Abstract
Aromatase is a particularly attractive drug target in the treatment of hormone-responsive breast cancer, and aromatase activity in breast cancer patients is greater in or near the tumor tissue compared with the normal breast tissue. Complex regulation of aromatase expression in human tissues involves alternative promoter sites that provide tissue-specific control. Previous studies in our laboratories suggested a strong association between aromatase (CYP19) gene expression and the expression of cyclooxygenase (COX) genes. Additionally, COX selective inhibitors can suppress CYP19 gene expression and decrease aromatase activity. Our current hypothesis is that pharmacological regulation of aromatase can act locally to decrease the biosynthesis of estrogen and may provide additional therapy options for patients with hormone-dependent breast cancer. Two pharmacological approaches are being developed, one approach utilizing small molecule drug design and the second approach involving mRNA silencing technology. The small molecule drug design approach focuses on the synthesis and biological evaluation of a novel series of sulfonanilide analogs derived from COX-2 selective inhibitors. Combinatorial chemistry approaches were used to generate diversely substituted novel sulfonanilides. The compounds suppress aromatase enzyme activity in SK-BR-3 breast cancer cells in a dose and time dependent manner, and structure activity analysis does not find a correlation between aromatase suppression and COX inhibition. Real-time PCR analysis demonstrates that the sulfonanilide analogs decrease aromatase gene transcription in breast cells. Furthermore, the sulfonanilide compounds selectively decrease aromatase gene expression in several breast cancer cells, without exhibiting cytotoxic or apoptotic effects at low micromole concentrations. A ligand based pharmacophore model for selective aromatase modulation (SAM) by the novel sulfonanilides identified an aromatic ring, two hydrogen bond acceptors, and a hydrophobic function as four key chemical features. In the second approach, short interfering RNAs (siRNA) were designed targeting human aromatase mRNA. Treatment of breast cancer cells with siRNAs targeting aromatase (siAROMs) completely masked the aromatase enzyme activity and resulted in suppression of CYP19 mRNA. Thus, these results suggest that the novel sulfonanilides and the siRNAs targeting aromatase expression may be valuable tools for selective regulation of aromatase in breast cancer.
Keywords: aromatase, COX-2 inhibitors, sulfonanilides, siRNA
1. Introduction
Estrogens are biosynthesized from androgens by the cytochrome P450 enzyme complex called aromatase [1;2]. The highest levels of enzyme are present in the ovaries of premenopausal women, in the placenta of pregnant women, and in the peripheral adipose tissues of postmenopausal women and of men. Aromatase activity has also been demonstrated in breast tissue in vitro [3–5], and expression of aromatase is highest in or near breast tumor sites [4;6]. Regulation of aromatase in various tissues is complex, and several tissue-specific promoter regions have been identified upstream from the CYP19 gene [2;7;8]. Promoter I.1 is the major promoter used in placental tissues and is the farthest upstream. The PII promoter is utilized in the ovary and in breast cancer tissues and contains a cAMP response element. Promoters I.3, I.4, I.6, and I.7 are the promoters used in extraglandular sites. Promoter PI.4 is the primary promoter used in normal adipose tissue and is responsive to glucocorticoids and cytokines such as IL-1β, IL-6 and TNFα.
In breast cancer tissues, an increased expression of aromatase cytochrome P450 is observed due to utilization of multiple promoter regions for gene expression. In the normal breast cells, aromatase expression is primarily derived by the tissue-specific promoter I.4 for transcription, whereas expression from breast cancer patients is primarily derived from the utilization of promoter I.3 and promoter II [9]. As a result of the use of the alternate promoters, the regulation of estrogen biosynthesis switches from one controlled primarily by glucocorticoids and cytokines to a promoter regulated through cAMP-mediated pathways, and the prostaglandin PGE2 increases intracellular cAMP levels and stimulates estrogen biosynthesis [9].
Research in our laboratories focuses on the development of novel therapeutic interventions targeting tissue-specific expression of aromatase. Our small molecule approach utilizes drug discovery technologies to develop novel sulfonanilides as selective agents regulating aromatase expression [10–14]. A second pharmacological approach involves the development of siRNA molecules that interfere with aromatase expression [10]. This manuscript reviews our recent publications in this area and presents preliminary results.
2. Novel Sulfonanilide Analogs for Suppression of Aromatase Expression and Activity
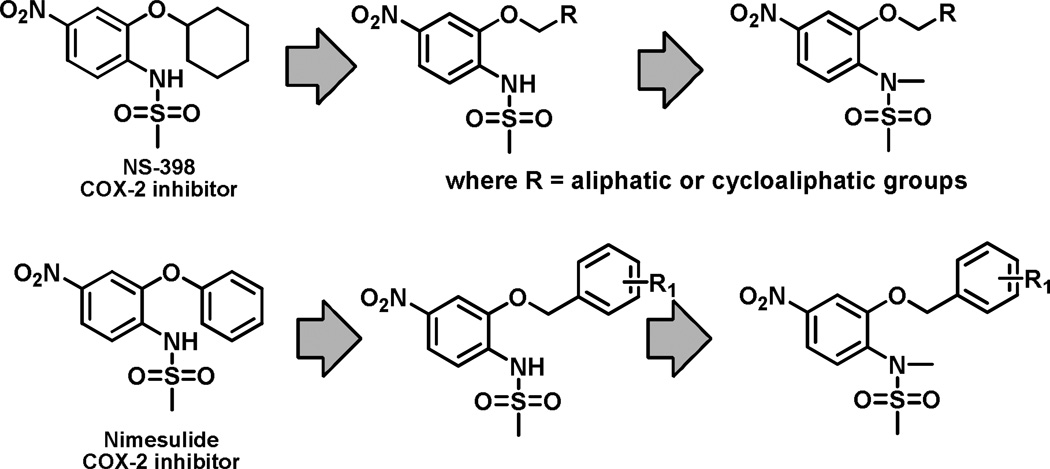
Previously, we demonstrated that COX-2 inhibitors nimesulide, N-(2-phenoxy-4-nitrophenyl)-methanesulfonamide, and NS-398, N-(2-cyclohexyloxy-4-nitrophenyl)-methanesulfonamide, suppress aromatase activity in breast cancer cells by suppressing aromatase transcription [15]. Interestingly, these two agents share very similar chemical structures (Figure 1). In addition, introduction of a methyl group at the N atom of the sulfonamide group to the COX-2 inhibitor nimesulide resulted in no COX-2 inhibitory activity [16]. This structural modification was utilized in our drug design of novel sulfonanilide derivatives of nimesulide and NS-398. The nitro group at the 4 position of the sulfonanilides was retained and modifications of the sulfonamide and of the 2 position group were made to generate the targeted libraries of new compounds (Figure 1) [11].
Figure 1.
Drug design approach for novel sulfonanilides as selective aromatase modulators.
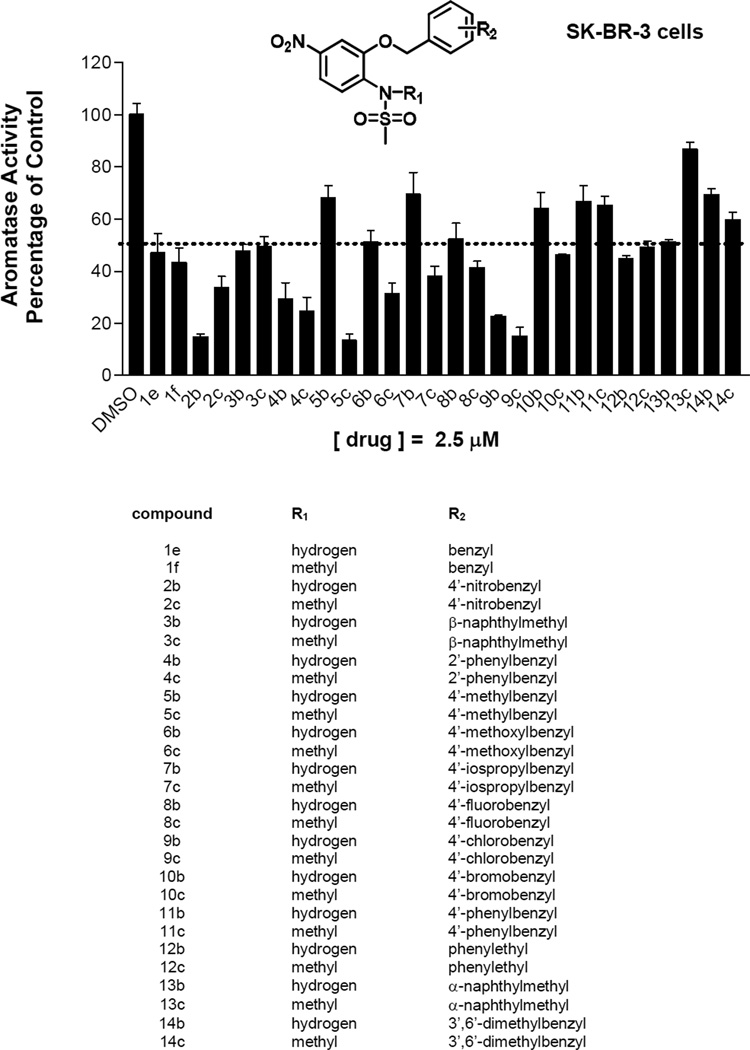
Evaluations of the synthetic compounds were performed in both SK-BR-3 breast cancer cells, which produce aromatase expression from promoter II and I.3 mediated by cAMP [12]. SK-BR-3 cells grown in defined media containing DMEM/F12 media, 1.0 mg/mL human albumin, 5.0 mg/L human transferin and 5.0 mg/L bovine insulin were treated with the synthetic compounds for 24 hours. Cells were then incubated for three hours with fresh media, and aromatase activity measured by the tritiated water release assay [12]. Many compounds significantly decreased aromatase activity in SK-BR-3 cells at 2.5µM (Figure 2), and those compounds with enhanced activities were further evaluated for selective regulation of aromatase expression.
Figure 2.
Suppression of aromatase activity by novel sulfonanilides in SK-BR-3 cells (adapted from [12]). SK-BR-3 cells were treated with compounds (2.5µM) and aromatase activity was measured. The value of 100% is equal to 0.03 pmol/hr/106 cells; n = 3.
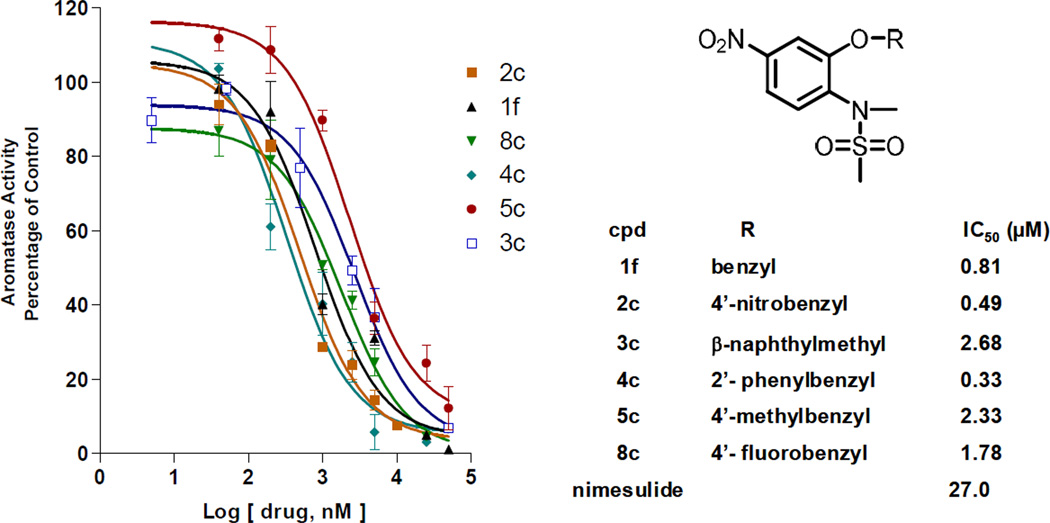
Dose-response studies of the effects on aromatase activity were performed in SK-BR-3 cells on six compounds. All six compounds exhibited dose-response curves for suppression of aromatase activity (Figure 3), and the corresponding IC50 values range from 0.33 to 2.68 µM. Furthermore, real time PCR experiments demonstrated that these compounds significantly decreased CYP19 gene expression at 5µM in SK-BR-3 cells [12]. The results of the cellular aromatase assay demonstrated that a one carbon extension at the 2-position of nimesulide results in significant increase in suppression of aromatase in breast cancer cells compared with nimesulide.
Figure 3.
Dose-response curves and IC50 values for selected novel sulfonanilides (adapted from [12]). The value of 100% is equal to 0.03 pmol/hr/106 cells. Each data point represents the mean results of three independent determinations, and the data were statistically analyzed by a nonlinear regression analysis method.
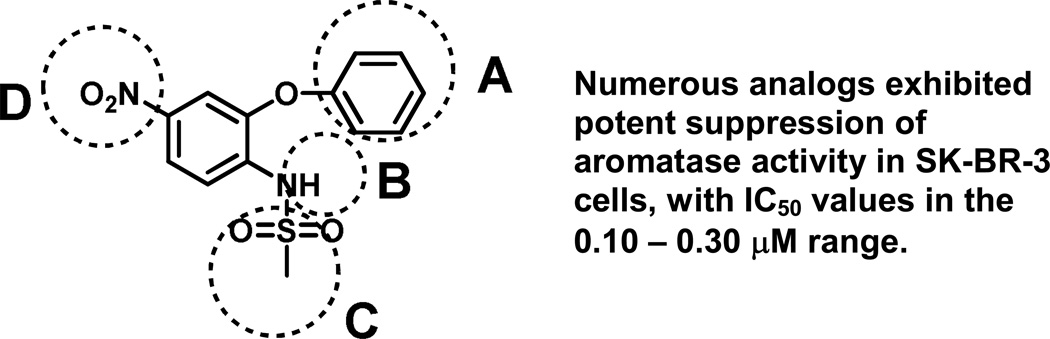
A combinatorial chemistry approach was used to generate diversely substituted nimesulide derivatives (Figure 4) by parallel synthesis [13;14]. This innovative technology of producing libraries of structurally related compounds is particularly beneficial in the step of lead optimization, facilitating structural modifications of a lead scaffold to enhance pharmacological activity, improve pharmacokinetic properties, and/or reduce unwanted side effects. Their pharmacological evaluation as agents for suppression of aromatase activity in breast cancer cells was further explored in the SK-BR-3 breast cancer cell lines and in MCF-7 cells transfected with the aromatase gene. A ligand-based pharmacophore model identified four chemical features critical for biological activity, namely, the central aromatic ring, one hydrophobic function (Figure 4, position A) and two hydrogen bond acceptors (Figure 4, positions C and D).
Figure 4.
Targeted libraries of novel sulfonanilides with modifications at positions A – D.
3. RNA Interference of Aromatase Expression
Newly developed RNA interference (RNAi) technology was utilized to further probe the interactions between aromatase and cyclooxygenases in breast cancer [10]. RNAi technology permits transient suppression of the levels of endogenous proteins in mammalian cells by enhancing the degradation of target mRNA. One RNAi approach involves transfection of 21–23 nucleotide double-stranded, short interfering RNAs (siRNA) into mammalian cells. These siRNAs are then incorporated into the RNA-inducing silencing complex (RISC), and RISC unwinds the siRNA duplex using ATP. The unwound, single-stranded antisense strand guides RISC to mRNA that has a complementary sequence, and the complex results in the endonucleolytic cleavage of the target mRNA.
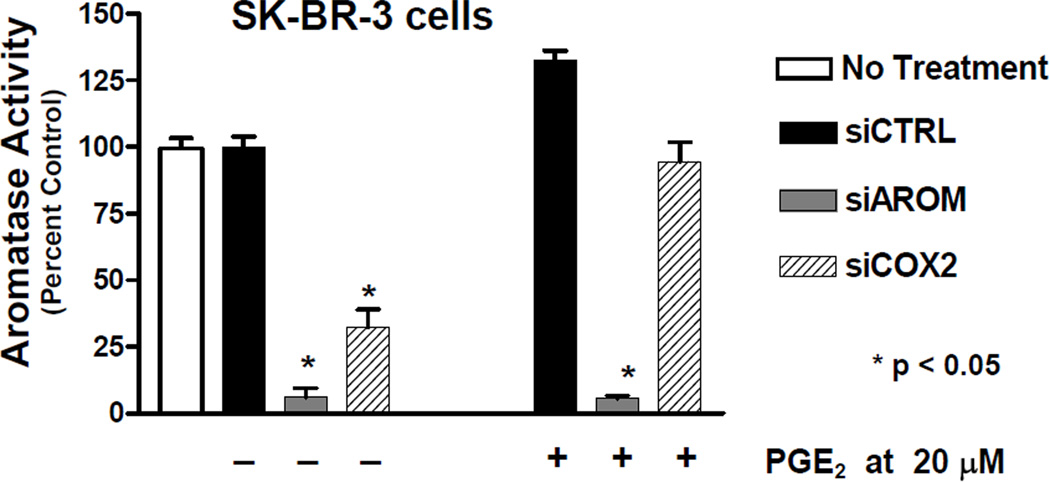
Short interfering RNAs (siRNA) were designed to target either human CYP19 mRNA or human COX-2 mRNA. The effects of siAROM and siCOX2 on aromatase enzyme activity in SK-BR-3 cells were examined (Figure 5). The transfection of siAROM resulted in suppression of basal levels of aromatase activity by greater than 90% compared with cells transfected with a nonspecific control siRNA or untreated cells. Also shown in this figure, the siCOX2 also resulted in suppression of aromatase activity by approximately 67%. Furthermore, the administration of PGE2 to cells treated with the siRNAs results in antagonism of only the siCOX2 and restores aromatase activity to untreated levels (Figure 5) [10].
Figure 5.
Effect of PGE2 on suppression of aromatase activity by siRNAs in SK-BR-3 cells [10]. The value of 100% is equal to 0.025 pmol/hr/106 cells; n = 4.
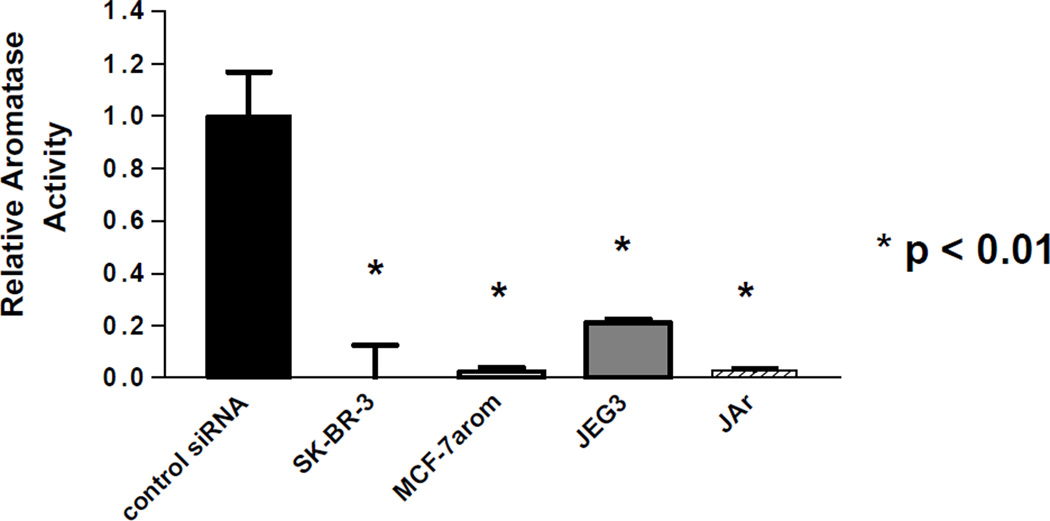
The effect of the siAROM on aromatase enzyme activity in several cancer cells were also examined (Figure 6). The siAROM silenced aromatase enzyme activity by greater than 90% in the SK-BR-3 human breast cancer cell line, in MCF-7 human breast cancer cells transfected with the aromatase gene, and in the JAr human choriocarcinoma cell line. The suppression of aromatase enzyme activity was greater than 80% in the JEG-3 human choriocarcinoma cell line.
Figure 6.
Suppression of aromatase activity by siRNAs in various human cancer cell lines. The value of 100% is equal to 0.025 pmol/hr/106 cells; n = 3.
4. Discussion
Two pharmacological approaches of novel therapeutic interventions targeting tissue-specific expression of aromatase have been recently developed in our laboratories. Our small molecule approach utilizes drug discovery technologies to develop novel sulfonanilides as selective agents regulating aromatase expression. Targeted synthetic libraries of novel sulfonanilide analogs, derived from COX-2 inhibitiors nimesulide and NS-398, decreased CYP19 mRNA expression and suppressed aromatase enzyme activity in SK-BR-3 breast cancer cells in a dose dependent manner. Furthermore, N-methyl sulfonanilide analogs without COX-2 inhibitory activity showed similar effects of decreased CYP19 mRNA expression and suppression of aromatase activity. Thus, these results suggest that the novel sulfonanilides targeting aromatase expression may be valuable tools for selective regulation of aromatase in breast cancer.
Investigations using RNA interference (RNAi) technology confirmed the interactions between aromatase and cyclooxygenases in breast cancer. Short interfering RNAs (siRNA) were designed against either human CYP19 mRNA or human COX-2 mRNA. Treatment of breast cancer cells with siAROM suppressed CYP19 mRNA and aromatase enzyme activity. Treatment with siCOX2 downregulated the expression of COX-2 mRNA and also resulted in suppression of aromatase mRNA. The administration of PGE2 to cells treated with the siRNAs results in antagonism of only the siCOX2 and restores aromatase activity to untreated levels. Finally, the siAROM was also very effective in silencing aromatase enzyme activity in human breast cancer cells in culture and in human choriocarcinoma cells in culture.
The regulation of aromatase in breast cancer involves complex autocrine and paracrine interactions, resulting in significant consequences on the pathogenesis of hormone-dependent breast cancer. The breast cancer tissue microenvironment can influence the extent of estrogen biosynthesis and metabolism, resulting in altered levels of hormonally active estrogens and therefore influencing breast tumor development and growth. Furthermore, novel sulfonanilides and various RNAi technologies targeting aromatase expression may be valuable tools for selective regulation of aromatase in breast cancer and could be developed into the first generation of Selective Aromatase Modulators for the treatment of hormone-dependent breast cancer.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) Grant R01 CA73698 (R.W.B.), the USAMRMC Breast Cancer Program Grant W81XWH-08-1-0521, and The Ohio State University Comprehensive Cancer Center Breast Cancer Research Fund.
Footnotes
Presented at the IX International Aromatase Conference, Shanghai, China, October 14–17, 2008.
References
- 1.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr. Rev. 1994;15:342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 2.Simpson ER, Clyne C, Rubin G, Boon WC, Robertson K, Britt K, Speed C, Jones M. Aromatase--a brief overview. Annu. Rev. Physiol. 2002;64:93–127. doi: 10.1146/annurev.physiol.64.081601.142703. [DOI] [PubMed] [Google Scholar]
- 3.James VH, McNeill JM, Lai LC, Newton CJ, Ghilchik MW, Reed MJ. Aromatase activity in normal breast and breast tumor tissues: in vivo and in vitro studies. Steroids. 1987;50:269–279. doi: 10.1016/0039-128x(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 4.Miller WR, O'Neill J. The importance of local synthesis of estrogen within the breast. Steroids. 1987;50:537–548. doi: 10.1016/0039-128x(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 5.Reed MJ, Owen AM, Lai LC, Coldham NG, Ghilchik MW, Shaikh NA, James VH. In situ oestrone synthesis in normal breast and breast tumour tissues: effect of treatment with 4-hydroxyandrostenedione. Int. J. Cancer. 1989;44:233–237. doi: 10.1002/ijc.2910440208. [DOI] [PubMed] [Google Scholar]
- 6.Bulun SE, Price TM, Aitken J, Mahendroo MS, Simpson ER. A link between breast cancer and local estrogen biosynthesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J. Clin. Endocrinol. Metab. 1993;77:1622–1628. doi: 10.1210/jcem.77.6.8117355. [DOI] [PubMed] [Google Scholar]
- 7.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Corbin CJ, Mendelson CR. Tissue-specific promoters regulate aromatase cytochrome P450 expression. Clin. Chem. 1993;39:317–324. [PubMed] [Google Scholar]
- 8.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Transcriptional regulation of CYP19 gene (aromatase) expression in adipose stromal cells in primary culture. J. Steroid Biochem. Mol. Biol. 1997;61:203–210. doi: 10.1016/s0960-0760(97)80013-1. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Estrogen biosynthesis proximal to a breast tumor is stimulated by PGE2 via cyclic AMP, leading to activation of promoter II of the CYP19 (aromatase) gene. Endocrinology. 1996;137:5739–5742. doi: 10.1210/endo.137.12.8940410. [DOI] [PubMed] [Google Scholar]
- 10.Brueggemeier RW, Su B, Sugimoto Y, Diaz-Cruz ES, Davis DD. Aromatase and COX in breast cancer: enzyme inhibitors and beyond. J. Steroid Biochem. Mol. Biol. 2007;106:16–23. doi: 10.1016/j.jsbmb.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su B, Diaz-Cruz ES, Landini S, Brueggemeier RW. Novel sulfonanilide analogues suppress aromatase expression and activity in breast cancer cells independent of COX-2 inhibition. J. Med. Chem. 2006;49:1413–1419. doi: 10.1021/jm051126f. [DOI] [PubMed] [Google Scholar]
- 12.Su B, Landini S, Davis DD, Brueggemeier RW. Synthesis and biological evaluation of selective aromatase expression regulators in breast cancer cells. J. Med. Chem. 2007;50:1635–1644. doi: 10.1021/jm061133j. [DOI] [PubMed] [Google Scholar]
- 13.Su B, Tian R, Darby MV, Brueggemeier RW. Novel Sulfonanilide Analogs Decrease Aromatase Activity in Breast Cancer Cells: Synthesis, Biological Evaluation, and Ligand-Based Pharmacophore Identification. J. Med. Chem. 2008;51:1126–1135. doi: 10.1021/jm701107h. [DOI] [PubMed] [Google Scholar]
- 14.Su B, Darby MV, Brueggemeier RW. Synthesis and biological evaluation of novel sulfonanilide compounds as antiproliferative agents for breast cancer. J. Comb. Chem. 2008;10:475–483. doi: 10.1021/cc700138n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Cruz ES, Shapiro CL, Brueggemeier RW. Cyclooxygenase inhibitors suppress aromatase expression and activity in breast cancer cells. J. Clin. Endocrinol. Metab. 2005;90:2563–2570. doi: 10.1210/jc.2004-2029. [DOI] [PubMed] [Google Scholar]
- 16.Julemont F, de L, Michaux XC, Damas J, Charlier C, Durant F, Pirotte B, Dogne JM. Spectral and crystallographic study of pyridinic analogues of nimesulide: determination of the active form of methanesulfonamides as COX-2 selective inhibitors. J. Med. Chem. 2002;45:5182–5185. doi: 10.1021/jm020920n. [DOI] [PubMed] [Google Scholar]