Abstract
Previous studies have suggested that certain genetic polymorphisms, specifically the Xeroderma pigmentosum group D (XPD) gene codon 751 and the X-ray repair cross-complementing group 1 (XRCC1) gene codon 399 polymorphisms, were associated with an increased risk of lung cancer, and, in some studies, with a greater risk for mutations in the p53 tumor suppressor gene in lung tumors. To evaluate whether these gene polymorphisms may be associated with an increased risk for bladder cancer or in association with p53 mutation status in bladder tumors, we screened for polymorphisms at XPD codons 751 and XRCC1 codon 399 in DNA isolated from blood of 194 bladder cancer patients and 313 healthy controls and for mutations in exons 4 to 8 of the p53 gene in bladder tumor DNA from 174 bladder cancer patients. There was a significantly higher prevalence of the XPD 751 Gln allele among the bladder cancer group, compared with the control group. No association was found between bladder cancer risk and the XRCC1 399 polymorphism. p53 mutations were found in 20.1% (35/174) patients. There was no difference in p53 mutation status among individuals with different genotypes. These results suggest that individuals who have the XPD 751 Gln allele may be at an increased risk for bladder cancer, although this may not lead to an increased risk for mutations in the p53 gene.
Keywords: bladder cancer, p53 mutations, DNA repair, Xeroderma pigmentosum group D, X-ray repair cross-complementing group 1
Introduction
Cigarette smoking is a primary risk factor for bladder cancer (1–4). Cigarette smoke contains a wide variety of chemical carcinogens, including polycyclic aromatic hydrocarbons, aromatic amines, N-nitroso and other compounds that can form bulky adducts and other types of damage on DNA of urothelial cells after activation by specific drug metabolism enzymes (5). Other metabolism enzymes function to repair damage to restore the integrity of the original DNA (6). This biological process prevents or at least minimizes the possibility that accumulated un-repaired damage irreversibly leads to genetic changes, including point mutations (7). For instance, benzo[a]pyrene can be bioactivated in vivo into benzo[a]-pyrene-diol epoxides (BPDE), which are well-known damaging metabolites and are related to a specific mutational spectrum in the p53 gene (7). Tobacco smoke and tobacco itself increase the production of radical oxygen species (ROS) in cells, resulting in the production of oxidative lesions in DNA. The accumulation of ROS may inflict oxidative DNA damage indirectly, by inactivation of enzymes that are involved in DNA repair, or directly, by generating DNA strand breaks and base damage that can lead to mutations in tumor suppressor genes or oncogenes (8,9).
Inactivation of the tumor suppressor gene p53 by mutations is common in a wide variety of human cancers, including bladder cancer (10). Within bladder tumors, the frequency of p53 mutations is between 15 and 60% (11–14). The origin(s) and mechanism of formation of these mutations in bladder cancer are unclear. However, the association of this cancer with tobacco smoke exposure suggests that some of the p53 mutations found in bladder tumors may derive from DNA damage caused directly or indirectly by tobacco smoke carcinogens. The removal or repair of DNA damage thus plays a key role in protecting the integrity of the genome from the insults of cancer-causing agents. DNA repair gene polymorphisms may result in altered function and/or efficiency of DNA repair, and may contribute to inter-individual variation of DNA repair capacity (6,15,16). Bulky adduct lesions induced by smoking chemical carcinogens are repaired through the nucleotide excision repair (NER) pathway (17). Xeroderma pigmentosum group D (XPD) is involved in the NER pathway by functioning as an ATP-dependent DNA helicase with its 5'->3' activity joint to the basal transcription factor IIH (TFIIH) (18). Several non-synonymous single nucleotide polymorphisms that induce amino acid changes have been found in the XPD gene including codon 751 (Lys/Gln) (19). In addition to NER, damaged bases and DNA single strand breaks can be repaired through the base excision repair (BER) pathway (20). The X-ray repair cross-complementing group 1 (XRCC1) protein is implicated in the BER processes by serving as a molecular scaffold interacting with poly (ADP-ribose) polymerase (PARP), DNA polymerase-β and DNA ligase IIIa (21–26). Multiple polymorphisms in the XRCC1 gene that lead to amino acid substitutions have also been described, including at codon 194 (Arg/Trp), codon 280 (Arg/His) and codon 399 (Arg/Gln) (19). The XRCC1 Gln399 polymorphism resulting in single base substitution may affect binding with PARP, leading to a deficiency of DNA repair (27). Epidemiological studies have indicated that these polymorphisms might modify the risk of bladder cancer (4,28–37).
There have been a number of studies investigating the prevalence of p53 mutations and their role in bladder cancer risk (10–12). A few studies have reported the relationship between polymorphisms of DNA repair genes, specifically XPD and XRCC1, and bladder cancer risk (28,29,31–35,38,39). These studies have suggested that there is an association between XPD and XRCC1 polymorphisms and risk for bladder cancer. Functional changes in repair capacity due to inheritance of certain polymorphisms could increase the chance that adducts produced from tobacco carcinogens resulting in p53 mutations. In this study, we explored the role of the XPD 751 and XRCC1 399 genetic polymorphisms as risk markers and investigated the relationship between these polymorphisms and presence of p53 mutations in bladder cancer in a UK population.
Materials and methods
Subjects and tissue specimens
All bladder cancer cases enrolled were among referrals to the ‘hematuria’ clinic at the Bristol Royal Infirmary in the UK. No patient had a prior confirmed diagnosis of bladder cancer. For study of the relationship between DNA repair polymorphisms and bladder cancer risk we analyzed 194 patients with incident transitional cell carcinoma of bladder from Bristol who had blood samples available for genotyping analysis (Table I). A fraction of these patients also had tumor tissues for p53 mutation analysis. However, only 44 healthy control subjects were available from Bristol. For this reason, 269 hospital-based clinic healthy subjects previously analyzed were also used for comparison (40). Information regarding age, gender, ethnicity, smoking history and other environmental factors was obtained by questionnaire. The 194 bladder cancer patients consisted of 154 males and 40 females, including 44 non-smokers and 150 smokers. The mean age of diagnosis for patients with bladder cancer was 71.2 years (range 39–100). The 44 Bristol controls consisted of 33 males and 11 females and included 19 non-smokers and 25 smokers with a mean age of 52.5 years (range 30–69). The 269 controls from Pittsburgh consisted of 119 males and 150 females, including 156 non-smokers and 113 smokers and had a mean age of 59.9 years (range 27–85). The age, gender distribution, and smoking status of these Pittsburgh control subjects were significantly different from those of the cancer cases from Bristol. They were adjusted for data analysis. Blood samples from all subjects and tumor specimens from bladder cancer patients were obtained with written consent.
Table I.
Genotype frequencies of DNA repair enzyme genes in cases and controls.
| Genotype | Cases | Controls-Bristol | Controls-Pittsburgh | ||
|---|---|---|---|---|---|
| N (%) | N (%) | OR (95% CI) | N (%) | OR (95% CI) | |
| XPD | |||||
| Lys751Lys | 64 (33.0) | 21 (47.7) | 1.0a | 153 (56.9) | 1.0 |
| Lys751Gln + Gln751Gln | 130 (67.0) | 23 (52.3) | 1.9 (1.0–3.6) | 116 (44.4) | 2.7 (1.8–3.9) |
| XRCC1 | |||||
| Arg399Arg | 85 (44.3) | 23 (52.3) | 1.0 | 113 (43.4) | 1.0 |
| Arg399Gln + Gln399Gln | 107 (55.7) | 21 (47.7) | 1.4 (0.7–2.7) | 156 (56.5) | 0.9 (0.6–1.4) |
Reference.
Analysis of p53 mutations and XPD and XRCC1 genotyping
Mutations in exons 4–8 of the p53 gene from 174 patients were analyzed using PCR+SSCP as described previously (41,42). Each mutant allele appearing on the gel was isolated and further characterized by sequencing, using an ABI PRISM 377 automatic sequencer. The XPD 751 Lys/Gln and XRCC1 399 Arg/Gln polymorphisms were analyzed in genomic DNA using the ABI Prism 7700 sequence detector (TaqMan, Applied Biosystems, Foster City, CA) as described previously (43–45).
Statistical analysis
Hardy-Weinberg equilibrium was tested by a goodness-of-fit χ2 test to compare the observed genotype frequencies with the expected frequencies. Logistic regression was used to estimate crude ORs and/or adjusted ORs and 95% CIs. Fisher's exact, and χ2 (Pearson's correlation) were employed to test the difference between cases and controls and the association between genotypes and p53 mutation frequency dichotomized as mutation negative and mutation positive. All statistical tests were two-sided. A p=0.05 was considered statistically significant. Analysis was performed using the STATA 9.0 software for Windows.
Results
Genotype and allele frequencies for XPD and XRCC1 polymorphisms
The genotypes and frequency distribution of alleles among the controls and cases are presented in Table I. There was a marginally significantly higher prevalence of the XPD 751 Gln allele among the bladder cancer group, compared with the Bristol control group (67.0 vs. 52.3%, OR=1.9, 95% CI=1.2–2.5, p=0.065). The significance was more evident as compared to the Pittsburgh control (67.0 vs. 43.1%, OR=2.7, 95% CI=1.8–3.9; adjusted OR=2.0, 95% CI=1.1–3.8, p=0.023), or the combined Bristol and Pittsburgh controls (67.0 vs. 44.4%, adjusted OR=1.9, 95% CI=1.1–3.7, p=0.021). No association was found between bladder cancer risk and either of the XRCC1 399 gene polymorphisms (p>0.05). Furthermore, analysis of the XPD 751 and XRCC1 399 alleles for the 44 controls from Bristol showed no significantly different allele frequencies for these two SNPs as compared to the Pittsburgh Caucasian population. The genotype frequencies were in Hardy-Weinberg equilibrium for both XPD 751 and XRCC1 399 in both the controls and cases (p>0.05), but not for the XPD 751 in the Pittsburgh control population (p<0.01).
p53 mutation in bladder tumors
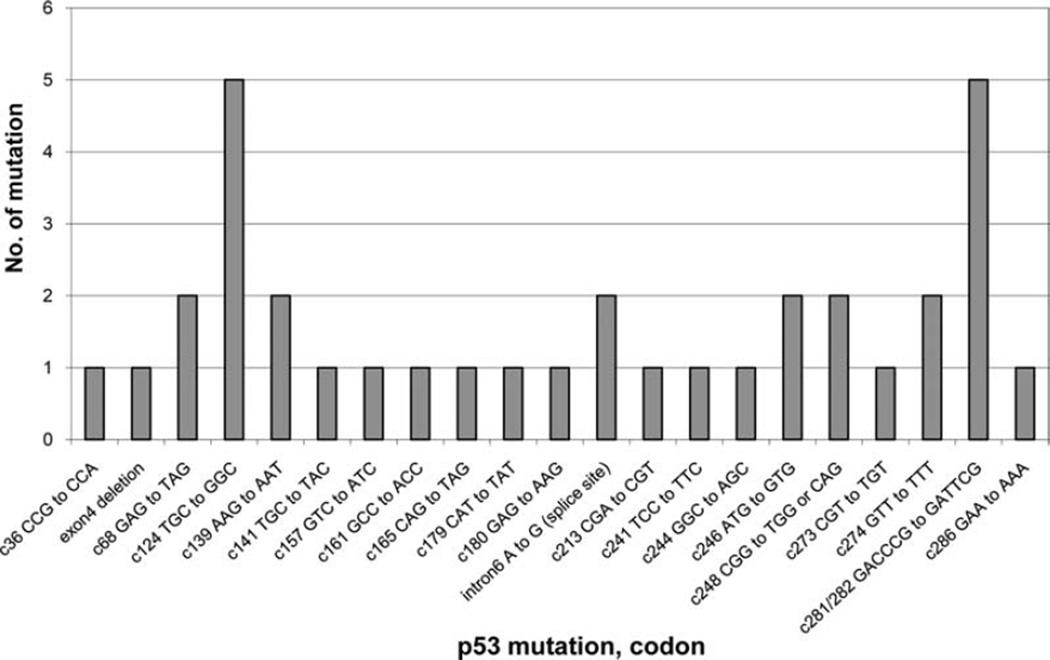
Tumor tissues from 174 bladder cancer patients were examined for mutations in exons 4–8 of the p53 gene. p53 mutations were found in 20.1% (35/174) patients. The distribution of p53 mutations is shown in Fig. 1. The p53 mutational events scattered throughout exons 4–8, with a preference for codons 124 (TGC to GGC) and 281/282 (GACCCG to GATTCG). The majority of p53 mutations was transitions (62.9%, 22/35, including 8 G to A, 5 C to T, 5 CC to TT double transitions, and 4 A to G), followed by 34.3% transversions (12/35, including 6 G to T, 5 T to G, and 1 A to T), and 2.9% (1/35) deletion.
Figure 1.
Distribution of p53 mutations in tumors from 35 of 174 bladder cancer patients.
Characterization of the roles of XPD and XRCC1 polymorphisms in p53 mutations
In order to investigate whether a variation in DNA repair capacity associated with XPD and XRCC1 polymorphisms affects the prevalence of mutations in the p53 gene, we compared the XPD and XRCC1 genotypes with p53 mutation frequencies in bladder tumors in 117 cases that had data available for genotypes and p53 mutations (Table II). Individuals with the XPD Lys751Lys, and Lys751Gln or Gln751Gln had a 13.9% (5/36), and 20.8% (16/77) p53 mutation frequency. Furthermore, individuals containing the XRCC1 Arg399Arg, and Arg399Gln or Gln399Gln had a 16.7% (9/54), and 20.6% (13/63) p53 mutation frequency. No evidence of an association between the presence of p53 mutations and the XPD 751 or XRCC1 399 genotypes was found (p>0.05).
Table II.
XPD and XRCC1 genotypes and p53 mutation.
| Genotypes | p53 mutations | ||||
|---|---|---|---|---|---|
| No. of patients | Frequency (%) | No. of patients | Frequency (%) | P-value | |
| XPD | |||||
| Lys751Lys | 36 | 31.9 | 5 | 13.9 | 0.38 |
| Lys751Gln + Gln751Gln | 77 | 68.1 | 16 | 20.8 | |
| XRCC1 | |||||
| Arg399Arg | 54 | 46.2 | 9 | 16.7 | 0.58 |
| Arg399Gln + Gln399Gln | 63 | 53.8 | 13 | 20.6 | |
Discussion
We investigated the role of polymorphisms in the DNA repair genes, XPD and XRCC1, and the status of p53 mutations in bladder cancer patients, as well as the relationship between these polymorphisms and p53 mutations in a UK population. Our results showed that there was a significantly higher prevalence of the XPD 751 Gln allele among the bladder cancer group, compared with the control groups. No association was found between bladder cancer risk and the XRCC1 399 gene polymorphism. Mutations in p53 were found in 20.1% (35/174) patients, with a majority of transition mutations (62.9%, 22/35). There was no difference in p53 mutation status among patients carrying different XPD 751 and XRCC1 399 genotypes.
In our previous studies, we hypothesized that if there were functional relevance for the polymorphic DNA repair enzymes in the removal of DNA damages, we would detect differences in p53 mutation frequencies in lung tumors of smoking and/or non-smoking lung cancer patients (43,45). We observed a significant association of p53 mutation frequencies and nucleotide excision repair polymorphisms of XPD in smoking non-small cell lung cancer patients (43) and with XRCC1 genotypes in non-small cell lung cancer patients from never-smokers (45). Furthermore, the p53 mutation frequency increased with an increasing number of combined genotypes associated with a lower DNA repair capacity of XPD and XRCC1 399 in lung cancer patients from both smokers and non-smokers (43,45). These results suggested that the presence of certain gene-gene combinations might influence the levels of p53 mutations in lung tumors.
There have been several studies investigating the relationship between polymorphisms of DNA repair genes, specifically XPD and XRCC1 (28,29,31–35,46,47), and the role of p53 mutations (10–12) in bladder cancer. Given the lack of direct functional measures of DNA repair capacity associated with the different polymorphisms, it is difficult to integrate the sometime divergent results of the existing studies on XPD and XRCC1. The meta-analyses for XPD and XRCC1 polymorphisms suggested that XPD 751 and XRCC1 399 polymorphisms might not be associated with bladder cancer susceptibility, except that XRCC1 399 Gln/Gln genotype decreased susceptibility of bladder cancer under recessive model and homozygote contract among ever-smokers (30,36,37). The results from our present study suggest that individuals who have the XPD 751 Gln allele may be at an increased risk for bladder cancer, consistent the results of a previous report (28). Nevertheless, conflicting findings from existing literature regarding XPD and XRCC1 polymorphisms in bladder cancer indicate that further larger studies are required to explore the roles of these polymorphisms in bladder cancer risk.
Only three studies have focused on the relationship between DNA repair polymorphisms and p53 mutations in bladder cancer (48–50). We did not find that the potential for altered repair capacity due to XPD 751 and XRCC1 399 genetic polymorphisms may influence the occurrence of p53 mutations in bladder cancer, consistent with the results of Ryk et al (48) and Sakano et al (49). However, the study by Stern et al suggested that the p53 mutation types in bladder cancer may differ according to the presence or absence of certain DNA repair gene variants (50). For instance, bladder cancer cases with the XRCC1 codon 399 Gln/Gln genotype were positively associated with the presence of p53 transversions while cases with the XPD codon 751 Gln/Gln genotype were positively associated with the presence of p53 transitions, in particular G:C-A:T transitions (50). We did not find any association between certain types of p53 mutations and XPD 751 or XRCC1 399 genetic polymorphisms (data not shown). In addition, compared with the results from our previous lung cancer studies (43,45), we did not find any association between the p53 mutation frequency in bladder tumor and an increasing number of combined genotypes associated with a lower DNA repair capacity of XPD 751, and XRCC1 399 (data not shown). The reasons for the discrepancies between the results of our study and those of Stern et al (50) are unclear. In our study, only 117 bladder cancer cases had data for both p53 mutations and DNA repair gene polymorphisms, which is smaller in comparison with the 139 bladder cancer cases that had both these data in the study by Stern et al (50). Further studies involving a larger number of subjects may help better understand the discrepancies among various studies regarding the relationship between the polymorphisms of these DNA repair genes and p53 mutations in bladder cancer risk.
While many studies showed that p53 mutations were frequently detected in bladder tumors, some studies reported these mutations mostly in the more malignant bladder cancer (51), suggesting they were involved in late events of bladder tumorigenesis. Furthermore, a few studies showed no obvious correlation between the types and/or frequencies of p53 mutations in bladder tumors and the patients' smoking history (52,53). In lines with several previous findings (5,10–13,50,54), the majority of p53 mutations in our study were G:C-A:T transitions (62.9%), followed by transversions (34.3%), and occurred throughout exons 4 to 8. Furthermore, eight of the mutations in our study occurred at the same codons as those reported by Stern et al (50), including a CGG-TGG transition at codon 282 and a CGG-CAG transition at codon 248 that were identified in both studies. The other common mutated codons (codons 141, 161, 244, 248 and 286) were the site for a different transition mutation in each study. In addition, in our study codons 124 (TGC-GGC) and 281/282 (GACCCG GATTCG) were preferentially mutated. The codon 124 mutation had not been reported previously. However, mutations in codons 280 through 287 had been reported in several studies although none of them corresponded to the CC to TT tandem mutation observed in our study (53–56). For instance, Spruck et al (53) found double mutations within the codons 280–287 sequence but in only tumors from current smokers and not in those from non-smokers. Another study reported similar frequencies and major types of p53 mutations in bladder tumors between arylamine-exposed workers and non-exposed workers (54). Overall, while bladder cancer risk has been strongly associated with environmental and occupational exposure, there have been no clear p53 mutational patterns linking bladder cancer with specific carcinogenic exposure. For comparison, our previous studies of lung tumors showed a predominance of G:C-T:A transversion, followed by G:C-A: T transition, many of them occurred at hotspot codons 248 and 249 in lung adenocarcinomas from smokers, suggesting they were primarily induced by adducts caused by polycyclic aromatic hydrocarbons and aromatic amines in tobacco smoke. These mutations were different from those found in lung tumors from non-smokers that consisted predominantly of transitions and were scattered throughout exons 5 to 8 (41,42). Nevertheless, in bladder cancer it has been suggested that the G:C-A:T mutations may be induced by alkyl adducts, such as those caused by exposure to nitrosamines (54). Furthermore, oxygen-free radicals that represent a constituent of tobacco smoke and are also produced endogenously by cellular processes may be bladder carcinogens capable of producing the types of mutations found in bladder tumors (53,57,58).
There are several limitations in our study. First, the number of controls from Bristol was relatively small. Secondly, the use of the Pittsburgh control population was not ideally designed for this study and the Hardy-Weinberg equilibrium indicated a possible selection bias for XPD 751 genotype. Nevertheless, we analyzed these subjects for polymorphisms at XPD codons 751 and XRCC1 codon 399. Our results showed no differences in allele frequencies for the two SNPs between the 44 Bristol control and the 269 Pittsburgh control subjects. Furthermore, the genotype data for the Bristol and Pittsburgh control subjects were comparable with those of other controls from previous studies using European population (31,32,38,39,59). These results are also consistent with those reported for these SNPs for Caucasian population in the database for Single Nucleotide Polymorphism (dbSNP, www.ncbi.nlm.nih.gov/projects/SNP/). Taken together, these results suggest that the use of a control population from either Bristol or Pittsburgh in this study may not affect our data inter-pretation regarding the relationship between XPD and XRCC1 polymorphisms and bladder cancer risk in Bristol. Another limitation is that only a subset of cases (117/194, 60.3%) had data available for both genotypes and mutations. However, this subset of cases seemed to be representative of all the cases in terms of p53 mutation frequency [22/117 (18.8%) vs. 35/174 (20.1%), p=0.78] and distribution of genotypes (p=0.84 for XPD 751; p=0.75 for XRCC1 399). Finally, given the small number of sample size, we were unable to perform combinative analyses of DNA repair polymorphisms, p53 mutations, smoking, and clinical parameters such as grade and stage.
Taken together, these results suggest that, unlike in lung cancer, the presence of p53 mutations is associated with advanced bladder cancer and defines an especially malignant subset of bladder tumors. Nevertheless, one cannot exclude the possibility that some of the p53 mutations found in bladder tumors may result from un-repaired damage induced by tobacco smoke carcinogens and represent early events in bladder tumorigenesis (60,61). Additional studies involving a larger number of bladder cancer patients with various tumor stages will help test this possibility and the association between polymorphisms of these DNA repair genes and the role of p53 mutations in bladder cancer.
Acknowledgements
This study was supported in part by NIH 2RO1 CA59834.
References
- 1.Clavel J, Cordier S, Boccon-Gibod L, Hemon D. Tobacco and bladder cancer in males: increased risk for inhalers and smokers of black tobacco. Int J Cancer. 1989;44:605–610. doi: 10.1002/ijc.2910440408. [DOI] [PubMed] [Google Scholar]
- 2.D'Avanzo B, Negri E, La Vecchia C, et al. Cigarette smoking and bladder cancer. Eur J Cancer. 1990;26:714–718. doi: 10.1016/0277-5379(90)90124-c. [DOI] [PubMed] [Google Scholar]
- 3.Hartge P, Silverman D, Hoover R, et al. Changing cigarette habits and bladder cancer risk: a case-control study. J Natl Cancer Inst. 1987;78:1119–1125. [PubMed] [Google Scholar]
- 4.Silverman DT, Rothman N, Devesa SS. Bladder Cancer. In: Syrigos KN, Skinner DG, editors. Bladder Cancer: biology, diagnosis, and management. New York: Oxford University Press; 1999. p. 11.p. 55. [Google Scholar]
- 5.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 6.Wei Q, Cheng L, Hong WK, Spitz MR. Reduced DNA repair capacity in lung cancer patients. Cancer Res. 1996;56:4103–4107. [PubMed] [Google Scholar]
- 7.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington DC: ASM Press; 1995. [Google Scholar]
- 9.Vineis P, Talaska G, Malaveille C, et al. DNA adducts in urothelial cells: relationship with biomarkers of exposure to arylamines and polycyclic aromatic hydrocarbons from tobacco smoke. Int J Cancer. 1996;65:314–316. doi: 10.1002/(SICI)1097-0215(19960126)65:3<314::AID-IJC6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 11.Husgafvel-Pursiainen K, Kannio A. Cigarette smoking and p53 mutations in lung cancer and bladder cancer. Environ Health Perspect. 1996;104(Suppl 3):S553, S556. doi: 10.1289/ehp.96104s3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannio A, Ridanpaa M, Koskinen H, et al. A molecular and epidemiological study on bladder cancer: p53 mutations, tobacco smoking, and occupational exposure to asbestos. Cancer Epidemiol Biomarkers Prev. 1996;5:33–39. [PubMed] [Google Scholar]
- 13.Ryk C, Berggren P, Kumar R, et al. Influence of GSTM1, GSTT1, GSTP1 and NAT2 genotypes on the p53 mutational spectrum in bladder tumours. Int J Cancer. 2005;113:761–768. doi: 10.1002/ijc.20650. [DOI] [PubMed] [Google Scholar]
- 14.Uchida T, Wada C, Ishida H, et al. p53 mutations and prognosis in bladder tumors. J Urol. 1995;153:1097–1104. [PubMed] [Google Scholar]
- 15.Mohrenweiser HW, Jones IM. Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat Res. 1998;400:15–24. doi: 10.1016/s0027-5107(98)00059-1. [DOI] [PubMed] [Google Scholar]
- 16.Wei Q, Cheng L, Amos CI, et al. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst. 2000;92:1764–1772. doi: 10.1093/jnci/92.21.1764. [DOI] [PubMed] [Google Scholar]
- 17.Benhamou S, Sarasin A. Variability in nucleotide excision repair and cancer risk: a review. Mutat Res. 2000;462:149–158. doi: 10.1016/s1383-5742(00)00032-6. [DOI] [PubMed] [Google Scholar]
- 18.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 19.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 20.Wilson DM, III, Thompson LH. Life without DNA repair. Proc Natl Acad Sci USA. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 23.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 24.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutat Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 27.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 2000;28:2049–2059. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrew AS, Nelson HH, Kelsey KT, et al. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 29.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 30.Lao T, Gu W, Huang Q. A meta-analysis on XRCC1 R399Q and R194W polymorphisms, smoking and bladder cancer risk. Mutagenesis. 2008;23:523–532. doi: 10.1093/mutage/gen046. [DOI] [PubMed] [Google Scholar]
- 31.Matullo G, Guarrera S, Sacerdote C, et al. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 32.Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 33.Stern MC, Johnson LR, Bell DA, Taylor JA. XPD codon 751 polymorphism, metabolism genes, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1004–1011. [PubMed] [Google Scholar]
- 34.Stern MC, Umbach DM, Lunn RM, Taylor JA. DNA repair gene XRCC3 codon 241 polymorphism, its interaction with smoking and XRCC1 polymorphisms, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:939–943. [PubMed] [Google Scholar]
- 35.Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:125–131. [PubMed] [Google Scholar]
- 36.Wang C, Sun Y, Han R. XRCC1 genetic polymorphisms and bladder cancer susceptibility: a meta-analysis. Urology. 2008;72:869–872. doi: 10.1016/j.urology.2007.12.059. [DOI] [PubMed] [Google Scholar]
- 37.Wang F, Chang D, Hu FL, et al. DNA repair gene XPD polymorphisms and cancer risk: a meta-analysis based on 56 case-control studies. Cancer Epidemiol Biomarkers Prev. 2008;17:507–517. doi: 10.1158/1055-9965.EPI-07-2507. [DOI] [PubMed] [Google Scholar]
- 38.Shen M, Hung RJ, Brennan P, et al. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case-control study in northern Italy. Cancer Epidemiol Biomarkers Prev. 2003;12:1234–1240. [PubMed] [Google Scholar]
- 39.Covolo L, Placidi D, Gelatti U, et al. Bladder cancer, GSTs, NAT1, NAT2, SULT1A1, XRCC1, XRCC3, XPD genetic polymorphisms and coffee consumption: a case-control study. Eur J Epidemiol. 2008;23:355–362. doi: 10.1007/s10654-008-9238-2. [DOI] [PubMed] [Google Scholar]
- 40.Buch S, Zhu B, Davis AG, et al. Association of polymorphisms in the cyclin D1 and XPD genes and susceptibility to cancers of the upper aero-digestive tract. Mol Carcinog. 2005;42:222–228. doi: 10.1002/mc.20086. [DOI] [PubMed] [Google Scholar]
- 41.Gao WM, Mady HH, Yu GY, et al. Comparison of p53 mutations between adenocarcinoma and squamous cell carcinoma of the lung: unique spectra involving G to A transitions and G to T transversions in both histologic types. Lung Cancer. 2003;40:141–150. doi: 10.1016/s0169-5002(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 42.Gealy R, Zhang L, Siegfried JM, Luketich JD, Keohavong P. Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol Biomarkers Prev. 1999;8:297–302. [PubMed] [Google Scholar]
- 43.Gao WM, Romkes M, Day RD, et al. Association of the DNA repair gene XPD Asp312Asn polymorphism with p53 gene mutations in tobacco-related non-small cell lung cancer. Carcinogenesis. 2003;24:1671–1676. doi: 10.1093/carcin/bgg115. [DOI] [PubMed] [Google Scholar]
- 44.Gao WM, Romkes M, Siegfried JM, Luketich JD, Keohavong P. No association between the XPD 312, 751, or XRCC1 399 polymorphisms and K-ras gene mutation in smoking non-small-cell lung cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:673–675. [PubMed] [Google Scholar]
- 45.Gao WM, Romkes M, Siegfried JM, Luketich JD, Keohavong P. Polymorphisms in DNA repair genes XPD and XRCC1 and p53 mutations in lung carcinomas of never-smokers. Mol Carcinog. 2006;45:828–832. doi: 10.1002/mc.20208. [DOI] [PubMed] [Google Scholar]
- 46.Narter KF, Ergen A, Agachan B, Gormus U, Timirci O, Isbir T. Bladder cancer and polymorphisms of DNA repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1) Anticancer Res. 2009;29:1389–1393. [PubMed] [Google Scholar]
- 47.Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Influence of XPD and APE1 DNA repair gene polymorphism on bladder cancer susceptibility in north India. Urology. 2009;73:675–680. doi: 10.1016/j.urology.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 48.Ryk C, Kumar R, Sanyal S, et al. Influence of polymorphism in DNA repair and defence genes on p53 mutations in bladder tumours. Cancer Lett. 2006;241:142–149. doi: 10.1016/j.canlet.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 49.Sakano S, Matsumoto H, Yamamoto Y, et al. Association between DNA repair gene polymorphisms and p53 alterations in Japanese patients with muscle-invasive bladder cancer. Pathobiology. 2006;73:295–303. doi: 10.1159/000099124. [DOI] [PubMed] [Google Scholar]
- 50.Stern MC, Conway K, Li Y, Mistry K, Taylor JA. DNA repair gene polymorphisms and probability of p53 mutation in bladder cancer. Mol Carcinog. 2006;45:715–719. doi: 10.1002/mc.20210. [DOI] [PubMed] [Google Scholar]
- 51.Fujimoto K, Yamada Y, Okajima E, et al. Frequent association of p53 gene mutation in invasive bladder cancer. Cancer Res. 1992;52:1393–1398. [PubMed] [Google Scholar]
- 52.Brockmoller J, Kaiser R, Kerb R, Cascorbi I, Jaeger V, Roots I. Polymorphic enzymes of xenobiotic metabolism as modulators of acquired P53 mutations in bladder cancer. Pharmacogenetics. 1996;6:535–545. doi: 10.1097/00008571-199612000-00007. [DOI] [PubMed] [Google Scholar]
- 53.Spruck CH, III, Rideout WM, III, Olumi AF, et al. Distinct pattern of p53 mutations in bladder cancer: relationship to tobacco usage. Cancer Res. 1993;53:1162–1166. [PubMed] [Google Scholar]
- 54.Taylor JA, Li Y, He M, et al. p53 mutations in bladder tumors from arylamine-exposed workers. Cancer Res. 1996;56:294–298. [PubMed] [Google Scholar]
- 55.Berggren P, Steineck G, Adolfsson J, et al. p53 mutations in urinary bladder cancer. Br J Cancer. 2001;84:1505–1511. doi: 10.1054/bjoc.2001.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroeder JC, Conway K, Li Y, Mistry K, Bell DA, Taylor JA. p53 mutations in bladder cancer: evidence for exogenous versus endogenous risk factors. Cancer Res. 2003;63:7530–7538. [PubMed] [Google Scholar]
- 57.Ames BN. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science. 1983;221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- 58.Kalra J, Chaudhary AK, Prasad K. Increased production of oxygen free radicals in cigarette smokers. Int J Exp Pathol. 1991;72:1–7. [PMC free article] [PubMed] [Google Scholar]
- 59.Matullo G, Dunning AM, Guarrera S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 60.Feng Z, Hu W, Rom WN, Beland FA, Tang MS. 4-amino-biphenyl is a major etiological agent of human bladder cancer: evidence from its DNA binding spectrum in human p53 gene. Carcinogenesis. 2002;23:1721–1727. doi: 10.1093/carcin/23.10.1721. [DOI] [PubMed] [Google Scholar]
- 61.Moore LE, Smith AH, Eng C, et al. P53 alterations in bladder tumors from arsenic and tobacco exposed patients. Carcinogenesis. 2003;24:1785–1791. doi: 10.1093/carcin/bgg136. [DOI] [PubMed] [Google Scholar]