Abstract
The practice of hybridization has greatly contributed to the increase in crop productivity. A major component that exploits heterosis in crops is the cytoplasmic male sterility (CMS)/nucleus-controlled fertility restoration (Rf) system. Through positional cloning, it is shown that heterozygous alleles (RsRf3-1/RsRf3-2) encoding pentatricopeptide repeat (PPR) proteins are responsible for restoring fertility to cytoplasmic male-sterile radish (Raphanus sativus L.). Furthermore, it was found that heterozygous alleles (RsRf3-1/RsRf3-2) show higher expression and RNA polymerase II occupancy in the CMS cytoplasmic background compared with their homozygous alleles (RsRf3-1/RsRf3-1 or RsRf3-2/RsRf3-2). These data provide new insights into the molecular mechanism of fertility restoration to cytoplasmic male-sterile plants and illustrate a case of overdominance.
Key words: Cytoplasmic male sterility, fertility restoration, heterozygous alleles, overdominance, radish.
Introduction
Plant cytoplasmic male sterility (CMS), a maternally inherited trait that prevents plants from producing functional pollen, has been identified in many higher plants, including rice, cotton, maize, and sorghum (Liu et al., 2001; Bentolila et al., 2002; Klein et al., 2005; Yin et al., 2006; Fujii et al., 2011). CMS restorer systems have been widely exploited to produce hybrids that outperform their inbred parents in yield, biomass, or other traits. CMS is usually attributed to an unusual chimeric gene in the mitochondrial genome (Schnable and Wise, 1998; Ivanov and Dymshits, 2007). In many cases, a nuclear-encoded fertility restorer gene (Rf) can restore fertility of the cytoplasmic male-sterile plants (Hanson and Bentolila, 2004). Therefore, the CMS/Rf system is an ideal model for dissecting the interaction between mitochondrial and nuclear genomes.
A variety of mechanisms of fertility restoration by the Rf genes have been reported for different CMS systems. T-urf13, a mitochondrial gene encoding a 13kDa protein, has been detected only in maize carrying T male-sterile cytoplasm (Dewey et al., 1987; Fujii and Toriyama, 2008). The first restorer allele cloned, the maize Rf2 gene, does not affect the expression of urf13 and encodes aldehyde dehydrogenase (ALDH), which is located in the mitochondrial matrix in a homotetrameric form (Cui et al., 1996; Liu et al., 2001; Liu and Schnable, 2002). The cloned petunia Rf-PPR592 gene is the first single dominant Rf gene encoding a pentatricopeptide repeat-containing (PPR) protein that can eliminate the CMS-associated protein PCF (Bentolila et al., 2002). The Rf-PPR592 protein contains 11 continuous PPR motifs and is a part of the mitochondrial inner membrane-associated, RNase-sensitive large soluble high molecular weight complex that interacts with pcf mRNA (Gillman et al., 2007). The radish Rfo/Rfk1 gene for Ogura/Kosena CMS also encodes a mitochondrially targeted PPR protein of 687 amino acids comprising 16 repeats of the 35 amino acid PPR motif (Brown et al., 2003; Desloire et al., 2003; Imai et al., 2003; Koizuka et al., 2003) that reduces the protein level of ORF138 without changing the level of mRNA (Bellaoui et al., 1999; Uyttewaal et al., 2008). Molecular components responsible for fertility restoration processes were deduced from various types of CMS/Rf systems in rice. Either of two tightly linked genes, Rf1a and Rf1b, can independently restore fertility to Boro II rice and have been identified as encoding PPR proteins, containing contiguous arrays of 18 and 11 PPR repeats, respectively (Komori et al., 2004; Wang et al., 2006). Rf1a blocks cytotoxic peptide ORF79 through endonucleolytic cleavage of the dicistronic B-atp6/orf79 mRNA, whereas Rf1b degrades B-atp6/orf79 mRNA (Wang et al., 2006; Kazama et al., 2008). Rf17 and Rf2 have been isolated as the Rf genes for CW-type and Lead Rice-type CMS, and have been shown to encode mitochondrial proteins, acyl-carrier protein synthase-like protein, and glycine-rich protein (GRP) that are not PPR proteins (Fujii and Toriyama, 2009; Itabashi et al., 2011). Interestingly, a recent work demonstrates that RF5, a PPR protein, physically interacts with GRP162, a GRP encoding 162 amino acids that was identified to bind to atp6-orfH79 (Hu et al., 2012).
Here the map-based cloning of heterozygous alleles controlling male fertility of CMS in radish is reported.
Materials and methods
Plant materials
9802A1 is a male-sterile strain. 9802B1 is a radish cultivar used as a maintainer of 9802A1. 9802A1 contains the same nuclear genome as 9802B1. 9606H is a Chinese radish cultivar. 0107H is a male-fertile strain carrying Rfob (EU163282). The cross between 9802A1 and 9606H yielded male-fertile F1 plants. Self-pollination of F1 plants yielded an F2 segregating population. The BC1F1 population was produced from the backcross between 9802A1 (the acceptor parent) and 9606H (the donor parent) (Supplementary Fig. S1 available at JXB online). Among the progeny involved in this work, two phenotypic classes were distinguished: male-fertile plants with full and dehiscent anthers and male-sterile plants with empty yellow anthers. To ensure the accuracy of the genetic position, the phenotypes of male-fertile recombinant F2 plants were confirmed by phenotyping the F3 progeny in a glasshouse.
Microscopic observation
Floral buds at different developmental stages were fixed overnight in FAA [ethanol 50% (v/v), acetic acid 5.0% (v/v), and formaldehyde 3.7% (v/v)]. Fixed floral buds were dehydrated with a 50–100% ethanol series and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim/Ts., Germany) according to the manufacturer’s manual. Transverse sections (1.5 μm thickness) were cut from the polymerized blocks on an ultramicrotome (Leica Ultracut R; Leica Microsystems) using glass knives, heat fixed to glass slides, and stained with 2% toluidine blue. Semi-thin sections were stained with 0.05% analine blue to detect callose. Slides were inspected and photographed using an Olympus BX61 microscope equipped with a colour CCD camera.
Macroarray
The radish bacterial artificial chromosome (BAC) library consists of 120 000 clones and represents the haploid radish genome at least 23 times over (Desloire et al., 2003). The macroarrays are produced by spotting thousands of bacterial clones on the nylon membrane with a high-throughput machine (Q-Bot). The macroarray in pre-hybridization buffer (6× SSC, 5×Denhardt’s, 100mg ml–1 salmon sperm DNA) was pre-hybridized for at least 2h at 68 ºC in a hybridization oven. The probe was denatured for 10min at 100 ºC and immediately placed on ice. The pre-hybridization buffer was removed from the macroarrays. Then, the probe was added to hybridization buffer and hybridized overnight at 68 °C. Hybridized macroarrays were washed twice for 30min in wash buffer (2× SSC, 0.1% SDS) at 50 ºC. After drying, macroarrays were wrapped in cling film, avoiding air bubbles. The level of radioactivity from a macroarray was checked, followed by fixation in a cassette. After scanning the macroarray with PhosphorImager, the analysis of the macroarray was carried out using high density filter reader (HDFR) software.
DNA extraction for genotyping
About 5 mm×5mm from young seedlings was harvested into 96-well PCR plates with 50 μl of NaOH solution (0.25M) in each well, and plates were sealed. Plates were incubated at 96° C for 10min in the PCR block, cooled on a bench, and spun down. A 50 μl aliquot of 0.25M HCl and 25 μl of buffer (0.5M TRIS-HCl, pH 8.0; 0.25% NP-40) were added. After mixing by inverting and spinning down, plates were incubated at 96° C for 1min in a PCR block. A 0.6 μl aliquot of DNA was used for each 12.5 μl PCR.
RT–PCR
Total RNA was isolated from floral buds using TRIzol reagent (Invitrogen Life Technologies) and first-strand cDNA was synthesized using a SMART™ RACE cDNA amplification kit (Clontech) according to the manufacturer’s instructions. After reverse transcription, the products were diluted 10-fold with distilled water. The PCR mixture and conditions as described above were followed. The primers used for 5’-rapid amplification of cDNA ends (RACE) and 3’-RACE are listed in Supplementary Table 1 at JXB online.
Real-time RT–PCR
The expression patterns of the PPR transcripts were examined through strand-specific RT–PCR, in which 1.5 μg of total RNA was used for the first-strand cDNA synthesis with the SuperScript III reverse transcriptase (Invitrogen) using the mixture of gene-specific primers. The cDNA reaction mixture was then diluted 10 times and 4 μl was used as a template in a 10 μl PCR with SYBR Green Supermix. PCR included a pre-incubation at 95 ºC for 5min followed by 45 cycles of denaturation at 95 ºC for 15 s, annealing at 60 ºC for 20 s, and extension at 72 ºC for 30 s. The comparative threshold (Ct) cycle method was used for determination of relative transcript levels with Actin 2/7 as an internal control.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described by Wierzbicki et al. (2008) with minor modifications. A 2g aliquot of above-ground tissue of seedlings was cross-linked with 1% formaldehyde by vacuum infiltration at room temperature three times, 5min each time, followed by the addition of glycine to 125mM vacuum infiltrate for an additional 5min. Plants were rinsed with distilled water, frozen in liquid nitrogen, ground into powder, suspended in 35ml of Honda buffer [20mM HEPES, 0.44M sucrose, 1.25% ficoll, 2.5% dextran T40, 10mM MgCl2, 0.5% Triton X-100, 5mM dithiothreitol (DTT), 1% plant protease inhibitors], filtered through two layers of Miracloth twice, and centrifuged at 3100 g for 20min. Nuclear pellets were resuspended in 1ml of Honda buffer, centrifuged at 3100 g for 10min at 4° C, resuspended in Nuclei Lysis Buffer (50mM TRIS-HCl pH 8.0, 10mM EDTA, 1% SDS, 1% plant protease inhibitors), and sonicated three times, 5min each time (30 s on/off intervals) at the ‘Middle’ setting. After centrifugation at 16 000 g for 7.5min, the supernatant was diluted 11-fold with ChIP dilution buffer (1.1% Triton X-100, 1.2mM EDTA, 16.7mM TRIS-HCl pH 8.0, 167mM NaCl, protease inhibitor cocktail). Immunoprecipitation was performed using 20 μl of Dynabeads protein G (Invitrogen) and 5 μl of Pol II antibody (Abcam ab5408). After reversion of cross-linking, samples were incubated with 40 μg of proteinase K (Invitrogen) at 50 ºC for 1h, followed by heat inactivation at 95 ºC for 10min. The resulting DNA was subjected to quantitative PCR in triplicate.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers JX521806 (RsRf3-1 gene) and JX521807 (RsRf3-2 gene).
Results
Histological analysis of male-sterile and male-fertile flowers of radish
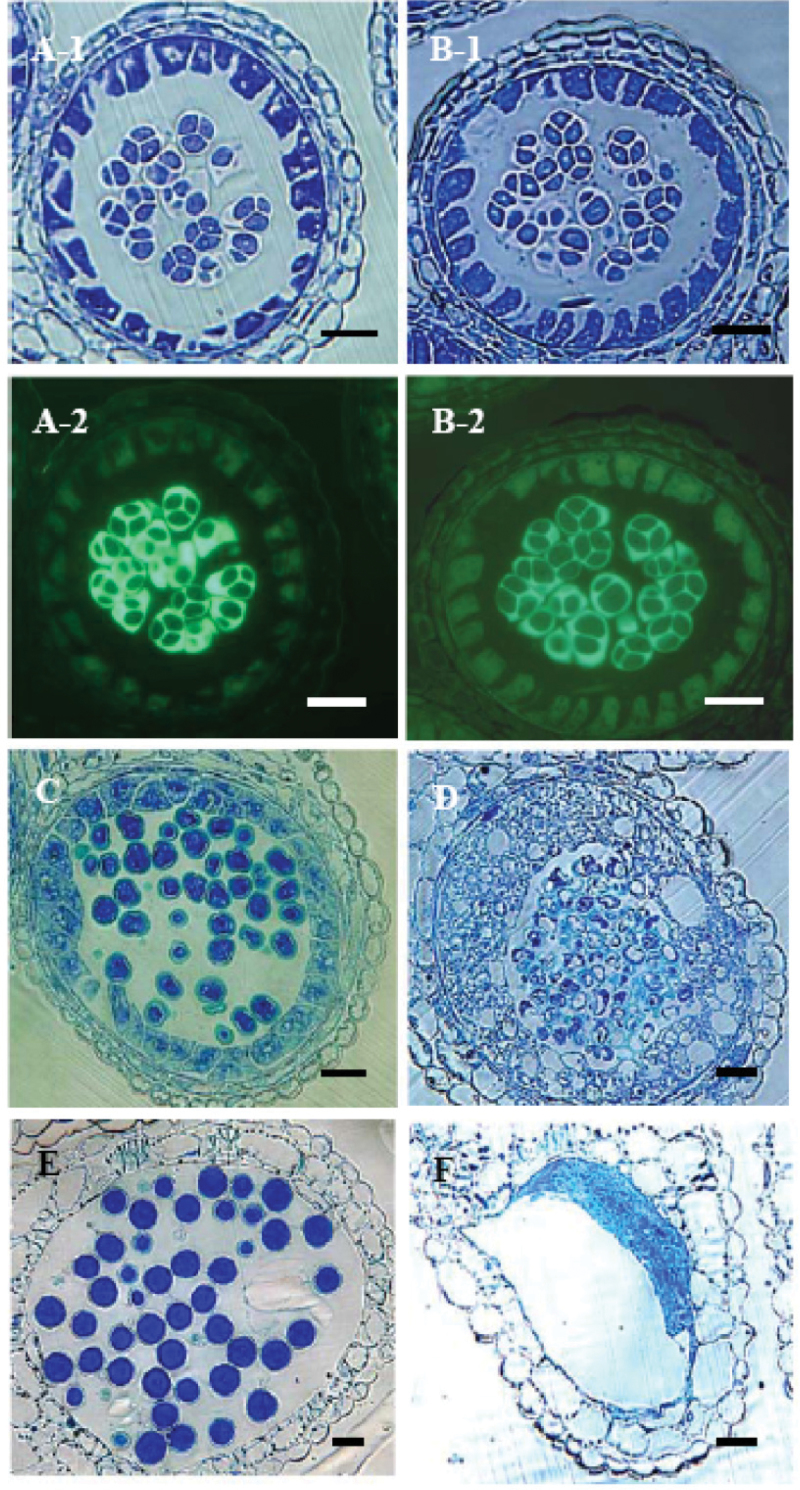
No cytological differences were observed between the fertile line 9802B1 and the male-sterile line 9802A1 in the anther tissues until the last stages of the tetrad (data not shown). At the tetrad stage, individual tetrads were completely encased by a thick callose wall in 9802B1 and 9802A1 (Fig. 1A, B). After the tetrad stage, the microspores from male-fertile plants increased in size and the microspore exine was quite evident (Fig. 1C). In contrast, although microspore exines were found in the male-sterile plants, microspores were malformed. At the same time, the tapetal cells grew abnormally large (Fig. 1D). At anthesis, the tapetal cells degenerated completely in both male-fertile and male-sterile anthers (Fig. 1E, F). Many fully mature pollen were observed in the locules of male-fertile anthers. In contrast, many remnants of microspore exine were found in the locules of male-sterile anthers (Fig. 1F).
Fig. 1.
Comparative study of anthers through different developmental stages in the male-fertile and male-sterile plants from 9802A1 and 9802B1. (A, C, E) Semi-thin sections from male-fertile anthers. (B, D, F) Semi-thin sections from male-sterile anthers. A-2 and B-2 were stained with aniline blue; the others were stained with toluidine blue. (This figure is available in colour at JXB online.)
The male fertility restorer is controlled by heterozygous alleles in an F2 segregating population
An F2 population was derived from a cross between 9802A1 and 9606H. In total, 600 plants were examined from the population, of which 296 male-fertile and 304 male-sterile plants were identified by investigation of male fertility. The segregation ratio of fertile to sterile plants in this F2 population best fitted a 1:1 ratio (P > 0.05), but not a 3:1 ratio (P < 0.05), indicating control by heterozygous alleles at a single locus (designated as RsRf3). In addition, the single locus control of male fertility was also strongly supported by a 1:1 BC1F1 ratio of male-fertile to male-sterile plants in the backcross population between 9802A1 (the acceptor parent) and 9606H (the donor parent) (Supplementary Fig. S1, Supplementary Table S2 at JXB online). The data also demonstrated that fertility of the cytoplasmic male-sterile plant was restored by heterozygous alleles in a sporophytic manner.
The RsRf3 locus was limited to a locus encoding PPR proteins
Most Rf genes cloned to date have PPR motifs (Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Imai et al., 2003; Koizuka et al., 2003; Komori et al., 2004; Wang et al., 2006). Rice Rf genes, Rf1a and Rf1b, share 70% identity between their protein sequences (Wang et al., 2006). It is possible that some radish Rf genes are descendents of a progenitor restorer PPR gene (Brown et al., 2003), and that these multiple genes arose in response to the appearance of new forms of CMS. These results offered compelling reasons for studies of possible genetic relationships between the PPR-coding genes and new Rf loci in radish. Consequently, a BAC radish library screening was performed by macroarray hybridization. This method allows fast access to homologues of the Rfo gene in the radish genome. Based on the radish Rfo gene (accession no. AJ535623), a pair of PCR primers (78F/79R) were designed (Supplementary Table S1 at JXB online) to amplify a PCR product of 469bp corresponding to the 1152–1620bp region of Rfob. One PCR product (~500bp) from the male-fertile line 0107H carrying Rfob (EU163282) was amplified using 78F/79R, purified, and used as a probe for a screening of the macroarray. Twenty-eight positive BAC clones were identified on macroarrays (Supplementary Fig. S2, Supplementary Table S3). Based on PCR by 78F/79R and sequencing, 18 clones corresponded to the Rfo locus. Primer pair 78F/79R did not amplify any product using the other 10 clones as templates, suggesting that these clones contain similar, but not identical, sequences to the Rfo gene. Then another 10 primer pairs were designed to amplify the corresponding Rfo amplicons from the 10 clones. The primer pair P1/P2 amplified ~300bp products in BAC clones 48H19 and 158K3. The sequencing result showed that the 296bp PCR product from 48H19 was identical to that from 158K3 and shared 96% identity with the corresponding region of Rfo. Pulsed field gel electrophoresis showed that 48H19 is ~145kb and 158K3 is ~130kb (Supplementary Fig. S3). Thus the BAC clone 48H19 was sequenced using 454 sequencing technology and 10 contigs were obtained. A contig, composed of 42 567bp, assembled by 8205 reads, contained two predicted PPR genes. One predicted PPR gene of 1440bp showed 90% identity with the Rfo gene. Based on previous genetic analysis, there should be polymorphism between the sterile line 9802A1 and the fertile line 9606H. Brief sequencing detected two polymorphic sites in predicted promoter and exon regions. Based on these polymorphic sites, two molecular markers were developed. Using the F2 population described above, linkage analysis showed that this PPR locus co-segregated with the RsRf3 locus (Supplementary Table S4). Based on the above contig sequence, a series of primers were designed to amplify the corresponding regions in 9802A1 and 9606H. The alignment of the corresponding sequences between 9802A1 and 9606H revealed a few more polymorphic sites. Additional molecular markers were developed and RsRf3 was resolved to a 4kb region containing only one gene using a large F2 population with 20 000 plants (Fig. 2). Mapping analysis using the BC1F1 population with 18 000 plants yielded the same result (data not shown).
Fig. 2.
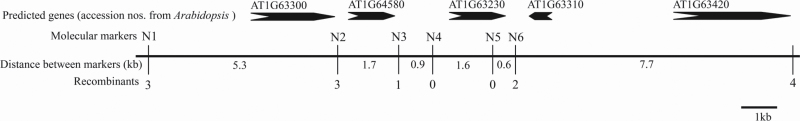
Genetic and physical maps of RsRf3. RsRf3 is mapped to the region between marker N3 and N6, which contains only one gene with homology to At1g63230.
Characterization of the RsRf3 locus
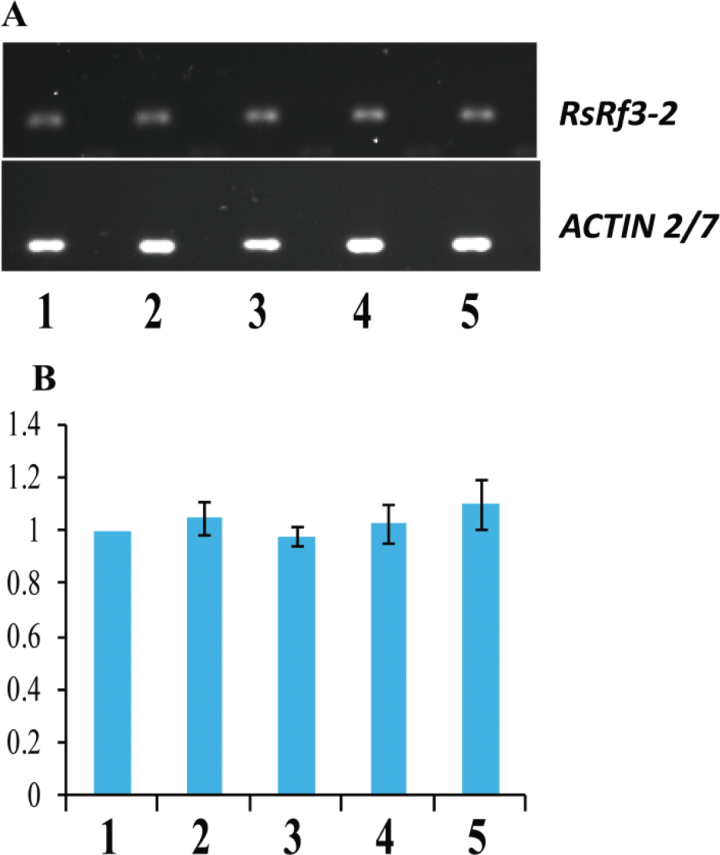
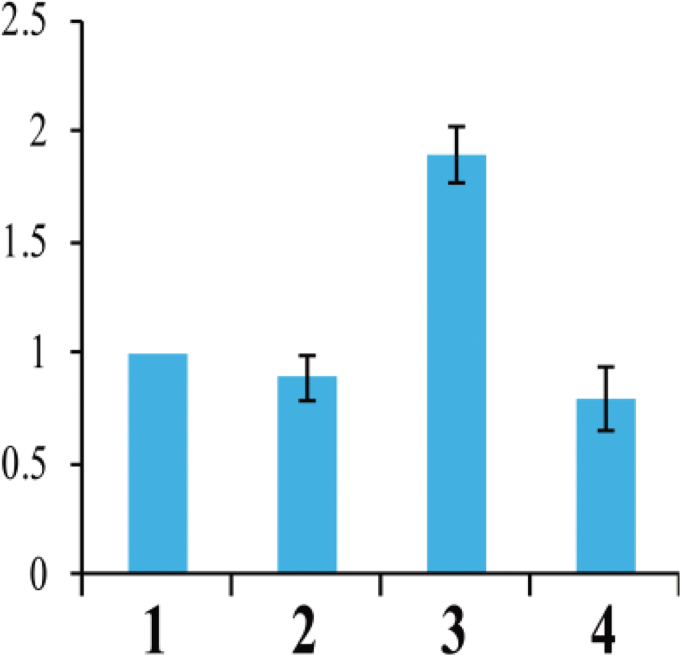
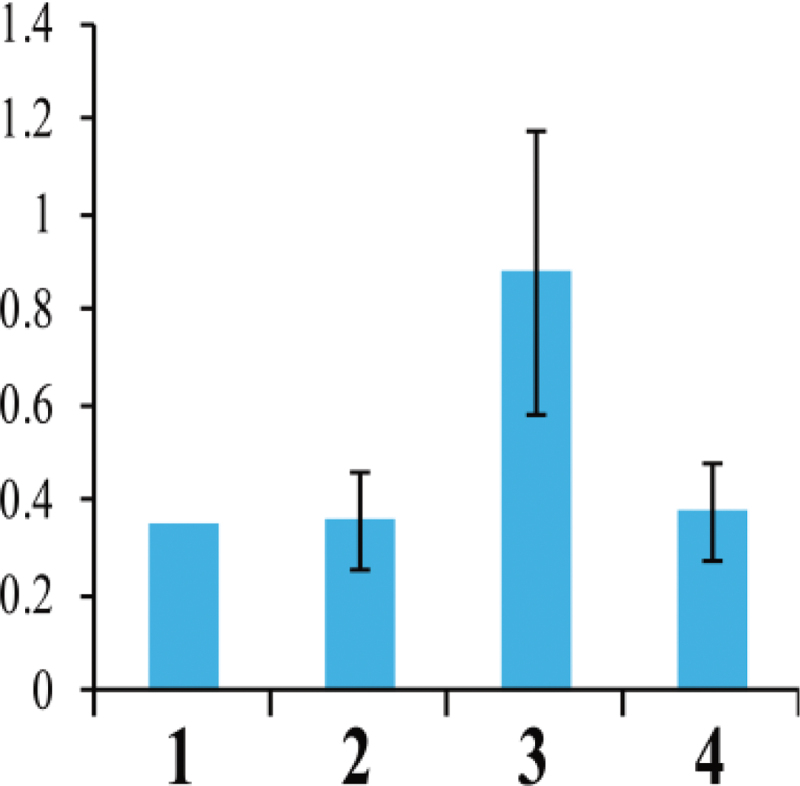
The 5’ and 3’ RACE experiments showed that the RsRf3 locus has an intron and encodes a PPR protein with 479 amino acids comprising 11 copies of the 35 amino acid PPR motif. The RsRf3 protein shows 85% similarity with the Rfo protein with 16 PPR domains (Fig. 3). Compared with the RsRf3-1 allele in 9802A1, the RsRf3-2 allele in 9606H contained one base substitution (C to T) and a CTT insertion at the promoter region. In the coding region, a single nucleotide polymorphism (SNP) results in a single amino acid replacement of arginine (AGG) in 9802A1 by lysine (AAG) in 9606H. Using a strand-specific RT–PCR method, the expression profile of RsRf3-2 in 9606H was assayed using RNA from five tissues. RsRf3-2 was detected in all of the tissues at the same expression level (Fig. 4). Interestingly, higher RsRf3 expression and RNA polymerase II (Pol II) enrichment were detected at the RsRf3 chromatin in F1 seedlings compared with its parents (Figs 5, 6). Furthermore, the increase in expression and Pol II enrichment was found only in plants carrying the cytoplasm from 9802A1 (Figs 5, 6).
Fig. 3.
Amino acid sequence alignment of the Rfo gene and the RsRf3-2 allele in 9606H.
Fig. 4.
Expression of the RsRf3-2 allele in different organs of the radish restorer line 9606H with specific primers F2/R5. (A) Expression profile of transcripts in different tissues. (B) Comparative expression analysis of the transcripts in different tissues. 1, 2, 3, 4, and 5: products with the first-strand cDNA of root, stem, leaf, flower, and young pod as template, respectively.
Fig. 5.
Quantitative RT–PCR analysis of Rf3 expression levels in the CMS/Rf system. The data are shown as mean ±SD (n=3). Columns 1, 2, 3, and 4 indicate the patterns of 9802A1, 9606H, F1 from 9802A1 (female parent) and 9606H, and F1 from 9802B1 (female parent) and 9606H, respectively.
Fig. 6.
ChIP analysis of Pol II on the RsRf3 locus. Columns 1, 2, 3, and 4 indicate the patterns of 9802A1, 9606H, F1 from 9802A1 (female parent) and 9606H, and F1 between from 9802B1 (female parent) and 9606H, respectively.
Discussion
In radish, several types of CMS/Rf systems have been reported. Ogura CMS is found to be controlled by orf138 (Bonhomme et al., 1992; Krishnasamy and Makaroff, 1993), and the Kosena CMS-associated gene orf125 shares extensive similarity with orf138 (Iwabuchi et al., 1999). Both Kosena and Ogura CMS are reversed by the same gene orf687 (Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003). The male-sterile line 9802A1 contains the full-length orf138 gene; the male-sterile line 9802A1 and its male fertility-restoring line 9606H contain the rfob gene that encodes the same predicted protein as the non-restoring allele rf (AJ535624) (Wang et al., 2008), suggesting that the CMS/Rf system used here (9802A1/9606H) is different from the Ogura/Kosena system. A recent report shows that the Ogura-type mitochondrial genome has been highly rearranged compared with the normal type genome and the rearrangement has resulted in four unique regions containing six open reading frames, including orf138 specific to the Ogura-type mitochondrial genome (Tanaka et al., 2012). After the tetrad stage, microspores of Ogura CMS began to degenerate, resulting in empty anthers with no pollen (Lee et al., 2008). In 9802A1, many remnants of microspore exine were found in anthers. Both 9802A1 and the fertility-restored hybrid (9802A1×9606H) show comparable levels of orf138 transcripts (Supplementary Fig. S4 at JXB online). The fact that the F1 plants of the cross between 9802A1 and 0107H, carrying Rfo and RsRf3-1, were male fertile indicates that male sterility in 9802A1 is due to orf138 but the restoration is achieved by another set of alleles (RsRf3). The male-sterile phenotype in 9802A1 is also different from other identified cytoplasmic male-sterile radish plants: a small number of pollen grains in the dehiscing anthers are visible in the NWB and DCGMS cytoplasmic male-sterile radish plants (Nahm et al., 2005; Lee et al., 2008). A new cytoplasmic male-sterile radish 805A has been identified that possesses two different abortion types in the locules during anther development (Shi et al., 2010).
To date, all the PPR-encoding Rf genes function dominantly against their non-functional rf allele. In this study, positional cloning of RsRf3 was performed and it was found that heterozygous PPR alleles are required for fertility restoration, suggesting that the rf allele in the CMS line is functional. How can heterozygous PPR alleles restore male fertility for CMS, but the homozygous allele cannot? Two possibilities are suggested to account for this observation. First, the two PPR proteins encoded by the different alleles at the same locus could show different functions, or a combination of their functions, probably forming a heterodimer, is sufficient to prevent the accumulation of CMS-associated gene products, which is supported by reports that PPR proteins might work as dimers (Nakamura et al., 2003; Okuda and Shikanai, 2012). The second hypothesis is based on the fact that the present work showed that the expression of homozygous alleles is lower than that of the heterozygous alleles. So, the elevated expression of heterozygous PPR alleles may be the reason for reversing the male-sterile phenotype. Both explanations warrant further investigation.
Alignment of the BAC clone Bac64 (127kb) containing the Rfo locus (rep) and the contig (42kb) carrying the RsRf3 locus from BAC clone 48H19 demonstrated that the Rfo and RsRf3 loci are located in different regions. A BLAST search against the Brassica rapa genome using the two clones indicated that the Rfo BAC clone and the RsRf3 contig correspond to 7059–7139kb and 7205–7256kb on chromosome 9 from B. rapa, respectively, suggesting tight linkage between the Rfo and RsRf3 loci (Supplementary Fig. S5 at JXB online).
The heterozygous RsRf3-1/RsRf3-2 alleles conditioning gain of function is a rare case which supports the ‘overdominance’ hypothesis of heterosis proposing that loci exhibit a heterozygote advantage (Crow, 1948). Recent research found that cis polymorphisms at the FLC locus and the LDMAR locus influence the accumulation of H3K27me3 and DNA methylation, respectively (Coustham et al., 2012; Ding et al., 2012). In this work, two polymorphic sites were detected between 9802A1 and 9606H, and higher expression and Pol II enrichment were found in F1 plants with the CMS background, suggesting that retrograde regulation, DNA or histone methylations could be involved in transcriptional regulation of RsRf3.
Supplementary data
Figure S1. Crossing scheme for the backcross.
Figure S2. Result of the macroarray hybridization. Positive BAC clones identified on the macroarray.
Figure S3. Pulsed field gel electrophoresis for BAC clones 48H19 (20) and 158K3 (22).
Figure S4. Expression of orf138 in 9802A1 (1) and F1 (2) from a cross between 9802A1 and 9606H.
Figure S5. BLAST of the RsRf3 contig (A) and the Rfo BAC clone (B) against the Brassica rapa genome.
Table S1. Primers used in this study.
Table S2. Backcross population of BC1F1.
Table S3. Positive BAC clones identified on macroarrays.
Table S4. F2 population crossed from 9802A1 and 9606H.
Acknowledgements
We thank members of Professor Caroline Dean’s lab at the John Innes Centre in Norwich, UK, for helpful suggestions and the anonymous reviewers for their valuable comments. This work was supported by the National Natural Science Foundation of China (31100231), the Natural Science Foundation of Hubei Province, China (2007ABA004, 2009CDB221, and 2011CDB408), the Youth Chenguang Project of Science and Technology of Wuhan, China (200850731402), and the fund of the CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture (Y152081t01).
References
- Bellaoui M, Grelon M, Pelletier G, Budar F. 1999. The restorer Rfo gene acts post-translationally on the stability of the ORF138 Ogura CMS-associated protein in reproductive tissues of rapeseed cybrids. Plant Molecular Biology 40, 893–902. [DOI] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR. 2002. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proceedings of the National Academy of Sciences, USA 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme S, Budar F, Lancelin D, Small I, Defrance MC, Pelletier G. 1992. Sequence and transcript analysis of the Nco2.5 Ogura-specific fragment correlated with cytoplasmic male sterility in Brassica cybrids. Molecular and General Genetics 235, 340–348. [DOI] [PubMed] [Google Scholar]
- Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS. 2003. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. The Plant Journal 35, 262–272. [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587. [DOI] [PubMed] [Google Scholar]
- Crow JF. 1948. Alternative hypotheses of hybrid vigor. Genetics 33, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wise RP, Schnable PS. 1996. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272, 1334–1336. [DOI] [PubMed] [Google Scholar]
- Desloire S, Gherbi H, Laloui W, et al. 2003. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Reports 4, 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey RE, Timothy DH, Levings CS. 1987. A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proceedings of the National Academy of Sciences, USA 84, 5374–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. 2012. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proceedings of the National Academy of Sciences, USA 109, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Bond CS, Small ID. 2011. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proceedings of the National Academy of Sciences, USA 108, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Toriyama K. 2008. Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility. Plant and Cell Physiology 49, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Toriyama K. 2009. Suppressed expression of RETROGRADE-REGULATED MALE STERILITY restores pollen fertility in cytoplasmic male sterile rice plants. Proceedings of the National Academy of Sciences, USA 106, 9513–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman JD, Bentolila S, Hanson MR. 2007. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. The Plant Journal 49, 217–227. [DOI] [PubMed] [Google Scholar]
- Hanson MR, Bentolila S. 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. The Plant Cell 16, S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang K, Huang WC, et al. 2012. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. The Plant Cell 24, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai R, Koizuka N, Fujimoto H, Hayakawa T, Sakai T, Imamura J. 2003. Delimitation of the fertility restorer locus Rfk1 to a 43-kb contig in Kosena radish (Raphanus sativus L.). Molecular Genetics and Genomics 269, 388–394. [DOI] [PubMed] [Google Scholar]
- Itabashi E, Iwata N, Fujii S, Kazama T, Toriyama K. 2011. The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. The Plant Journal 65, 359–367. [DOI] [PubMed] [Google Scholar]
- Ivanov MK, Dymshits GM. 2007. Cytoplasmic male sterility and restoration of pollen fertility in higher plants. Russian Journal of Genetics 43, 354–368. [PubMed] [Google Scholar]
- Iwabuchi M, Koizuka N, Fujimoto H, Sakai T, Imamura J. 1999. Identification and expression of the kosena radish (Raphanus sativus cv. Kosena) homologue of the ogura radish CMS-associated gene, orf138 . Plant Molecular Biology 39, 183–188. [DOI] [PubMed] [Google Scholar]
- Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. 2008. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. The Plant Journal 55, 619–628. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein P, Mullet J, Minx P, Rooney W, Schertz K. 2005. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theoretical and Applied Genetics 111, 994–1012. [DOI] [PubMed] [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J. 2003. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. The Plant Journal 34, 407–415. [DOI] [PubMed] [Google Scholar]
- Komori T, Ohta S, Murai N, Takakura Y, Kuraya Y, Suzuki S, Hiei Y, Imaseki H, Nitta N. 2004. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). The Plant Journal 37, 315–325. [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Makaroff CA. 1993. Characterization of the radish mitochondrial Orfb locus—possible relationship with male-sterility in Ogura radish. Current Genetics 24, 156–163. [DOI] [PubMed] [Google Scholar]
- Lee YP, Park S, Lim C, Kim H, Lim H, Ahn Y, Sung SK, Yoon MK, Kim S. 2008. Discovery of a novel cytoplasmic male-sterility and its restorer lines in radish (Raphanus sativus L.). Theoretical and Applied Genetics 117, 905–913. [DOI] [PubMed] [Google Scholar]
- Liu F, Cui XQ, Horner HT, Weiner H, Schnable PS. 2001. Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. The Plant Cell 13, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Schnable PS. 2002. Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiology 130, 1657–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm SH, Lee HJ, Lee SW, Joo GY, Harn CH, Yang SG, Min BW. 2005. Development of a molecular marker specific to a novel CMS line in radish (Raphanus sativus L.). Theoretical and Applied Genetics 111, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Meierhoff K, Westhoff P, Schuster G. 2003. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. European Journal of Biochemistry 270, 4070–4081. [DOI] [PubMed] [Google Scholar]
- Okuda K, Shikanai T. 2012. A pentatricopeptide repeat protein acts as a site-specificity factor at multiple RNA editing sites with unrelated cis-acting elements in plastids. Nucleic Acids Research 40, 5052–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Wise RP. 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends in Plant Science 3, 175–180. [Google Scholar]
- Shi S, Ding D, Mei S, Wang J. 2010. A comparative light and electron microscopic analysis of microspore and tapetum development in fertile and cytoplasmic male sterile radish. Protoplasma 241, 37–49. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tsuda M, Yasumoto K, Yamagishi H, Terachi T. 2012. A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics 13, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttewaal M, Arnal N, Quadrado M, Martin-Canadell A, Vrielynck N, Hiard S, Gherbi H, Bendahmane A, Budar F, Mireau H. 2008. Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. The Plant Cell 20, 3331–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zou Y, Li X, et al. 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. The Plant Cell 18, 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Zhang YJ, Xiang CP, Mei SY, Zhou Y, Chen GP, Wang T. 2008. A new fertility restorer locus linked closely to the Rfo locus for cytoplasmic male sterility in radish. Theoretical and Applied Genetics 117, 313–320. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. 2008. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Guo W, Yang L, Liu L, Zhang T. 2006. Physical mapping of the Rf1 fertility-restoring gene to a 100kb region in cotton. Theoretical and Applied Genetics 112, 1318–1325. [DOI] [PubMed] [Google Scholar]