1:2). (B and C) Quantification of root parameters in 5-week-old wild-type (black bars) and RNAi plants (line P16 at T1 generation, grey bars). Data are presented as means ±SD (n > 6). Asterisks indicate statistically significant differences for *P < 0.05 and **P < 0.01. (This figure is available in colour at JXB online.)
Effect of LjCCD7 silencing on reproduction
Lotus japonicus ecotype ‘Gifu’ flowers profusely and, in subsequent rounds, yields thousands of seeds within months after pollination (Handberg and Stougaard, 1992). We observed that seed production was severely impaired in RNAi plants compared with controls, particularly after two rounds of seed settings (representative individuals are shown in Supplementary Fig. S4A at JXB online). For a statistical assessment, all flowers from the third round on were counted and pods were harvested separately. An at least 7-fold reduction of flower and pod numbers was observed in RNAi lines relative to the NC (Fig. 5). After the above-ground part of 11-month-old plants was lopped off, ~40 times fewer pods were counted on the emergent branches of RNAi plants than on controls (not shown). Impairment of flower and seed setting was less severe for line P16 than lines PG and P9, which is why the first was used for most analyses. There were no significant changes with respect to size, shape, or colour of the flowers, pods, and seeds. Given the intimate cross-talk occurring between SLs and auxin during reproductive development (Kohlen et al., 2012), the levels of free indole acetic acid (IAA) were measured in flowers collected at anthesis. Only a mild but statistically non-significant reduction was observed in RNAi line P16 compared with controls (Supplementary Fig. S4B at JXB online).
Fig. 5.
LjCCD7 transcript correlates with flower and pod number. Quantification of flowers and pods in 11-month-old transgenic negative control (NC, black bars) and RNAi lines PG, P9, and P16 at the T1 generation (grey bars). Values are means ±SD (n=3 independent plants per genotype). The asterisks indicate statistically significant differences for P < 0.01.
Effect of decreased LjCCD7 transcript levels on plant senescence
Lotus is a deciduous perennial displaying progressive leaf senescence and abscission in temperate climates. Since a role for SLs in senescence had been proposed (Snowden et al., 2005; Ledger et al., 2010), the leaves of the transgenic plants were monitored for any obvious colour change compared with control, and chlorophyll in leaves of comparable physiological age was quantified. All three RNAi lines remained green for a longer time than the control (see Supplementary Fig. S5A at JXB online) and accumulated significantly more chlorophyll a (Chla) and Chlb than the NC, with a cumulative 2-fold increase in total chlorophyll content when ~8 months old (Fig. 6A). At a younger stage (~3 months old), RNAi lines PG and P9 showed a significant increase in Chla and Chlb, respectively; however, total chlorophyll content was not significantly different from that of the NC (Fig. 6A). When the top and internal part of 8-month-old RNAi plants left to senesce spontaneously had slightly started to fade in colour, controls were already severely senescing throughout their aerial parts (Supplementary Fig. S5B at JXB online). Also, more new shoots and branches kept emerging in RNAi lines compared with NCls (Supplementary Fig. S3C at JXB online). Finally, the RNAi line had higher biomass relative to control plants when ~3.5 months old (Fig. 6B); the trend was maintained with plant ageing. However, the shoot-to-root ratio was largely unchanged (average ratio ±SD: 2.796±0.204 for RNAi lines versus 2.591±0.094 for controls; data in Fig. 6B).
Fig. 6.
LjCCD7 influences age-related senescence progression and biomass in Lotus. (A) Total chlorophyll (Chla, striped bars; Chlb, solid bars) in leaves of transgenic negative controls (NC, black bars) and RNAi lines PG, P9, and P16 at the T1 generation (grey bars), 3 or 8 months old. Values are means ±SD (n=8 for each RNAi line and n=7 for NC). The asterisks indicate significant differences of Chla and Chlb between RNAi plants and NCs of the same age for *P < 0.05 and **P < 0.01. (B) Fresh weight of roots and aerial parts in 3.5-month-old wild-type plants (black bars) compared with RNAi line P16, T1 generation (grey bars). Values are means ±SD (n≥ 6). The asterisks indicate statistically significant differences for *P < 0.05 and **P < 0.01.
LjCCD7-silenced Lotus nodulates less but is colonized by AMF as much as control plants
The influence of SLs on Lotus determinate root nodules was explored. There were no significant differences in root fresh weight of RNAi plants compared with the wild-type and NC plants (not shown). However, their adventitious roots were significantly more branched also in the specific culture conditions used for this test (Fig. 7A, B). Although this situation potentially offers a larger surface to rhizobial infection for the same root biomass, RNAi roots carried significantly fewer nodules per gram of fresh weight than the NC 2 weeks post-inoculation (~20% reduction; Fig. 7C). In contrast, when controls and RNAi lines P16 and P9 were inoculated with spores of the AMF G. margarita, no significant differences in the colonization parameters evaluated were detected (Fig. 7D). SL-depleted roots supported both extra- and intraradical mycelium, which did not differ in quantity and developmental pattern from control roots. Also, arbuscules were morphologically indistinguishable (not shown).
Fig. 7.
SL-depleted Lotus plants nodulate less than controls but are not affected in AM symbiosis. (A) Representative picture of the differences in nodule number and root development detected between the wild type (left), RNAi line P16 (middle), and the transgenic negative control (NC, right), T1 generation, 2 weeks after inoculation in perlite. (B) Root branching and (C) nodule number per gram of fresh root weight of 4-week-old roots in the wild type and NCs (black bars), and RNAi line P16, T1 generation (grey bar). All values are displayed as means ±SD (n=17 for the wild type, n=6 for NC, and n=14 for the RNAi line). Asterisks indicate significant differences for *P < 0.05. (D) Results of mycorrhization tests on controls (wild-type and NC plants, black bars) and RNAi lines P9 and P16, T2 generation (grey bars). Trouvelot tests were performed on n=3 root apparatuses for each genotype, sandwich-inoculated with Gigaspora margarita spores. F%, frequency of mycorrhization; M%, intensity of mycorrhization; a%, percentage of arbuscules within the infected areas. A%, percentage of arbuscules in the whole root system. (This figure is available in colour at JXB online.)
Discussion
The role of SLs in the regulation of plant architecture is conserved in L. japonicus, but with some peculiarities
The phenotype of the SL-depleted L. japonicus plants characterized in this study was overall expected, on the basis of previously published work. RNAi plants were in fact stunted and their shoots more branched than controls, as were root apparatuses. More adventitious roots (not shown) were also observed, consistent with published data on Arabidopsis and pea (Rasmussen et al., 2012). Notwithstanding, the shoot morphology of L. japonicus is quite different from that of other model plant species, which makes it an attractive model to study SL function. In fact, in wild-type Lotus, the axillary buds in the cotyledonary node or in the leaf axils are already visible during the early vegetative stage. Once these buds are initiated, they develop directly into lateral branches without being inhibited by any factors released by the shoot apical meristems. Thereafter, numerous accessory meristems will potentially develop lateral branches in the cotyledonary node zone, but in the aerial part only a single bud in the leaf axis develops into a branch while the others remain dormant (Alvarez et al., 2006). This peculiar developmental pattern was reflected in some specific shoot features in the SL-depleted plants. In contrast to most SL-related phenotypes described so far, in fact, branching of secondary shoots was highly increased compared with controls. This phenotype has been reported, though to a milder extent, only in CCD7- and CCD8-silenced tomato plants (Vogel et al., 2010; Kohlen et al., 2012). In transgenic Lotus, this increase was most probably due to several of the accessory buds in the aerial axils developing into lateral shoots, in contrast to what happens in the wild type (Alvarez et al., 2006). Also, many more basal branches than in the wild type were produced from the activity of accessory buds in the cotyledonary axil, resulting in more numerous primary shoots (Supplementary Fig. S3C at JXB online). Primary and secondary shoots were shorter in CCD7-silenced plants compared with control plants because of a reduced internode length (as in petunia and maize) (Snowden et al., 2005; Guan et al., 2012) rather than number (as in tomato) (Kohlen et al., 2012).
LjCCD7-silenced plants differed slightly but significantly from other published SL mutants at the root level as well, in that they consistently exhibited longer primary roots than the controls. Results in Arabidopsis, rice, and maize point instead to a role for SLs in the promotion of primary root growth (Koltai et al., 2010a ; Ruyter-Spira et al., 2011; Arite et al., 2012; Guan et al., 2012). In these species, SLs were proposed to alter the auxin gradient in the root tips, thus changing both the number and length of the cells located in the meristem and transition zone of primary roots. The results suggest that the role of SLs in primary root development differs from species to species, possibly in relation to varying endogenous hormonal levels.
Finally, the above- and below-ground mass of SL-depleted plants was increased, so that the shoot-to-root biomass ratio was comparable with that of controls. This observation differs from what has been reported for the petunia ccd7/dad3 mutant, which displays a higher shoot-to-root ratio than the wild type (Snowden et al., 2005). This discrepancy may be due to the different species, growth conditions (hydroponic for petunia), and/or physiological age of the plants.
CCD7 affects reproduction and age-dependent senescence in L. japonicus
A role for SLs was postulated in reproduction, considering the high expression of Arabidopsis MAX3 in siliques and seeds (Booker et al., 2004; Mashiguchi et al., 2009), of tomato SlCCD7 in green immature fruits (Vogel et al., 2010), of AcCCD7 and AcCCD8 in young kiwifruits and seeds (Ledger et al., 2010), and of ZmCCD8 in maize shank and ear shoots of female inflorescences (Guan et al., 2012). To date, however, CCD7 activity has never been proven to affect reproduction, and, more specifically, the reduced flower, fruit, and seed numbers observed in L. japonicus have never been reported for SL-depleted plants of any species. Rather, the petunia dad1/ccd8 mutant is delayed in the setting of flowers, which also weigh less and are smaller than those of the wild type (Snowden et al., 2005). SlCCD8-silenced tomato plants also have smaller floral organs, fruits, and seeds, as well as 60% fewer seeds relative to controls (Kohlen et al., 2012). Also a significant reduction in the size and diameter of ear and shank was observed in maize ZmCCD8 mutants (Guan et al., 2012). In contrast, abnormal floral organs, altered size and colour of flowers, pods, and seeds, or seed patterning inside the pod were not observed in transgenic Lotus consistent with the fact that no significant differences in free IAA content could be detected in flowers of RNAi and wild-type plants. Therefore, it is proposed that the observed reduction in seed production in transgenic plants is mainly due to the reduced number of flowers. To form its pseudoraceme inflorescences, the shoot main apex of L. japonicus produces a flower branch, which is subtended in the compound leaves and is terminated when one or two flowers are developed (Guo et al., 2006). A possible explanation for the reduced number of flowers in RNAi compared with transgenic control plants, in spite of their higher number of branches, is that their potential to form new buds that can produce flower branches may be reduced compared with controls. This, in turn, could be linked to the fact that the dormancy of many accessory buds was broken to give higher order branches, before transition from vegetative growth to flowering. Alternatively, or additionally, fewer resources could be available to reproduction in the SL-depleted plants because of the more vigorous vegetative growth (see below).
LjCCD7-silenced Lotus lines lagged in leaf senescence and abscission; this the first report linking senescence to a functional defect in a CCD7 homologue. A correlation between SLs and senescence was instead reported in Arabidopsis, rice, and petunia plants lacking MAX2 (Woo et al., 2001; Yan et al., 2007; Drummond et al., 2012), and in the ccd8 mutants of petunia and kiwifruit (Snowden et al., 2005; Ledger et al., 2010). Transcripts of the Arabidopsis MAX3/CCD7 and MAX4/CCD8 genes accumulate during age-dependent leaf senescence, further suggesting that the role of SLs in the process may be direct (Breeze et al., 2011). However, mutation of the MAX2 orthologue in pea (RMS4) does not affect leaf senescence (Johnson et al., 2006), implying that this is not a conserved feature of SLs across plant species.
A link may be seen between the delayed leaf senescence and impaired flower and seed production in the RNAi plants used here. Reproductive development controls the timing of leaf senescence, with photosynthates translocated from leaves to reproductive organs before death. The negative correlation between senescence and reproduction could be even more evident in polycarpic plants such as L. japonicus, which sets numerous inflorescences in subsequent rounds. The iterative production of reproductive organs and seeds may require more resources at the expense of vegetative fitness (Munné-Bosch, 2008). In this scenario, LjCCD7-silenced plants would allocate fewer resources than control plants to reproduction, thus delaying senescence and allowing higher biomass. This interpretation agrees with the observation that chlorophyll content was higher in the leaves of the RNAi lines used here compared with control plants, particularly at and after reproduction. It also agrees with the fact that total biomass was higher in RNAi plants than in controls. Whether the higher resource allocation to vegetative growth is the cause of the observed impairment of reproduction, or rather its effect, cannot be determined at this stage.
Role of SL in the regulation of root symbiosis in L. japonicus
The present data prove for the first time that SLs promote the formation of determinate nodules. The correlation between SLs and nodulation was previously investigated in M. sativa and pea, both forming indeterminate nodules (Soto et al., 2010; Foo and Davies, 2011; Foo et al., 2013). In L. japonicus as well, LjCCD7-silenced plants carried 20% fewer nodules than controls. As in the pea rms1 mutant, alterations in nodule development and morphology were not detected. SLs apparently have no direct influence on the growth of rhizobia, but they do have a quantitative effect on nodulation, in spite of their negative effect on the total root length and surface available to infection (Foo and Davies, 2011). Even if nodules and lateral roots form from different founder cells (being initiated by cortical versus pericycle cell divisions, respectively), a balance was proposed long ago to exist between nodule and lateral root formation, with nodule primordia initiation dependent on the suppression of lateral root emergence (Nutman, 1949). Later, it was proposed that the balance of auxin, cytokinins, and abscisic acid (ABA) decides the fate of pericycle cells and cortical cells, dictating whether lateral roots or nodules will be initiated (Ding and Oldroyd, 2009). Given the proven cross-talk of SLs with auxin (Crawford et al., 2010; Domagalska and Leyser, 2011; Ruyter-Spira et al., 2011; Kohlen et al., 2012) and ABA (Lechat et al., 2012; Toh et al., 2012), and the importance of both these hormones for normal frequency and development of determinate nodules in Lotus (Ding and Oldroyd, 2009), the phenotype observed in the SL-depleted plants could be auxin and/or ABA mediated, and be the outcome of an obviously defective suppression mechanism on lateral root emergence.
As proven for 5-deoxystrigol in root exudates of L. japonicus in the first place, SLs induce hyphal branching of AMF, increasing the chances of successful root colonization (Akiyama et al., 2005). A MAX2/D3-dependent, D14-independent effect on AMF colonization of roots was also observed very recently in rice (Yoshida et al., 2012). In an SlCCD8-knockdown line of tomato, ~50% reduction of total SLs in roots correlated with a 27% decrease in colonization by Glomus intraradices (Kohlen et al., 2012). Interestingly, the RNAi lines used in the present study did not show a significant reduction in AMF colonization for any of the parameters tested. Among all natural and synthetic SLs, 5-deoxystrigol (the only SL found in Lotus) is the most active, inducing AMF branching at subnanogram concentrations (Akiyama et al., 2010; Xie et al., 2010). Therefore, it is possible that the residual 20–30% of 5-deoxystrigol in the RNAi lines used here is still above the required threshold to induce fairly normal hyphal branching of G. margarita in our inoculation system (clean roots placed in close contact with germinating AMF spores). Very recently, protocols enabling highly efficient insertion mutagenesis were set up for Lotus (Fukai et al., 2012; Urbanski et al., 2012). The availability of such reverse genetic tools, through which complete knockout of genes would be achievable, will allow this point to be solved.
In summary, this is the first report of an SL-related phenotype in the perennial herbaceous model L. japonicus. Evidence was presented that LjCCD7 is crucial for SL synthesis in Lotus, where the role of SLs is conserved in the regulation of root and shoot architecture, with some peculiarities; among them, the effects on primary root growth, internode length, number of higher order branches, and root-to-shoot biomass ratio. These apparent discrepancies are most probably specific features of SL action in the model plant used here, and, thus, just the reflection of biological diversity. Additional effects on reproduction and senescence were for the first time linked to an impairment in the expression of a CCD7 enzyme, and an effect of SLs on determinate nodulation was described. Further work is under way to unravel cross-talk of SLs with other hormones and their relative contributions to the observed phenotypes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic analysis of CCD7 protein homologues.
Figure S2. LjCCD7 has carotenoid cleavage activity.
Figure S3. Representative comparison of high-order and basal shoot emergence between SL-depleted (RNAi) and negative transgenic control (NC) plants.
Figure S4. LjCCD7 influence on reproduction.
Figure S5. LjCCD7 impact on age-related senescence processes.
Table S1. List of primers used in this work.
Table S2. List of protein sequences used in phylogenetic analysis.
Acknowledgements
The authors wish to thank M. Chiurazzi (CNR, Italy) for suggestions about plant transformation, J. Stougaard (Aarhus University, Denmark) for Gifu B-129 seeds; S. Karunairetnam (Plant & Food Research, New Zealand) and R. Geurts (Wageningen University, The Netherlands) for vector pTKO2 and Mesorhizobium loti, respectively; W. Kohlen and I. Haider for their guidance during JL’s. stay in Wageningen. Research was funded by the BioBITs Project [Piedmont Region, Converging Technologies 2007] to FC, AC, CL. and PB, and by the Netherlands Organization for Scientific Research [Vici grant, 865.06.002, 834.08.001] to HJB; JL was funded by the China Scholarship Council [2008108168].
References
- Agusti J, Herold S, Schwarz M, et al. 2011. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences, USA 108, 20242–20247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Akiyama K, Ogasawara S, Ito S, Hayashi H. 2010. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant and Cell Physiology 51, 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. [DOI] [PubMed] [Google Scholar]
- Alvarez ND, Meeking RJ, White DWR. 2006. The origin, initiation and development of axillary shoot meristems in Lotus japonicus . Annals of Botany 98, 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite T, Kameoka H, Kyozuka J. 2012. Strigolactone positively controls crown root elongation in rice. Journal of Plant Growth Regulation 31, 165–172. [Google Scholar]
- Barbulova A, D’Apuzzo E, Rogato A, Chiurazzi M. 2005. Improved procedures for in vitro regeneration and for phenotypic analysis in the model legume Lotus japonicus . Functional Plant Biology 32, 529–536. [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Kyozuka J. 2009. New genes in the strigolactone-related shoot branching pathway. Current Opinion in Plant Biology 13, 1–6. [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser C. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. 2005. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Developmental Cell 8, 443–449. [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Roux C, López-Ráez JA, Bécard G. 2007. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends in Plant Science 12, 224–230. [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, et al. 2011. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. The Plant Cell 23, 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. 1971. Control of leghaemoglobin synthesis in snake beans. Biochemical Journal 125, 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Muller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- Cui HL, Wang YC, Qin S. 2012. Genomewide analysis of carotenoid cleavage dioxygenases in unicellular and filamentous cyanobacteria. Comparative and Functional Genomics 2012, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux PM, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N. 2012. Origin of strigolactones in the green lineage. New Phytologist 195, 857–871. [DOI] [PubMed] [Google Scholar]
- Ding Y, Oldroyd GE. 2009. Positioning the nodule, the hormone dictum. Plant Signaling and Behavior 4, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nature Reviews Molecular Cell Biology 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Drummond RS, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. 2009. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE 7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology 151, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RS, Sheehan H, Simons JL, Martinez-Sanchez NM, Turner RM, Putterill J, Snowden KC. 2012. The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Frontiers in Plant Science 2, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Davies NW. 2011. Strigolactones promote nodulation in pea. Planta 234, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Foo E, Yoneyama K, Hugill C, Quittenden L, Reid J. 2013. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Molecular Plant 6, 76–87. [DOI] [PubMed] [Google Scholar]
- Fukai E, Soyano T, Umehara Y, Nakayama S, Hirakawa H, Tabata S, Sato S, Hayashi M. 2012. Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1 . The Plant Journal 69, 720–730. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- Guan JC, Suzuki M, Wu S, Latshaw SP, Petruff T, Goulet C, Klee HJ, Koch KE, McCarty DR. 2012. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific sub-network. Plant Physiology 160, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhao Z, Chen J, Hu X, Luo D. 2006. A putative CENTRORADIALIS/TERMINAL FLOWER 1-like gene, LjCEN1, plays a role in phase transition in Lotus japonicus . Journal of Plant Physiology 163, 436–444. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. 1992. Lotus japonicus, an autogamous, diploid legume species for classical and molecular-genetics. The Plant Journal 2, 487–496. [Google Scholar]
- Hoagland A, Arnon D. 1950. The water-culture method for growing plants without soil. California Agriculture Experiment Station Circular No. 347.
- Hu Z, Yan H, Yang J, Yamaguchi S, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. 2010. Strigolactones negatively regulate mesocotyl elongation in rice during germination and growth in darkness. Plant and Cell Physiology 51, 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Muller B. 2012. Cytokinin signaling networks. Annual Review of Plant Biology 63, 353–380. [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology 142, 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Lammers M, et al. 2012. The tomato CAROTENOID CLEAVAGE DIOXYGENASE 8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytologist 196, 535–547. [DOI] [PubMed] [Google Scholar]
- Koltai H. 2011. Strigolactones are regulators of root development. New Phytologist 190, 545–549. [DOI] [PubMed] [Google Scholar]
- Koltai H. 2012. Strigolactones activate different hormonal pathways for regulation of root development in response to phosphate growth conditions. Annals of Botany (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H, Dor E, Hershenhorn J, et al. 2010. a Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. Journal of Plant Growth Regulation 29, 129–136. [Google Scholar]
- Koltai H, LekKala SP, Bhattacharya C, et al. 2010. b A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. Journal of Experimental Botany 61, 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat MM, Pouvreau JB, Peron T, et al. 2012. PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. Journal of Experimental Botany 63, 5311–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger SE, Janssen BJ, Karunairetnam S, Wang T, Snowden KC. 2010. Modified CAROTENOID CLEAVAGE DIOXYGENASE 8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytologist 188, 803–813. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhao L, Challis R, Leyser O. 2010. Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum). Journal of Experimental Botany 61, 3069–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, et al. 2011. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. The Plant Cell 23, 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Bird DM. 2003. Lotus japonicus: a new model to study root-parasitic nematodes. Plant and Cell Physiology 44, 1176–1184. [DOI] [PubMed] [Google Scholar]
- Lombari P, Ercolano E, El Alaoui H, Chiurazzi M. 2003. A new transformation–regeneration procedure in the model legume Lotus japonicus: root explants as a source of large numbers of cells susceptible to Agrobacterium-mediated transformation. Plant Cell Reports 21, 771–777. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Kohlen W, Charnikhova T, et al. 2010. Does abscisic acid affect strigolactone biosynthesis? New Phytologist 187, 343–354. [DOI] [PubMed] [Google Scholar]
- Marasco EK, Vay K, Schmidt-Dannert C. 2006. Identification of carotenoid cleavage dioxygenases from Nostoc sp PCC 7120 with different cleavage activities. Journal of Biological Chemistry 281, 31583–31593. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T. 2009. Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Bioscience, Biotechnology, and Biochemistry 73, 2460–2465. [DOI] [PubMed] [Google Scholar]
- Matusova R, Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ. 2005. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiology 139, 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, Lekkala SP, Resnick N, Wininger S, Bhattacharya C, Lemcoff JH, Kapulnik Y, Koltai H. 2010. Strigolactones are positive regulators of light-harvesting genes in tomato. Journal of Experimental Botany 61, 3129–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Turnbull CGN, Murfet IC, Beveridge CA. 2001. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiology 126, 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Annals of Botany 107, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné-Bosch S. 2008. Do perennials really senesce? Trends in Plant Science 13, 216–220. [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Chen ZJ. 2009. Chlorophyll and starch assays. Protocol Exchange DOI: 10.1038/nprot.2009.1012 [Google Scholar]
- Novero M, Faccio A, Genre A, Stougaard J, Webb KJ, Mulder L, Parniske M, Bonfante P. 2002. Dual requirement of the LjSym4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New Phytologist 154, 741–749. [DOI] [PubMed] [Google Scholar]
- Nutman PS. 1949. Physiological studies on nodule formation: I. The relation between nodulation and lateral root formation in red clover. Annals of Botany 13, 261–283. [Google Scholar]
- Proust H, Hoffmann B, Xie XN, Yoneyama K, Schaefer DG, Yoneyama K, Nogué F, Rameau C. 2011. Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens . Development 138, 1531–1539. [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, et al. 2012. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiology 158, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Frankel LK, Bricker TM. 2011. Developmental defects in mutants of the PsbP domain protein 5 in Arabidopsis thaliana . PLoS One 6, e28624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, et al. 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiology 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. 2005. Auxin’s role as an example of the mechanisms of shoot/root relations. Plant and Soil 268, 13–19. [Google Scholar]
- Schwartz SH, Qin XQ, Loewen MC. 2004. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. Journal of Biological Chemistry 279, 46940–46945. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. 2005. The Decreased apical dominance 1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE 8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell 17, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, et al. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes and Development 17, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MJ, Fernández-Aparicio M, Castellanos-Morales V, García-Garrido JM, Ocampo JA, Delgado MJ, Vierheilig H. 2010. First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biology and Biochemistry 42, 383–385. [Google Scholar]
- Sugimoto Y, Ueyama T. 2008. Production of (+)-5-deoxystrigol by Lotus japonicus root culture. Phytochemistry 69, 212–217. [DOI] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y. 2012. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant and Cell Physiology 53, 107–117. [DOI] [PubMed] [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V. 1986. Mesure du taux de mycorrhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle. Paris: INRA Press, 217–222. [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P. 2010. A small-molecule screen identifies new functions for the plant hormone strigolactone. Nature Chemical Biology 6, 741–749. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Urbanski DF, Malolepszy A, Stougaard J, Andersen SU. 2012. Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus . The Plant Journal 69, 731–741. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Walter MH, Giavalisco P, et al. 2010. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. The Plant Journal 61, 300–311. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. 2001. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. The Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XN, Yoneyama K, Yoneyama K. 2010. The strigolactone story. Annual Review of Phytopathology 48, 93–117. [DOI] [PubMed] [Google Scholar]
- Yan H, Saika H, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. 2007. Rice tillering dwarf mutant dwarf3 has increased leaf longevity during darkness-induced senescence or hydrogen peroxide-induced cell death. Genes and Genetic Systems 82, 361–366. [DOI] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, et al. 2009. Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti . The Plant Cell 21, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Awad AA, Xie XN, Yoneyama K, Takeuchi Y. 2010. Strigolactones as germination stimulants for root parasitic plants. Plant and Cell Physiology 51, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kameoka H, Tempo M, Akiyama K, Umehara M, Yamaguchi S, Hayashi H, Kyozuka J, Shirasu K. 2012. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytologist 196, 1208–1216. [DOI] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal 48, 687–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.