Abstract
Environmental stresses such as drought, salinity, and cold are major adverse factors that significantly affect agricultural productivity. Protein phosphorylation/dephosphorylation is a major signalling event induced by osmotic stress in higher plants. Sucrose non-fermenting 1-related protein kinase 2 (SnRK2) family members play essential roles in the response to hyperosmotic stresses in plants. In this study, the TaSnRK2.3 gene, a novel SnRK2 member was cloned, and three copies located on chromosomes 1A, 1B, and 1D were identified in common wheat. TaSnRK2.3 was strongly expressed in leaves, and responded to polyethylene glycol, NaCl, abscisic acid, and cold stresses. To characterize its function, transgenic Arabidopsis overexpressing TaSnRK2.3–GFP controlled by the cauliflower mosaic virus 35S promoter was generated and subjected to severe abiotic stresses. Overexpression of TaSnRK2.3 resulted in an improved root system and significantly enhanced tolerance to drought, salt, and freezing stresses, simultaneously demonstrated by enhanced expression of abiotic stress-responsive genes and ameliorative physiological indices, including a decreased rate of water loss, enhanced cell membrane stability, improved photosynthetic potential, and significantly increased osmotic potential and free proline content under normal and/or stressed conditions. These results demonstrate that TaSnRK2.3 is a multifunctional regulator, with potential for utilization in transgenic breeding for improved abiotic stress tolerance in crop plants.
Key words: expression pattern, morphological character, osmotic stress, physiological trait, stress response.
Introduction
Plants are constantly confronted by environmental stresses, including drought, high salinity, and extreme temperatures, that affect both biomass and grain yield of crops. Plants have evolved a range of molecular and physiological mechanisms to cope with multiple adverse stresses. Numerous lines of evidence indicate that protein kinases/protein phosphatases play essential roles in the response to environmental stimuli (Hong et al., 1997).
In eukaryotes, reversible protein phosphorylation is a mechanism to perceive and respond to environmental stresses, and constitutes a means to maintain cellular functions when responding to environmental stimuli, pathogens, and various phytohormones (Cohen, 1988). Genetic evidence shows that protein phosphatases 2C and 2A play crucial roles in the early abscisic acid (ABA) signalling pathway (Hauser et al., 2011; Lee and Luan, 2011). Various stress-inducible protein kinase families such as mitogen-activated protein kinase (Wrzaczek and Hirt, 2001), calcium-dependent protein kinase (CDPK) (Ludwig et al., 2004), and sucrose non-fermenting 1 (SNF1)-related protein kinase (SnRK), are activated by ABA and diverse stress signals.
Yeast SNF1 protein kinase, mammalian AMP-activated protein kinase (AMPK), and plant SnRK protein are highly conserved and play pivotal roles in growth and metabolic responses to cellular stresses. Plant SnRKs are grouped into three subfamilies: SnRK1, SnRK2, and SnRK3. The SnRK1 and SnRK2 subfamilies are similar in their catalytic domains but divergent in their C-terminal domains, whereas the SnRK3 protein subfamily forms a unique group (Hardie et al., 1998). SnRK1 kinase is well characterized at the molecular and biochemical levels, and evidence shows that SnRK1s play a role in the regulation of carbon and nitrogen metabolism, whereas SnRK2 and SnRK3 function mainly in stress signalling (Hrabak et al., 2003). Recent studies suggest that SnRK2 and SnRK3 originated by duplication of SnRK1 and then diverged rapidly during plant evolution to fulfil new roles that enabled plants to develop networks linking abiotic stress and ABA signalling with metabolic signalling (Hrabak et al., 2003; Hauser et al., 2011). The SnRK2 and SnRK3 subfamilies are unique to plants (Halford et al., 2003). One of best-studied kinases in the SnRK3 family is SOS2, which is vital for Na+ and K+ homeostasis and abiotic stress tolerances (Liu et al., 2000). The activities of SnRK3 kinases are regulated in a calcium-dependent manner, and they may function as cross-talk nodes in complex signalling networks (Kim et al., 2003; Nolan et al., 2006).
Accumulated evidence shows that SnRK2s are a merging point of ABA-dependent and -independent pathways for osmotic stress response and may be involved in diverse developmental processes in plants (Fujii et al., 2011; Kulik et al., 2011). Ten SnRK2s have been identified in Arabidopsis, among which SnRK2.2–3 and SnRK2.6–8 are activated by ABA, and SnRK2.2–10 is activated by hyperosmotic and salinity stresses, whereas none is activated by cold stress (Boudsocq et al., 2004, 2007). Overexpression of SnRK2.8 or NTL6, an NAC transcription factor linked directly with SnRK2.8-mediated signalling, leads to enhanced drought tolerance (Umezawa et al., 2004; Kim et al., 2012). Furthermore, SnRK2.4 and SnRK2.10 are involved in the maintenance of root system architecture during salt stress (McLoughlin et al., 2012). Similarly, ten SnRK2s, designated SAPK1–10, have been identified in rice. All are activated by hyperosmotic stress, and SAPK8–10 are also activated by ABA (Kobayashi et al., 2004). Overexpression of SAPK4 significantly enhances tolerance to salt in rice (Diedhiou et al., 2008). Recently, ten maize SnRK2 members were cloned, and most ZmSnRK2s were induced by one or more abiotic stresses (Huai et al., 2008). In wheat, the first SnRK2 member, PKABA1 (also named TaW55), was induced by ABA and hyperosmotic stress, and was involved in the response to multiple environmental stresses (Anderberg and Walker-Simmons, 1992; Xu et al., 2009). In previous studies, we observed dynamic expression of TaSnRK2.4 and TaSnRK2.7–8 under various abiotic stresses; their overexpression resulted in enhanced tolerance to multi-environmental stresses (Mao et al., 2010; Zhang et al., 2010, 2011). Thus, solid evidence shows that the SnRK family of protein kinases is involved in multi-environmental stress responses and all have potential use for improvement of abiotic stress tolerance and yield enhancement (Piattoni et al., 2011).
As a world staple crop, wheat production is severely constrained by abiotic stresses, such as drought, salinity, and extreme temperatures. Understanding the molecular basis of abiotic stress response, especially the role of specific SnRK2s in response to adverse stresses, is a prerequisite for genetic improvement of abiotic stress tolerance. To this end, we cloned TaSnRK2.3, determined its expression patterns under various abiotic stresses and in different wheat tissues, and characterized its function in Arabidopsis. Transgenic experiments indicated that overexpression of TaSnRK2.3 in Arabidopsis enhanced tolerance to drought, salt, and freezing stresses without growth retardation. Therefore, TaSnRK2.3 could be utilized to improve abiotic stress tolerance in plants.
Materials and methods
Plant materials and stress experiments
Wheat (Triticum aestivum L.) genotype ‘Hanxuan 10’ with a significant degree of drought tolerance was used in this study. Growth conditions and stress treatment assays were as described previously (Mao et al., 2010). To study the expression of target genes at different developmental stages, seedling leaves and roots, spindle leaves at jointing, and young emerging ears were sampled as described previulsy (Mao et al., 2010).
To investigate the genomic origin of the target gene, 20 accessions of various wheat species, including four A genome accessions (Triticum urartu), four S genome accessions (Aegilops speltoides, the putative B genome donor), four diploid D genome accessions (Aegilops tauschii), four AB genome accessions (three Triticum dicoccoides and one Triticum dicoccum), and four hexaploid wheat accessions were selected for PCR (Table S1, at JXB online). Forty-one nulli-tetrasomic lines of Chinese Spring were deployed to identify the chromosome origin of the target gene. A doubled haploid (DH) population (Hanxuan 10×Lumai 14) with 150 lines was used to map TaSnRK2.3.
Arabidopsis thaliana (ecotype Columbia) for transgenic analysis was grown in a controlled environment chamber at 22 °C, with a 12h/12h photoperiod, a light intensity of 120 mmol m–2 s–1, and 70% relative humidity. Four T3 homozygous transgenic lines with relatively higher expression levels of the target gene were selected for phenotypic assays, and all seeds used for phenotypic assays were from the same harvest and stored under the same conditions. To identify the expression patterns of abiotic stress-responsive genes in transgenic Arabidopsis, 7-d-old Arabidopsis seedlings germinated on MS plates were transferred to a stainless steel sieve and subjected to polyethylene glycol (PEG-6000, –0.5MPa) stress for 3h.
Cloning the full-length cDNA and sequence analysis of TaSnRK2.3
Tissues from wheat seedlings at various stages and from mature plants were collected to extract total RNA with TRIzol reagent (Invitrogen), and mRNA was isolated with oligo(dT)–cellulose (Qiagen). Several full-length cDNA libraries of wheat in λ Zap II (Stratagene) were constructed by an optimized cap-trapper method (Mao et al., 2005). A full-length wheat cDNA database was generated with the 3’ end and 5’ sequencing data of full-length cDNA clones. To obtain the cDNA sequence of TaSnRK2.3, the amino acid sequence of rice SAPK3 was used as a query probe to screen the full-length wheat cDNA database. Three candidate clones were obtained by tBLASTn, and the full-length cDNA of TaSnRK2.3 was identified by sequencing the ends.
Database searches of the nucleotide and deduced amino acid sequences were performed using BLAST. Sequence alignment and similarity analyses were conducted by multiple sequence alignment programs in DNASTAR. Signal sequence and transmembrane regions were predicted with SignalP (http://genome.cbs.dtu.dk/services/SignalP) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Subcellular localization was predicted online (http://wolfpsort.org/). Secondary structure was revealed with PREDATOR (http://bioweb.pasteur.fr/seqanal/protein/intro-uk.html). The functional region and activity sites of target proteins were predicted with InterProScan (http://www.ebi.ac.uk/Tools/InterProScan) and ScanProsite (http://www.expasy.ch/tools/scanprosite). To reveal the relationship between TaSnRK2.3 and SnRK2 members in other plant species, a maximum likelihood tree was constructed with the PHYLIP soft package (version 3.69).
Genetic characterization of TaSnRK2.3
To analyse the structure of TaSnRK2.3, several pairs of primers flanking the ORF were designed, and two pairs of primers amplified the target fragments were obtained. One specifically amplified the B genomic allele (GBF/GBR), and the other simultaneously amplified the A, B, and D genomic alleles (GTF/GTR) (Table S2 at JXB online). The genomic fragments of TaSnRK2.3 were amplified using TransStart FastPfu Taq DNA polymerase and ligated into a pEASY-Blunt cloning vector (TransGen Biotech), and then sequenced with a DNA analyser (ABI 3730XL). To further probe the chromosome origin of different alleles and simplify the PCR results, three allele-specific primer pairs flanking the polymorphism-enriched regions of the three alleles were obtained (Table S2). After nucleotide sequence polymorphism assays, another pair of primers was designed to develop a cleaved amplified polymorphic sequence (CAPS) marker, and a DH population (Hanxuan 10×Lumai 14) with 150 lines was used for gene mapping. MapMaker (version 3.0) was used to determine the location on the chromosome.
To isolate the promoter of TaSnRK2.3, the genomic DNA sequence of TaSnRK2.3 was compared with a genome sequence database of A. tauschii (DD) (unpublished data), the diploid D genome donor species of common wheat in BLASTn assays. The hit scaffold with the highest similarity to the query sequence was selected, and gene-specific primers (covering the promoter and partial coding regions) were designed according to the sequence. The promoters of three genomic alleles were isolated by PCR, and their cis-acting regulatory elements were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Quantitative real-time PCR (qRT-PCR)
The cDNA samples were obtained as described above and qRT-PCR was performed in triplicate with an ABI PRISM® 7900 system using a SYBR Green PCR Master Mix kit (Applied Biosystems) according to the manufacturer’s instructions. Tubulin transcripts of wheat were used to quantify relative transcript levels. Oligonucleotides of qRT-PCR primers for TaSnRK2.3 wae listed in Table S2. Relative gene expression levels were detected using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Actin transcripts of Arabidopsis were used to quantify the expression levels of TaSnRK2.3 and abiotic stress responsive genes in transgenic Arabidopsis plants. The oligonucleotides for abiotic stress-responsive genes have been described elsewhere (Ding et al., 2009).
Subcellular localization of the TaSnRK2.3 protein
The ORF of TaSnRK2.3 was fused upstream of the green fluorescent protein (GFP) gene and place under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter in the pJIT163-GFP expression vector to construct a 35S::TaSnRK2.3–GFP fusion protein. Restriction sites were added to the 5’ and 3’ ends of the coding region by PCR; the oligonucleotides for fusion GFP subcloning were: forward, 5’-CCCAAGCTTATGGAGGAGAGGTACGAGGC-3’ (HindIII site in bold italics), reverse, 5’-GAGAGTCGACGTAGGTCTCCC CCTCGGCT-3’ (SalI site in bold italics). The PCR product was cloned into the pJIT163-GFP plasmid for expression of the fusion protein. The subcellular location of TaSnRK2.3 was detected as described previously (Mao et al., 2010).
Generation of transgenic plants
The coding region of TaSnRK2.3 cDNA was amplified by primers: forward, 5’-CTCCCGGGATGGAGGAGAGGTACGAGGCG-3’ (SmaI site in bold italics); reverse, 5’-CTGTCGACGTAGGTCTCC CCCTCGGCT-3’ (SalI site in bold italics), and cloned into a pPZP211 vector as a GFP-fused fragment driven by the CaMV 35S promoter (Hajdukiewicz et al., 1994). The transformation vectors harbouring 35S::GFP and 35S::TaSnRK2.3–GFP were introduced into Agrobacterium and transferred into wild-type (WT) Arabidopsis plants by floral infiltration. Positive transgenic lines were screened on kanamycin plates and identified by RT-PCR, and the expression level of TaSnRK2.3 was determined by qRT-PCR and the protein level evaluated by Western blotting.
Protein level assays for TaSnRK2.3
Total protein was extracted from approximately 0.1g of Arabidopsis seedling tissue (Mao et al., 2010). The concentration of total protein was tested with a spectrophotometer (NanoDrop 2000C; Thermo Scientific). Protein samples were separated electrophoretically on 12.5 % polyacrylamide gel with a visible protein marker. The gel was stained with Ponceau S, and the proteins were subsequently transferred to polyvinylidene fluoride membranes (Amersham) by semi-dry electroblotting (Mini-Protean II system; Bio-Rad). The membrane was blocked with 5% skimmed milk and blotted with a commercial GFP-tag rabbit monoclonal antibody. After extensive washings, the bound primary antibody was detected with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody according to the manufacturer’s recommendations (Amersham).
Morphological characterization of transgenic plants
Transgenic plants were characterized for morphological changes under short-day (12h/12h light/dark) photoperiods in a growth chamber with a constant temperature of 22 °C. Root morphology was examined on MS medium solidified with 1.0% agar. Briefly, T3 homozygous transgenic and WT seeds were germinated on MS medium and grown vertically for primary root length measurement (10 d) and determination of the number of lateral roots (14 d). For biomass measurement, transgenic plants and two controls were planted in sieve-like plates filled with mixed soil (vermiculite:humus, 1:1) and cultured under well-watered conditions.
Physiological characterization of TaSnRK2.3 transgenics
To probe the potential effects of TaSnRK2.3 overexpression in Arabidopsis, four transgenic lines, as well as WT and GFP controls, were cultured in soil for specified periods and subjected to salt, drought stress, or no stress to measure abiotic stress-related physiological indices, including relative water loss rate, chlorophyll content, water potential (WP), chlorophyll fluorescence (CF), cell membrane stability, osmotic potential, free proline content, and H+, Na+, and K+ flux rates.
Water loss rates were measured using ten plants each for TaSnRK2.3 transgenic and control plants. Four-week-old plants were detached from the roots and immediately weighed to determine the fresh weight (FW); the plants were then left on a laboratory bench (humidity, 45–50%, 20–22 °C) and weighed at designated time intervals. The proportion of FW loss was calculated on the basis of the initial plant weight. Plants were finally oven dried to a constant dry weight (DW) at 80 °C over a 24h period. Water retention ability (WRA) was measured according to the formula: WRA (%)=(desiccated weight – DW)/(FW – DW)×100 (Xu et al., 2007).
Chlorophyll content was measured with a chlorophyll meter (SPAD 502 Plus; Konica Minolta Sensing). Measurements of chlorophyll content under stress conditions were performed after applying a treatment of 300mM NaCl for 2 d or when moderate drought stress occurred. One measurement for fully expanded leaves was made for each plant, and 20 plants of each line were used for chlorophyll content assays.
CF was measured with a portable CF meter (OS 30P; Opti-Sciences). CF measurements were performed after applying a treatment of 300mM NaCl for 12h or when moderate drought stress occurred. Forty fully expanded leaves were selected to determine the CF parameters. The maximum efficiency of photosystem α (PSα) photochemistry, F v/F m=(F m – F 0)/F m, was deployed to assess changes in the primary photochemical reactions of the photosynthetic potential after exposure to stress.
Plant cell-membrane stability (CMS) was determined with a conductivity meter (Orion 3 Star portable conductivity meter; Thermo Scientific) as: CMS (%)=(1 – initial electrical conductivity/electrical conductivity after boiling)×100. For CMS under salt stress, soil-grown seedlings were exposed to NaCl (300mM) stress from the bottom of the container. When signs of salt stress began to appear on WT plants (after about 20h), the harvested Arabidopsis seedlings were rinsed completely and immersed in 20ml of ddH2O at room temperature for CMS measurement. For CMS under drought stress, measurements were carried out when moderate drought stress occurred (after about 4 weeks) and when symptoms of drought stress were evident.
Osmotic potential (OP) was measured with a Micro-Osmometer (Fiske® Model 210; Fiske® Associates) as described (Mao et al., 2010). Free proline was extracted and quantified from fresh tissues of seedlings (0.5g) as described previously (Hu et al., 1992).
The WP of seedlings was measured with a WP meter (WP4; Decagon Devices). Measurements were taken in dew point mode at room temperature. Arabidopsis plants growing in the same container as described in Materials and methods were selected for WP assays when moderate drought stress occurred. Five plants of each line were collected as a sample for WP measurement.
Ion fluxes were measured non-invasively under conditions of salt shock and salt pre-treatment. For salt-shock treatment, net H+ and K+ fluxes were measured in the root apices of 5-d-old seedlings of WT and TaSnRK2.3 plants. The seedlings were pre-incubated in buffer (0.5mM KCl, 0.1mM MgCl2, 0.1mM CaCl2, 0.2mM Na2SO4, and 0.3mM MES, pH 6.0) for 30min and assayed in the same buffer containing 100mM NaCl at pH 6.0. The transmembrane H+ and K+ fluxes in the roots of TaSnRK2.3 transgenic plants (100 µm to root apex) were compared with that of the WT. For salt pre-treatment experiments, 4-d-old seedlings were grown for 24h on MS medium to which 100mM NaCl had been added and the transmembrane Na+ fluxes were then measured as described. Ionic fluxes were calculated using the Mageflux program developed by the Xuyue Company (http://www.xuyue.net/).
Abiotic stress-tolerance assays
To measure the germination rates of transgenic plants under normal and stressed conditions, same-batch-harvested seeds of homozygous TaSnRK2.3 T3 plants, as well as WT and GFP controls, were sown on MS medium, supplemented with abiotic stress factors, including mannitol (200mM), ABA (0.5 μM), and NaCl (100mM). Germination rates were recorded after 2 weeks; seeds with green euphylla were regarded as germinated.
Drought-tolerance assays were performed on seedlings. After germination on MS plates, 7-d-old seedlings (including transgenics, WT, and GFP control) were planted in sieve-like rectangular plates (3cm deep) fully filled with a soil mixture (vermiculite:humus, 1:1) and well watered. Seedlings were then cultured in a greenhouse (22 °C, 70% humidity, 120 µmol m–2 s–1, 12h/12h light/dark cycle) withholding watering.
Seedlings for salt-tolerance assays were grown as for the drought-tolerance assays. Water was withheld for 3 weeks before being irrigated with NaCl solution (250mM) applied from the bottom. When the soil was completely saturated with salt water, the free NaCl solution was removed and the plants were cultured normally.
For cold-tolerance assays, four 1-week-old seedlings were grown in identical pots and cultured normally as described above. Four weeks later, the seedlings were stressed in a –8±1°C freezer for 1h, and then cultured at 15 °C for 24h to facilitate recovery before growing under normal conditions.
Results
Molecular characterization of TaSnRK2.3
TaSnRK2.3 was obtained by screening full-length wheat cDNA libraries. TaSnRK2.3 cDNA was 1447bp and consisted of a 145bp of a 5′-untranslated region (5′-UTR), 1029bp ORF and 273bp 3′-UTR. The ORF encoded a polypeptide of 342 aa with a predicted molecular mass of 45.6kDa and pI of 5.55. The deduced amino acid sequence showed high homology with counterpart monocot SnRK2 family members, i.e. Oryza sativa and Zea mays, and relatively lower homology with SnRK2s from dicot species, such as Glycine max and A. thaliana. TaSnRK2.3 had 87.4% identity to O. sativa SAPK3 (BAD17999), 86.2% to ZmSnRK2.3 (ACG50007), 75.3% to Ricinus communis (XP_002517501), and a relatively lower similarity (64.0%) to AtSnRK2.3 (NP_201489).
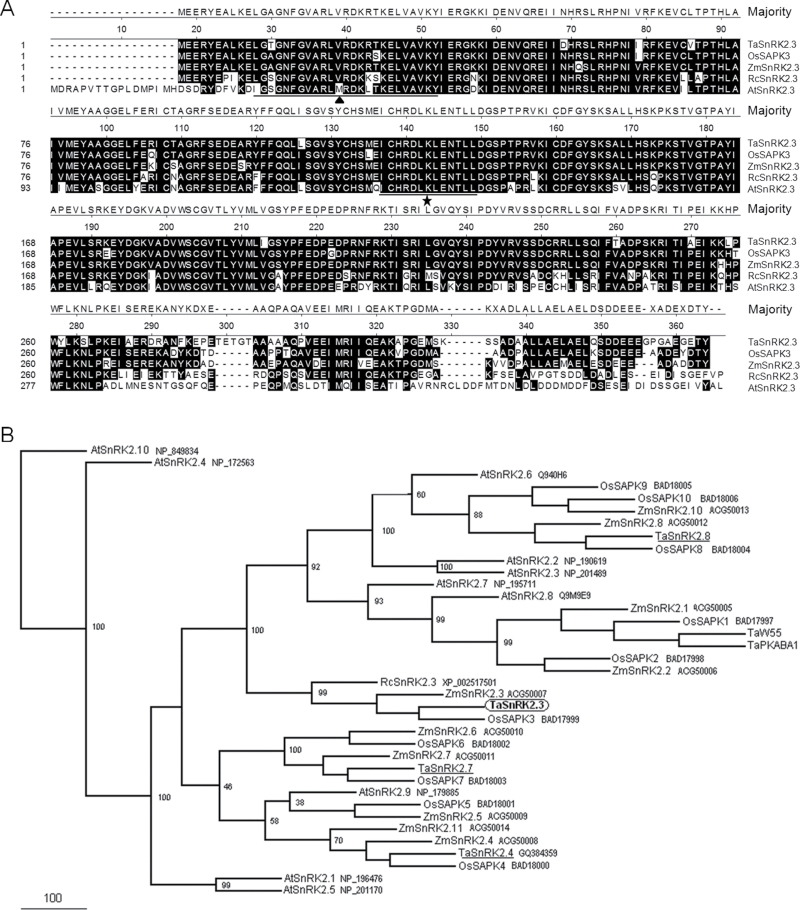
Scansite analysis indicated that TaSnRK2.3 has the potential for serine/threonine and tyrosine kinase activities and, like other SnRK2s, has two domains in its N- and C-terminal regions. The N-terminal catalytic domain (aa 5–261) was highly conserved, containing an ATP-binding site (aa 11–34) and a protein kinase-activating site (aa 120–132) (Fig. 1A). One potential transmembrane helix (aa 184–201) was identified, but no signal peptide was detected. The secondary structure prediction revealed that TaSnRK2.3 protein formed ten α-helixes and nine β-pleated sheets.
Fig. 1.
Sequence alignment of TaSnRK2.3 and SnRK2s in other plant species. (A) Amino acid alignment of TaSnRK2.3 and other SnRK2 family members from selected plant species. Numbers on the left indicate amino acid position. Shared amino acid residues have a black background. Gaps, indicated by dashed lines, were introduced for optimal alignment. The filled triangle indicates an ATP-binding region signature; the a star indicates a serine/threonine protein kinase activating signature. Alignments were performed using the Megalign program of DNASTAR. (B) Phylogenetic tree of TaSnRK2.3 and SnRK2 members from other plant species. At, Arabidopsis thaliana; Os, Oryza sativa; Rc, Ricinus communis; Ta, Triticum aestivum; Zm, Zea mays. The phylogenetic tree was constructed with the full-length amino acid sequences of SnRK2s using the PHYLIP 3.69 package; bootstrap values are in percentages.
A phylogenetic tree was constructed with the full-length putative amino acid sequences of TaSnRK2.3 and SnRK2 subfamily members of Arabidopsis, rice, maize, and wheat. Phylogenetic assays showed that TaSnRK2.3 clustered in the same clade as OsSAPK3, ZmSnRK2.3, and RcSnRK2.3 (Fig. 1B).
Genetic characterization of TaSnRK2.3
To investigate the structure and genomic origin of TaSnRK2.3, 20 accessions of various wheat species were subjected to PCR. The target fragments could be amplified in all 20 accessions (using primer pair GTF/GTR), indicating that there were three copies of TaSnRK2.3 in common wheat, originating from the A, B, and D genomes. The amplified fragments were about 2.7kb, consisting of nine exons and eight introns, with all splicing sites complying with the GT–AG rule. Phylogenetic assays showed that the obtained cDNAs were clustered in the same clade of D genomic fragments, suggesting that the cDNA was the transcript of the D genomic allele in common wheat.
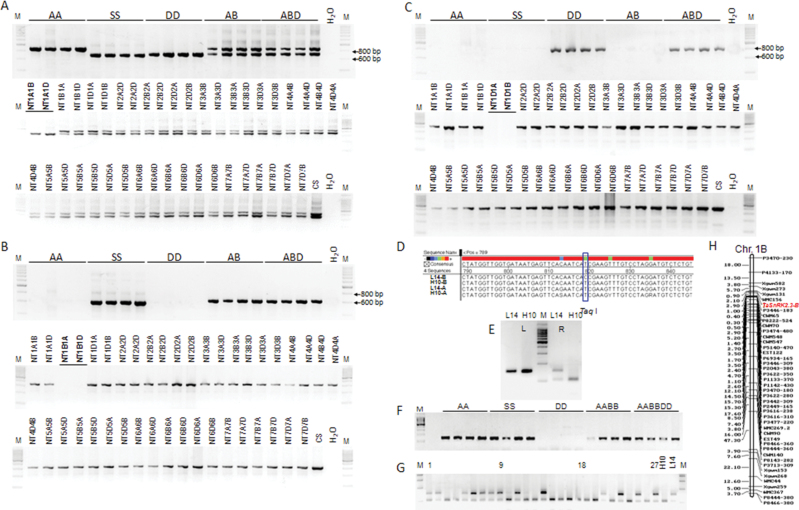
To further probe the chromosome origin of different alleles and simplify the PCR results, three allele-specific primer pairs were obtained. Primer pair AGSF/GSR1 could amplify the target regions of the three alleles in all 20 accessions, whilst the A genomic fragments were evidently longer than those of the other two alleles (Fig. 2A). Primer pair BGSF/GSR2 could specifically amplify the B genomic allele (Fig. 2B), and DGSF/GSR2 the D genomic allele (Fig. 2C). Furthermore, the three alleles were located on chromosomes 1A, 1B, and 1D with a set of nulli-tetrasomic lines of Chinese Spring (Fig. 2A–C).
Fig. 2.
Genetic characterization of TaSnRK2.3 in wheat. (A–C) Chromosome identification of TaSnRK2.3-A (A), TaSnRK2.3-B (B), and TaSnRK2.3-D (C) in common wheat. AA, T. urartu; SS, A. speltoides; DD, A. tauschii; AB, T. dicoccoide and T. dicoccum; ABD, T. aestvium; NT, nulli-tetrasomic lines of Chinese Spring; M, 200bp DNA ladder. (D) SNP of TaSnRK2.3-B between the two parents of the DH population. A SNP (T→C) occurring at 819bp of the B genomic allele of Lumai 14 resulted in the absence of a cutting site for TaqI. (This part is available in colour at JXB online.) (E) A cleaved amplified polymorphic sequence (CAPS) marker was developed with restriction enzyme TaqI. L, before digestion; R, after digestion with TaqI; H10, Hanxuan 10; L14, Lumai 14. (F) The CAPS marker was detected in species with A and/or B genomes. (G) Cleaved amplified sequence polymorphisms identified in the DH population. 1–27, Lines of the DH population, followed by the two parents. (H) The B genomic allele TaSnRK2.3-B was mapped on chromosome 1B flanked by wmc156 and P3346-183.
To identify the chromosome locations of different genomic alleles, a DH population (Hanxuan 10×Lumai 14) was used for single-nucleotide polymorphism (SNP) analysis and gene mapping, and a single-nucleotide polymorphic site was identified at nt 819 of the B genomic allele between the two parents of the DH population (amplified by B allele-specific primer pair GBF/GBR; Fig. 2D and Table S2). The single-nucleotide change resulted in the absence of a TaqI cutting site in Lumai 14. To simplify the restriction digest results, another pair of primers (CAPSF/CAPSR) flanking the SNP was designed to specifically amplify the target regions of the A and B alleles (Fig. 2E and Table S2). From this, a cleaved amplified polymorphic sequence (CAPS) marker was developed (Fig. 2F). Using the DH population, the B genomic allele TaSnRK2.3-B was mapped on chromosome 1B flanked by wmc156 (2.1 cM) and P3346-183 (2.9 cM) (Fig. 2G, H), co-located with a quantitative trait locus controlling total root length and plant height (Liu et al., 2013; Wu et al., 2010).
A scaffold (no. 27623) with the highest identity to the query TaSnRK2.3 genomic sequence was obtained by BLASTn assays with a D genomic sequence database of A. tauschii. Three pairs of gene-specific primers were designed according to the sequence and one pair of primers (PF/PR) that amplified the target promoter sequence in the 20 wheat species accessions was obtained (Table S2). The target fragments were about 2200bp and were designated ProA, ProB, and ProD based on their genomic origins. Cis-acting regulatory element analysis showed the presence of some basic components and stress-responsive element-binding motifs in the three promoters (1500bp), including TATA boxes, CAAT boxes, abiotic stress-response cis-elements, i.e. MYB-binding site (involved in drought inducibility), C-repeat/DRE regulatory element (involved in cold and dehydration response), ABA response elements, and multiple biotic stress response elements, including salicylic acid and MeJA-responsive elements. However, the category, position and number of cis-regulation elements varied remarkably in the three promoters (Table S3 and Fig. S1 at JXB online).
Dynamic expression of TaSnRK2.3 in different tissues and under abiotic stresses
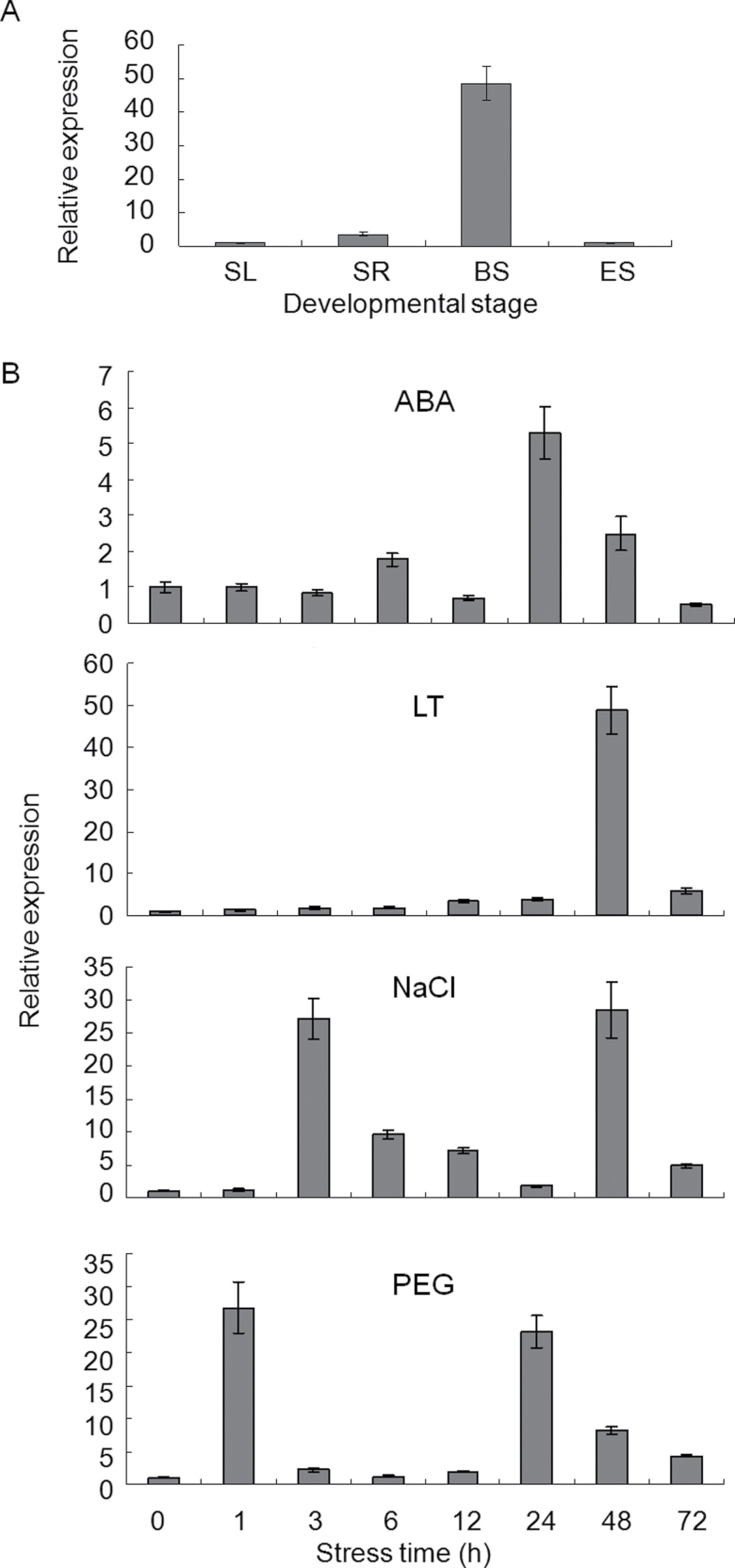
The combined expression patterns of TaSnRK2.3 were identified by qRT-PCR with a pair of primers that could simultaneously amplify the three alleles in common wheat. As shown in Fig. 3A, TaSnRK2.3 was strongly expressed in booting spindles and less so in seedling roots, seedling leaves, and emerging spikes. Various expression patterns were observed under water deficit, salt, low temperature, and ABA treatments (Fig. 3B). TaSnRK2.3 was significantly activated by salt, low temperature, and water-deficit stresses, and relatively weakly by ABA. Among the four stimuli, TaSnRK2.3 was extremely sensitive to PEG stress at an early stage (responding at 1h), followed by NaCl, cold, and ABA. The expression patterns and maximum expression levels differed remarkably for each type of stress. The expression levels peaked at 1 and 24h for PEG, 3 and 48h for NaCl, 24h for ABA, and 48h for cold, and the corresponding maxima were 27 and 23, 27 and 28, 5, and 48 times greater than the control.
Fig. 3.
Expression patterns of TaSnRK2.3 revealed by qRT-PCR analysis. (A) Expression patterns of TaSnRK2.3 in tissues at different developmental stages. SL, seedling leaf; SR, seedling root; BS, booting spindle; ES, emerging spike. (B) Expression patterns of TaSnRK2.3 under ABA, salt (NaCl), PEG, and low temperature (LT) treatments. Two-leaf seedlings of common wheat cv. Hanxuan 10 were exposed to abiotic stresses. The 2–ΔΔCT method was used to measure the relative expression level of the target gene, and the expression of TaSnRK2.3 in non-stressed seedling leaves was used as the control. Means were generated from three independent measurements; bars indicate standard error (SE).
Subcellular localization of TaSnRK2.3
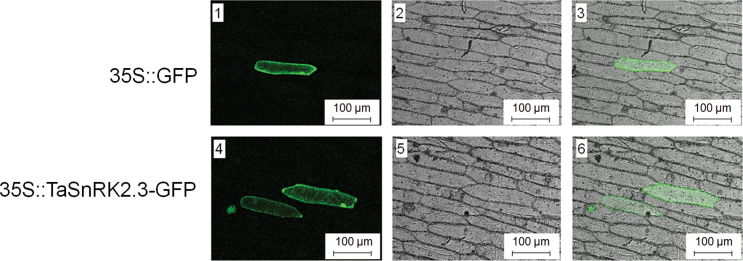
Protein kinases localize to specific cell compartments for proper function, and scanning sequences often specify their intracellular locations. We examined the subcellular distribution of TaSnRK2.3 in onion epidermal cells by transient expression of proteins fused with GFP by fluorescence microscopy. As shown in Fig. 4, TaSnRK2.3–GFP was present in the cell membrane, cytoplasm, and nucleus.
Fig. 4.
Subcellular localization of TaSnRK2.3 protein in onion epidermal cells. Onion epidermal cells were bombarded with constructs carrying GFP or TaSnRK2.3–GFP. GFP and TaSnRK2.3–GFP fusion proteins were transiently expressed under the control of the CaMV 35S promoter and observed with a laser-scanning confocal microscope. Images were taken in a dark field for green fluorescence (1, 4), and the cell outline (2, 5) and ombination (3, 6) were photographed in a bright field. Bar, 100 µm. Each construct was bombarded into at least 30 cells.
Gene expression and protein levels in TaSnRK2.3 transgenics
Six transgenic lines were randomly selected for detection of gene expression, and line 6 was selected to quantify the expression level of TaSnRK2.3 because it had the lowest expression. The expression levels of TaSnRK2.3 varied in different transgenic lines; the highest expression occurred in line 2, followed by lines 4, 5, 3 and 1 (Fig. S2). The four lines with higher expression levels were selected for further analysis. Western blotting assays showed that the protein levels of TaSnRK2.3 in lines 2 and 4 were quite similar, and both were much higher than those in lines 3 and 5, with the lowest protein level occurring in line 3. As shown in Figs S2 and S3 (at JXB online), the protein levels were not exactly consistent with the gene expression levels, although the general trends in gene expression and protein levels were quite similar in the different transgenic lines.
TaSnRK2.3 transgenics have a larger root system
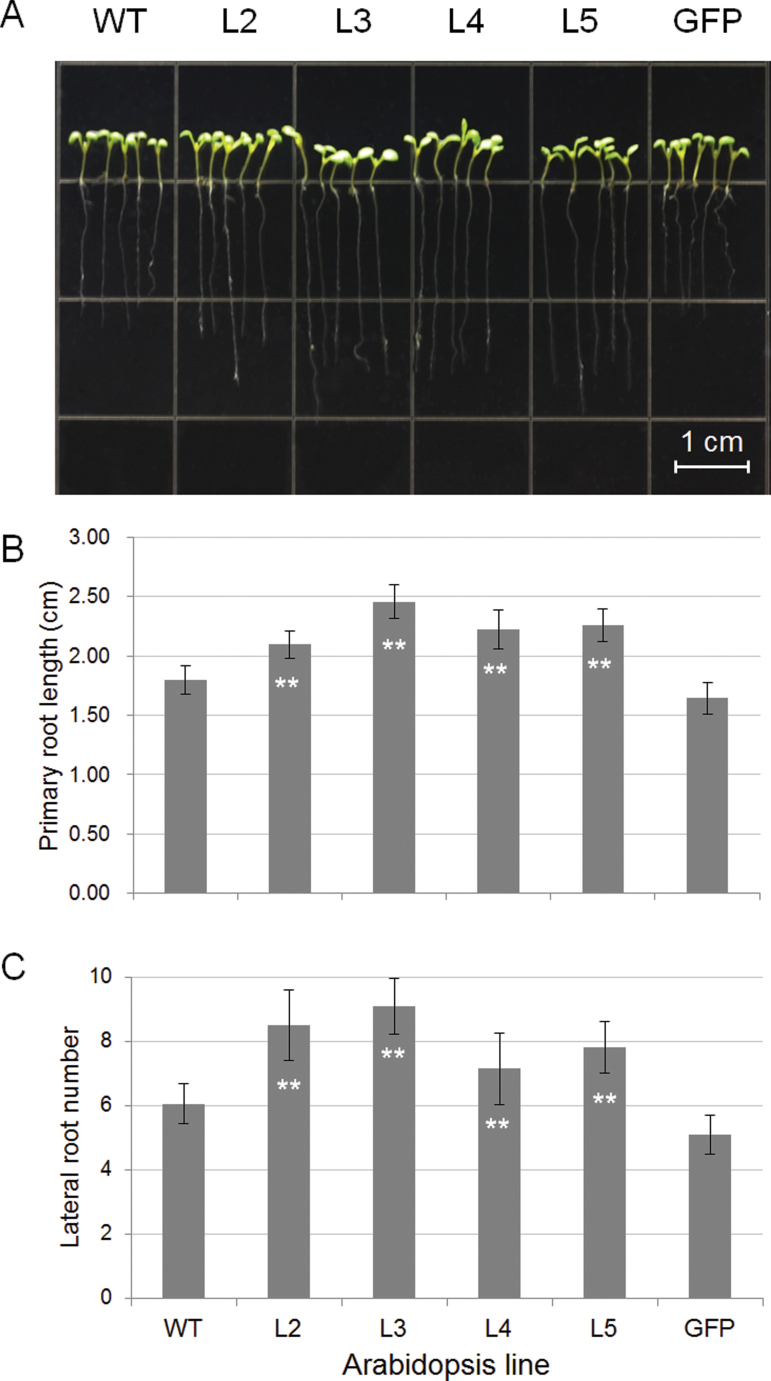
To evaluate the applicability of TaSnRK2.3 in transgenic breeding for abiotic stress tolerance, the phenotypes of TaSnRK2.3 Arabidopsis were characterized at the seedling stage. The primary roots of the transgenics were significantly longer than those of the WT and GFP plants (Fig. 5A, B), and the number of lateral roots was larger than that of the two controls (Fig. 5C). No evident differences were observed between seedlings grown on MS medium and soil. There were also no differences in FW and DW between the transgenics and WT plants (data not shown).
Fig. 5.
Comparison of primary root lengths and lateral root numbers for TaSnRK2.3 plants. Seeds of four TaSnRK2.3 transgenic Arabidopsis lines and two controls were planted on MS agar plates and cultured under short-day conditions. Ten seeds of each line were planted in triplicate and root lengths were measured after 7 d. The primary root lengths are shown in (A). Actual measurements of primary root length and lateral root numbers are compared in (B) and (C). **, Significantly different from WT at P <0.01. Values (and error bars) in (B) and (C) were calculated from 20 plants. (This figure is available in colour at JXB online.)
TaSnRK2.3 transgenics have improved physiological traits for abiotic stress
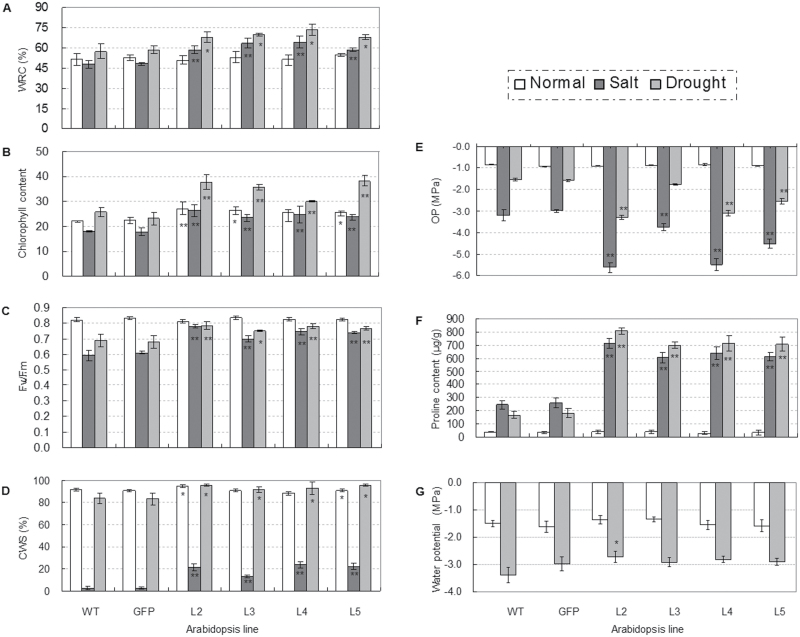
To ascertain the physiological changes in TaSnRK2.3 transgenics, the four selected homozygous T3 transgenic lines, as well as WT and GFP plants, were subjected to physiological assays. Eight physiological parameters related to abiotic stress tolerance were assayed. There was no difference in WRA between transgenic lines and the two controls under normal growth conditions, but the WRA of TaSnRK2.3 transgenics was significantly higher than that of the two controls under salt- and drought-stress conditions (P <0.01; Fig. 6A).
Fig. 6.
Comparisons of physiological indices related to abiotic stress responses for TaSnRK2.3 plants under stress or no-stress conditions. (A–F) TaSnRK2.3 transgenics had a higher water retention ability (WRA) (A), chlorophyll content (B), photosynthetic potential (C), cell membrane stability (CMS) (D), proline content (E), and water potential (F) under drought and/or salt stress conditions. * and ** indicate significant differences at P <0.05 and P <0.01, respectively. WT, wild type; GFP, GFP transgenic line; L2–L5, four TaSnRK2.3 transgenic lines. The values are means ±SE. For (A–C), n=10, n=20, and n=40, and for (E) and (F), n=3. (G) Transgenic TaSnRK2.3 plants had lower osmotic potential (OP). Four TaSnRK2.3 transgenic lines, as well as the WT and GFP plants, cultured under well-watered conditions, were subjected to OP assays. Five plants of each line were collected as a sample; three replications were set for each line. The values are means ±SE (n=3).
Clear differences in chlorophyll content under both normal and salt- and drought-stress conditions were evident. The chlorophyll contents for three of the four transgenic lines were higher (although not significantly) than the controls under normal growth conditions, whereas the chlorophyll content in all transgenic plants was significantly higher than the WT and GFP controls under moderate drought- and salt-stress conditions (P <0.5 or P <0.01; Fig. 6B).
For CF, no differences were identified between the TaSnRK2.3 lines and WT in the F v/F m ratio under normal growth conditions, whereas the ratios for the transgenics were significantly higher than the controls under both drought- and salt-stress conditions, suggesting that stresses to the control plants were more damaging than to the TaSnRK2.3 transgenics (P <0.5 or P <0.01; Fig. 6C).
Under normal growth conditions, the CMS of transgenic plants was higher than that of the controls, and differences for two transgenic lines reached significance levels (P <0.05). Under drought- and salt-stress conditions, the CMS of all transgenic lines was significantly higher than the controls, and the differences reached significance levels for both drought (P <0.05) and salt (P <0.01) (Fig. 6D).
Free proline content was determined under salt and drought stress, as well as under normal growth conditions. No differences were identified under normal growth conditions, whereas the free proline content of all transgenics was significantly higher than that of both controls under salt and drought stresses (P<0.01; Fig. 6E).
For WP assays, there were no differences between controls and transgenics under normal growth conditions, whereas the WP for the transgenic lines was higher than that of the controls under moderate drought stress (when the rosette leaves became dark), and the difference for L2 reached the significance level (P <0.05), indicating that the transgenics probably retained more water than the controls (Fig. 6F).
There was no difference in OP between the transgenics and control plants under normal growth conditions, but the OP of all TaSnRK2.3 transgenic plants was significantly lower than that of the WT and GFP controls under salt and drought stresses, whereas no difference was identified between WT and GFP plants (Fig. 6G).
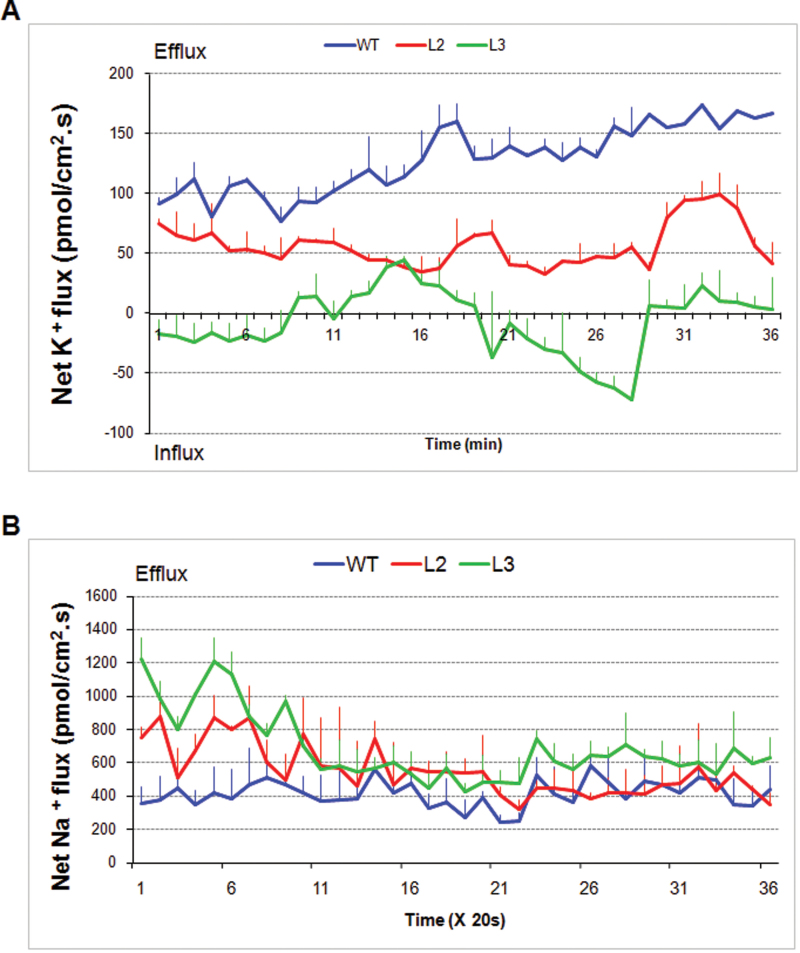
To decipher the mechanism of salt tolerance for TaSnRK2.3 plants, H+ and K+ were measured after a 100mM NaCl shock. Evidently lower K+ ion efflux rates were identified in the transgenic lines compared with in the WT (Fig 7A). However, there was no difference in H+ flux rate (data not shown). To further disentangle the physiological mechanism, Na+ ion fluxes were measured in plants pre-treated with salt. The Na+ ion efflux rates were significantly higher in the transgenic lines than in the WT control at the early stage of measurement (about 5min), but subsequently declined and approached the level of the control. The final Na+ ion efflux rate of L3 remained higher than that in the WT (Fig 7B).
Fig. 7.
TaSnRK2.3 transgenics have stronger K+-retaining and Na+-extruding capabilities relative to WT Arabidopsis. Two transgenics had lower K+ ion efflux rates (A) and higher Na+ ion extruding rates (B) than WT plants. For K+ ion flux rate measurement, the Arabidopsis seedlings were pre-incubated in buffer (0.5mM KCl, 0.1mM MgCl2, 0.1mM CaCl2, 0.2mM Na2SO4, and 0.3mM MES, pH 6.0) for 30min and assayed in the same buffer containing 100mM NaCl. For Na+ ion efflux rate measurement, the Arabidopsis seedlings were pre-treated on MS medium supplemented with 100mM NaCl for 24h. Five plants were measured for each line, and the values are means ±SE (n= 5).
TaSnRK2.3 transgenics have a pronounced tolerance to multiple abiotic stresses
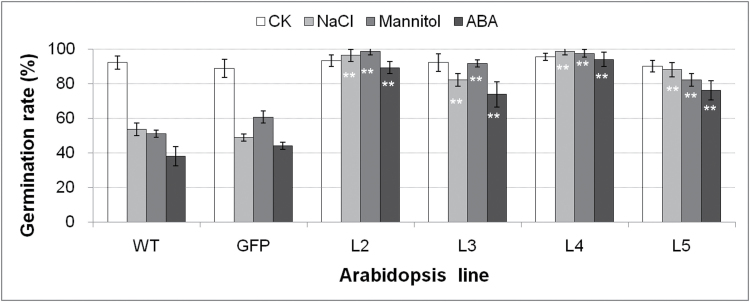
To examine the function of TaSnRk2.3 under osmotic stress, seeds of transgenics and controls were germinated on MS medium supplemented with mannitol, ABA, or NaCl. The germination rates were recorded after 2 weeks. There were no differences in germination rates between the transgenic lines and controls on normal MS medium (no stress), whereas the rates for all transgenics were significantly higher than the controls when exposed to mannitol, NaCl, or ABA stress, suggesting that the TaSnRk2.3 transgenics had enhanced tolerances to multi-abiotic stresses (Fig. 8).
Fig. 8.
Comparison of germination rates of transgenic and control lines under stress conditions. MS, normal MS medium; NaCl, 100mM NaCl; Mannitol, 200mM mannitol; ABA, 0.5 µM ABA; WT, wild-type; GFP, GFP transgenic line; L2–L5, individual transgenic lines. Values are means ±SE (n=3). ** indicates a significant difference at P <0.01.
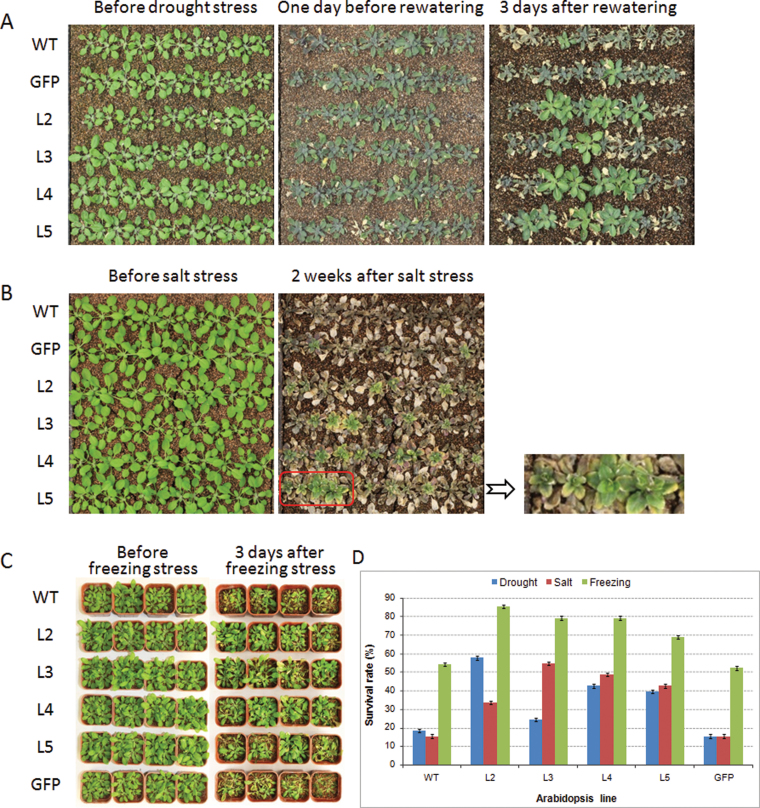
To assess the performance of TaSnRk2.3 plants in soil, seedlings of the transgenic lines were subjected to drought treatments. The lower rosette leaves of WT and GFP plants showed strong wilting after a 40-d water-withholding period compared with much less wilting for the TaSnRk2.3 transgenics. After watering for 3 d, only 15–18% of the WT and GFP plants had survived, compared with 24–57% for the TaSnRK2.3 transformants (Fig. 9A, D).
Fig. 9.
TaSnRK2.3 transgenics have enhanced tolerance to drought, salt, and freezing stress. (A–C) Phenotypes of the four TaSnRK2.3 transgenics and the WT and GFP controls following drought stress (A), salt stress (B), and freezing stress (C). (D) Survival rates of the transgenics and the control plants after abiotic stresses. For drought and salinity stresses, three separate identical plates were used. For freezing stress, normally pot-cultured transgenic seedlings at 4 weeks were divided into three groups, and each group was stressed at –10 °C for 1.5h. Twenty plants (five pots) of each line represented each experiment.
To determine whether TaSnRK2.3 overexpression could enhance tolerance to salt stress, Arabidopsis seedlings grown in soil were exposed to 250mM NaCl solution. The leaf tips of WT Arabidopsis plants began to crimple 20h later, but no evident crimpling was observed on the transgenics. Three days later, the rosette leaves of WT plants began to bleach and after a further 2 weeks, differences between transgenics and controls were quite evident; only 15% of the WT and GFP plants survived compared with 33–54% of the TaSnRK2.3 transformants (Fig. 9B, D).
The survival rates of WT and GFP plants subjected to freezing stress were 52–55% compared with 68–85% for the TaSnRK2.3 transgenics (Fig. 9C, D).
Enhanced expression of abiotic stress responsive genes in TaSnRK2.3 plants
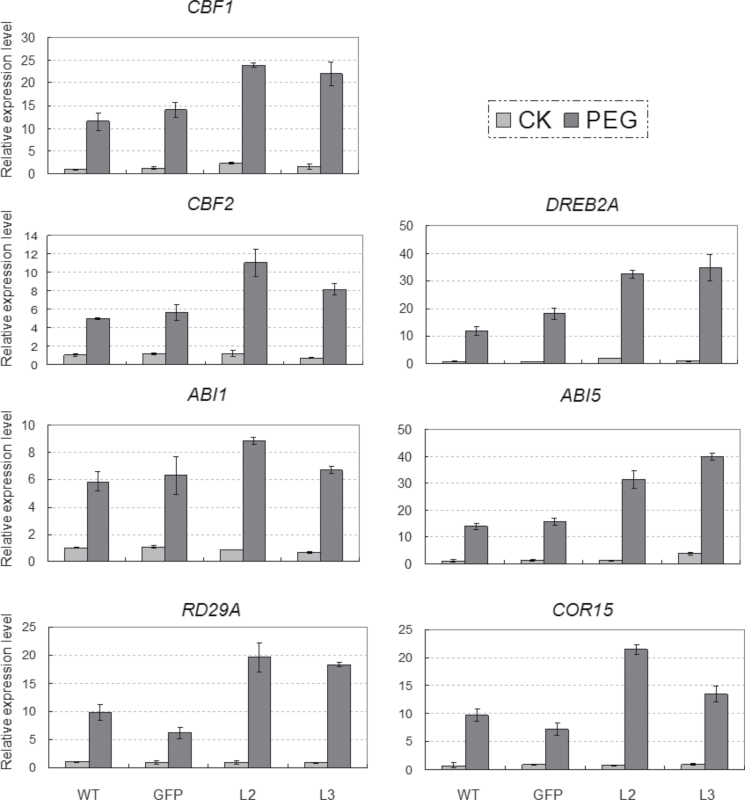
Morphological assays indicated that the TaSnRK2.3 transgenics had enhanced tolerance to drought, salt, and freezing stress. To reveal the underlying molecular mechanisms, transgenic lines L2 and L3 with the highest and lowest expression levels of TaSnRK2.3 among the four lines were selected for expression pattern assays using ten typical abiotic stress-responsive genes, DREB1A, DREB2A, CBF1, CBF2, RD29A, RD29B, RD22, COR15, COR47, and Rab18, and six ABA synthesis or response genes, ABA1, ABI1, ABI2, ABI3, ABI4, and ABI5, under normal and water-deficit conditions. Transcripts of two genes (CBF1 and DREB2A) were consistently and significantly higher in both normal and stressed conditions, whereas expression levels of five genes (CBF2, COR15, RD29A, ABI1,and ABI5) were significantly higher in PEG-stressed plants (Fig. 10). Transcript levels of the other nine genes (DREB1A, RD29B, RD22, COR47, Rab18, ABI2, ABI3, ABI4, and ABA1) were not significantly changed (data not shown).
Fig. 10.
Comparisons of relative transcript levels of CBF1, CBF2, DERB2A, ABI1, ABI5, RD29A, and COR15 in the control plants and TaSnRK2.3 transgenics treated with PEG-6000 (–0.5MPa). Columns indicate relative transcript levels. Ten seedlings were pooled as a sample, and three samples were collected for each line and three duplications were performed for each sample in qRT-PCR. The values (±SE) were calculated from three independent experiments.
Discussion
TaSnRK2.3 possesses typical features of the SnRK2 subfamily
Growing evidence supports a role for the SnRK2 family in response to multi-environmental stresses. Kobayashi et al. (2004) observed upregulation of SAPK3 by ABA in the blades and sheaths and downregulation by mannitol treatment in the roots of rice. Huai et al. (2008) identified a strong response of ZmSAPK3/ZmSnRK2.3 to NaCl in maize seedlings. In this study, we detected expression of TaSnRK2.3 under diverse environmental stresses. Significant differences in transcription levels and response times indicated that TaSnRK2.3 is very sensitive to PEG and NaCl stress (Fig. 3B), and the expression patterns under water deficit and NaCl stress were similar to those of TaSnRK2.4, while they varied significantly compared with TaSnRK2.7–8 (Mao et al., 2010; Zhang, et al., 2010, 2011).
Previous research has indicated that overexpression of TaSnRK2.4 leads to delayed seed germination in transgenic Arabidopsis (Mao et al., 2010). A similar event did not occur in TaSnRK2.3 transgenics, suggesting TaSnRK2.3 might not participate in regulating seed dormancy. Root length assays indicated that TaSnRK2.3 plants had longer primary roots relative to control plants, a result similar to the effects of other SnRK2 family members, including AtSnRK2.4, AtSnRK2.10, TaSnRK2.4, and TaSnRK2.7–8 (Mao et al., 2010; McLoughlin et al., 2012; Zhang et al., 2010, 2011), and suggesting that promotion of root growth might be a common feature of the SnRK2 subfamily. Additionally, the TaSnRK2.3-B locus was co-located with the quantitative trait locus for plant height and total root length (Wu et al., 2010; Liu et al., 2013), suggesting that the gene could be used to improve root systems and biomass in crops subjected to drought.
Physiological changes in transgenic TaSnRK2.3 plants under various conditions
Environmental stresses often cause physiological changes in plants. Indices such as CMS, OP, WRA, chlorophyll content, CF, free proline content, and WP are typical physiological parameters for evaluating abiotic stress tolerance and resistance in crops.
WRA and detached-leaf water loss rate are essential parameters of water status in plants and have been proposed as important indicators of water status (Clarke et al., 1989; Dhanda and Sethi, 1998). WRA is closely related to cell volume and may more closely reflect the balance between water supply to the leaves and transpiration rate (Farquhar et al., 1989). In our work, the detached-leaf water loss rate of TaSnRK2.3 Arabidopsis was lower than that of the the WT and GFP controls, and the WRA for TaSnRK2.3 seedlings was significantly higher than that of the controls (Fig. 6A), strongly indicating that transgenic lines had a higher water-retention ability.
Chlorophyll content, a determination factor for accumulation of biomass and grain yield, as well as an important parameter for assessment of environmental stress resistance, has been widely used in drought-, heat-, and salt-tolerance assays. Significant increases in chlorophyll content were observed in transgenic plants under normal conditions and when subjected to drought and salt stresses (Fig. 6B), revealing that the TaSnRK2.3 transgenics had higher photosynthetic capacities.
CF from intact leaves, especially the fluorescence induction pattern, is a reliable, non-invasive method for monitoring photosynthetic events and reflects the physiological status of the plant (Strasser et al., 2002). The ratio of variable to maximal fluorescence is an important parameter used to assess the physiological status of the photosynthetic apparatus. Environmental stresses that affect PS Ⅱ efficiency are known to cause decreases in the F v/F m ratio (Krause and Weis, 1991). In the present research, significantly higher F v/F m ratios were evident in the transgenic plants (Fig. 6C), clearly indicating that the TaSnRK2.3 transgenics had more robust photosynthetic abilities than the controls under drought- and salt-stress conditions.
Cell membranes are among the first targets of many plant stresses, and the maintenance of membrane integrity and stability under water-stress conditions is a major component of environmental stress tolerance in plants (Levitt, 1980). In most studies, CMS exhibits a positive correlation with water-use efficiency (Franca et al., 2000), stomatal resistance, OP and leaf-rolling index, K+ concentration, and/or WRA (Munns, 2002). In this study, the CMS of TaSnRK2.3 plants under both osmotic and salinity stresses was higher than that of the WT and GFP controls, clearly demonstrating that CMS enhancement was attributable to overexpression of TaSnRK2.3 (Fig. 6D), and predicting that TaSnRK2.3 plants might have a strong capacity for adapting to environmental stresses, as verified by the morphological assay results in Arabidopsis (Figs 8 and 9).
Osmotic adjustment is an essential cell tolerance response to osmotic stress. OP is a direct reflection of osmotic adjustment capability at the physiological level, and has been used as an effective index to screen germplasm for osmotic stress tolerance. Our research indicated that OP in the TaSnRK2.3 transgenics was significantly lower than that in the WT and GFP controls under both drought- and salt-stress conditions (Fig. 6E). Decreased OP is primarily attributed to accumulation of osmoprotectants, including amino acids, quaternary amines, and various sugars. It is well documented that proline is the most widely distributed multifunctional osmolyte, playing important roles in enhancing osmotic stress tolerance (Bartels and Sunkar, 2005). Increases in free proline were detected in the drought- and salt-stressed TaSnRK2.3 plants (Fig. 6F), suggesting that proline was contributing to OP reduction, and that TaSnRK2.3 might be directly or indirectly involved in the pathway of proline metabolism under osmotic stress. Lower OP commonly predicts a higher water retention capacity and a lower rate of water loss, as well as higher water-use efficiency. The results of OP analysis were consistent with the above WRA results (Fig. 6A), and partially explain the enhanced tolerances to drought, salt, and cold stresses.
The ion flux measuring technique provides a unique possibility to link genetic/genomic data and cellular physiological behaviour because of its non-invasive, high spatial and temporal resolution features (Shabala, 2006). NaCl-induced K+ efflux has been demonstrated as a physiological ‘marker’ for salt tolerance in several species, including maize (Pandolfi et al., 2010; Wakeel et al., 2011), barley (Chen et al., 2005, 2007a , b ), wheat (Cuin et al., 2008, 2011) and Arabidopsis (Shabala et al., 2003, 2006). The ability to retain K+ most effectively (i.e. to minimize K+ efflux when Na+ was applied) was strongly correlated with an ability to thrive at high salt concentrations in barley, and K+ flux measurement has been recommended as a screen for salt tolerance in crop species (Chen et al., 2005). In this study, we measured K+ efflux under salt-shock conditions and found that the K+ efflux rate of the TaSnRK2.3 transgenics was significantly lower than that of WT Arabidopsis at the early stage of measurement (Fig. 7A), suggesting that the transgenics had a stronger capacity to retain K+, consistent with the evident salt-tolerant phenotypes of the transgenics (Figs 8 and 9B, D). To further decipher the physiological mechanism of enhanced salt tolerance in the transgenic plants, Na+ ion efflux rates were measured under conditions of salt pre-treatment, and the Na+ efflux rates in transgenics were significantly higher than in the WT control (Fig. 7B). The TaSnRK2.3 plants also had a stronger capacity to extrude Na+ ions. Based on the phenotypes and ion flux measurement results, we propose that the enhanced tolerance to high salinity is mainly attributable to strengthened K+-retaining and Na+-excluding capabilities in the transgenics.
Overexpression of TaSnRK2.3 enhances the multi-environmental stress responses in Arabidopsis
It is well established that the SnRK2 family plays critical roles in the responses to hyperosmotic stress and ABA treatment. Ten SnRK2s have been identified in Arabidopsis, rice, and maize (Boudsocq et al., 2004, 2007; Kobayashi et al., 2004). Several studies have shown that OST1/SnRK2E/SRK2.6 and Vicia faba AAPK are involved in ABA-dependent stomatal regulation (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). Overexpression of AtSnRK2C/AtSnRK2.8 increases the expression of stress-related genes and thus enhances drought tolerance in Arabidopsis (Boudsocq et al., 2004; Shin et al., 2007). Transgenic rice overexpressing SAPK4 has enhanced salt tolerance by regulating genes involved in ion homeostasis and the oxidative stress response (Diedhiou et al., 2008). Transgenic expression of TaSnRK2.4 and TaSnRK2.7–8 confers enhanced tolerance to multiple abiotic stresses (Mao et al., 2010; Zhang et al., 2010, 2011). In this study, overexpression of TaSnRK2.3 led to enhanced tolerance to drought, salinity, and freezing stress, simultaneously supported by morphological and physiological evidence. As far as this point is concerned, TaSnRK2.3 is quite similar to TaSnRK2.4 and TaSnRK2.7–8 in enhancing tolerances to abiotic stresses. However, the involved molecular mechanisms might be different. For instance, TaSnRK2.3 was involved in the regulation of Rd29A, COR15, and DREB2A (Fig. 9), whereas TaSnRK2.8 was involved in the regulation of RD29B, RD20, ABI2, ABI3, and ABI4 (Zhang et al., 2010), although both were involved in upregulation of CBF1, CBF2, ABI1, and ABI5, suggesting the presence of functional diversity between different TaSnRK2 members.
In the present research, morphological and physiological evidence strongly demonstrated that the transgenic TaSnRK2.3 plants acquired strengthened tolerances to multiple abiotic stresses. We speculate that the enhanced tolerances to abiotic stresses are mainly attributable to consistently and/or significantly increased expression of abiotic stress-responsive genes, including CBF1, DREB2A, ABI1, ABI5, COR15, and RD29A. CBF1 encodes an AP2 domain-containing transcriptional activator binding to the low-temperature-responsive element CCGAC, which induces the expression of cold-regulated genes and increases plant freezing tolerance through an ABA-independent pathway (Medina et al., 1999). DREB2A is a crucial regulatory element involved in drought response (Liu et al., 1998). Consistent upregulation of CBF1 and DREB2A undoubtedly increases the expression levels of downstream freezing and drought stress-responsive genes, and enhanced tolerance to drought and/or other abiotic stresses due to widespread ‘cross-talk’ between various environmental stresses (Xiong et al., 1999; Seki et al., 2002). ABI1 encodes a type 2C protein phosphatase involved in ABA signalling (Chak et al., 2000), and ABI5 encodes a basic leucine zipper transcription factor, involved in altering the expression of ABA-regulated genes (Finkelstein and Lynch, 2000). Their high levels of expression probably lead to upregulation of genes controlled by ABI1 and ABI5 in an ABA-dependent pathway, and possibly enhance integrative tolerance to multiple abiotic stresses. Additionally, we witnessed significant increases in expression of RD29A and COR15. These genes encode low-molecular-weight hydrophilic proteins (Yamaguchi-Shinozaki and Shinozaki, 1993; Zhou et al., 2009), and their significant enhancement in transcription undoubtedly leads to increased solute levels in tissue sap, resulting in decreased OP of cells and reduced rates of water loss under stressed conditions. However, the phenotypic data were not exactly consistent with the expression and protein levels of target genes in the transgenics under adverse conditions; we presume this inconsistency might be attributable to the difference in insertion sites for TaSnRK2.3 in the transgenics.
This study was mainly concerned with the morphological and physiological features of TaSnRK2.3 overexpression in Arabidopsis under normal and adverse conditions, as well as the potential molecular mechanisms for dynamic expression patterns of abiotic responsive genes. The results will be helpful in understanding the mechanisms of environmental stresses on plants. Further ongoing research on transgenic wheat will enable us to validate the functions of TaSnRK2.3 in enhancing tolerance to abiotic stresses in crops.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Plant materials used for identification of genomic origins.
Table S2. Oligonucleotides for genetic assays.
Table S3. Comparison of cis-acting elements identified in the promoter regions of TaSnRK2.3s.
Fig. S1. Sequence alignment of the promoter regions for TaSnRK2.3s.
Fig. S2. Gene expression levels of TaSnRK2.3 in different transgenic lines.
Fig. S3. Protein levels for TaSnRK2.3 in transgenic lines.
Acknowledgements
We thank Robert A. McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia) for critical reading and comments on the manuscript. This study was supported by the National Natural Science Foundation of China (31040089), and the National High-tech R&D Program (863 Program, 2011AA100501).
Glossary
Abbreviations:
- ABA
abscisic acid
- CAPS
cleaved amplified polymorphic sequence
- CF
chlorophyll fluorescence
- CMS
cell membrane stability
- DH
doubled haploid
- GFP
green fluorescent protein
- OP
osmotic potential
- PEG
polyethylene glycol
- qRT-PCR
quantitative real-time PCR
- WRA
water retention ability
- SnRK
sucrose non-fermenting 1-related protein kinase
- WP
water potential
- WT
wild type.
References
- Anderberg RJ, Walker-Simmons MK. 1992. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proceedings of the National Academy of Sciences, USA. 89, 10183–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. 2005. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 24, 23–58. [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. 2004. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana . Journal of Biological Chemistry. 279, 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C. 2007. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Molecular Biology. 63, 491–503. [DOI] [PubMed] [Google Scholar]
- Chak RK, Thomas TL, Quatrano RS, Rock CD. 2000. The genes ABI1 and ABI2 are involved in abscisic acid- and drought-inducible expression of the Daucus carota L, Dc3 promoter in guard cells of transgenic Arabidopsis thaliana (L.) Heynh. Planta. 210, 875–883. [DOI] [PubMed] [Google Scholar]
- Chen Z, Newman I, Zhou M, Mendham N, Zhang G, Shabala S. 2005. Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant, Cell & Environment. 28, 1230–1246. [Google Scholar]
- Chen Z, Pottosin II, Cuin TA, et al. 2007. a Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiology. 145, 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhou M, Newman IA, Mendham NJ, Zhang GP, Shabala S. 2007. b Potassium and sodium relations in salinised barley tissues as a basis of differential salt tolerance. Functional Plant Biology. 34, 150–162. [DOI] [PubMed] [Google Scholar]
- Clarke J, Romagosa M, Jana I, Srivastava JP, McCaig TN. 1989. Relationship of excised-leaf water loss rate and yield of durum wheat in diverse environments. Canadian Journal of Plant Science. 69, 1075–1081. [Google Scholar]
- Cohen P. 1988. Protein phosphorylation and hormone action. Proceedings of the Royal Society B: Biological Sciences. 234, 115–144. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Betts SA, Chalmandrier R, Shabala S. 2008. A root’s ability to retain K+ correlates with salt tolerance in wheat. Journal of Experimental Botany. 59, 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S. 2011. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant, Cell & Environment. 34, 947–961. [DOI] [PubMed] [Google Scholar]
- Dhanda SS, Sethi GS. 1998. Inheritance of excised-leaf water loss and relative water content in bread wheat (Triticum aestivum). Euphytica. 104, 39–47. [Google Scholar]
- Diedhiou CJ, Popova OV, Dietz KJ, Golldack D. 2008. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biology. 8, 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D. 2009. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana . Journal of Genetics and Genomics. 36, 17–29. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, Wong SC, Evans JR, Hubic KT. 1989. Photosynthesis and gas exchange. In: Jones HG, Flowers TJ, Jones MB, eds. Plant under stress. Cambridge: Cambridge University Press, 47–69. [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca MGC, Thi ATP, Pimentel C, Rossiello R OP, Zuily FY, Laffray D. 2000. Differences in growth and water relations among Phaseolus vulgaris cultivars in response to induced drought stress. Environmental and Experimental Botany. 43, 227–237. [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J. 2011. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proceedings of the National Academy of Sciences, USA. 108, 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y. 2003. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. Journal of Experimental Botany. 54, 467–475. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annual Review of Biochemistry. 67, 821–855. [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. 2011. Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology. 21, R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Jon JH, Kwak JM, Nam HG. 1997. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopis thaliana . Plant Physiology. 113, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, et al. 2003. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiology. 132, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CA, Delauney AJ, Verma DP. 1992. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proceedings of the National Academy of Sciences, USA. 89, 9354–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, Zhao J, Wang G. 2008. Cloning and characterization of the SnRK2 gene family from Zea mays . Plant Cell Reports. 27, 1861–1868. [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S. 2003. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis . Plant Cell. 15, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Park MJ, Seo PJ, Song JS, Kim HJ, Park CM. 2012. Controlled nuclear import of NTL6 transcription factor reveals a cytoplasmic role of SnRK2.8 in drought stress response. Biochemistry Journal. 448, 353–363. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. 2004. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 16, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GH, Weis E. 1991. Chlorophyll fluorescence and photosynthesis: the basics. Annual Review of Plant Physiology and Plant Molecular Biology. 42, 313–349. [Google Scholar]
- Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. 2011. SnRK2 protein kinases—key regulators of plant response to abiotic stresses. OMICS. 15, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Luan S. 2011. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant, Cell & Environment. 35, 53–60. [DOI] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. 1980. Responses of plants to environmental stresses. In: Water, radiation, salt and other stresses. , vol. II New York: Academic Press, 3–211. [Google Scholar]
- Li J, Wang X, Watson MB, Assmann SM. 2000. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 287, 300–303. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. 2000. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proceedings of the National Academy of Sciences, USA. 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis . Plant Cell. 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li R, Chang X, Jing R. 2013. Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica. 189, 51–66. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JD. 2004. CDPK-mediated signalling pathways: specificity and cross-talk. Journal of Experimental Botany. 55, 181–188. [DOI] [PubMed] [Google Scholar]
- Mao X, Kong X, Zhao G, Jia J. 2005. Construction of a full-length cDNA library of Aegilops speltoides Tausch with optimized cap-trapper method. Acta Genetica Sinica. 32, 811–817. [PubMed] [Google Scholar]
- Mao X, Zhang H, Tian S, Chang X, Jing R. 2010. TaSnRK2.4, an SNF1-type serine-threonine protein kinase of wheat (Triticum aestivum L.) confers enhanced multi-stress tolerance in Arabidopsis . Journal of Experimental Botany. 61, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin F, Galvan-Ampudia CS, Julkowska MM, Caarls L, van der Does D, Lauriere C, Munnik T, Haring MA, Testerink C. 2012. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. The Plant Journal. 72, 436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. 1999. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiology. 119, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. 2002. Comparative physiology of salt and water stress. Plant, Cell & Environment. 25, 239–250. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. 2002. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KE, Saeed NA, Rose RJ. 2006. The stress kinase gene MtSK1 in Medicago truncatula with particular reference to somatic embryogenesis. Plant Cell Reports. 25, 711–722. [DOI] [PubMed] [Google Scholar]
- Pandolfi C, Pottosin I, Cuin T, Mancuso S, Shabala S. 2010. Specificity of polyamine effects on NaCl-induced ion flux kinetics and salt stress amelioration in plants. Plant Cell Physiology. 51, 422–434. [DOI] [PubMed] [Google Scholar]
- Piattoni CV, Bustos DM, Guerrero SA, Iglesias AA. 2011. Nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase is phosphorylated in wheat endosperm at serine-404 by an SNF1-related protein kinase allosterically inhibited by ribose-5-phosphate. Plant Physiology. 156, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, et al. 2002. Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Functional & Integrative Genomics. 2, 282–291. [DOI] [PubMed] [Google Scholar]
- Shabala S. 2006. Non-invasive microelectrode ion flux measurements in plant stress physiology. In: Volkov A, ed. Plant electrophysiology—theory and methods. Berlin: Springer-Verlag, 35–71. [Google Scholar]
- Shabala S, Demidchik V, Shabala L, Cuin TA, Smith SJ, Miller AJ, Davies JM, Newman IA. 2006. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+ -permeable channels. Plant Physiology. 141, 1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Shabala L, Van, Volkenburgh E. 2003. Effect of calcium on root development and root ion fluxes in salinised barley seedlings. Functional Plant Biology. 30, 507–514. [DOI] [PubMed] [Google Scholar]
- Shin R, Alvarez S, Burch AY, Jez JM, Schachtman DP. 2007. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proceedings of the National Academy of Sciences, USA. 104, 6460–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser RJ, Srivastava A, Tsimilli-Michae MI. 2002. The fluorescence transient as a tool to characterize and screen photosynthetic samples. In: Mohanty P, Yunus U, Pathre M, eds. Probing photosynthesis: mechanism, regulation and adaptation. London: Taylor and Francis, 443–480. [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. 2004. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA. 101, 17306–17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeel A, Sumer A, Hanstein S, Yan F, Schubert S. 2011. In vitro effect of different Na+/K+ ratios on plasma membrane H+-ATPase activity in maize and sugar beet shoot. Plant Physiology and Biochemistry. 49, 341–345. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Hirt H. 2001. Plant MAP kinase pathways: how many and what for? Biology of the Cell. 93, 81–87. [DOI] [PubMed] [Google Scholar]
- Wu X, Wang Z, Chang X, Jing R. 2010. Genetic dissection of the developmental behaviours of plant height in wheat under diverse water regimes. Journal of Experimental Botany. 61, 2923–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Zhu JK. 1999. Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis . Plant Physiology. 119, 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Jing R, Mao X, Jia X, Chang X. 2007. A wheat (Triticum aestivum) protein phosphatase 2A catalytic subunit gene provides enhanced drought tolerance in tobacco. Annals of Botany. 99, 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Liu L, Ni Z, Liu P, Chen M, Li L, Chen Y, Ma Y. 2009. W55a encodes a novel protein kinase that is involved in multiple stress responses. Journal of Integrative Plant Biology. 51, 58–66. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Molecular and General Genetics. 236, 331–340. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. 2002. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis . Plant Cell Physiology. 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Zhang H, Mao X, Jing R, Chang X, Xie H. 2011. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. Journal of Experimental Botany. 62, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mao X, Wang C, Jing R. 2010. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis . PLOS ONE. 5, e16401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhou J, Meng L, Wang Q, Xie H, Guan Y, Ma Z, Zhong Y, Chen F, Liu J. 2009. Duplication and adaptive evolution of the COR15 genes within the highly cold-tolerant Draba lineage (Brassicaceae). Gene. 441, 36–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.