Abstract
A growing body of evidence suggests an important role for fibrinogen-like proteins in innate immunity in both vertebrates and invertebrates. It has been shown that fibrinogen-related proteins (FREPs), plasma proteins present in the freshwater snail Biomphalaria glabrata, the intermediate host for the human blood fluke Schistosoma mansoni, are diverse and involved in snail innate defense responses. To gain further insight into the functions of FREPs, recombinant FREP proteins (rFREPs) were produced in Escherichia coli and antibodies (Abs) were raised against the corresponding rFREPs. We first show that most FREP proteins exist in their native conformation in snail hemolymph as multimeric proteins. Western blot analyses reveal that expression of multiple FREPs including FREP4 in plasma from M line and BS-90 snails, which are susceptible and resistant to S. mansoni infection, respectively, is up-regulated significantly after infection with the trematode Echinostoma paraensei. Moreover, our assays demonstrate that FREPs are able to bind E. paraensei sporocysts and their secretory/excretory products (SEPs), and a variety of microbes (Gram-positive and Gram-negative bacteria and yeast). Furthermore, this binding capability shows evidence of specificity with respect to pathogen type; for example, 65–75-kDa FREPs (mainly FREP4) bind to E. paraensei sporocysts and their SEPs whereas 95-kDa and 125-kDa FREPs bind the microbes assayed. Our results suggest that FREPs can recognize a wide range of pathogens, from prokaryotes to eukaryotes, and different categories of FREPs seem to exhibit functional specialization with respect to the pathogen encountered.
Keywords: Fibrinogen-related protein (FREP), invertebrate innate immunity, mollusc, Biomphalaria glabrata, Echinostoma paraensei, Schistosoma mansoni
Introduction
Many human infectious diseases such as schistosomiasis and malaria are caused by parasites that are dependent on invertebrates (e.g. molluscs and arthropods) as the intermediate hosts for perpetuating their life cycles. The freshwater planorbid snail Biomphalaria glabrata (Gastropoda, Planorbidae) is one of the prominent intermediate hosts for the human parasite Schistosoma mansoni, one of the most common causative agents of schistosomiasis. It is estimated that more than 600 million people live in schistosomiasis-endemic areas and that more than 200 million people are still infected.1
Snails like other invertebrates have efficient innate immune systems and one of the potential approaches for controlling schistosomiasis is to exploit these defense responses to block schistosomiasis development and transmission. This depends on an in-depth understanding of the molecular interactions between schistosomes and snails. Although B. glabrata is the most intensively studied schistosome intermediate host, and largely for this reason is the subject of a genome sequencing program, one of the few initiated for any mollusc,2 much remains to be learned about snail innate immunity.
Fibrinogen-related proteins (FREPs) have been identified from the plasma of B. glabrata.3,4 It has been shown that FREPs have a unique molecular structure, with one or two N-terminal immunoglobulin superfamily (IgSF) domain(s) separated from a C-terminal β/γ-fibrinogen (FBG) domain by an interceding region (ICR).5–7
FREPs are of particular interest from an immunological viewpoint because they contain two domains, IgSF and FBG, both of which are involved in innate defense, in both vertebrates and invertebrates. IgSF defense functions are noted with vertebrate antibodies (Abs),8 molluscan defense molecule (MDM),9 amphioxus immunoglobulin-type variable region-containing chitin-binding proteins (VCBPs),10 insect hemolin11 and Down’s syndrome cell adhesion molecule (DSCAM).12,13
With respect to FBG domains, extensive functional studies have been performed on three vertebrate ficolins, L-, H-, and M-ficolin, for which the FBG-like domain comprises a major part.14–16 FBG-encoding genes or molecules have been recently identified in a variety of invertebrates, and almost all are implicated in innate immune-related function although the detailed functions are not fully understood. They include horseshoe crab tachylectin,17 ascidian ficolins,18 FBG-like immunolectins from mosquito species,19–23 sponges,24 and slugs.25 Additionally, recent genome sequencing projects from different invertebrates have revealed that FBG-encoding genes are surprisingly abundant. For example, the on-going genome project for the gastropod mollusc Lottia gigantea (Gastropoda, Lottiidae) reveals at least 70 FBG-encoding genes to be present in its genome (<http://supfam.cs.bris.ac.uk/SUPERFAMILY/cgi-bin/gen_list.cgi?genome=gy>). A comparable number of FBG sequences have been uncovered from mosquitoes.26 More surprisingly, the Florida amphioxus Branchiostoma floridae, a cephalochordate invertebrate, possesses 378 putative genes encoding FBG domain(s) (<http://www.jpi.doe.gov>). The heavy representation of FBG-encoding genes in both deuterostome and protostome invertebrates suggests they have considerable functional significance, and that they will become an important research subject, especially in the context of innate immunity.
It has been shown that snail FREPs belong to a complex gene family. At present, we know that the B. glabrata genome contains about 14 FREP subfamilies,6,27 which can be classified into two types – those with one or two IgSF domains. All FREPs thus far observed have a single FBG-like domain. Additionally, the B. glabrata FBG domain has been found to be associated with non-IgSF domains,27 making FBG-containing molecules even more complicated in this snail species. Our previous study uncovered 42 unique FBG-like sequences from 180 clones sequenced, and all were derived from PCR products amplified from a single M line snail, a snail strain maintained in our laboratory for more than two decades.7 The number of FBG-like sequences in the B. glabrata genome may prove to be comparable to that in the L. gigantea or mosquito genome.
The finding of alternatively spliced forms of FREP transcripts and FREP retrosequences suggests that FREP diversification occurs at both genomic and transcriptional levels.6,7 We have also shown that B. glabrata generates a surprising diversity of FREP genes through point mutation and recombination.28 This along with analogous discoveries from different invertebrate animals has provoked a general reconsideration of the capacity of invertebrates to generate diverse molecules for defense responses.12,13,28–30 Recent studies of invertebrates, jawless vertebrates and higher plants suggest that innate immune systems are much more complex than previously thought. Snail FREPs may serve as one of the candidate molecules used to understand invertebrate immune mechanisms better.8,29,31–33
One of the most important groups of pathogens to infect snails routinely are digenetic trematodes, including schistosomes, like S. mansoni. Another trematode commonly employed in laboratory-based studies is Echinostoma paraensei. Previous studies have suggested functional roles for FREPs in defense against trematodes because they bind to sporocysts, and their encoding genes exhibit up-regulated expression after infection.3 These earlier studies were undertaken before the complex array of FREPs had come to light, and did not employ Abs specific to particular FREPs. More recently, Hertel et al.34 found that FREP genes, as measured by quantitative PCR (qPCR), display complex responses in schistosome-resistant (BS-90 strain) or schistosome-susceptible (M line strain) snails after exposure to trematodes. These previous expression studies were performed at the messenger RNA (mRNA) level, leaving unexplored, for lack of the necessary tools, the responses of individual FREP proteins in snail blood. In general, much of the previous work with B. glabrata–schistosome interactions has been confined to genomic DNA and transcriptional studies due to the lack of specific Abs to probe particular snail proteins and their functions. Some commercially available Abs raised against proteins from model organisms have been employed,3,35,36 as have Abs raised against purified snail blood proteins, but the specific identity of the purified proteins was not known at the time the Abs were raised. For example, Hertel et al.37 produced Abs against affinity purified snail lectins that were likely to be FREPs, though FREPs were unknown at the time.
To overcome these problems, we have developed Abs raised against purified recombinant FREP proteins (rFREPs) which, in turn, were based on specific cDNA sequences cloned from snails. Using these newly-developed Abs, we have addressed several function-related questions, with focus on the B. glabrata–E. paraensei system. E. paraensei normally develops in B. glabrata and has been used as a convenient model to elicit strong FREP responses from this snail.3 Here, we ask the following questions using these antibodies. How do FREP protein levels change following exposure to E. paraensei, the parasite that provokes the strong responses from the snail immune system? Are any differences in response evident between strains of B. glabrata either susceptible or resistant to S. mansoni? Do FREPs bind to potential pathogens of different major categories, including to E. paraensei, to Gram-positive or Gram-negative bacteria, or to the yeast Saccharomyces cerevisiae? Does any of the binding observed exhibit specificity with respect to the categories of organisms bound? By addressing these questions, we gain novel insights into FREP functions.
Materials and Methods
Biological samples
M line and BS-90 strains of the snail B. glabrata are commonly used for snail immunological studies. The two snail strains and digenetic trematodes E. paraensei and S. mansoni were maintained in the laboratory.38 Both trematodes use the snail B. glabrata as an intermediate host for their life cycles. M line and BS-90 snails are susceptible and resistant to S. mansoni, respectively. The Gram-positive bacterium Staphylococcus aureus, the Gram-negative bacterium Escherichia coli, and the yeast S. cerevisiae used in this study were derived from the teaching collection of the Department of Biology, University of New Mexico, USA.
Construction of expression vectors
Full-length cDNAs of FREP2, FREP3 and FREP4 that were cloned from M line snails were used to generate full-length or truncated forms of the corresponding genes. The primers that were used for generating expression constructs are listed in Table 1 and the corresponding restriction enzyme site is indicated in each primer. The expression vector pET23b(+) (Novagen) was used for the construction of expression vectors.
Table 1.
The primers used to generate cDNA sequences for construction of expression vectors
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| FREP2 | pETbF2Ig-F | CTACATATGAGTTCATGGCTAAATTTTACAG |
| pETbF2Ig-R | GTGCTCGAGGTCATCTGTACTTGA | |
| pETbF2FG-F | CTCCATATGTCTTGTCGTGACGTCATC | |
| pETbF2FG-R | GTGCTCGAGGTTTAGCTCTATTTCTCTAATT | |
| FREP3 | pETbF3Ig1-F | CAGGCTAGCGGTTCAGAACTTGTCATAGA |
| pETbF3Ig1-R | GTCCTCGAGTAAAGCGGTCGAGTTTGTTT | |
| pETbF3Ig2-R | GTCCTCGAGCAAATCAAACGTTGGTC | |
| pETbF3FBG-F | CTCGCTAGCTCTTGTCGTGACGTCAACA | |
| pETbF3FBG-R | GTGCTCGAGGATTTCACGGATTTTCATCTCGGTA | |
| FREP4 | pET23bF4Ig-F | CAGGCTAGCTTGTCTTTTAATGCAAATGTG |
| pET23bF4Ig-R | AGCCTCGAGAAGATCAGCAATCTCTAGTTTC | |
| pET23bF4FG-F | CAGGCTAGCTCCTGTCGTGACGTTATCAG | |
| pET23bF4FG-R | AGCCTCGAGGTCAATTTCTCTTATTTTTATTTC |
Sequences underlined in the primers CATATG, GCTAGC and CTCGAG are restriction enzyme sites for Nde I, Nhe I and Xho I, respectively. Those restriction enzyme sites are also present in the pET23b(+) vector. The letter F or R at the end of each primer name indicates the primer is forward (F) or reverse (R). The size of peptide to be expressed and primer combination used for generation of the peptide are provided in the caption to Figure 1.
PCR products that contained restriction sites and the pET23b(+) vector were cleaved by the appropriate restriction enzymes at 37°C overnight (see Table 1), and run on a 1% agarose gel to separate the expected PCR products. The expected PCR band and pET23b(+) vector were cut from the gel and purified using QIAquick gel extraction kit (Qiagen). The purified PCR product that contained an open reading frame (ORF) with a restriction site at either end was then ligated to the pET23b(+) vector (Novagen). The resultant construct contained a conventional six-histidine (6xHis) sequence at the C-terminus. The expression vector was transformed into the E. coli BL21(DE3) strain, or the toxin-resistant BL21(DE3)pLysS strain (Novagen). After confirming by sequencing that the vector contained the correct ORF sequence, expression was performed as below.
Expression and purification of rFREP proteins
To optimize expression conditions for each protein, expressions in small scale were conducted prior to large scale expressions. The expression clones carrying the coding sequence of interest were cultured in 2 ml Luria–Bertani (LB) medium at 37°C until A600= 0.6–1.0. Expression was induced by addition of iso-propyl-β-1-D-thiogalactopyranoside (IPTG) at different concentrations (0.5–1.0 mM) for various times (2–6 h) at 37°C. Protein expression was assessed on a pre-cast SDS-PAGE gel (10–20% acrylamide gradient; BioRad) under reducing condition using 10 μl of a total bacterial lysate, and further confirmed by Western blot using an anti-6xHis monoclonal Ab (details of Western blotting are presented below).
All expressed polypeptides were tagged with a C-terminal 6xHis that facilitated subsequent purification using Ni-NTA agarose (Qiagen). Prior to accumulation of a given recombinant protein, a small aliquot of purified protein was used for size verification using Western blotting. Once the purified protein was accumulated, it was sent to Zymed Laboratories Inc. (Invitrogen) for generation of Abs in rabbits. The specificity of each Ab was tested by the company using ELISA method, and further testing was performed in our laboratory as described in the Results section.
Preparation of plasma samples from snails after exposure to trematode parasites
M line or BS-90 snails (6–8 mm shell diameter) were exposed individually to 20 miracidia of E. paraensei or S. mansoni. At 2- or 4-days post-exposure, snails exposed to trematodes were bled by cardiac puncture to collect hemolymph. Plasma was separated from hemocytes by centrifugation (1000 g for 5 min). For the expression studies, plasma samples were obtained from individual snails at different days post-exposure. For the binding assay, plasma from ~60 snails was collected at 4 days post-exposure to E. paraensei.
Preparation of snail plasma samples after injection with microbes
Gram-positive and Gram-negative bacteria were cultured in LB medium overnight. Yeast were grown in yeast medium (10 g Bacto yeast, 20 g Bacto peptone, 20 g dextrose, final volume after adding water 1000 ml; for making agar plates, 20 g agar was added to 1000 ml medium) overnight. After collection, microbes were used for the binding assay or suspended in phosphate buffered saline (PBS; all PBS used in this study was adjusted to 50% strength of mammalian PBS, to be suitable for snails) for injection (5 μl volume for injection). Snail injection was performed according to our established procedure.39 At the same time, a small amount of each type of microbe was diluted 10-, 100-, and 1000-fold and plated onto agar LB plates to determine the number of microbes injected. In this study, the number injected per snail for Staph. aureus, E. coli and S. cerevisiae was estimated to be 1.2 × 106, 1.5 × 106 and 4.4 × 104, respectively. For each treatment, plasma was collected from ~60 snails at 4-days post-injection.
Collection of sporocysts and their secretory–excretory products from E. paraensei
In vitro transformation of E. paraensei sporocysts was performed according to Adema et al.40 Briefly, E. paraensei eggs collected from adult worms were incubated for 3–4 weeks at 27°C, washed in the 50% (v/v) 199-medium (Sigma) containing (4% gentamycin + 1% penicillin) 7 times and finally washed with 50% 199-medium (4% gentamycin). The eggs were then exposed to light and allowed to hatch into miracidia. Miracidia together with culture media were transferred into tubes and incubated at 27°C for 2–4 days. About 100–200 eggs were incubated in 500 μl medium. Secretory–excretory products (SEPs) were collected and filtered by Utrafree-MC centrifugal filter units (Millipore) to remove debris. SEPs were further concentrated (30-fold) using Centricon-3 filters (retaining molecules > 3 kDa; Amicon). Sporocysts were rinsed three times with 50% (v/v) 199-medium containing 4% gentamycin before use in binding assays and three times after the assays.
Binding of FREPs from snail plasma to E. paraensei secretory–excretory products
For all binding experiments, 40-μl aliquots of plasma obtained from a pool of plasma derived from ~40 snails (either controls or subject to different challenges) were used. In the experiment in which snail plasma and SEPs were mixed, 40 μl plasma was mixed with 8 μl SEPs (30-fold concentrated), and the mixture was kept for 2 h at room temperature. The pellet formed by subsequent centrifugation at 13,000 g for 15 min was transferred to a clean Eppendorf tube, dissolved in 40 μl gel sample buffer and loaded onto a 5–20% gradient SDS-PAGE gel, the type of gel used throughout all experiments in this study.
Binding of plasma FREPs to E. paraensei sporocysts
About 150 sporocysts suspended in 40 μl half-strength 199-medium were added to 40 μl plasma, mixed and kept at room temperature for 2 h. The sporocysts were gently pelleted (1000 g for 5 min), and unbound plasma supernatant was removed. After washing with half-strength 199-medium (containing gentamycin) 3 times, all sporocysts were transferred into a tube and mixed with 40 μl gel sample buffer, boiled and an aliquot of this solution was loaded onto the gel.
Binding of plasma FREPs to three types of microbes
Each type of microbe was grown to mid-log phase in culture media overnight, and collected by centrifugation. Normally, 80 μl of the resultant medium was collected, suspended in PBS, and washed 3 times with PBS. Bacteria suspended in 40 μl PBS were mixed with 40 μl plasma. The mixture was kept at room temperature for 2 h and washed 3 times with PBS. The washed sample was dissolved in 40 μl sample buffer in preparation for electrophoresis.
Western blotting
Since FREPs are plasma proteins, and no housekeeping secretory proteins or antibodies are available for the snail, an equal volume of plasma (2 μl) that was diluted 10-fold in Laemmli sample buffer (BioRad) was loaded into each lane in all experiments. For reducing conditions which were used unless otherwise specified, 2-mercaptoethanol (2%) was present in sample buffer. Diluted samples (10 μl) and 5 μl of protein molecular markers were loaded onto separate lanes of a SDS-PAGE gel (5–20%). Following electrophoresis, separated bands were transferred to a nitrocellulose membrane at 75 V for 1 h. Blots were washed with TBS twice, and then blocked with 1.5% gelatin (dissolved in TBS). After washing with TBS with 0.05% Tween 20, blots were exposed to an anti-rFREP primary Ab (1/5000 dilution for anti-rFREP4 and anti-rFBG Abs) overnight at 4°C, and then, following rinsing to remove primary Ab, placed in anti-rabbit IgG Ab conjugated to alkaline phosphatase (Sigma; secondary AB, 1/500 dilution) at room temperature for 1 h. Finally, the blot was developed in a solution containing 0.3 mM nitroblue tetra-zolium and 0.4 mM bromo-4-chloro-3-indoyl phosphate.37
Proteomics analysis
To confirm the specificity of antibodies generated, a pro-teomics approach was applied. After running SDS-PAGE gel electrophoresis, the protein bands at expected sizes were cut and sent to the University of New Mexico Mass Spectrometry Facility (<http://hsc.unm.edu/envirohealth/Facility>). Peptide mass spectra were obtained on a MALDI-TOF/TOF mass spectrometer (4700 Proteomics Analyzer, Applied Biosystems). Spectra were processed and analyzed by the Global Protein Server Workstation (Applied Biosystems), which uses internal Mascot (Matrix Science, UK) software for searching the peptide mass fingerprints and MS/MS data. Searches were performed against the NCBI non-redundant protein database.
Data collection and analysis
For quantitation of a particular protein of interest, intensity of the protein band on the Western blot was quantified densitometrically using Kodak Molecular Imaging Software (v.4.0.3). An area covering the full width of all visible bands was selected for each lane to determine the relative density of the lane. Net intensity was measured as a way to correct for background, and the net intensity value of non-exposed control lanes was set to a value of 1. All lanes for exposed snails were then compared to this value.
After the relative intensity from individual snail plasma samples was obtained, one-way ANOVA analysis using post-hoc multiple comparisons was performed using SPSS (v.13.0). Data are represented as mean ± SE, and difference was considered statistically significant at P < 0.05.
Results
Expression and purification of recombinant FREP proteins (rFREPs)
While attempting to express rFREPs, we discovered that only the IgSF domains of all three FREPs could be expressed in the normal BL21(DE3) bacterial strain. Both isolated FBG domains and full-length FREPs bearing FBG domains were not expressed. We later transformed the same vectors into BL21(DE3)pLysS, a toxin-resistant strain, which expressed both FBG domains as well as full-length FREPs successfully. This suggests that the FBG domain of FREPs may have antimicrobial properties. Using a bacterial expression system, we have produced 10 rFREPs, including truncated forms from the best known FREPs, FREP2, FREP3, and FREP4 (Fig. 1A, B). These are important reagents to enable us to explore FREP biology further.
Fig. 1.
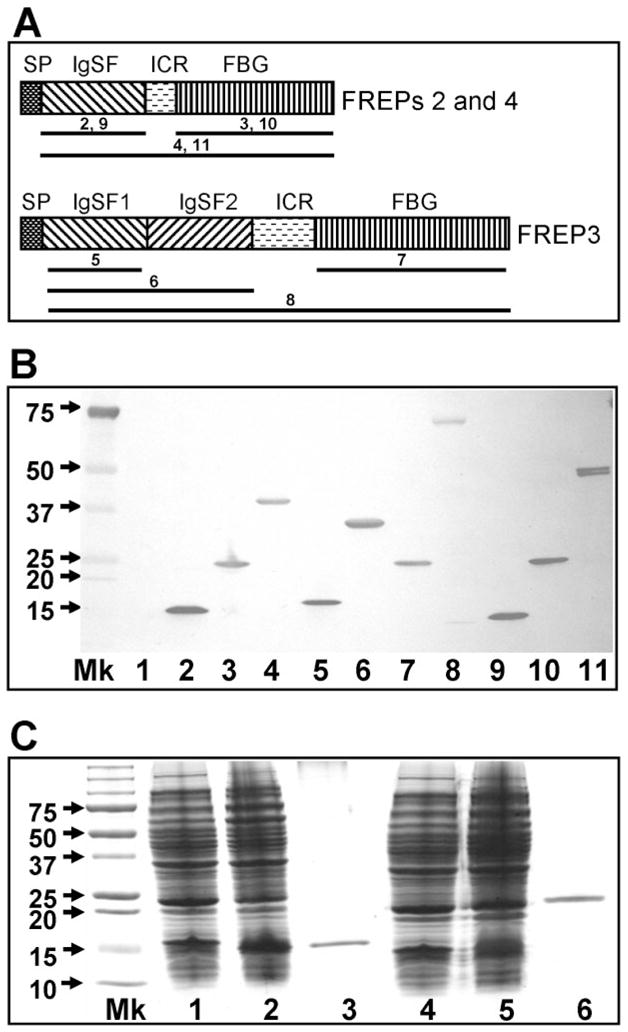
In vitro expression of full-length and truncated forms of FREPs. (A) Schematic diagram of gene products of FREPs with one IgSF domain (FREPs 2 and 4) or two IgSF domains (FREP3). SP, signal peptide; ICR, interceding region; IgSF, immunoglobin superfamily; FBG, fibrinogen. The regions underlined for each FREP were cloned into vectors and expressed in E. coli. The number above the line represents the number of the gel lane in (B). The diagrams are not drawn to scale. (B) Western blot showing expression of 10 rFREP proteins including truncated forms. Blots were probed with anti-6xhis monoclonal antibody because all rFREPs are tagged with 6 histamines at the C-terminus. Mk, protein marker; Lane 1 is loaded with control bacteria lacking expression constructs. Lanes are numbered as follows: (2) rIgSF-FREP2 (142 aa; pETbF2Ig-F+pETbF2Ig-R), (3) rFBG-FREP2 (210 aa; pETbF2FG-F+pETbF2FG-R), (4) rFREP2 (374 aa; pETbF2Ig-F+pETbF2FG-R), (5) rIgSF1-FREP3 (141 aa; pETbF3Ig1-F+pETbF2Ig1-R), (6) rIgSF1+2-FREP3 (301 aa; pETbF3Ig1-F+pETbF3Ig2-R), (7) rFBG-FREP3 (212 aa; pETbF3FG-F+pETbF3FG-R), (8) rFREP3 (648 aa; pETbF3Ig1-F+pETbF3FG-R), (9) rIgSF-FREP4 (141 aa; pETbF4Ig-F+pETbF4Ig-R), (10) rFBG-FREP4 (266 aa; pETbF4FG-F+pETbF4FG-R), and (11) rREPF4 (408 aa; pETbF4Ig-F+ pETbF4FG-R). The size of the peptide generated and the primer combination used for generation of the peptide are provided in parentheses. Primer sequences are listed in Table 1. (C) SDS-PAGE gel showing the expression and purification of IgSF and FBG domains of FREP4 from E. coli stained by Coomassie blue. Lane Mk is protein size markers; Lanes 1 and 4 are proteins expressed by E. coli lacking expression vectors; Lanes 2 and 5 are proteins expressed by bacteria transformed with expression vectors (lane 2, IgSF vector; lane 5, FBG vector); Lanes 3 and 6 show the purified rIgSF and rFBG proteins obtained using the nickel column.
In this study, we have concentrated on providing a broad overview of multiple FREPs, and on FREP4 as a representative FREP. The reasons for singling out FREP4 are as follows. The first FREP sequence to be generated, based on information derived from actual peptide sequence, was FREP4.3,5 A recent qPCR study showed that FREP4 responds strongly to trematode infection34 and our experience with FREP4 suggests that it is the FREP subfamily that is most consistently responsive to trematode infection. As demonstrated by previous studies, the IgSF regions of individual FREPs are more divergent than the relatively conserved FBG domain,7,40 so we probed FREP4 responses and functions using the Ab raised to the rIgSF of FREP4. To assess responses and functions of a broad repertoire of FREPs, the Ab raised against the rFBG domain of FREP4 was used as a probe. This also allowed us to compare the two Abs raised from recombinant proteins derived from same gene, but from different regions. Figure 1C shows the successful expression and purification of rIgSF and rFBG of FREP4.
The rIgSF and rFBG of FREP4 were purified using a nickel-chelating resin column and polyclonal Abs were produced commercially. The specificity of the Abs generated was checked by comparing the molecular weights of bands bound on Western blots with the known or predicted sizes of FREPs. Under the same experimental conditions (same amount of plasma and exposure time), no positive bands could be seen on Western blots when pre-immune serum was used as source of the primary antibody. Snail plasma proteins of 65–75 kDa, which can precipitate E. paraensei SEPs and that are known to be FREP4,3,5 were bound by the anti-rIgSF-FREP4 Ab (called anti-FREP4 Ab hereafter). Our recent proteomics analysis confirmed that the 65–75 kDa band was FREP4. FREP2 which has also been directly identified as a band of ~50 kDa was not bound by the anti-FREP4 Ab (not shown). Regarding the anti-rFBG-FREP4 Ab (called anti-FBG Ab hereafter), as this Ab was expected to detect all potential FREPs, it is difficult to characterize the specificity fully. However, as expected, a wide range of bands was revealed by the Ab raised by rFBG-FREP4, including FREP4 and FREP2. This pattern is broadly supported by Southern blot studies which revealed FBG-encoding molecules to be abundant in the snail genome (IgSF and FBG).7
Most FREP proteins are multimeric proteins
To determine whether FREP polypeptides aggregate into high molecular-weight multimers, we first used the anti-FBG Ab to probe Western blots of snail plasma obtained from M line or BS-90 snails under both reducing and non-reducing conditions (Fig. 2A). In addition to the bands observed under reducing condition, multiple bands with high molecular-weights have also been observed, suggesting that most FREPs form multimeric proteins in snail hemolymph. It is also possible that other large proteins are present, and share antigenic determinants for this antibody, but the diffuse nature of these higher molecular-weight bands suggests they are FREPs.
Fig. 2.
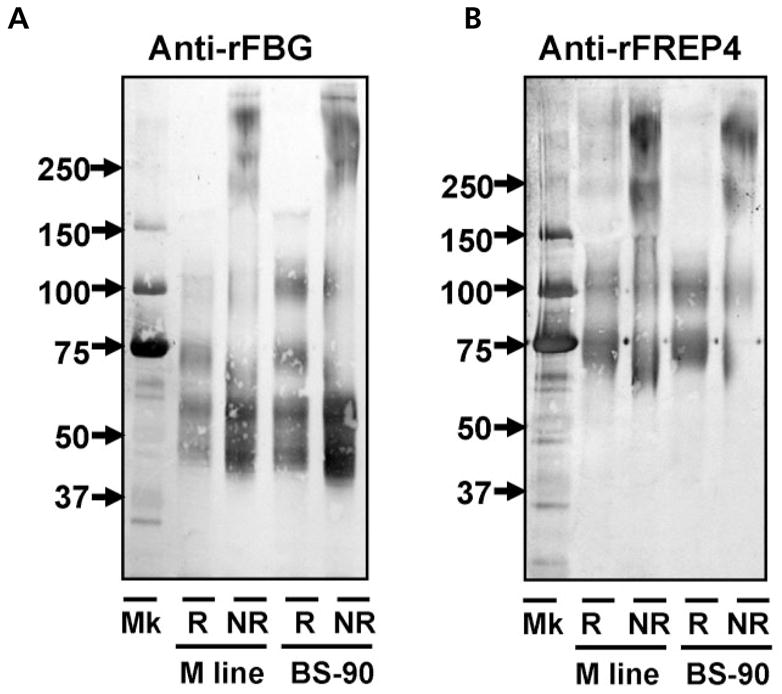
Detection of multiple FREPs and FREP4 under reducing and non-reducing conditions. Western blots shown were probed with anti-rFBG Ab (A) and anti-rFREP4 Ab (B). Lanes R and NR show the plasma from M line or BS-90 snails at 4-days post-exposure to E. paraensei under reducing and non-reducing conditions, respectively.
Next we used anti-FREP4 Ab to probe a blot loaded with the same two types of plasma samples (Fig. 2B). Under non-reducing conditions, in addition to the two bands (65–75 kDa strong band, and a weaker 90–100 kDa band) seen under reducing conditions, two additional diffuse bands with high molecular weights were detected. The smaller of these is ~250 kDa. Regarding the 90–100 kDa band, as we currently have no information about its nature, we have designated it as ‘FREP4-like protein’. Although the nature of the 90–100 kDa band requires further investigation, this study nevertheless has shown that classic FREP4 of 65–75 kDa, like most FREPs, is multimeric in its native configuration, comprised of at least four FREP4 subunits (tetramers).
Finally, we examined the snail FREP2 protein using anti-rIgSF-FREP2 Ab. Interestingly, no difference has been observed between reducing and non-reducing conditions, suggesting FREP2 does not form a multimer (data not shown).
Significant up-regulation of FREP expression in M line and BS-90 snails after exposure to E. paraensei
Figure 3A shows a Western blot loaded with the plasma samples from individual M line snails at different days post-exposure to the trematode E. paraensei, and probed with anti-FBG Ab. As expected, a wide range of FREPs were detected, including the 65–75 kDa region corresponding to FREP4 and the 50-kDa FREP2 band (see below). Significant up-regulation has been observed from 2 up to 8 days post-exposure with a gradual return to control levels after 10 days post-exposure. The overall highest expression level observed was at 6 days post-exposure (~18-fold, P < 0.05), and the next highest level was at 4 days post-exposure (~14-fold, P < 0.05).
Fig. 3.
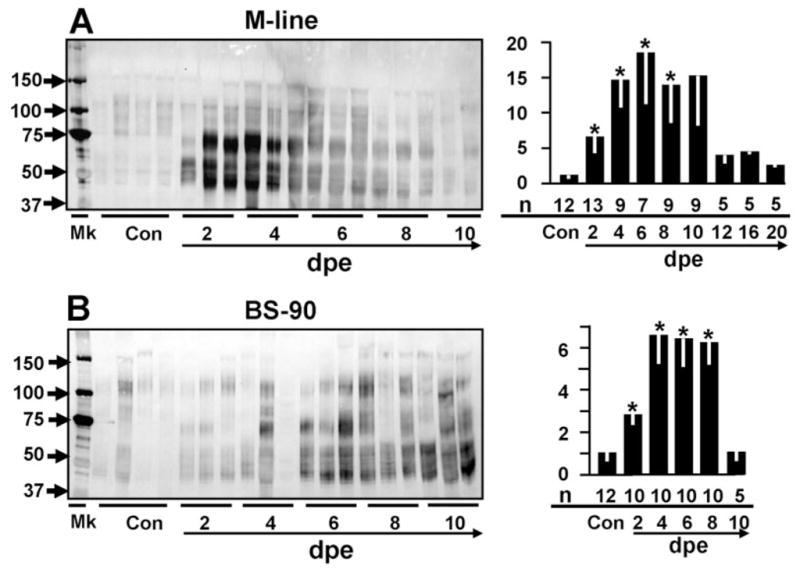
As detected by probing with anti-rFBG, expression of putative multiple FREP proteins in the plasma of M line (A) and BS-90 snails (B) in unexposed control snails (Con) or after exposure to E. paraensei at different days post-exposure (dpe). The left panel shows the Western blots from relatively few random samples; the days post-exposure for each lane is indicated by a solid line under the lane. The right panel shows the expression of FREPs relative to the non-exposed snails based on a statistical analysis of densitometric scans. The Y-axis shows the relative expression level and the X-axis shows the different days post-exposure (below line). The numbers indicated above the line are the numbers of all snail plasma samples that were scanned and used for the statistical analysis at each time point. The asterisks indicate statistical significance (P < 0.05) as compared to unexposed control snails. The numbers above the arrowed line show the total number of random samples used for the Western blots as shown in the right panel and those samples were also used for the statistical analysis.
Figure 3B shows a similar blot with plasma from BS-90 snails after exposure to E. paraensei. It is again apparent that E. paraensei can activate FREP expression in BS-90 snails in the same general pattern as noted with M line snails. Significant up-regulation was observed from 2 to 8 days post-exposure, with the fold increase in expression level generally being lower, ~50% of that seen in M line snails.
Expression of FREP4 in M line and BS-90 snails after exposure to E. paraensei
We next investigated the responses of an individual FREP, FREP4, to E. paraensei in both snail strains. FREP4 expression is strongly up-regulated (~20-fold; P < 0.05) in M line snails after exposure to E. paraensei (Fig. 4A). Instead of the broad range of bands detected by anti-FBG Ab (Fig. 3), only a major band of 65–75 kDa was noted when probing with anti-rFREP4 Ab. In infected snails, this band strongly increased in intensity, and also showed as a characteristic tendency to be diffuse rather than tightly focused, spanning a molecular weight range of ~60–80 kDa. Note that the actual range of molecular weights represented varies somewhat from one snail (lane) to the next. Additionally, a weaker 90–100 kDa band can be seen Figure 4A.
Fig. 4.
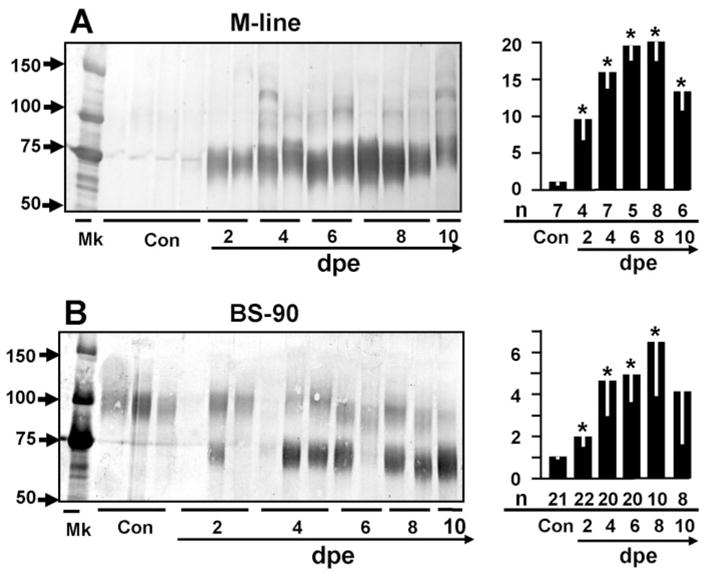
As detected by probing with anti-rFREP4 Ab, expression of putative FREP4 proteins in plasma of M line (A) and BS-90 snails (B) after exposure to E. paraensei. Abbreviations, lane designations and statistical analysis are as described in the caption to Figure 3.
With respect to FREP4 expression in BS-90 snails, a 65–75-kDa band was also up-regulated, but less so than seen with M line snails (~3-fold). Different from M line snails, the 90–100-kDa protein is constitutively expressed at a relative high level in BS-90 snails (Fig. 4B).
The 65–75-kDa FREPs prominently bind to E. paraensei sporocysts and their products
To examine the ability of FREPs to bind various antigen presentations, we used the anti-FBG Ab because of its ability to bind multiple FREPs. Snail blood (plasma) was collected from snails under seven different conditions: non-challenged controls, exposed to either trematode, or injected with PBS or with one of three types of microbes (for details, see the caption to Fig. 5A). For the purpose of comparison, these replicate plasma samples were used for all treatments in the binding study.
Fig. 5.
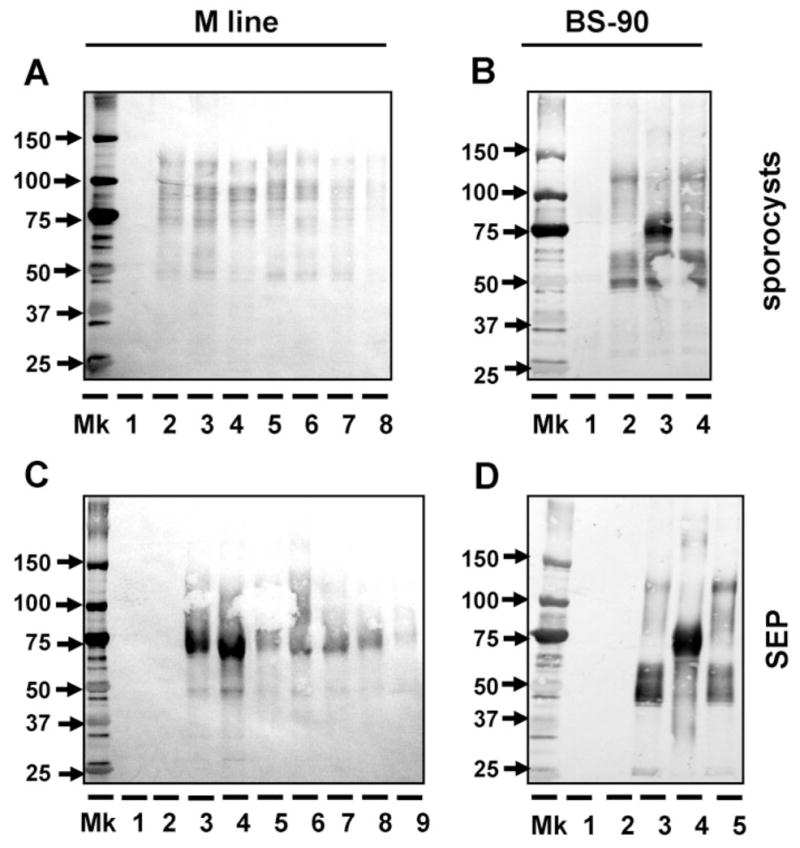
Use of anti-rFBG antibody to detect the binding of putative multiple FREPs from two snail strains (M line and BS-90) to E. paraensei sporocysts (A, B) and E. paraensei sporocyst SEPs (C, D). (A) Binding of M line plasma to E. paraensei sporocysts. Lane 1 is loaded with solubilized sporocysts. Lanes 2–8 are loaded with M line plasma components that bound sporocysts, from non-exposed controls snails (lane 2); snails exposed to E. paraensei (lane 3) or S. mansoni (lane 4); or snails injected with PBS (lane 5), Staph. aureus (lane 6), E. coli (lane 7) or S. cerevisiae (lane 8). All plasma was collected at 4 days post-infection or post-injection for all binding studies throughout this work. (B) Binding of BS-90 plasma to E. paraensei sporocysts. Lane 1 is loaded with sporocyst antigens only. Plasma components binding SEPs are derived from non-exposed control snails (lane 2), or snails exposed to E. paraensei (lane 3) or S. mansoni (lane 4). (C) Binding of M line plasma to SEPs. All SEPs used in this study was derived from E. paraensei sporocysts at 2–4 days post-transformation in vitro. Lane 1 received SEPs only. Lane 2 was loaded with half-strength 199-medium. Treatments for lanes 3–9 are the same, and in the same order, as described for lanes 2–8 in Figure 6A. (D) Binding of BS-90 plasma to SEPs. Lanes 1 and 2 were loaded the same as lanes 1 and 2 as described in the caption to Figure 5C. Treatments for lanes 3 to 5 are the same, and in the same order, as lanes 2 to 4 in Figure 5B.
The first binding experiment was performed to examine whether FREPs bind to E. paraensei sporocysts. Figure 5A shows that in vitro transformed sporocysts are bound by a broad range of FREPs from M line snails, from 50 kDa to 125 kDa. No consistent difference was observed among the seven sources of M line plasma used.
When plasma from BS-90 snails (unexposed controls or snails exposed to either trematode) was used, the results were quite different. Sporocysts were weakly bound by 50 kDa and 125 kDa FREPs if the plasma was from unexposed controls or snails exposed to S. mansoni, but when the plasma was from snails exposed to E. paraensei, sporocysts were predominantly bound by 65–75-kDa FREPs. Notably, 65–75-kDa proteins from controls or S. mansoni-exposed snails bind to E. paraensei sporocysts weakly or not at all (Fig. 5B).
Next the binding of FREPs to SEPs of E. paraensei sporocysts was tested, using the same plasma samples as described above for the sporocyst-binding experiments. SEPs were bound mostly by 65–75-kDa FREPs (Fig. 5C, D). Echinostome sporocysts and their secretory products provoke different FREP binding patterns, with the soluble secretory products revealing binding capacities in M line snails that the sporocyst bodies do not. The results show clearly when using secretory products as targets that exposure of snails to trematodes, particularly to E. paraensei, has induced the production of FREPs with distinct binding specificities (lane 3, Fig. 5D) or with enhanced amounts of binding (lanes 3 and 4, Fig. 5C).
High molecular-weight FREPs bind to Gram-positive and Gram-negative bacteria and to yeast
The Gram-positive bacterium Staph. aureus, the Gram-negative bacterium E. coli, and the yeast S. cerevisiae were used as three representative microbe targets in another binding study. Plasma samples from the seven specified sources for M-line snails, and three sources for BS-90 snails were again used. Neither the experimental source of the plasma nor the snail strain strongly influenced the results. The major FREPs binding to all three microbes are of ~95 kDa and ~125 kDa. The two snail strains differed slightly with respect to relative staining intensity of the two major bands (Fig. 6A–F). It is clear that exposure to a particular microbe did not engender production of FREPs that were specifically up-regulated as a consequence of that exposure.
Fig. 6.
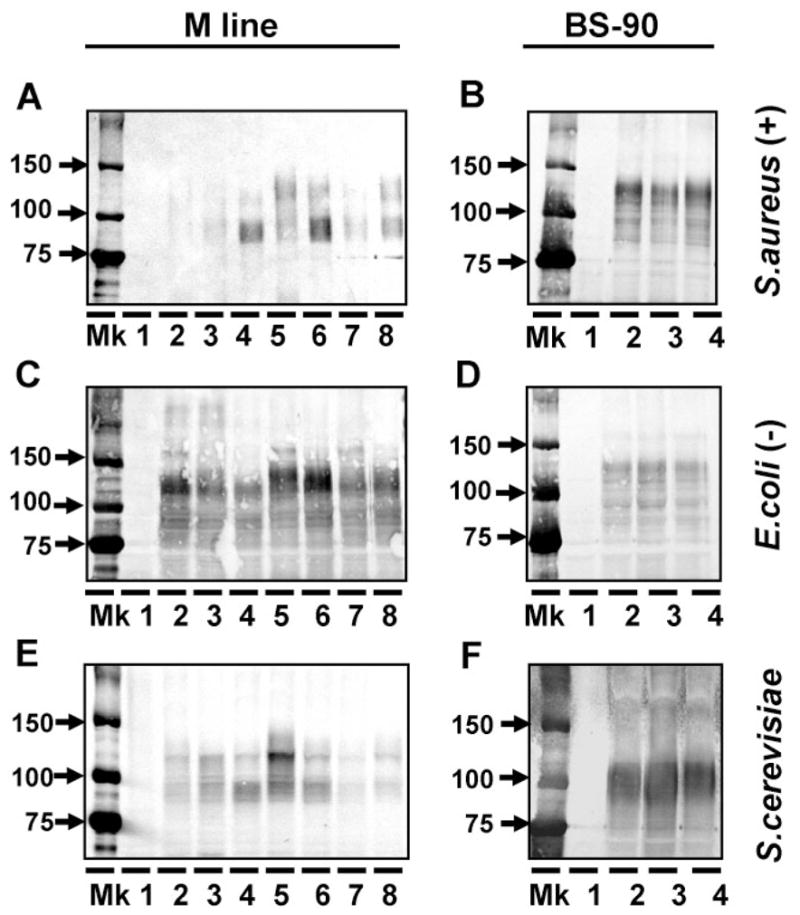
Use of anti-rFBG antibody to examine the binding of putative multiple FREPs from two snail strains to Staph. aureus (A, B), E. coli (C, D) and S. cerevisiae (E, F). The left and right panels show binding of plasma components from M line and BS-90 snails, respectively. Lane 1 was loaded with putative antigen that was tested for binding assay. The M line plasma treatments for lanes 2–8 shown in the left panels are the same as lanes 2–8 in Figure 5A and plasma from lanes 2 to 4 in (BS-90 snails: on the right) are same as lanes 2 to 4 in Figure 5B.
Discussion
FBG-containing molecules, often in surprising diversity with respect to numbers of putative coding sequences, have been documented in a wide variety of invertebrates and vertebrates; their functions, however, are not well understood.15,16 Accumulating data suggest that the primary function of FBG-bearing proteins is in innate immune responses. In the sponge Suberites domuncula, a protein containing a FBG-like domain binds carbohydrates and is up-regulated by curdlan.24 The ascidian ficolins, which are FBG-bearing proteins, bind to N-acetyl-D-glucosamine (GlcNAc) and N-acetyl-D-galactosamine (GalNAc) and activate immune responses.18 The horseshoe crab tachylectins have been found to possess antibacterial activities and non-self recognition capabilities.17 In mosquitoes of the genera Anopheles and Armigeres, FBG-like proteins have been implicated in immune responses to microbes or malaria parasites because their transcript levels are up-regulated after infection.19–23 Sialic-acid-binding lectins which contain FBG domains have been identified from the slug Limax flavus and found to be down-regulated after parasitism.25 With respect to human ficolins, which are the best-studied FBG-bearing proteins in terms of structure and function, three types of ficolins (L-, H-, and M-ficolin) have lectin activity and can activate the lectin complement pathway.14–16
The molecular nature of FBG-bearing proteins (FREPs) from the snail B. glabrata was discovered about 10 years ago and considerable progress has since been made in terms of revealing gene structure, mRNA expression and diversification.4 However, one of the shortcomings in our current knowledge of FREPs is that we need a more precise understanding of their functional roles.
To help redress this shortcoming, we have expressed a number of rFREP proteins, generated Abs to them, and applied these reagents to functional assays. Although we have only discussed two such Abs (anti-rFBG and anti-rFREP4 Abs) in this study, we have generated 10 different rFREP proteins, each of which provides a plausible target for a specific antibody which can, in turn, be used to dissect the function of a specific FREP subfamily.
While interpreting the results of this paper, it is noteworthy that many of the ‘bands’ from snail hemolymph detected by anti-rFREPs are broad and ‘fuzzy’; therefore, we represent their presence as a range of molecular weights (e.g. 60–80 kDa in some E. paraensei-infected individuals). We have long noted this tendency of several snail hemolymph components and, as discussed elsewhere,4 believe this fundamentally to be a consequence of the fact that the various FREP components identified exist as diversified clusters of related molecules that form broad rather than sharply focused bands.
Our previous studies based on DNA sequence and Southern blot analyses revealed that probes from the FREP IgSF region could detect member(s) of a single FREP subfamily, and that probes from the FREP FBG-encoding region, because the latter is more conserved, hybridize with many FREP encoding genes, potentially from all FREP subfamilies.6,7,41 Based on this result, it was predicted that Abs raised against the IgSF region of FREP of a given subfamily (e.g. FREP4 in this study) would potentially detect FREPs of this subfamily. In contrast, Abs raised against the rFBG domain of any FREPs (e.g. FREP4) should be able to react with multiple FREPs. This prediction has essentially been verified. For example, the anti-rFBG Ab recognizes complex profiles of plasma proteins relative to the simpler profiles recognized by anti-FREP4 Ab. In addition to FREP4, some additional bands bound by anti-rFBG are known to be FREPs, including a band known to be comprised of a different subclass, FREP2 (data not shown).
Regarding the two main bands (65–75 kDa and 90–100 kDa) detected by anti-rFREP4 Ab, there is confirmatory evidence that the 65–75-kDa band is the expected target. A peptide fragment originally derived from a band of the same molecular weight provided the starting point for the eventual cloning and sequencing of the gene first identified as FREP4.3,5 Moreover, we recently directly confirmed that the 65–75-kDa band identified by our Ab was FREP4 using similar proteomic approaches. One explanation for the relatively weak 90–100-kDa band bound by anti-rFREP4 Ab is that it is a variant form possessing tandem FREP4 IgSF domains, or it may represent cross-reaction of anti-FREP4 with another FREP. The responsiveness of the two bands detected by the anti-rFREP4 Ab in the binding studies is clearly different, as discussed further below.
Previous studies demonstrated that two main protein populations/bands designated G1M (150–200 kDa) and G2M (75–130 kDa) could be harvested from the plasma of unexposed control or E. paraensei-infected M line snails using a mixture of monosaccharide-conjugated agarose beads.42,43 Further study using antisera raised against these affinity purified plasma components revealed that G1M and G2M were able to bind to E. paraensei sporocysts and bacteria.37 In the present study, the size of snail plasma proteins detected by our current Abs was found in the range of G2M, but typically not in the range of G1M. This supports our assertion that G2M molecules collected previously are FREPs. These results also suggest that G1M proteins may not be FREPs because the snail proteins detected by our FREP Abs are less than 150 kDa. One hypothesis is that G1M molecules are composed of multiple IgSF domains, making them similar to molluscan defense molecules (MDM), described by Hoek et al.9
As demonstrated in the study, some major FREPs, including FREP4, likely have multimeric constitution in their native configuration. Two diffuse bands, 65–75 kDa and 90–100 kDa, were found in reducing conditions, as detected by anti-FREP4 Ab (see Figs 2B and 4A). Under non-reducing conditions, two higher molecular-weight bands were also observed, consistent with the interpretation that FREP4 exists in its native configuration in snail hemolymph as tetramers or even higher level multimers. Previous study also revealed that snail plasma molecules of 65–75 kDa, now known to be FREP4, exist as high molecular weight multimers, and actively precipitate E. paraensei SEPs from solution. It was also suggested that FREP4 forms covalently bound hexamers of ~400 kDa, which are further non-covalently associated into tetramers of ~1600 kDa.44
Formation of multimers in molecules containing FBG domains has also been observed in human, horseshoe crab and ascidian ficolins.15,17,18,45 A recent crystal structure study suggests that actual interfaces after association of three ficolin subunits expand greatly, implying the overall pattern recognition capabilities of ficolins are much greater than previously thought.46 In the case of mosquito FBG-like proteins, it has been hypothesized that different FBG-containing monomers are joined as multimers, potentially resulting in large numbers of different combinations which would further increase their recognition capability (Dimopoulos G., personal communication). Although this hypothesis presents a daunting challenge, if it can be shown to apply to FREPs, then the overall recognition capability of molecules of the FREP family could be even further increased given that each member of FREPs possesses two functional domains (IgSF and FBG) and that this family already is known to be comprised of multiple subfamilies which are also diversified by other means.28 Unlike most of FREPs, including FREP4, FREP2 maintains a monomeric form. It was shown that FREP4 and FREP2 have the same molecular structure (one IgSF and one FBG domain),5 but our finding suggests that they differ with respect to multimeric formation. This suggests that even although their molecular structures are very much the same, they differ in surprising ways with respect to how they associate with one another and their functional capacities.
One of the important goals of this study was to learn if our newly-generated anti-rFREP antibodies could be used to detect the presence and changes in abundance of potentially all or of specific FREPs in M line or BS-90 snail plasma following exposure to E. paraensei, because early studies suggested that FREPs or putative FREPs sharply increase in abundance following E. paraensei exposure. The significance of the present approach is that it offers the possibility of documenting which plasma proteins are FREPs, thus helping to fulfil a long-standing need to identify more thoroughly the component molecules comprising snail hemolymph. Our study first showed with the use of anti-rFBG Ab that the plasma of B. glabrata contains many polypeptides with FBG domains. As we have noted previously, many of these molecules migrate on standard denaturing SDS-PAGE gels as broad diffuse bands. Also, use of either anti-rFBG or of specific anti-FREP Abs like anti-rFREP4 Ab showed that many FREPs are elevated in response to trematode infections, and present complicated response patterns. This study, using non-glycosylated recombinant proteins known to be FREPs or portions thereof, provides both general and more specific confirmation of previous studies, showing the dynamic changes of both multiple FREP proteins and of specific FREP (e.g. FREP4) in snail plasma after exposure to trematodes.3
Further insight into FREP function has been gained by the binding assay we used which suggests that FREPs have the ability to recognize a wide range of potential pathogens including bacteria, yeast, and trematode, and associated molecules such as SEPs. This suggests that FREPs may play a role in defense responses to a variety of pathogens, potentially explaining why there are several subfamilies of FREPs. Some capacity for specificity in FREP responses is exhibited in that the microbes examined were bound mainly by 95-kDa and/or 125-kDa FREPs whereas E. paraensei or its products are bound predominantly by FREPs of 65–75 kDa.
One intriguing possibility pointed out in this study that requires further investigation is the possibility that exposure to trematode infection can promote production of molecules with binding capacities not seen in unexposed snails. For example, the 65–75-kDa FREP4 molecules from the plasma of BS-90 snails exposed to E. paraensei are able to bind to sporocysts or SEPs from this trematode, whereas there is no evidence to suggest that plasma from unexposed controls or S. mansoni-exposed snails has the same capacity. Although there are likely differences in the abundance of FREP4 among the three groups of snails, this alone does not seem sufficient to account for the pronounced binding differences. Up-regulation of FREPs provoked by exposure to some infections may be accompanied by production of molecules with binding capabilities not seen in other snails. The range of mass of the FREP4 band up-regulated in infected snails seen in Figure 4 is further suggestive of this possibility.
As proposed previously, one of the important functions of FBG-containing proteins is carbohydrate recognition.14,15 Our binding assay suggests that FREPs are able to bind to a wide range of pathogens displaying carbohydrates on their surfaces. Another interesting aspect of FREP molecules is that they have two domains, FBG and IgSF, that potentially could bind to pathogens. One hypothesis is that the IgSF domain is responsible for binding to non-self objects such as trematodes or microbial surfaces. The FBG-domain, similar to other invertebrate FBG-containing molecules, has lectin activity and, as proposed for molluscan lectins by Renwrantz et al.,47 could bind to a carbohydrate-bearing ligand on a hemocyte membrane. In this view, FREPs could serve as bringing molecules that when engaged activate effector function such as phagocytosis or encapsulation responses.4
Although our binding assay should be considered preliminary, several supportive lines of evidence also suggest that FREPs are able to bind to the parasites and microbes. For example, a previous study using antibody raised against purified snail plasma proteins suggested that molecules likely to be FREPs were able to bind parasites and microbes.37 The first partial FREP peptide sequence was obtained from snail plasma proteins that had precipitated E. paraensei SEPs, and a recent study using a proteomics approach has reconfirmed that the major plasma protein binding to SEPs is FREP4. Finally, the binding assay employed suggests all FREPs do not indiscriminately bind to all foreign objects; microbes bound different suits of FREPs than trematodes and different microbes bound different FREP profiles. To study FREP functions further, we have expressed recombinant FREP proteins in insect cell expression systems. Such proteins will be more similar to their natural counterparts including glycosylation, and thus will be well-suited for our functional studies because snail FREPs are glycosylated.3
Conclusions
This study demonstrates the feasibility of expressing recombinant B. glabrata proteins and of making antibodies to recombinant FREPs as valuable ways to probe FREP function. Our results confirm that snails mount very complex responses manifested as visibly altered quantities of FREP polypeptides in snail plasma, and further show that at least some FREPs exist as multimers (e.g. FREP4). Only selected stimuli elicit an obvious plasma FREP response, and snails of two strains respond differently to the same stimulus. Evidence is presented that microbes bind different suites of FREPs than trematode larvae. However, many questions await further study. For example, how do other individual FREPs not studied here respond to trematode infection? Do FREPs influence compatibility between snail and schistosome? Do FREPs play a role in defense against microbes? What are the functions of the two domains found in FREPs? Many of these questions are amenable to further study using the technique of RNAi which has been developed for B. glabrata39 and will be assisted by the use of the antibody probes we have developed here.
Acknowledgments
This work was supported by National Institutes of Health Grant RO1 AI067686 (to S-MZ) and Grant Number RR-1P20RR18754 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources. The journal’s contributor publishing agreement complies with the NIH Public Access Policy.
References
- 1.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 2.Raghavan N, Knight M. The snail (Biomphalaria glabrata) genome project. Trend Parasitol. 2005;22:148–151. doi: 10.1016/j.pt.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stout B, Adema CM, Zhang S-M, Loker ES. The biology of FREPs: diversified lectins with fibrinogen-related domains from the freshwater snail B. glabrata. In: Vasta GR, Ahmed H, editors. Animal Lectins: A Functional View. Cleveland, OH: CRC Press Taylor & Francis; 2008. In press. [Google Scholar]
- 5.Léonard PM, Adema CM, Zhang S-M, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S-M, Loker ES. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Dev Comp Immunol. 2003;27:175–187. doi: 10.1016/s0145-305x(02)00091-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S-M, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the snail Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–266. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 9.Hoek RM, Smit AB, Frings H, et al. A new Ig-superfamily member, molluscan defence molecule (MDM) from Lymnaea stagnalis, is down-regulated during parasitosis. Eur J Immunol. 1996;26:939–944. doi: 10.1002/eji.1830260433. [DOI] [PubMed] [Google Scholar]
- 10.Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- 11.Sun SC, Lindstrom I, Boman HG, et al. Hemolin: an insect-immune protein belonging to the immunoglobulin superfamily. Science. 1990;250:1729–1732. doi: 10.1126/science.2270488. [DOI] [PubMed] [Google Scholar]
- 12.Watson FL, Puttmann-Holgado R, Thomas F, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1978. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 13.Dong YM, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biology. 2006;4:137–146. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmskov U, Thiel S, Jenseius JC. Collectins and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 15.Fujita T, Matsushita M, Endo Y. The lectin-complement pathways- its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 16.Endo Y, Matsushita M, Fujita T. Role of ficolin in innate immunity and its molecular basis. Immunobiology. 2007;212:371–379. doi: 10.1016/j.imbio.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Gokudan S, Muta T, Tsuda R, et al. Horseshoe crab acetyl group-recognizing lectins involved in innate immunity are structurally related to fibrinogen. Proc Natl Acad Sci USA. 1999;96:10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenjo A, Takahashi M, Matsushita M, et al. Cloning and characterization of novel ficolins from the solitary ascidian, Halocynthia roretzi. J Biol Chem. 2001;276:19959–19965. doi: 10.1074/jbc.M011723200. [DOI] [PubMed] [Google Scholar]
- 19.Dimopoulos G, Casavant TL, Chang SR, et al. Anopheles gambiae pilot gene discovery project: identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc Natl Acad Sci USA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartholomay LC, Cho WL, Rocheleau TA, et al. Description of the transcriptomes of immune response-activated hemocytes from the mosquito vectors Aedes aegypti and Armigeres subalbatus. Infect Immun. 2004;72:4114–4126. doi: 10.1128/IAI.72.7.4114-4126.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan P, Abraham EG, Ghosh AK, et al. Analysis of the Plasmodium and Anopheles transcriptomes during oocyst differentiation. J Biol Chem. 2004;279:5581–5587. doi: 10.1074/jbc.M307587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Rocheleau TA, Fuchs JF, et al. A novel lectin with a fibrinogen-like domain and its potential involvement in the innate immune response of Armigeres subalbatus against bacteria. Insect Mol Biol. 2004;13:273–282. doi: 10.1111/j.0962-1075.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- 23.Dong YM, Aguila R, Xi E, et al. Anopheles gambiae immune response to human and rodent Plasmodium parasite species. PLoS Pathogens. 2006;6:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perovic-Ottstadt S, Adell T, Proksch P, et al. A (1–3)-β-D-glucan recognition protein from the sponge Suberites domuncula: mediated activation of fibrinogen-like protein and epidermal growth factor gene expression. Eur J Biochem. 2004;271:1924–1937. doi: 10.1111/j.1432-1033.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 25.Kurachi S, Song ZW, Takagaki M, et al. Sialic-acid-binding lectin from the slug Limax flavus: cloning, expression of the polypeptide, and tissue location. Eur J Biochem. 1998;254:217–222. doi: 10.1046/j.1432-1327.1998.2540217.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Zhao Q, Christernsen BM. Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaste, genomes. BMC Genom. 2005;6:114. doi: 10.1186/1471-2164-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S-M, Nian H, Zeng Y, DeJong RJ. Fibrinogen-bearing protein genes in the snail Biomphalaria glabrata: characterization of two novel genes and expression studies during ontogenesis and trematode infection. Dev Comp Immunol. 2008 doi: 10.1016/j.dci.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S-M, Adema CM, Kepler TB, Loker ES. Diversification of Ig genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 29.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nat Rev Immunol. 2005;11:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger DP, Buckley KM, Mehta D, et al. Unexpected diversity displayed in cDNA expressed in the immune cells of the purple sea urchin, Strongylocentrotus purpuratus. Physiol Genom. 2006;26:134–144. doi: 10.1152/physiolgenomics.00011.2006. [DOI] [PubMed] [Google Scholar]
- 31.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Little TJ, Hultmark D, Read AE. Invertebrate immunity and the limits of mechanistic immunology. Nat Immunol. 2006;6:651–654. doi: 10.1038/ni1219. [DOI] [PubMed] [Google Scholar]
- 33.Litman GW, Cooper MD. Why study the evolution of immunity? Nat Immunol. 2007;8:547–548. doi: 10.1038/ni0607-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Enchinostoma paraensei. Dev Comp Immunol. 2005;29:295–303. doi: 10.1016/j.dci.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Humphries JE, Yoshino TP. Schistosoma mansoni excretory-secretory products stimulate a p38 signalling pathway in Biomphalaria glabrata embryonic cells. Int J Parasitol. 2006;36:37–46. doi: 10.1016/j.ijpara.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Zelck UE, Gege BE, Schmid S. Specific inhibitors of mitogen-activated protein kinase and P13-K pathways impair immune responses by hemocytes of trematode intermediate host snails. Dev Comp Immunol. 2007;31:321–331. doi: 10.1016/j.dci.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Hertel LA, Stricker SA, Monroy FP, et al. Biomphalaria glabrata hemolymph lectins – binding to bacteria, mammalian erythrocytes, and to sporocysts and rediae of Echinostoma paraensei. J Invertebr Pathol. 1994;64:52–61. doi: 10.1006/jipa.1994.1068. [DOI] [PubMed] [Google Scholar]
- 38.Loker ES, Hertel LA. Alternations in Biomphalaria glabrata plasma induced by infection with the digenetic trematode Echinostoma paraensei. J Parasitol. 1987;75:505–513. [PubMed] [Google Scholar]
- 39.Jiang Y, Loker ES, Zhang S-M. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Comp Immunol. 2006;30:855–866. doi: 10.1016/j.dci.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adema CM, Hertel LA, Loker ES. Evidence from two planorbid snails of a complex and dedicated response to digenean (echinostome) infection. Parasitology. 1999;119:395–404. doi: 10.1017/s0031182099004850. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S-M, Léonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria glabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. 2001;53:684–694. doi: 10.1007/s00251-001-0386-8. [DOI] [PubMed] [Google Scholar]
- 42.Monroy F, Hertel LA, Loker ES. Carbohydrate-binding plasma proteins from the gastropod Biomphalaria glabrata: strain specificity and the effect of trematode infection. Dev Comp Immunol. 1992;16:355–366. doi: 10.1016/0145-305x(92)90038-e. [DOI] [PubMed] [Google Scholar]
- 43.Monroy F, Loker ES. Production of heterogeneous carbohydrate-binding proteins by the host snail Biomphalaria glabrata following exposure to Echinostoma paraensei and Schistosoma mansoni. J Parasitol. 1993;79:416–423. [PubMed] [Google Scholar]
- 44.Adema CM, Hertel LA, Loker ES. Infection with Echinostoma paraensei (Digenea) induces parasite-reactive polypeptides in the hemolymph of the gastropod host Biomphalaria glabrata. In: Beckage NE, editor. Parasites and Pathogens: Effect on the Host Hormones and Behavior. New York: Chapman & Hall; 1997. [Google Scholar]
- 45.Le Y, Tan SM, Lee SH, et al. Human L-ficolin: plasma levels, sugar specificity, and assignment of its lectin activity to the fibrinogen-like (FBG) domain. FEBS Lett. 1998;425:367–370. doi: 10.1016/s0014-5793(98)00267-1. [DOI] [PubMed] [Google Scholar]
- 46.Garlatti V, Belloy N, Martin L, et al. Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 2007;26:623–633. doi: 10.1038/sj.emboj.7601500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renwrantz L, Schancke W, Harm H, et al. Discriminative ability and function of the immunobiological recognition system of the snail Helix pomatia. J Comp Physiol. 1981;141:477–488. [Google Scholar]


