Abstract
In yeasts and animals, premature entry into mitosis is prevented by the inhibitory phosphorylation of cyclin-dependent kinase (CDK) by WEE1 kinase, and, at mitosis, WEE1 protein is removed through the action of the 26S proteasome. Although in higher plants WEE1 function has been confirmed in the DNA replication checkpoint, Arabidopsis wee1 insertion mutants grow normally, and a role for the protein in the G2/M transition during an unperturbed plant cell cycle is yet to be confirmed. Here data are presented showing that the inhibitory effect of WEE1 on CDK activity in tobacco BY-2 cell cultures is cell cycle regulated independently of the DNA replication checkpoint: it is high during S-phase but drops as cells traverse G2 and enter mitosis. To investigate this mechanism further, a yeast two-hybrid screen was undertaken to identify proteins interacting with Arabidopsis WEE1. Three F-box proteins and a subunit of the proteasome complex were identified, and bimolecular fluorescence complementation confirmed an interaction between AtWEE1 and the F-box protein SKP1 INTERACTING PARTNER 1 (SKIP1). Furthermore, the AtWEE1–green fluorescent protein (GFP) signal in Arabidopsis primary roots treated with the proteasome inhibitor MG132 was significantly increased compared with mock-treated controls. Expression of AtWEE1–YFPC (C-terminal portion of yellow fluorescent protein) or AtWEE1 per se in tobacco BY-2 cells resulted in a premature increase in the mitotic index compared with controls, whereas co-expression of AtSKIP1–YFPN negated this effect. These data support a role for WEE1 in a normal plant cell cycle and its removal at mitosis via the 26S proteasome.
Key words: Arabidopsis thaliana, bimolecular fluorescence complementation (BiFC), BY-2 cell line, CDKA/B, cell cycle, F-box, green fluorescent protein (GFP), mitosis, Nicotiana tabacum, 26S proteasome SKIP1, WEE1.
Introduction
The cell cycle is conserved in all eukaryotes, with G1/S and G2/M being regulated by cyclin-dependent kinases (CDKs). In plants, >160 CDK-related genes have been cloned from >20 higher plant species and are apportioned into classes A–G and CDK-like (Dudits et al., 2007). The CDKB family is unique to plants. In Arabidopsis, CDKA;1 activity peaks at G1/S and G2/M, whereas CDKB2;1 peaks at G2/M (Joubes et al., 2000). In tobacco BY-2 cells, CDKA activity is relatively constant from S-phase to mitosis, whereas B-type activity peaks in mid-G2 (Porceddu et al., 2001; Sorrell et al., 2001).
In Schizosaccahromyces pombe, the CDK, Cdc2, is phosphoregulated in G2, negatively by SpWee1 kinase and positively by SpCdc25 phosphatase (Nurse, 1990). A partial WEE1 homologue was cloned in maize and inhibits CDK activity in vitro (Sun et al., 1999) whilst a full-length Arabidopsis WEE1 is highly expressed in meristems (Sorrell et al., 2002). T-DNA insertional lines are hypersensitive to DNA-damaging agents, demonstrating that AtWEE1 participates in the DNA damage and replication checkpoints (De Schutter et al., 2007). However, the role of WEE1 in a normal cell cycle remains uncertain since T-DNA insertional lines grow and develop normally. This is in contrast to Wee1–/– mice that die during embryogenesis (Tominaga et al., 2006). However, a systematic analysis of cell cycle gene expression during Arabidopsis development shows tight regulation of WEE1 expression during the cell cycle in plants, as indicated by patchy expression patterns in parts of the young and mature leaves, shoot apical meristem, and young roots (de Almeida Engler et al., 2009). Furthermore, there may be a role for WEE1 during endoreduplication given that WEE1 transcript levels were high during this process both in the endosperm of Zea mays (Sun et al., 1999) and in tomato fruit (Gonzalez et al., 2004, 2007). The latter authors concluded that tomato WEE1 negatively regulates CDKA activity to control cell size acting through a regulation of cell expansion and/or endoreduplication. However, in these systems, neither WEE1 protein nor the ability of WEE1 from plant protein extracts to inhibit CDK activity was measured and linked to the cell cycle phase.
Timely degradation of regulatory proteins has a critical effect on cell cycle progression. The most widely studied method of proteolysis in eukaryotes is the ubiquitin–26S proteasome pathway. Its mechanism is widely conserved among eukaryotes, including plants (reviewed by Sullivan et al., 2003). One of the components of the ubiquitin cascade is E3 ligase, which provides the specificity to the target protein. Several types of E3 ligase exist in eukaryotes. The SCF E3 ligase complex consists of four subunits. S PHASE KINASE-ASSOCIATED PROTEIN 1 (SKP1) and CULLIN provide the structural backbone, while RBX is a ring finger protein which binds the E2, or ubiquitin-conjugating, enzyme. Finally an F-box protein binds to SKP1, providing specificity to the target protein (Skowyra et al., 1997; Deshaies et al., 1999; Zheng et al., 2002). In Arabidopsis, 692 F-box genes have been identified through homology searches (Xu et al., 2009), although only a relatively small number of these proteins have been studied functionally.
In budding yeast and mammalian cells, two E3 ligases, SCF and APC, are important components of the degradatory pathways that remove unwanted cell cycle proteins such as WEE1 kinase and cyclins. Their timely removal is followed by normal G2/M and metaphase/anaphase progression (King et al., 1995; Feldman et al., 1997; Skowyra et al., 1997). In Xenopus laevis, the degradation of WEE1 via the 26S proteasome is required for the correct timing of mitosis, and was blocked by inhibition of DNA replication, clearly indicating a link between the completion of S-phase and the progression of mitosis (Michael and Newport, 1998). The Saccharomyces cerevisiae WEE1 homologue, SWE1, is targeted for degradation by a SUMO (small-ubiquitin modifier protein, similar to ubiquitin) protein, SMT3, via the E3 ligase SIZ1 (Simpson-Lavy and Brandeis, 2010). F-box proteins including MET30 are implicated in WEE1 degradation, in S. cerevisiae (Kaiser et al., 1998), as is TOME-1 in Xenopus (Ayad et al., 2003). The most comprehensive study of WEE1 kinase activity during the cell cycle was carried out for HeLa cells, where WEE1 kinase activity was detected during interphase but not in mitosis (McGowan and Russell, 1995). In these assays, native WEE1 was pulled-down using human WEE1 antibody and then used to inhibit CDK activity in histone H1 kinase assays (McGowan and Russell, 1995).
The aim of this work was to test the hypothesis that plant WEE1 action is under cell cycle control, and investigate the mechanism by which CDKs are released from WEE1 inhibition as cells enter mitosis. Data presented here show, for the first time in plant cells, cell cycle regulation of WEE1 at the protein level and a drop in WEE1 inhibition of CDK activity during G2 that remained low as cells entered mitosis. Data presented here indicate that WEE1 protein degradation in plants is 26S proteasome dependent and targeted via the protein’s physical interaction with the 26S proteasome F-box protein, SKIP1.
Materials and methods
Cell cycle measurements
Tobacco (Nicotiana tabacum) BY-2 cells were subcultured every 7 d and synchronized as described previously (Francis et al., 1995). At hourly intervals following removal of aphidicolin, the mitotic index was derived from scoring ≥300 Hoechst-stained cells per slide in random transects using fluorescence microscopy (Olympus BH2, UV, λ=420nm).
Cloning Nicta;WEE1
Degenerate primers were designed based on the maize ZmWEE1 (accession no. AAD52983) and AtWEE1 (accession number: CAD28679) (Supplementary Table S2 available at JXB online) and used to amplify a 339bp fragment of NtWEE1 from N. tabacum var. Samsun genomic DNA. The PCR product was cloned in pGEM T-Easy (Promega, Southampton, UK) and sequenced. One cycle of 3’ rapid amplification of cDNA ends (RACE) and two cycles of 5’ RACE (using the BD SMART™ RACE cDNA amplification Kit, Clontech) furbished the whole open reading frame (ORF) (EMBL database accession nos: AJ866274, AJ866275, AJ866276, and AJ866277). The entire ORF was amplified (primers are given in Supplementary Table S2) from BY-2 cDNA and cloned into pTA7002 by digestion with XhoI/SpeI, creating pTA7002 NtWEE1. The ORF was fully sequenced (EMBL database accession no. AM408785). Clustal W within DNAstar (Lasergene), BIOEDIT version 7.0.1 (Hall, 1999), and MEGA software version 3.1 (Tamura et al., 2007) were used to compare the tobacco ORF with other wee1 sequences. pTA7002 NtWEE1 was transformed into Agrobacterium tumefaciens LBA4404 and GV3101 and used to transform BY-2 cells and Arabidopsis var. Columbia, respectively, as described previously (An, 1985; Clough and Bent, 1998; Orchard et al., 2005).
Semi-quantitative RT–PCR
Total RNA was extracted from BY-2 cells using TRI reagent (Sigma Aldrich, Gillingham, UK) and residual genomic DNA removed by DNase treatment (Ambion, Austin, TX, USA). RNA (5 µg) was reacted with Superscript II reverse transcriptase (GIBCO, Paisley, UK) and used for semi-quantitative reverse transcription–PCR (RT–PCR) of NtWEE1 expression in synchronized cells (primers are listed in Supplementary Table S2 at JXB online).
Histone H4 primers (Supplementary Table S2) were used to verify cell cycle stage, and 18S rRNA primers for normalization (Orchard et al., 2005). For all semi-quantitative RT–PCRs, the cycle number was optimized such that the amount of product was proportional to the amount of input total RNA, verified using a dilution series of cDNAs in each PCR. Products of three replicate PCRs were quantified using ethidium bromide-stained agarose gels and GeneGenius software (Syngene, Cambridge, UK).
Protein extractions from BY-2 cells and western blotting
Protein extraction from N. tabacum BY-2 cell cultures was essentially performed as described in Cockcroft et al. (2000). Protein extract concentrations were determined using a Bradford assay (Bradford, 1976; Bradford Reagent, Sigma, Dorset, UK) to ensure equal loading on SDS–gels and western blots (20 µg). Loading was also verified using replicate Coomassie brilliant blue-stained gels. The WEE1 antibody used was as described in Lentz Grønlund et al. (2009). Western blotting was as described in Lentz Grønlund et al. (2009) using a WEE1 antibody dilution of 1:1000 followed by α-rabbit IgG (1:2500) (Sigma, Dorset, UK). Proteins were visualized by western blotting using ECL reagents (Amersham Biosciences, Amersham, UK) and quantified using an internal control to normalize across different gels and GeneGenius software (Syngene, Cambridge, UK). Quantified data presented are the means of three independent western blots for protein levels and two gels for the kinase assays (±SE).
Recombinant protein expression and purification
The coding sequences of NtWEE1 and At14-3-3ω were PCR amplified (primers are listed in Supplementary Table S2 at JXB online) using Pfu polymerase and cloned into the pET15B vector system using NdeI/BamHI. The insertions were verified by sequencing and the plasmids were transformed into Escherichia coli DE3 Rosetta pLysS cells. Recombinant protein was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) and the purity of the recombinant proteins was analysed by SDS–PAGE.
Immunoprecipitation and kinase assay
The CDK substrate for the kinase assays was pulled down from N. tabacum BY-2 cells using a p13SUC1 agarose conjugate (Upstate) from 100–250 µg of protein extract. WEE1 protein was immunoprecipitated from 100 µg of protein extracts from N. tabacum BY-2 cells at different times following synchronization using WEE1 antibody raised as described in Lentz Grønlund et al. (2009). The histone H1 assay was essentially as described in Cockroft et al. (2000) using 5 µl of NtWEE1 antibody. Samples were subjected to SDS–PAGE. Products were quantitated from autoradiographs using GeneGenius software (Syngene). Results are expressed as the percentage inhibition of CDK activity normalized to the control CDK activity without addition of recombinant WEE1. Results are based on two independent replicates.
Two-hybrid analysis
The bait plasmid pBD-Gal4-cam AtWEE1 was constructed as described in Lentz Grønlund et al. (2009). An Arabidopsis seedling root primary cDNA library was constructed in the HybriZAP-2.1 lambda vector (Stratagene) (Sorrell et al., 2003). The primary library was amplified and converted by in vivo excision into a GAL4 transcriptional activation domain pAD-GAL4-2.1 library according to the manufacturer’s protocol (Stratagene). Both bait and cDNA library were transformed into S. cerevisiae strain YRG-2 [MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu 2–3, 112 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4 17mers(x3)-TATACYC1-lacZ] (Stratagene). Approximately 1–2×106 transformants were plated onto His– synthetic dextrose minimal medium and screened as described in Sorrell et al. (2003) using both the HIS3 and LacZ reporter genes. Interacting proteins were identified by colony PCR and sequenced. Sequences were identified using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
Bimolecular fluorescence complementation (BiFC)
The AtWEE1 ORF was amplified and cloned into the BiFC vector containing the C-terminal portion of yellow fluorescent protein (YFP), pkanII-SPYCE(M) (Waadt et al., 2008) as described in Lentz Grønlund et al. (2009). The AtSKIP1 ORF was PCR amplified (primers are listed in Supplementary Table S2 at JXB online) and cloned using the AscI/XmaI sites into the BiFC vector fusing the SKIP1 ORF in-frame with the N-terminal portion of YFP, pSPYNE (Walter et al., 2004). The constructs were transformed into A. tumefaciens strain EHA105 and used to co-transform transiently (as described in Lentz Grønlund et al., 2009) and transform stably BY-2 cells as described previously (Orchard et al., 2005). Cells were monitored for fluorescence using fluorescence microscopy (Olympus BH2, UV, λ=420nm).
AtWEE1 and AtWEE1–GFP transgenic BY-2 and Arabidopsis lines
AtWEE1 under the 35S promoter in the BIN HYG TX vector was assembled as described in Spadafora et al. (2012). A WEE1–green fluorescent protein (GFP) fusion protein construct was created by amplifying the AtWEE1 ORF (using the primers listed in Supplementary Table S2 at JXB online) and cloned into the Gateway (Invitrogen) vector system to create an entry clone in pDonr207. The insert was then transferred to pGFP-N-Bin (Invitrogen) to create an N-terminal fusion. The constructs were transformed into A. tumefaciens strain LBA4404 for transformation into BY-2 cells and A. tumefaciens GV3101 for transformation into Arabidopsis var. Columbia as described above. The presence of the transgene was checked by PCR (for primers, see Table S2; data not shown).
The 35S::GFP BY-2-transformed line was kindly donated by Dr Lukás Fischer (Nocarova and Fischer, 2009).
Both cell lines were synchronized with aphidicolin (Orchard et al., 2005) and samples were stained with Hoechst. GFP and Hoechst were visualized using an Olympus BX61 microscope at λ=380nm or 530nm. Where GFP signal was absent following Hoechst staining (35S::GFP line), further images were taken using differential interference contrast (DIC) microscopy.
Arabidopsis lines expressing WEE1–GFP were selected based on GFP fluorescence visualized as above and crossed with a line expressing AtSUN1–mRFP (monomeric red fluorescent protein) (Graumann et al., 2010). Root tips of 5- to 7-day-old seedlings were imaged using an oil immersion ×40 lens on a Zeiss LSM510 confocal microscope. GFP was excited with a 488nm argon laser and fluorescence was captured with a 505–530nm bandpass filter; mRFP was excited with a 543nm helium–neon laser and fluorescence was captured with a 560–615nm bandpass filter. Images were captured with the Zeiss LSM software and exported in TIF format.
Propidium iodide staining and confocal imaging of Arabidopsis seedlings
AtWEE1–GFP localization was observed using a Leica TCS SP2 AOBS spectral confocal microscope employing 5-day-old seedlings grown on MS medium (Murashige and Skoog, 1962). Cell walls were counterstained using propidium iodide. GFP fluorescence was excited using a 488nm argon ion laser line and detected between 500nm and 550nm. Propidium iodide fluorescence was excited using a 543nm helium–neon ion laser line and detected between 600nm and 650nm. Images were captured using Leica confocal software. The H2B-YFP Arabidopsis line was kindly donated by Professor J. Murray, School of Biosciences, Cardiff University, Wales, UK.
MG132 treatment of Arabidopsis seedlings
Five-day-old seedlings grown as above were carefully removed from the surface of the agar and incubated with MG132 [stock solution 25mg ml–1 in dimethylsulphoxide (DMSO) diluted to 50 µM in liquid MS medium or, for the mock treatment, an equal volume of DMSO instead of MG132 stock] and incubated with occasional gentle agitation in the light for 6h at 21 °C. Seedlings were then transferred to fresh liquid MS medium and used for confocal imaging as above. Fluorescence was quantified using ImageJ software.
Results
Recombinant N. tabacum WEE1 inhibits CDK activity in vitro
The WEE1 antibody recognized a single 56kDa band in proteins extracted in lag (day 1), exponential (day 3), and stationary phase (day 5) of a 7 day subculture of BY-2 cells, indicating good antibody specificity as shown previously with Arabidopsis (Lentz Grønlund, 2009) (Supplementary Fig. S1 at JXB online).
The CDK inhibitory activity of NtWEE1 was investigated using a WEE1 kinase inhibition assay based on that used with recombinant Z. mays and Solanum lycopersicum WEE1 proteins (Sun et al., 1999; Gonzalez et al., 2007). This assay tests whether recombinant NtWEE1 can inhibit CDK phosphorylation of histone H1 in vitro. Total CDK activity in this assay is interpreted as the inverse of WEE1 activity, as previously demonstrated for Homo sapiens Wee1 by McGowan and Russell (1995). Induction of NtWEE1 in E. coli was tested by western blotting, showing good specificity of the NtWEE1 antibody; purification of the recombinant WEE1 protein using His-beads resulted in a single band of the expected 56kDa on a Coomassie-stained gel (Supplementary Fig. S2 at JXB online).
Addition of recombinant NtWEE1 resulted in a 5-fold decrease in CDK activity (Fig. 1A) compared with CDK alone. This shows that recombinant NtWEE1 protein produced in E. coli can negatively regulate CDK activity in vitro as do other plant WEE1 kinases. This is taken to indicate that the WEE1 is inhibiting CDK activity through its kinase activity (see Sun et al., 1999).
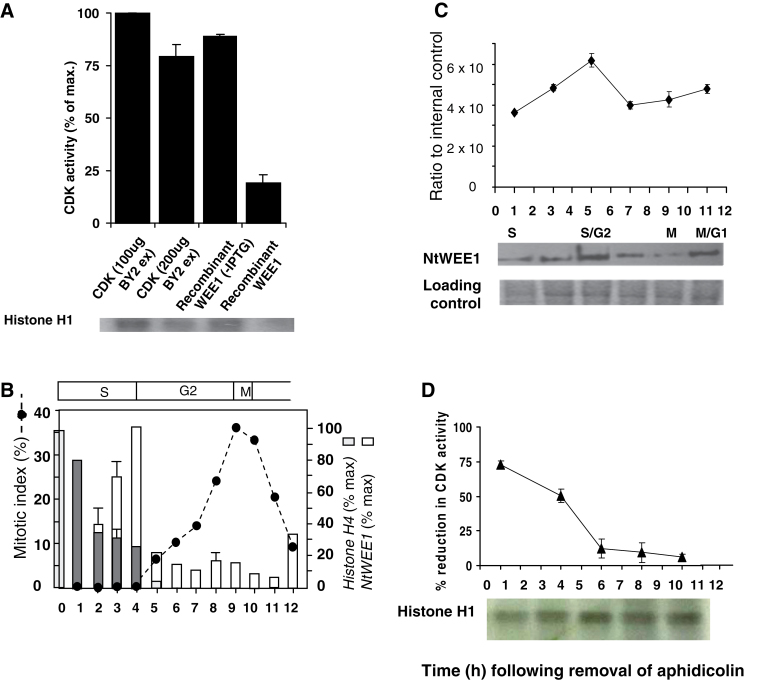
Fig. 1.
(A) Reduction in CDK activity elicited by the addition of recombinant NtWEE1. HIS6-NtWEE1 fusion protein, induced in E. coli (+IPTG) [compared with uninduced (–IPTG) control] was purified by affinity purification, and added to a kinase assay reaction containing CDKs (200 µg) purified from wild-type N. tabacum BY-2 cell culture protein extracts using p13SUC1 beads and histone H1 substrate; compared with the CDK alone (100 or 200 µg). A representative autoradiograph is shown below the histogram. The incorporation of 32P was assayed by quantification of the bands on the autoradiograph. (B–D) Cell cycle regulation of WEE1 mRNA, protein, and kinase activity in a synchronized wild-type N. tabacum BY-2 cell culture following removal of aphidicolin. (B) Mitotic index profile (dotted line) calculated as the sum of prophase, anaphase, metaphase, and telophase mitotic figures as a percentage of a minimum of 300 cells; mean histone H4 expression level (% max ±SE) (grey bars), mean NtWEE1 expression level (% max ±SE) (white bars), error bars absent when variation about the mean was <5% (n=3). The corresponding cell cycle phases are shown above the mitotic index graph. (C) Immunodetection of NtWEE1 protein extracted from synchrony samples (10 µg of protein per lane) and subjected to western blotting using the NtWEE1 antibody. The histogram displays mean (±SE) WEE1 protein levels as a ratio to an internal control (n=3). Representative western blot and a Coomassie stain loading control are shown below the histogram. (D) Inhibition of CDK activity by NtWEE1. The incorporation of 32P into histone H1 was assayed by quantification of the bands on the autoradiographs and expressed as the percentage reduction in CDK activity (± SE). CDK was pulled-down from 100 µg of BY-2 proteins. A representative autoradiograph is shown below the histogram.
WEE1 activity drops during G2 and remains low as cells enter mitosis
Tobacco BY-2 cells were synchronized with aphidicolin and, following removal of this reversible inhibitor of DNA polymerase α (Nagata et al., 1992), semi-quantitative RT–PCR generated histone H4 profiles showed an S-phase duration of 4h (Fig. 1B). Mitotic indices peaked at 9h (and also at 23h), giving a cell cycle duration of 14h (later part of the curve not shown). The phase durations (shown above Fig. 1B) are highly comparable with previously published cell cycle data for BY-2 cells (see Nagata et al., 1992; Sorrell et al., 2001; Orchard et al., 2005). Expression of NtWEE1 peaked at 4h (S/G2; Fig. 1B) and is also highly comparable with published data (Gonzalez et al., 2004).
WEE1 protein level was highest at 5h (early G2, Fig. 1C), 1h following the WEE1 mRNA peak (Fig. 1B). WEE1 levels dropped significantly (P < 0.05) at 7h (mid-G2) and remained low at 9h (mid-G2 to mitosis) as more cells entered mitosis, before showing a slight rise at 11h (early G1) (Fig. 1C).
WEE1 inhibition of CDK activity (Fig. 1D) was maximal when WEE1 was pulled-down from proteins extracted from BY-2 cells at 1h (early S-phase), decreased significantly (P < 0.05) by 4h (S/G2) and again (P <0.05) from 4h to 6h (early G2), and then remained at this level throughout G2 and M phase. The data are thus consistent in showing a drop in WEE1 kinase activity during G2 that remained low when cells entered mitosis.
WEE1–GFP signal essentially disappears at metaphase of mitosis
Data for WEE1 protein and WEE1 inhibition of CDK activity suggest strongly that WEE1 is cell cycle regulated in BY-2 cells. However, neither total protein nor activity levels disappeared. Since only ~40% synchrony was achieved (Fig. 1B), residual WEE1 kinase activity may derive from incomplete shut down throughout mitosis or persisted because of unsynchronized interphase cells. To test more precisely the levels of WEE1 protein during the cell cycle, GFP signal was monitored during the synchronized cell cycle of a 35S::AtWEE1–GFP cell line compared with a 35S::GFP-expressing control line. The AtWEE1–GFP signal was detected mainly in nuclei and chromosomes, with background/residual GFP signal in the cytoplasm (Fig. 2A).
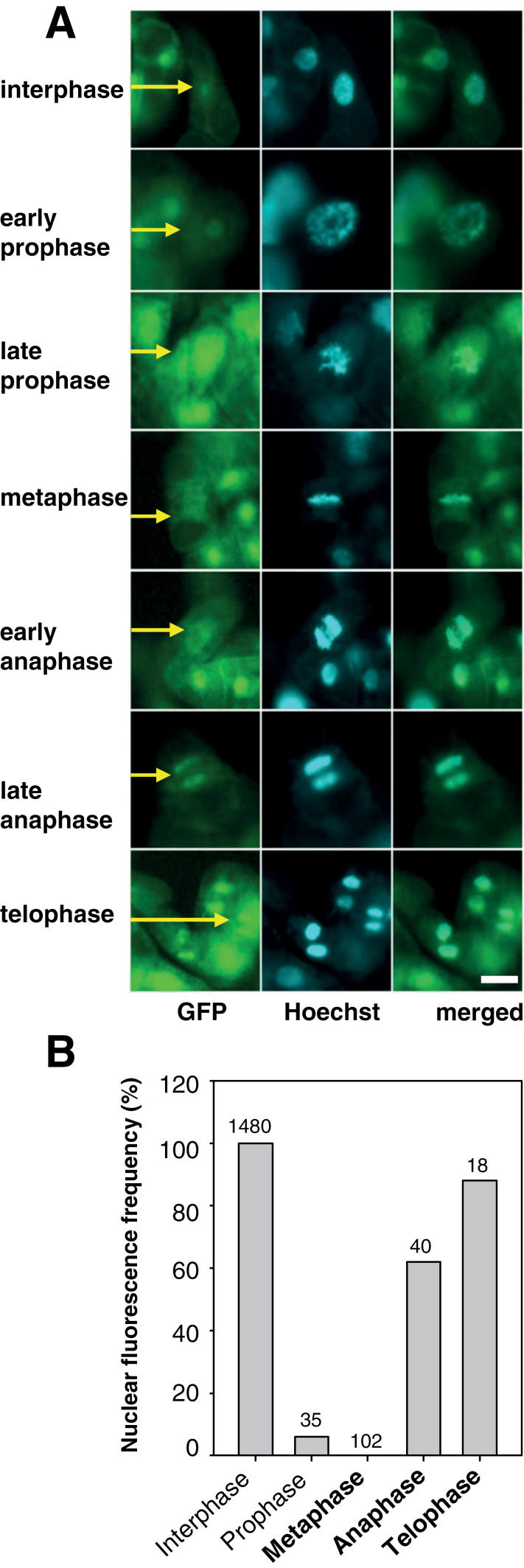
Fig. 2.
In the AtWEE1–GFP line, GFP signal is greatly reduced in metaphase and early anaphase. (A) 35S::AtWEE1–GFP (GFP, Hoechst, and merged GFP–Hoechst; Hoechst λ=420nm GFP λ=530nm). Yellow arrows indicate a representative cell; scale bar=50 µm for all images. (B) Nuclear fluorescence frequency (%) of cells sampled from the synchronized 35S::AtWEE1–GFP cell line. n values are indicated on each bar. Contingency χ2=1511 df 4, P < 0.001.
A clear nuclear AtWEE1–GFP signal was seen in interphase; however, signal associated with the chromosomes of early and late prophase cells was weaker, followed by an almost complete absence of signal from chromosomes at metaphase and early anaphase but its re-establishment in late anaphase and telophase. This pattern of alteration of AtWEE1–GFP intensity contrasts with a constant GFP signal regardless of cell cycle stage for the 35S::GFP line (Supplementary Fig. S3 at JXB online). Quantification of nuclear fluorescence from >1500 cells in both lines confirmed that in the 35S::AtWEE1–GFP line, GFP signal was undetectable in metaphase cells and only very few prophase cells emitted signal (Fig. 2B). A contingency χ2 indicated a highly significant difference in the relative frequency of fluorescence between stages for the 35S::AtWEE1–GFP line. In contrast, in the 35S::GFP line all cells (interphase and all mitotic stages) emitted a GFP signal and hence would be quantified as 100% were it to be shown in Fig. 2B.
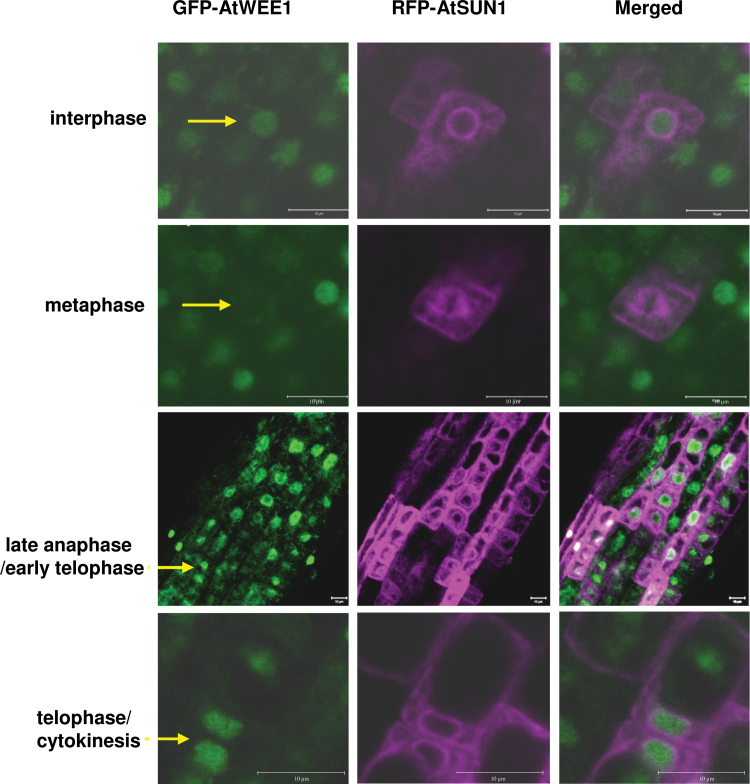
The dispersal of AtWEE1 during metaphase was also confirmed in Arabidopsis roots. An Arabidopsis line transformed with the WEE1–GFP construct was crossed with a line transformed with the nuclear envelope marker, AtSUN1–mRFP (Graumann et al., 2010; Graumann and Evans, 2011). In the resultant line, the pattern of WEE1–GFP signal during the cell cycle is similar to that seen in BY-2 cells (Fig. 3). In interphase, a clear nucleoplasmic AtWEE1–GFP signal is visible, surrounded by nuclear envelope labelling of AtSUN1–mRFP (Fig. 3). During metaphase AtWEE1–GFP signal is essentially absent but a clear AtSUN1–mRFP signal can be observed in mitotic spindle membranes (Fig. 3). AtWEE1–GFP signal reappears in late anaphase/early telophase cells, while AtSUN1–mRFP is present in the reforming nuclear envelope, and, finally, strong GFP and RFP signals were observed during cytokinesis. Hence, there is a remarkably precise cell cycle regulation of WEE1 with presumed degradation or destabilization of WEE1 when chromosomes align at the metaphase plate for both BY-2 cells and Arabidopsis root cells.
Fig. 3.
AtWEE1–GFP and AtSUN1–mRFP expression in different cell cycle phases in roots of transgenic Arabidopsis expressing both genes. Green colouring indicates AtWEE1–GFP expression, while purple colouring indicates AtSUN1–mRFP expression. Scale bar=10 µm. Yellow arrows indicate a representative cell.
WEE1 protein is not detectable in lateral root primordial cells
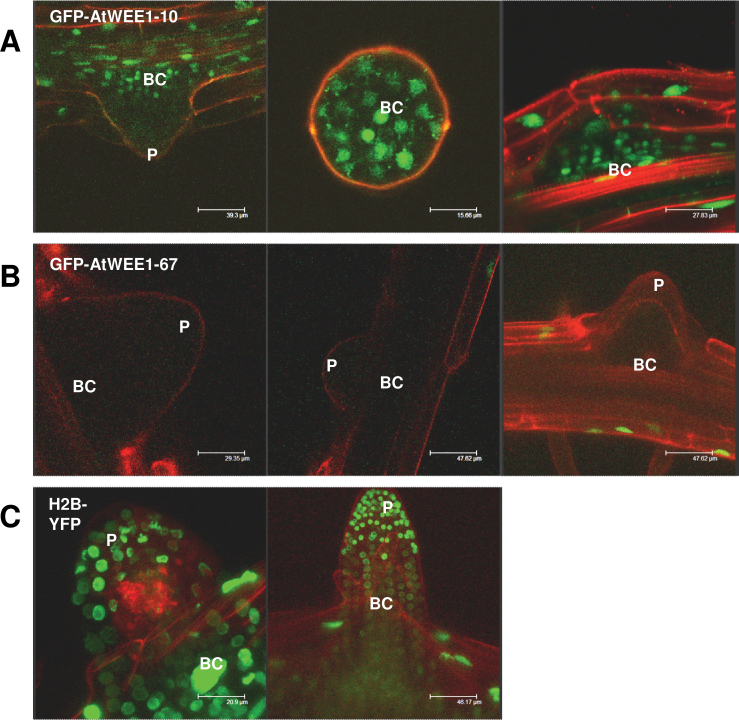
The stability of WEE1 protein was also examined in lateral roots of two independent 35S::AtWEE1–GFP-expressing Arabidopsis lines, #10 and #67. In line #10, a weak GFP signal was observed in the nuclei of the basal cells of the lateral root primordia (Fig. 4A), but fluorescence was not detected in the rest of the primordium. In line #67, a fluorescent signal could not be detected in any of the cells of the lateral root primordia (Fig. 4B). In contrast, in the 35S::H2B–YFP line, strong YFP expression was observed throughout the lateral root primordia (Fig. 4C).
Fig. 4.
Lateral root primordia of AtWEE1–GFP lines #10 and #67, and H2B–YFP seedlings. (A and B) Green colouring indicates AtWEE1–GFP expression in two independent lines, #10 and #67. (C) Green indicates H2B expression. Red colouring is propidium iodide counterstain for the cell walls. Representative images of at least five seedlings examined. P, lateral root primordium; BC, basal cells of lateral root primordium. Note that the middle image of (A) is a primordium seen from above, and demarcated by propidium iodide counterstaining.
AtWEE1 is degraded via the 26S proteasome degradation pathway
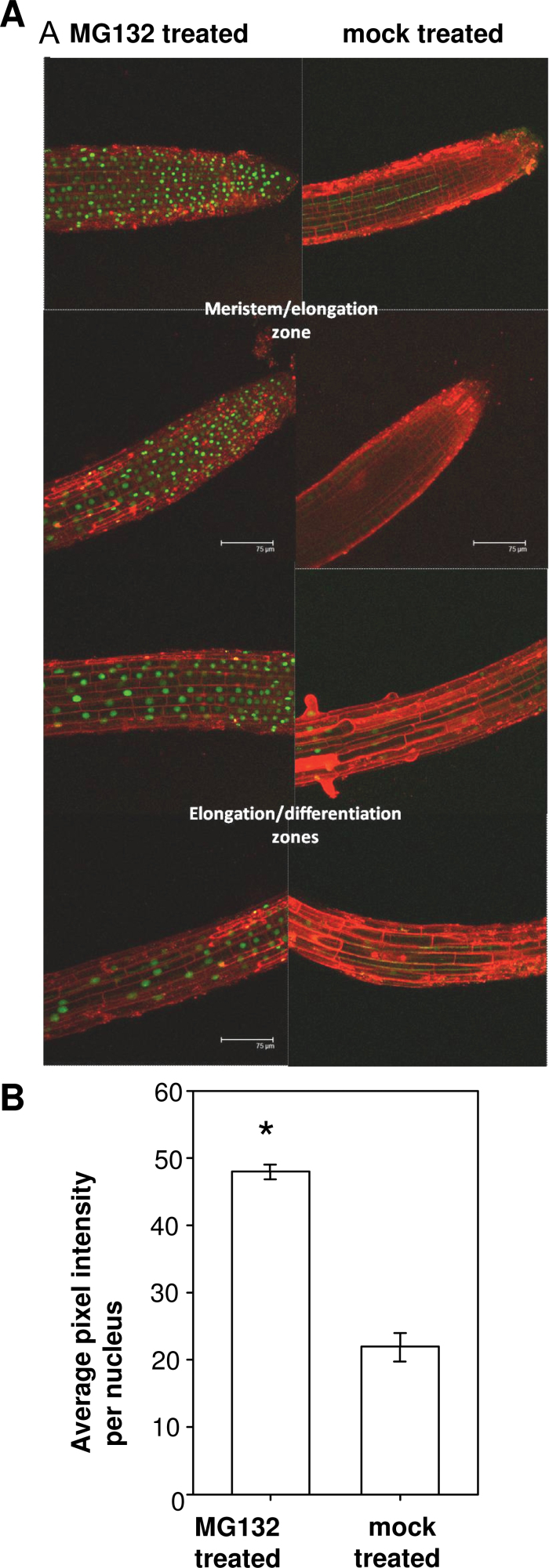
The proteasome inhibitor MG132 was used to determine whether the reduced fluorescent signal observed in the AtWEE1–GFP lines was caused by proteasome-mediated protein degradation. GFP signal clearly increased in AtWEE1–GFP seedlings treated with MG132 relative to the mock-treated seedlings (Fig. 5A) both in the root tips and further up the root. Quantification of the fluorescent signal showed that there was a significant (P < 0.05) 2-fold increase in GFP signal in the MG132-treated seedlings compared with the mock-treated seedlings (Fig. 5B). This demonstrates that the AtWEE1–GFP protein persists when the degradation route via the 26S proteasome is blocked.
Fig. 5.
MG132- and mock-treated 5-day-old AtWEE1–GFP line # 67 seedlings. (A) Confocal images of root tips and more basal regions: green indicates AtWEE1–GFP expression and red is propidium iodide counterstain for the cell walls. Representative images of three seedlings examined for each treatment. To allow for accurate and direct comparison between the two treatments, the confocal settings were not altered between the imaging of the MG132-treated and mock-treated seedlings. (B) GFP signal intensity (±SE). *Significant difference between treatments (P < 0.05; n= ≤466 to ≥2121).
In a yeast two-hybrid screen AtWEE1 interacts with components of the proteasome machinery
AtWEE1 was used as a bait in a yeast two-hybrid screen to search for interacting proteins that might play a regulatory role in its turnover. Approximately 1×107 transformants were screened from a library generated using Arabidopsis primary and secondary root tips (Sorrell et al., 2003). Over 900 interactors were detected by an ability to grow on His– medium and, in a second screen for β-galactosidase activity, 82 of these were confirmed. Sequencing of plasmid insertions revealed 60 different AtWEE1 interaction partners, of which 11 were identified multiple times (Supplementary Table S1 at JXB online). Functionally the interacting proteins could be divided into seven groups (Table 1). Of direct relevance to this work, four proteins associated with ubiquitin-mediated degradation were detected (Supplementary Table S1); each one was only detected once. One of these is a regulatory subunit of the 26S proteasome, while the other three are F-box proteins including SKIP1 (SKP1 INTERACTING PARTNER 1).
Table 1.
Functional groups of proteins that interacted with AtWEE1 in a yeast two-hybrid library screen
| Functional group | No. of proteins |
|---|---|
| Transcription factors/DNA- or RNA-binding proteins, histone modifications | 9 |
| Plant growth regulation and signal transduction | 3 |
| Stress responses/detoxification/pathogen responses | 13 |
| Cell division/cell size/cell wall and cell growth | 8 |
| Ribosomes/protein biosynthesis | 4 |
| Ubiquitin-mediated degradation | 4 |
| Other | 19 |
A protein–protein interaction between AtWEE1 and AtSKIP1 was confirmed in vivo through BiFC
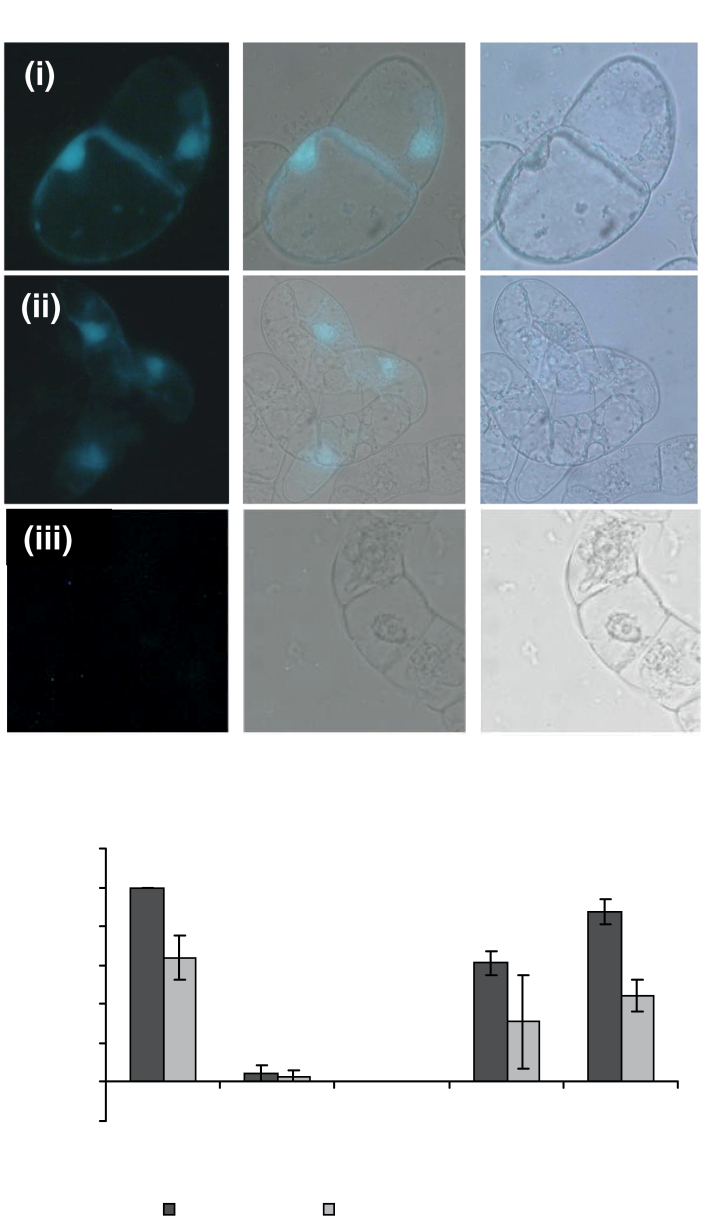
BiFC in BY-2 cells was used to verify the AtSKIP1–AtWEE1 interaction in plant cells, initially by transient transformation. Interacting proteins were mostly localized in the nucleus; however, the interaction was also detected at the cell wall, especially at the junctions between adjacent cells (Fig. 6A). The AtWEE1–YFPC (in the SPYCE vector) and AtSKIP1–YFPN (in the SPYNE vector) constructs were stably co-transformed into BY-2 cells to study the dynamics of the interaction during the cell cycle. The frequency with which an interaction between AtWEE1 and AtSKIP1 was observed in each cell cycle phase was similar to that of AtWEE1–GFP, with a drop in interactions observed between interphase and prophase, and interactions were not observed during metaphase. Interactions were again observed during anaphase and telophase (Fig. 6B).
Fig. 6.
(A) Tobacco BY-2 cells co-transformed with (i) AtWEE1–YFPC in the vector pSPYCE and AtSKIP1–YFPN in the vector pSPYNE; (ii) AtBZIP63 in both pSPYCE and pSPYNE (positive control); and (iii) AtWEE1–YFPC and AtBZIP63–YFPN (negative control); under UV light (left), white light (right), and the two merged (centre). Blue colouring indicates a positive interaction between the two proteins (representative images). (B) Mean nuclear fluorescence frequency (%; ±SE, n=3) in each cell cycle phase in cells from the following transgenic BY-2 lines: AtWEE1–GFP line # 4 (GFP–WEE1) and AtWEE1–YFPC/AtSKIP1–YFPN, line # 6 (WEE1–C/SKIP1-N6).
Expression of AtSKIP1 and AtWEE1 in BY-2 cells restores mitotic timing to wild-type levels compared with cultures expressing AtWEE1 alone
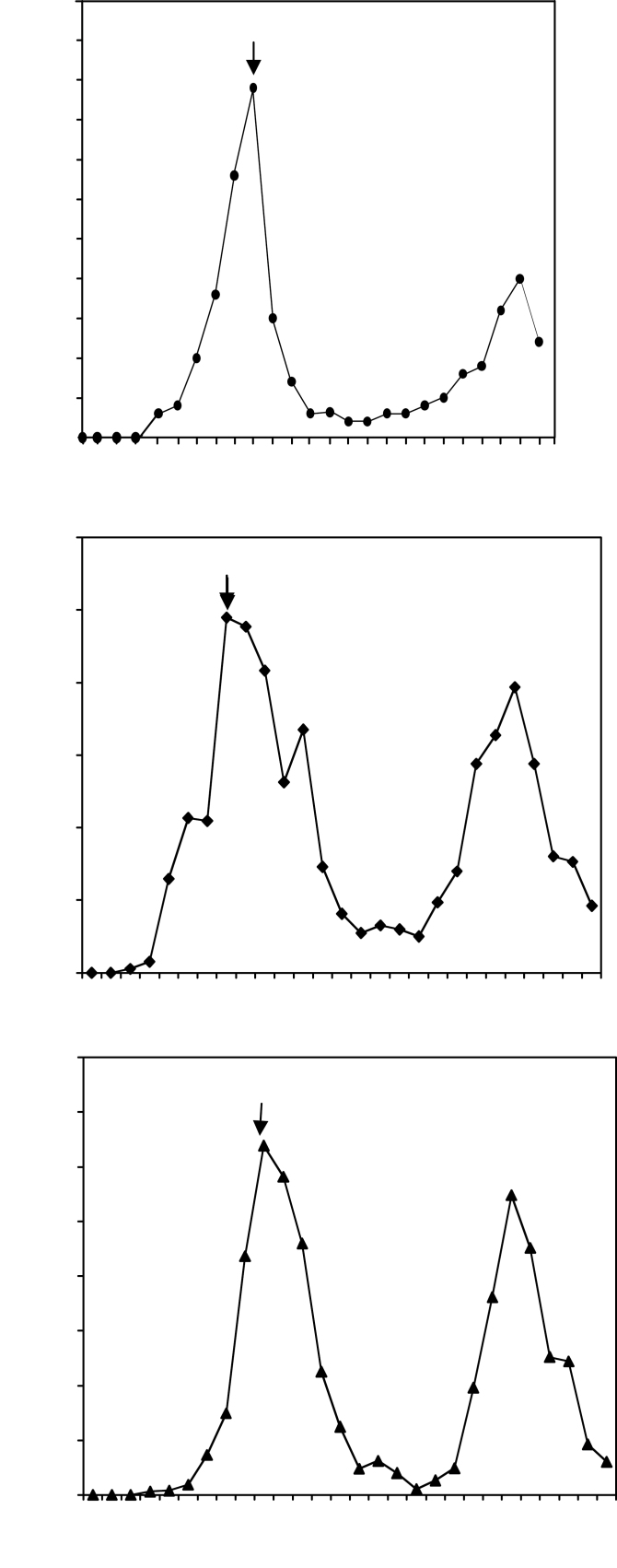
Given an interaction between AtWEE1 and AtSKIP1, the extent to which these genes affected aphidicolin-induced synchronized cell cycles in BY-2 cells was examined. Expression of AtWEE1–YFPC resulted in a mitotic peak at 7h compared with 9h in wild-type BY-2 cells (Fig. 7A, B). The mitotic peak was also earlier (at 4h) when AtWEE1 was expressed in BY-2 cells in the BIN HYG TX vector under an attenuated 35S promoter (Supplementary Fig. S4 at JXB online). However, when AtSKIP1–YFPN was co-expressed with AtWEE1–YFPC, there was an interaction between the two proteins, and the mitotic peak was restored to wild-type timing of 9h (Fig. 7C).
Fig. 7.
Mitotic index (±SE) of BY-2 cell lines: (A) The wild type. (B) AtWEE1–YFPC and (C) AtWEE1–YFPC/AtSKIP1–YFPN line #6 synchronized with aphidicolin compared with the wild type. The timing of the first mitotic peak in each line is shown with an arrow.
Discussion
In the work reported here, the inhibitory effect of the WEE1 protein pulled-down from BY-2 cells on CDK activity drops as cells traverse G2 and enter mitosis. Remarkably, a GFP signal essentially disappeared at metaphase in both BY-2 cells and Arabidopsis lines expressing WEE1–GFP. The GFP signal was then restored towards the end of mitosis at late anaphase/early telophase. Previous work in Arabidopis and tobacco BY-2 cells established that the G2/M transition is regulated by both CDKA and CDKB. While CDKA activity is generally constant during S-phase and G2, CDKB peaks in mid- to late G2 phase (Porceddu et al., 2001; Sorrell et al., 2001; Orchard et al., 2005). In other words, WEE1 activity data are the opposite of the typical CDKB activity profile reported for BY-2 cells (Sorrell et al., 2001; Orchard et al., 2005). Such CDK activity is required until metaphase when partner mitotic cyclins are degraded via a proteolytic pathway that deploys the anaphase-promoting complex (APC-Cdh1; Peters, 2002). BY-2 cells transformed with non-degradable mitotic B1 cyclin exhibited normal prophase and metaphase, but from there on was a mitotic catastrophe (Weingartner et al., 2004). In other words, a normal mitosis is finely tuned and depends on CDK kinase activity persisting until metaphase but then finishing abruptly.
Another notable feature of the profile reported here is that NtWEE1 kinase-mediated inhibition of CDK activity is highest in early S-phase. Recently, it was shown in Arabidopsis that following hydroxyurea treatment, roots exhibited rapid accumulation of WEE1 transcripts at the start of S-phase, leading to the suggestion that WEE1 kinase has a role during S-phase following the induction of the DNA replication checkpoint (Cools et al., 2011). Thus data here support a role for plant WEE1 in S-phase although, of course, it is still possible that there may also be other negative regulators of CDK activity in the in vivo plant cell cycle (e.g. ICK1/2), as proposed by Boudolf et al. (2006).
Unlike WEE1 kinase in the human cell cycle (McGowan and Russell, 1995), NtWEE1 activity never drops to zero. While the use of aphidicolin is an established and proven method for cell cycle synchrony (Nagata et al., 1992), it is not possible to achieve 100% synchronization of mitosis. Hence the residual protein and kinase activity is most probably due to WEE1 protein in interphase cells. Furthermore, BY-2 and Arabidopsis lines expressing AtWEE1–GFP exhibited WEE1 protein localization in mitotic phases other than metaphase. This is another reason why WEE1 kinase activity did not drop to zero during mitosis in synchronized BY-2 cells.
The 35S promoter normally confers strong, constitutive expression in BY-2 and Arabidopsis root tip cells. So the dramatic reduction of GFP signal in metaphase cells in both BY-2 and Arabidopsis root cells expressing WEE1 is consistent with the removal of the WEE1 protein at this stage of mitosis. In the 35S::GFP control BY-2 cells, GFP signal persisted in 100% of cells, regardless of mitotic phase.
A previous study of the interactions between rice cyclins and CDKs in BY-2 cells showed that there was a tight association of CDKB2;1–GFP and CycB2;2–GFP with chromosomes of transgenic BY-2 cells during an aphidicolin-induced synchronous cell cycle (Lee et al., 2003). This conclusion was supported by the finding that treatment with Triton X-100 resulted in a complete loss of GFP fluorescence in cells expressing GFP alone whereas GFP fluorescence was unaffected by the Triton treatment in the lines transformed with the CDK–GFP or cyclin–GFP fusion proteins. Exactly the same phenomenon was seen here when the GFP-expressing line was stained with Hoechst; this staining reaction depends on a pre-treatment with Triton X-100. Hence, in the 35S::AtWEE1–GFP line, the data suggest that the loss of GFP signal is due to WEE1 degradation at metaphase and its re-appearance due to re-synthesis at anaphase. Moreover the same pattern of alteration of GFP signal was seen in root meristems of 35S::AtWEE1–GFP crossed with AtSUN1–RFP.
The pattern of 35S::AtWEE1–GFP signal was also examined in lateral root primordia in two independent Arabidopsis lines. AtWEE1–GFP levels were reduced in both lines, to the point of being undetectable in line #67, whereas 35S::H2B–YFP was strongly and constitutively expressed in the lateral root primordia (Boisnard-Lorig et al., 2001). This implies a high turnover of AtWEE1 in lateral root primordia, supporting a role for AtWEE1 degradation in the initiation of lateral root growth. This is further supported by the down-regulation of AtWEE1 in the pericycle, when roots were stimulated to produce laterals by 1-naphthaleneacetic acid (NAA) treatment (de Almeida Engler et al., 2009). Since pericycle cells re-enter the cell cycle for the initiation of laterals, this supports a role for inhibition of the cell cycle via WEE1 and its removal in pericycle cells primed to divide.
Timely degradation of plant cell cycle proteins, including cyclins, is frequently achieved via the 26S proteasome degradation pathway (Genschik et al., 1998). WEE1 degradation is also achieved via the 26S proteasome degradation pathway in budding yeast (Simpson-Lavy and Brandeis, 2010), Xenopus (Michael and Newport, 1998), and humans (Watanabe et al., 2004). However, the mechanism for the degradation of WEE1 protein in plants is yet to be described. When a 26S proteasome inhibitor, MG132 (Rock et al., 1994), was supplied to seedlings of an AtWEE1–GFP-expressing line, the AtWEE1–GFP signal was significantly enhanced (Fig. 5A, B). These results strongly indicate that AtWEE1 protein is degraded via the 26S proteasome degradation pathway; note that GFP is unaffected by MG132 treatment (Song and Wu, 2005).
The finding that AtWEE1 interacts with several proteasome-related proteins in a two-hybrid screen further supports this route for its degradation. BiFC was used to confirm the interaction between AtWEE1 and the F-box protein AtSKIP1. AtSKIP1 interacts with the SKP1/ASK1 subunit of SCF ubiquitin ligase (Risseeuw et al., 2003). It is not closely related to any other proteins in Arabidopsis; the closest homologue is a member of the RNI-superfamily, the Arabidopsis putative F-box/leucine-rich repeat protein 19 (At4g30640), which shares 40% gene sequence identity with AtSKIP1.
This interaction could provide the specificity for the targeted removal of AtWEE1, although the finding that AtWEE1 interacts with several F-box proteins indicates that different F-box proteins may mediate the specificity of WEE1 removal in different tissues or under different cellular conditions. If AtSKIP1 promotes AtWEE1 degradation, then co-expression of the two proteins might tend to destabilize the AtWEE1. This hypothesis was tested using synchronized BY-2 cells. Expressing AtWEE1 in tobacco BY-2 cells under the 35S promoter led to a shortening of G2 and a premature rise in the mitotic peak, as was the case in the AtWEE1–YFPC line. This was an unexpected result since expression of AtWEE1 in fission yeast (Sorrell et al., 2002) and Arabidopsis (Spadafora et al., 2012) tends to delay mitosis, resulting in a larger cell phenotype. This result is probably due to an interaction between AtWEE1 and the tobacco cellular machinery resulting in an interference in the normal functions of NtWEE1. However, co-expression of AtSKIP1–YFPN with AtWEE1–YFPC clearly delayed the mitotic peak compared with AtWEE1–YFPC alone, restoring the timing back to the wild type. This suggests that the removal of AtWEE1 by SKIP1 prevented a premature rise in the mitotic index in synchronized cells. Thus the hypothesis that the interaction between AtWEE1 and AtSKIP1 is indeed functional was supported.
In conclusion, a drop in WEE1 activity was discovered when synchronized BY-2 cells traverse G2 and enter mitosis. This is linked to a fine-tuned regulation of WEE1 protein, which essentially disappeared from chromosomes at metaphase in both BY-2 cells and Arabidopsis roots. Given what is known about CDK activity at G2/M and during mitosis, this suggests an inverse relationship between WEE1 and CDKB activity in the normal BY-2 cell cycle. The removal of WEE1 protein from lateral root primordial cells also supports the idea that WEE1 is degraded, enabling cell division in the pericyle and in developing lateral root primordia. Furthermore, the data here demonstrate for the first time that WEE1 protein is degraded via the 26S proteasome in plants, and that this may be mediated by an interaction with specific F-box proteins including SKIP1.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Immunodetection of WEE1 in protein extracts from wild-type N. tabacum BY-2 cell culture.
Figure S2. Expression and affinity purification of recombinant NtWEE1 and AtGF14ω proteins in E. coli.; immunodetection of the recombinant NtWEE1 protein by western blotting
Figure S3. 35S::GFP (GFP, DIC, merged GFP–DIC) images showing that GFP signal is retained throughout the cell cycle.
Figure S4. Mitotic index (± SE) in synchronized BY-2 cell lines of two independent lines transformed with AtWEE1 compared with empty vector.
Table S1. Proteins that interacted with AtWEE1 in a yeast two-hybrid library screen.
Table S2. Primer sequences for PCR analysis and vector construction.
Acknowledgements
IS and NS thank the University of Calabria for an international research award (borsa di specializzazione all’estero per giovani ricercatori), GSC, IS, ALG, and NS thank Cardiff University and University of Worcester (UW) for research studentships, and RJH thanks UW for sabbatical leave to work in Cardiff. KG thanks the Leverhulme Trust for a research grant and fellowship. We also thank Dr L. Fischer (Charles University, Prague, CZ) for BY-2 callus transformed with a 35S::GFP construct, and Professor Jim Murray (Cardiff University) for provision of the H2B-YFP Arabidopsis line.
References
- An GH. 1985. High-efficiency transformation of cultured tobacco cells. Plant Physiology 79, 568–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayad NG, Rankin S, Murakami M, Jebanathirajah J, Gygi S, Kirschner MW. 2003. Tome-1, a trigger of mitotic entry, is degraded during G1 via the APC. Cell 113, 101–113. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F. 2001. Dynamic analyses of the expression of the HISTONE::YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. The Plant Cell 13, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Inze D, De Veylder L. 2006. What if higher plants lack a CDC25 phosphatase? Trends in Plant Science 11, 474–479. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JM, Murray JA. 2000. Cyclin D control of growth rate in plants. Nature 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, Van den Daele H, Isterdale GV, Schnittger A, De Veylder L. 2011. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. The Plant Cell 23, 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J, De Veylder L, De Groodt R, et al. 2009. Systematic analysis of cell-cycle gene expression during Arabidopsis development. The Plant Journal 59, 645–660. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Joubes J, Cools T, et al. 2007. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. The Plant Cell 19, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annual Review of Cell and Developmental Biology 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dudits D, Cserháti M, Miskolczi P, Horváth V. 2007. The growing family of plant cyclin-dependent kinases with multiple functions in cellular and developmental regulation. In: Inze D, ed. Cell cycle control and plant development Oxford: Blackwell Publishing; 1–30. [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Francis D, Davies MS, Braybrook A, James NC, Herbert RJ. 1995. An effect of zinc on M-phase and G1 of the plant cell cycle in the synchronous TBY-2 tobacco cell suspension. Journal of Experimental Botany 46, 1887–1894. [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. 1998. Cell cycle-dependent proteolysis in plants. Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. The Plant Cell 10, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A. 2007. The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. The Plant Journal 51, 642–655. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Hernould M, Delmas F, Gevaudant F, Duffe P, Causse M, Mouras A, Chevalier C. 2004. Molecular characterization of a WEE1 gene homologue in tomato (Lycopersicon esculentum Mill.). Plant Molecular Biology 56, 849–861. [DOI] [PubMed] [Google Scholar]
- Graumann K, Evans DE. 2011. Nuclear envelope dynamics during plant cell division suggest common mechanisms between kingdoms. Biochemical Journal 435, 661–667. [DOI] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE. 2010. Characterization of SUN-domain proteins at the higher plant nuclear envelope. The Plant Journal 61, 134–144. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98. [Google Scholar]
- Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, Renaudin JP. 2000. CDK-related protein kinases in the plant cell cycle. Plant Molecular Biology 43, 607–620. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Sia RA, Bardes EG, Lew DJ, Reed SI. 1998. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes and Development 12, 2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81, 279–288. [DOI] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, Hashimoto J, Tsutsumi N, Uchimaya H, Umeda M. 2003. Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. The Plant Journal 34, 417–425. [DOI] [PubMed] [Google Scholar]
- Lentz Grønlund A, Dickinson JR, Kille P, Herbert RJ, Francis D, Rogers HJ. 2009. Plant WEE1 kinase interacts with a 14-3-3 protein but a mutation of WEE1 at S485 alters their spatial interaction. Open Plant Science Journal 3, 40–48. [Google Scholar]
- McGowan CH, Russell P. 1995. Cell cycle regulation of human WEE1. EMBO Journal 14, 2166–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Newport J. 1998. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science 282, 1886–1889. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–479. [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. 1992. Tobacco BY-2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. International Review of Cytology 132, 1–30. [Google Scholar]
- Nocarova E, Fischer L. 2009. Cloning of transgenic tobacco BY-2 cells; an efficient method to analyse and reduce high natural heterogeneity of transgene expression. BMC Plant Biology 9, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. 1990. Universal control mechanism regulating onset of M-phase. Nature 256, 547–551. [DOI] [PubMed] [Google Scholar]
- Orchard CB, Siciliano I, Sorrell DA, et al. 2005. Tobacco BY-2 cells expressing fission yeast cdc25 bypass a G2/M block on the cell cycle. The Plant Journal 44, 290–299. [DOI] [PubMed] [Google Scholar]
- Peters JM. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Molecular Cell 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld JP, Segers G, De Veylder L, Barroco RP, Casteels P, Van Montagu M, Inze D, Mironov V. 2001. A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. Journal of Biological Chemistry 276, 36354–36360. [DOI] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. 2003. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. The Plant Journal 34, 753–767. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Brandeis M. 2010. Cdk1 and SUMO regulate Swe1 stability. PLoS One 5, e15089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Song Z, Wu M. 2005. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene 24, 2723–2734. [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Marchbank AM, Chrimes DA, Dickinson JR, Rogers HJ, Francis D, Grierson CS, Halford NG. 2003. The Arabidopsis 14-3-3 protein, GF14ω, binds to the Schizosaccharomyces pombe Cdc25 phosphatase and rescues the DNA replication checkpoint in the rad – 24 mutant. Planta 218, 50–57. [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Marchbank A, McMahon K, Dickinson JR, Rogers HJ, Francis D. 2002. A WEE1 homologue from Arabidopsis thaliana . Planta 215, 518–522. [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Menges M, Healy JMS, et al. 2001. Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar bright yellow-2 cells. Plant Physiology 126, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora N, Perotta L, Nieuwland J, et al. 2012. Gene dosage effect of WEE1 on growth and morphogenesis from Arabidopsis hypocotyl explants. Annals of Botany110, 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW. 2003. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nature Reviews Genetics 4, 948–958. [DOI] [PubMed] [Google Scholar]
- Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jug R, Gordon-Kamm WJ, Larkins BA. 1999. Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proceedings of the National Academy of Sciences, USA 96, 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tominaga Y, Tominaga Y, Cuiling L, Rui-Hong C, Wang R-H, Deng C-X. 2006. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. International Journal of Biological Sciences 2, 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. The Plant Journal 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proceedings of the National Academy of Sciences, USA 101, 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Meszaros T, Binarova P, Schmit AC, Helfer A, Derevier A, Erhardt M, Bogre L, Genschik P. 2004. Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. The Plant Cell 16, 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H. 2009. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proceedings of the National Academy of Sciences, USA 106, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, et al. 2002. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.