Abstract
A chemoenyzmatic method for direct glycosylation of polypeptides is described. The method consists of two site-specific enzymatic glycosylation steps: introduction of a glucose moiety at the consensus N-glycosylation sequence (NXS/T) in a polypeptide by an N-glycosyltransferase (NGT) and attachment of a complex N-glycan to the glucose primer by an endoglycosidase (ENGase)-catalyzed transglycosylation. Our experiments demonstrated that a relatively small excess of the UDP-Glc (the donor substrate) was sufficient for an effective glucosylation of polypeptides by the NGT, and different high-mannose and complex type N-glycans could be readily transferred to the glucose moiety by ENGases to provide full-size glycopeptides. The usefulness of the chemoenzymatic method was exemplified by an efficient synthesis of a complex glycoform of polypeptide C34, a potent HIV inhibitor derived from HIV-1 gp41. A comparative study indicated that the Glc-peptide was equally efficient as the natural GlcNAc-peptide to serve as an acceptor in the transglycosylation with sugar oxazoline as the donor substrate. Interestingly, the Glc–Asn linked glycopeptide was completely resistant to PNGase F digestion, in contrast to the GlcNAc–Asn linked natural glycopeptide that is an excellent substrate for hydrolysis. In addition, the Glc–Asn linked glycopeptide showed at least 10-fold lower hydrolytic activity toward Endo-M than the natural GlcNAc–Asn linked glycopeptide. The chemoenzymatic glycosylation method described here provides an efficient way to introducing complex N-glycans into polypeptides, for gain of novel properties that could be valuable for drug discovery.
Keywords: Glycopeptide, Glycoprotein, Chemoenzymatic synthesis, N-glycosyltransferase, Endoglycosidase, Transglycosylation, Enzymatic glycosylation
1. Introduction
Protein glycosylation is one of the most common post-translational modifications involved in protein folding, intracellular trafficking, secretion, and many other important biological recognition processes.1–5 However, a detailed understanding of the effects of glycosylation on the structure and function of proteins is hampered by the lack of well-defined homogeneous glycopeptides and glycoproteins, which are difficult to obtain from natural sources. To overcome this hurdle, many laboratories worldwide are dedicated to the development of various chemical, enzymatic, chemoenzymatic, and bioengineering methods for the synthesis of homogeneous glycopeptides and glycoproteins.6–10 As part of these efforts, several groups including Wang and co-workers have developed a convergent chemoenzymatic method that permits site-specific enzymatic ligation between an activated glycan oxazoline and a GlcNAc-peptide/protein to give the homogeneous glycopeptide/glycoprotein.11–30 This chemoenzymatic method is based on the transglycosylation activity of a class of endo-β-N-acetylglucosaminidases (ENGases)—their native function is to hydrolyze N-glycans from glycoproteins. A big advantage of the endoglycosidase-based method is its high convergence—ligation of a large intact oligosaccharide en bloc to a GlcNAc-polypeptide in a single step and in a regio- and stereo-specific manner. Moreover, the discovery of synthetic glycan oxazolines as the donor substrates and the generation of novel glycosynthases that can use glycan oxazolines of different types for transglycosylation but lack the product hydrolysis activity, has significantly expanded the scope and efficiency of the method for glycopeptide and glycoprotein synthesis.7,9,10,16,31 Nevertheless, this chemoenzymatic method requires a facile preparation of a monosaccharide-containing polypeptide or protein as the precursor. While a monosaccharide moiety can be introduced during the polypeptide synthesis via automated solid-phase peptide synthesis, a method that allows efficient site-specific attachment of a monosaccharide moiety to free natural or synthetic polypeptides or proteins would be highly desirable.20 It has been previously reported that a class of bacterial N-glycosyltransferases (NGTs) (also called HMW1C proteins) are responsible for glycosylation of HMW1A proteins (important for cell adhesion) by transferring a monosaccharide moiety (Glc or Gal) from the corresponding sugar-nucleotide (UDP-Glc or UDP-Gal) to the Asn side chain in the consensus N-glycosylation sequence (NXS/T) of the proteins.32–35 Recently the Aebi group has reported that a recombinant NGT from Actinobacillus pleuropneumoniae (ApNGT) can perform in vitro glycosylation on various polypeptides having the NXS/T sequence.36 In this paper, we report an efficient method for direct glycosylation of free polypeptides with a defined complex oligosaccharide that combines the action of two enzymes, the NGT for initial glycosylation of the polypeptide and the ENGase for transfer of an N-glycan to the installed glucose moiety. We also show that the Glc–Asn linkage is resistant to PNGase cleavage and has much reduced susceptibility toward ENGase-catalyzed hydrolysis.
2. Results and discussion
2.1. NGT-catalyzed glucosylation of polypeptides at the consensus NXS/T N-glycosylation site
Previous work has shown that the glycosyl transferase from Actinobacillus pleuropneumoniae (ApNGT) was able to transfer a glucose moiety from UDP-Glc to the Asn side chain in a consensus sequence NXS/T of a polypeptide, where X is any natural amino acid except proline.36 Initial studies used a large excess of UDP-Glc (100:1, donor/acceptor) to drive the reaction. To establish a practical synthetic procedure, we first examined the effects of the ratios of donor/acceptor substrates on the efficiency of glucosylation, using a 16-mer peptide (compound 1) derived from the yeast glycoprotein lysophospholipase 2 (Plb2) as the acceptor (Scheme 1). Recombinant ApNGT and acceptor (1) were kept at a constant concentration while the donor UDP-Glc concentration was varied in a Tris buffer (pH 8.0). The reaction was monitored by reverse-phase HPLC and the results were shown in Figure 1. It was found that the change in donor/acceptor ratio from 100:1 to 10:1 had no apparent effect on the rate of glucose transfer to the peptide (Fig. 1A). The enzymatic reaction slowed down when the donor/acceptor ratio was dropped from 10:1 to 5:1, but the glycosylation still proceeded smoothly to give the product (2) over the time course (Fig. 1A). To examine whether product inhibition (caused by the released UDP) was a factor for the slowing down of the enzymatic reaction when the donor/acceptor ratio was reduced, we included Calf intestinal phosphatase37 in the reaction to perform in situ hydrolysis of UDP. However, no change in the Glc-peptide yield (2) or the reaction rate was observed (data not shown). Thus, an appropriate excess of the donor substrate (e.g., 5- to 10-fold excess) seems sufficient for an efficient enzymatic transformation. The glycosylation product (2) was readily purified by RP-HPLC and its identity was confirmed by ESI-MS analysis (calcd for glycopeptide 2, M = 1924.08; found, 1925.00 [M+H], 963.30 [M+2H]2+, 642.60 [M+3H]3+) (Fig. 1B). It should be mentioned that the enzymatic glycosylation was scalable and the yield could reach over 90% when the reaction was performed with elongated time (overnight incubation) with excess of donor substrate (5- to 10-fold excess).
Scheme 1.
NGT-catalyzed glucosylation of polypeptides.
Figure 1.
Effects of donor/acceptor ratios on ApNGT catalyzed glucosylation. (A) Glucosylation of peptide 1 with varying donor/acceptor ratio: 100:1 (○), 50:1 (■), 25:1 (▲), 10:1 (◇), and 5:1 (□). Reaction was run at RT using constant enzyme (ApNGT) and acceptor Plb2 (1). Aliquots were taken at intervals and analyzed by RP-HPLC. (B) ESI-MS spectrum of purified product, Glc-peptide 2. Inset: HPLC profile of purified Glc-peptide 2.
Previous study indicated that the ApNGT was able to glycosylate a series of synthetic peptides derived from yeast glycoproteins carrying the conserved NXS/T N-glycosylation site.36 We next examined the glycosylation of a large 34-mer polypeptide, C34, a potent HIV fusion inhibitor derived from HIV-1 gp41 envelope gly-coprotein (amino acid sequence 628–661).38–40 C34 contains a conserved N-glycosylation site at the N637 position (HXB2 gp120 numbering). It was found that ApNGT was equally efficient at introducing a glucose moiety in C34 at the consensus N-glycosylation site (N637) (Scheme 1). As monitored by HPLC (Fig. 2A), the glycosylation product (4), Glc–C34, appeared earlier than C34 under the RP-HPLC condition, and the product was easily purified. The identity of the purified product (4) was confirmed by ESI-MS analysis (Fig. 2B). Deconvolution of the MS data gave an observed molecular mass of 4453.84, which matches well with the calculated molecular mass of Glc–C34 (M = 4453.67).
Figure 2.
HPLC and ESI-MS analysis of HIV-gp41 C-peptide (C34). (A) HPLC trace of glucose transfer reaction to C34; peak (a) is the starting peptide 3, where peak (b) is glucosylated peptide 4. (B) ESI-MS profile of Glc–C34. Inset: HPLC profile of the product.
2.2. ENGase-catalyzed transglycosylation of glucose-containing polypeptides
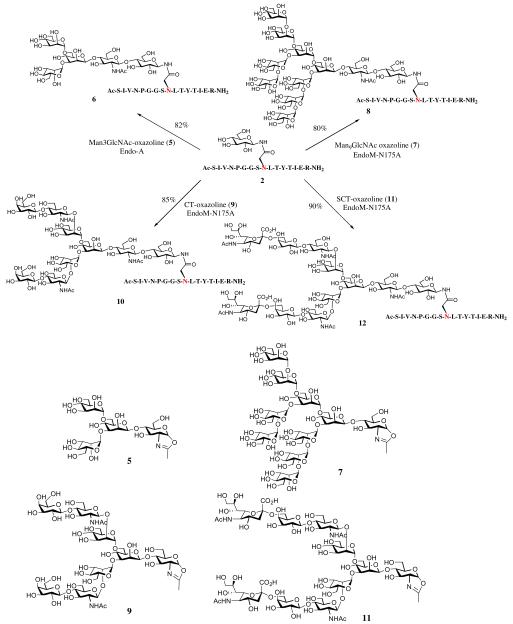
The efficient glycosylation of polypeptides by ApNGT permits site-specific introduction of a glucose moiety at the consensus N-glycosylation site, which provides a primer for potential extension of the sugar chain. We have previously shown that the endoglycosidases from Arthrobacter protophormiae (Endo-A) and Mucor hiemalis (Endo-M) were able to recognize the glucose moiety on various natural products for transglycosylation and could introduce an N-glycan onto the natural products using a glycan oxazoline donor substrate.41 An earlier study has also demonstrated that Endo-A could transglycosylate a glucose moiety in a short peptide.42 To test the feasibility of ENGase-catalyzed transglycosylation of large Glc-peptides for making complex glycopeptides, we first tested the Man3GlcNAc oxazoline (5) corresponding to the N-glycan core for transglycosylation (Scheme 2). Incubation of Man3GlcNAc oxazoline (5) and Glc-peptide (2) (donor/acceptor, 4:1) along with Endo-A in a phosphate buffer (pH 7.4) resulted in the formation of a new product (6) that was eluted earlier than Glc-peptide (2) under a RP-HPLC condition. The reaction was monitored by RP-HPLC, and additional sugar oxazoline and enzyme were added to drive the reaction to reaching over 90% yield (based on the acceptor). The transglycosylation product (6) was isolated by RP-HPLC and its identity was confirmed by ESI-MS analysis (Fig. 3A). The deconvolution of the MS data showed a peak at 2613.60, which was in good agreement with the calculated molecular mass of 6 (M = 2613.68). It should be mentioned that the regio- and stereo-specificity of the endoglycosidase-catalyzed transglycosylation on glucose-containing acceptors was previously examined by a detailed NMR (TOCOSY and NOESY) analysis of the transglycosylation product.41 The NOESY analysis unambiguously confirmed that the transferred glycan was attached to the 4-OH of the glucose moiety to form a β-1,4-glycosidic linkage between the GlcNAc and Glc residues, which showed exactly the same regio- and stereo-specificity as in the case of the enzymatic transglycosylation with the GlcNAc-peptide acceptors.11,41 Thus, it is assumed that the newly formed glycosidic linkage in glycopeptide 6 would be the expected β-1,4-glycosidic linkage.
Scheme 2.
ENGase-catalyzed transglycosylation of Glc-containing polypeptide with different glycan oxazolines.
Figure 3.
ESI-MS profiles of the transglycosylation products. (A) Man3GlcNAc–Glc-peptide (6); (B) Man9GlcNAc–Glc-peptide (8); (C) asialylated complex type glycan-containing glycopeptide (10); and (D) asialylated complex type glycan-containing glycopeptide (12).
With this success, we sought to expand the attached glycan from the core to more complex glycan types, including a high-mannose type, an asialylated complex type, and a fully sialylated bi-antennary complex type N-glycan. Incubation of Man9GlcNAc oxazoline (7)43 and the Glc–Plb2 peptide (2) (donor/acceptor, 4:1, molar ratio) along with EndoM-N175A43 in a phosphate buffer (pH 7.4) resulted in the formation of high-mannose type glycopeptide 8 in over 50% within 1 h. Addition of a new portion of EndoM-N175A and sugar oxazoline (2 mol equiv) and incubation of the mixture for another 3 h led to the formation of ca. 80% of the product (8). The transglycosylation product was purified by RP-HPLC and its identity was confirmed by ESI-MS analysis (Fig. 3B) (calcd for glycopeptide 8, M = 3586.56; found = 1794.10 [M+2H]2+, 1196.50 [M+3H]3+; deconvoluted molecular mass, M = 3586.50). Gratifyingly, the excess glycan oxazoline was efficiently recovered in the form of free reducing glycan during HPLC purification of the transglycosylation product, and it could be readily converted into the glycan oxazoline (7) for re-use via an efficient single step transformation using a 2-chloroimidazolinium salt as the activating reagent.18,44 The transglycosylations of the Glc-peptide (2) with the asialylated complex type glycan oxazoline (CT-oxazoline, 9)17 and the sialylated complex type glycan oxazoline (SCT-oxazoline 11)18 using EndoM-N175A as the catalyst were performed in the similar manner as in the use of the Man9GlcNAc oxazoline (7), leading to the formation of the corresponding complex type glycopeptides 10 and 12, respectively (Scheme 2). The transglycosylation gave a satisfactory yield (80–90%) when an excess sugar oxazoline and sufficient enzyme were used. The respective transglycosylation product was purified by RP-HPLC and characterized by ESI-MS [Fig. 3C for glycopeptide 10, calculated, M = 3344.38; found, M = 3344.70 (deconvoluted data); Fig. 3D for glycopeptide 12, calculated, M = 3926.89; found, M = 3927.00 (deconvolution data)]. Again, the excess glycan oxazoline was recovered as reducing glycan after the completion of the reaction and was re-used for transglycosylation after re-activated. These results suggest that both high-mannose type and complex type N-glycans could be efficiently transferred to the glucose moiety in the Glc-peptide under the catalysis of an appropriate glycansynthase to provide a large glycopeptide.
2.3. Synthesis of a complex type glycoform of C34
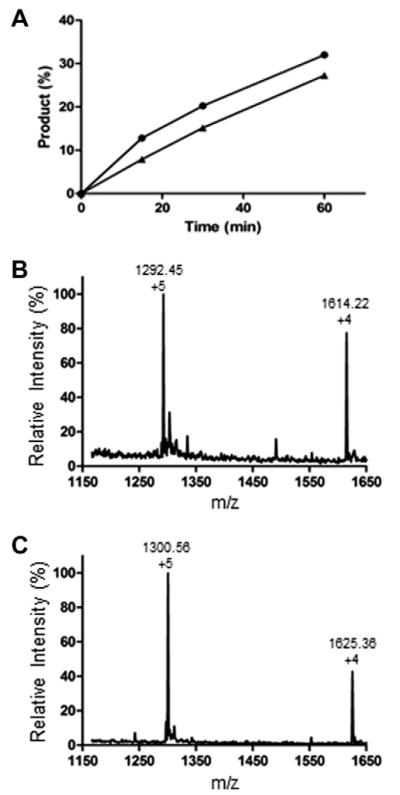
After showing the feasibility of enzymatic glucosylation and sugar chain extension by the action of the two enzymes, we further expanded the approach to the synthesis of more complex glycopeptides derived from C34. The transglycosylation of a complex type N-glycan to the Glc–C34 and GlcNAc–C34 was compared (Scheme 3). The Glc–C34 was synthesized as described above, and the GlcNAc–C34 was prepared by solid-phase peptide synthesis using the Fmoc-Asn(Ac3GlcNAc)-OH as the building block to install the GlcNAc–Asn moiety according to our previously described procedure.40 Transfer of the sialylated complex type N-glycan from the SCT-glycan oxazoline (11) to Glc–C34 (4) and GlcNAc–C34 (14) was performed in a phosphate buffer (pH 7.4) containing 20% DMSO in the presence of EndoM-N175A mutant. DMSO was added to enhance the solubility of the C34 polypeptide acceptor. Reactions were monitored by RP-HPLC at dual wavelengths (214 and 280 nm). The Glc- and GlcNAc-peptide acceptors (4 and 14) showed comparable reactivity in the enzymatic transglycosylation with the active sugar oxazoline and gave the corresponding transglycosylation products, 13 and 15, respectively (Fig. 4A). The yield was driven to over 80% when additional enzyme was added with a prolonged incubation time (5 h). The products (SCT–Glc–C34 and SCT–GlcNAc–C34) were purified by preparative HPLC and their identity was confirmed by ESI-MS analysis (Fig. 4B and C): ESI-MS of SCT–Glc–C34 (13), calcd, M = 6456.67; found, M = 6456.30 (deconvolution data). ESI-MS of SCT–GlcNAc–C34 (15), calcd, M = 6497.70; found, M = 6497.80 (deconvolution data). These experiments demonstrate that the combined use of ApNGT-catalyzed glucosylation and ENGase-catalyzed transglycosylation provides a feasible approach to introducing complex N-glycans into free polypeptides in a site-specific manner with natural glycosidic linkages, in which the GlcNAc–Asn linkage found in mammalian natural N-glycoproteins is replaced with a Glc–Asn moiety.
Scheme 3.
ENGase-catalyzed trasnglycosylation of Glc–C34 and GlcNAc–C34.
Figure 4.
Analysis of the transglycosylation of Glc–C34 and GlcNAc–C34. (A) Transglycosylation of GlcNAc–C34 (●) and Glc–C34 (▲) with sialylated glycan (SCT) oxazoline (11); (B) ESI-MS spectrum of SCT–Glc–C34 (13). (C) ESI-MS spectrum of SCT–GlcNAc–C34 (15).
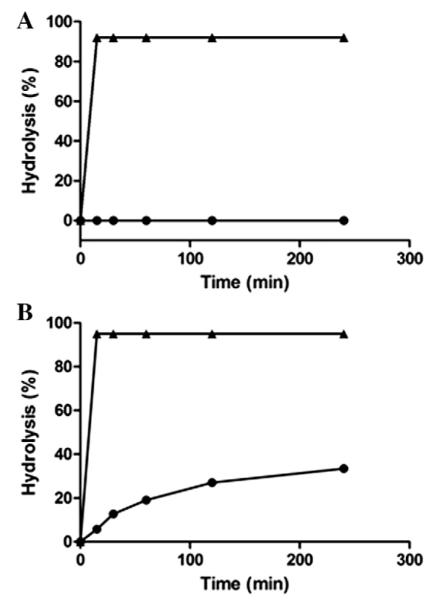
2.4. Hydrolysis of Glc–Asn and GlcNAc–Asn linked glycopeptides by PNGase F and Endo-M
To examine how the replacement of the Asn linked GlcNAc by a Glc moiety in a complex N-glycopeptide affects the stability of the glycopeptide toward glycan-hydrolyzing enzymes, we tested the hydrolysis of the synthetic Glc- and GlcNAc-containing glycoforms (13 and 15, respectively) of C34 by PNGase F and Endo-M. PNGase F is a peptide-N4-(N-acetyl-β-d-glucosaminyl) asparagine amidase that cleaves the glycosylaminde bond in Asn linked glycopeptides and glycoproteins releasing the glycans and converts the asparagine residue to an aspartic acid. The SCT–Glc–C34 (13) and SCT–GlcNAc–C34 (15) were treated with PNGase F and the reaction was monitored by RP-HPLC. It was found that PNGase F could rapidly hydrolyze the natural glycopeptide SCT–GlcNAc–C34 (15) within 15 min under the conditions, but the Glc–Asn linked glycopeptide (13) was completely resistant to PNGase hydrolysis (Fig. 5A). This result was consistent with previous reports showing that PNGase F was unable to hydrolyze Glc–Asn linked synthetic glycopeptides.42,45 Next we tested the ability of Endo-M to recognize and hydrolyze the synthetic Glc-glycopeptide. Endo-M is an endoglycosidase that cleaves β-1,4-glycosidic linkage between the two GlcNAc residues in the chitobiose core of natural N-glycans for deglycosylation of glycoproteins. As expected, treatment of the natural GlcNAc–Asn linked C34 glycopeptide (15) with Endo-M led to a fast hydrolysis of the glycopeptide. It was found that more than 90% of 15 was hydrolyzed within 15 min under the reaction conditions (Fig. 5B). The Endo-M was found to be able also to hydrolyze the Glc–Asn linked glycopeptide (13) but, interestingly, the enzymatic hydrolysis was at a much slower rate, with less than 10% hydrolysis within the first 15 min under the same reaction conditions (Fig. 5B). Given the fact that ENGases can transglycosylate both the Glc- and GlcNAc-containing polypeptide C34 with sugar oxazoline donor substrate at comparable reaction rates (Fig. 4), the big difference in reactivity of glycopeptides 13 and 15 toward Endo-M catalyzed hydrolysis suggests that the Asn linked GlcNAc serves as an important element for the enzyme to recognize for the hydrolysis of the ground-state substrates.
Figure 5.
Digestion of Glc–Asn linked (13) and GlcNAc–Asn linked glycopeptide (15) by PNGase F and Endo-M. (A) SCT–Glc–C34 (●) and SCT–GlcNAc–C34 (▲) were digested by PNGaseF; (B) SCT–Glc–C34 (●) and SCT–GlcNAc–C34 (▲) were digested by Endo-M.
3. Conclusion
Described in this paper is an efficient chemoenzymatic method for direct glycosylation of free polypeptides with a variety of complex glycans. This method combines the action of two enzymes, an N-glycosyltransferase (NGT) for initial site-specific introduction of a monosaccharide primer at the conserved N-glycosylation site (NXS/T) and an endoglycosidase or its mutant for subsequent extension of the sugar chain into complex N-glycan. We have shown that small or large synthetic polypeptides can be glycosylated and that the resulting glycopeptides possess interesting properties toward digestion by PNGase F and ENGases. A major advantage of the chemoenzymatic method is its convergence and flexibility, which provides a solution to the long-standing problem of incompatibility of the chemistry for the synthetic manipulations of oligosaccharide and polypeptide portions in chemical synthesis of complex glycopeptides. The next step is to test an expanded library of synthetic polypeptides as well as natural or recombinant proteins to define the scope (and limitation) of the chemoenzymatic method.
4. Experimental
4.1. Materials and general methods
N-glycosyltransferase from Actinobacillus pleuropneumoniae (ApNGT) was cloned, expressed, and purified as demonstrated previously.36 Wild type Endo-A and mutant EndoM-N175A were expressed and purified according to the previously reported procedures.43,46 Peptide (1) (Ac-SIVNPGGSNLTYTIER-NH2) derived from the yeast glycoprotein lysophospholipase 2 (Plb2), C34 (a potent HIV inhibitor derived from HIV-1 gp41), and GlcNAc–C34 were synthesized on an automated peptide synthesizer using Fmoc chemistry, following our previously described procedures.40 The synthesis of Man3GlcNAc oxazoline (5) was previously described.11 Man9GlcNAc oxazoline (7) was synthesized according to our previously described procedure.43 The asialo- and sialylated complex glycan oxazolines (CT-oxazoline 9 and SCT-oxazoline 11, respectively) were prepared from the sialyl glycopeptide (SGP) isolated from chicken egg yolks, following our previously described procedure.17,18 UDP-glucose was purchased from Calbiochem. Calf intestinal phosphatase and PNGase F were purchased from New England Biolabs (NEB). All other reagents were purchased from Sigma–Aldrich and used as received. Analytical HPLC was carried out on a Waters 626 HPLC instrument equipped with a Waters Nova-Pak C18 column (3.9 × 150 mm) at 40 °C. The column was eluted with a linear gradient of 0–90% MeCN containing 0.1% TFA at a flow rate of 1 mL/min over 20 min. Peptides and glycopeptides were detected at dual wavelengths (214 and 280 nm). Preparative HPLC was performed on a Waters 600 HPLC instrument with a Waters C18 preparative column (Symmetry 300, 19 × 300 mm). The column was eluted with a suitable gradient of aq MeCN containing 0.1% TFA at 10 mL/min. Glycopeptides were quantified by HPLC analysis through integration of the peptide/glycopeptide peaks using standard Plb2 peptide solution as the reference.
4.2. Analysis of effects of changes in donor/acceptor ratios on the rate of apNGT-catalyzed glucosylation of polypeptide (1)
A solution of the donor substrate (UDP-Glc) and the acceptor substrate (peptide 1) at varied ratios (from 100:1 to 5:1, at a fixed concentration of 0.6 mM for peptide 1) in a buffer (150 mM NaCl buffered with 25 mM Tris, pH 7.9) was incubated with enzyme ApNGT at a fixed concentration of 9 μM). The reaction was run at 30 °C for 4 h taking time points by quenching an aliquot of the reaction in a 0.1% TFA solution. Samples were then injected and run over a Waters 626 HPLC, Symmetry™ 300 C18 (5.0 μm, 4.6 × 250 mm), with a gradient from 0% to 90% acetonitrile containing 0.1% TFA over a 30 min period while monitoring at 214 nm wavelength. Peaks were integrated using analysis software and percent glycosylation was plotted as a function of time. Peak identification was determined using the Waters Micromass ZQ-4000 single quadrupole electron spray ionization-mass spectrometer (ESI-MS).
4.3. Synthesis of Glc-peptide (2)
A solution of UDP-Glc (3.9 mg, 6.9 μmol) and peptide 1 (3.0 mg, 0.70 μmol) in a Tris buffer (25 mM, pH 7.9, 3 mL) containing 20% DMSO (to increase the solubility of the peptide) was incubated at 30 °C for 32 h in the presence of ApNGT (10 μM). The glucosylation product was purified by preparative RP-HPLC on a Waters 626 HPLC instrument with a Symmetry Prep™ C18-column (7 μm, 19 × 300 mm) (gradient: 0–90% acetonitrile containing 0.1% trifluoracetic acid over 30 min). The fractions containing pure product were pooled and lyophilized to give Glc-peptide 2 (2.8 mg) in 90% yield. ESI-MS of Glc-peptide 2: calcd, M = 1924.08; found, 1925.00 [M+H], 963.52 [M+2H]2+, 642.60 [M+3H]3+.
4.4. Synthesis of Glc–C34 (4) via glucosylation of peptide C34
ApNGT-catalyzed glucosylation of C34 (3) (1 mg) was performed under the same conditions described for the preparation of Glc-peptide (2) using an excess amount of UDP-Glc (UDP-Glc/C34, 10:1). The product (4) was purified by preparative HPLC to give Glc–C34 (0.9 mg) in 88% yield. ESI-MS: calcd for Glc–C34, M = 4453.67; found, 1114.46 [M+4H]4+, 1485.55 [M+3H]3+; deconvolution data, M = 4453.84.
4.5. Synthesis of the Man3GlcNAc–Glc-peptide (6) through Endo-A catalyzed transglycosylation
A mixture Man3GlcNAc–oxazoline (5) (1.2 mg, 1.8 μmol, 3.6 mM) and Glc-peptide (2) (1.2 mg, 0.55 μmol, 1.2 mM) in a phosphate buffer (500 μL, 100 mM, pH 7.4) was incubated with Endo-A (2 μg) at 30 °C. Reaction progression was monitored using analytical RP-HPLC. After 2 h, additional sugar oxazoline (0.66 mg, 1.0 μmol) was added and the solution was incubated for another 2 h when HPLC indicated the completion of the enzymatic reaction. The reaction mixture was subject to preparative HPLC purification to afford the glycopeptide product (6) (1.0 mg, 82%). ESI-MS of glycopeptide 6: calcd, M = 2613.68; found, 1307.80 [M+2H]2+; deconvolution data, M = 2613.60.
4.6. Synthesis of the Man9GlcNAc–Glc-peptide (8), the asialylated complex type glycopeptide (10) and the sialylated complex type glycopeptide (12) through EndoM-N175A catalyzed transglycosylation
A mixture of Man9GlcNAc–oxazoline (7) (260 μg, 156 nmol) and Glc–Plb2 (2) (100 μg, 52 nmol) in a phosphate buffer (10 μL, 100 mM, pH 7.4) was incubated with EndoM-N175A (10 μg) at 30 °C. Reactions were monitored by RP-HPLC. After 2 h, additional Man9GlcNAc oxazoline (7) (100 μg) and EndoM-N175A (5 μg) were added and the reaction was continued for another 3 h. The product was purified by RP-HPLC to give the Man9GlcNAc–Glc-peptide (8) (149 μg, 80% yield). The amount of the product was quantified by RP-HPLC analysis using a standard concentration of the Plb2 peptide (1) as the reference. The synthesis of the asialylated glycopeptide (10) and the sialylated glycopeptide (12) was performed in the same way as for the synthesis of the Man9GlcNAc–Glc-peptide (8), using the corresponding glycan oxazolines (9 and 11) as the donor substrates, respectively. The products were purified by RP-HPLC and analyzed by ESI-MS. Glycopeptide 8: yield, 80%; ESI-MS, calcd, M = 3586.56; found, 1794.10 [M+2H]2+, 1196.50 [M+3H]3+; deconvolution data, M = 3586.50. Glycopeptide 10, yield, 85%. ESI-MS, calcd, M = 3344.38; found, 1673.60 [M+2H]2+, 1115.90 [M+3H]3+; deconvolution data, M = 3344.70. Glycopeptide 12: yield, 90%; ESI-MS, calcd, M = 3926.89; found, 1964.50 [M+2H]2+, 1310 [M+3H]3+, 982.90 [M+4H]4+; deconvolution data, M = 3926.60.
4.7. Synthesis of sialylated complex type C34 glycopeptide (13)
A solution of SCT–oxazoline (11) (1.0 mg, 560 nmol) and Glc–C34 (4) (500 μg, 112 nmol) in a phosphate buffer (100 μL, 100 mM, pH 7.4) was incubated with EndoM-N175A (10 μg) at 30 °C. Reactions were monitored by RP-HPLC. After 2 h, additional SCT–oxazoline (11) (200 μg) and EndoM-N175A (5 μg) were added and the reaction was continued for another 3 h. The product was purified by RP-HPLC to give the SCT–Glc-peptide (13) (600 μg, 83% yield). ESI-MS of SCT–Glc–C34 (13): calcd M = 6456.67; found, 1292.20 [M+5H]5+, 1615.22 [M+4H]4+.
4.8. Comparative analysis on the enzymatic transglycosylation of Glc–C34 and GlcNAc–C34
A solution of Glc–C34 (4) (100 μg, 22.4 nmol) or GlcNAc–C34 (14) (100 μg, 22.2 nmol) and the sialylated glycan oxazoline (11) (224 μg, 112 nmol) (molar ratio, donor/acceptor ratio,5:1) in a phosphate buffer (10 μL, 100 mM, pH 7.4) was incubated at 30 °C in the presence of EndoM-N175A (5 μg). Reactions were monitored by HPLC analysis of the aliquots taken at intervals over the time course. The aliquots were treated with a 0.1% TFA solution to quench the reaction before HPLC analysis. The transglycosylation yields at time points were calculated by dividing the product peak area by the total peak areas of product and the acceptor substrate. The transglycosylation product was confirmed by ESI-MS analysis. ESI-MS of SCT–GlcNAc–C34 (15): calcd M = 6497.04; found, 1300.56 [M+5H]5+, 1625.36 [M+4H]4+.
4.9. Digestion of Glc–Asn and GlcNAc–Asn linked glycopeptides by PNGase F and Endo-M
SCT–Glc–C34 (13) or SCT–GlcNAc–C34 (15) (10 μg) in a G7 buffer containing 10% NP-40 was treated with PNGase F (500 NEB units, equivalent to 7.7 milli IUB unit) at 30 °C for 4 h. The digestion progress was monitored by analytical RP-HPLC. The digestion of SCT–Glc–C34 (13) or SCT–GlcNAc–C34 (14) (10 μg) with Endo-M (7.2 μg) in a phosphate buffer (50 mM, pH 6.5) was performed at 30 °C for 4 h, and the reaction was monitored in a similar way by RP-HPLC. Hydrolysis was determined by integration of the peaks corresponding to starting material and product.
Acknowledgments
We thank Dr. Wei Huang and other members of the Wang Lab for technical assistance. A.N. is a member of the Zurich PhD program in molecular life science. This work was supported in part by the US National Institutes of Health (NIH Grant R01 GM080374 to L.-X.W.) and the Swiss National Science Foundation (Grant CRSII3_127333 to M.A.).
References and notes
- 1.Dwek RA. Chem. Rev. 1996;96:683. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 2.Helenius A, Aebi M. Science. 2001;291:2364. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 3.Hart GW, Copeland RJ. Cell. 2010;143:672. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marth JD, Grewal PK. Nat. Rev. Immunol. 2008;8:874. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jefferis R. Nat. Rev. Drug Disc. 2009;8:226. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 6.Gamblin DP, Scanlan EM, Davis BG. Chem. Rev. 2009;109:131. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 7.Rich JR, Withers SG. Nat. Chem. Biol. 2009;5:206. doi: 10.1038/nchembio.148. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Y, Chen J, Wan Q, Wilson RM, Danishefsky SJ. Biopolymers. 2010;94:373. doi: 10.1002/bip.21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmaltz RM, Hanson SR, Wong CH. Chem. Rev. 2011;111:4259. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- 10.Wang LX, Lomino JV. ACS Chem. Biol. 2012;7:110. doi: 10.1021/cb200429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Zeng Y, Hauser S, Song H, Wang LX. J. Am. Chem. Soc. 2005;127:9692. doi: 10.1021/ja051715a. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Li B, Song H, Breydo L, Baskakov IV, Wang LX. J. Org. Chem. 2005;70:9990. doi: 10.1021/jo051729z. [DOI] [PubMed] [Google Scholar]
- 13.Li B, Song H, Hauser S, Wang LX. Org. Lett. 2006;8:3081. doi: 10.1021/ol061056m. [DOI] [PubMed] [Google Scholar]
- 14.Wei Y, Li C, Huang W, Li B, Strome S, Wang LX. Biochemistry. 2008;47:10294. doi: 10.1021/bi800874y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochiai H, Huang W, Wang LX. J. Am. Chem. Soc. 2008;130:13790. doi: 10.1021/ja805044x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang LX, Huang W. Curr. Opin. Chem. Biol. 2009;13:592. doi: 10.1016/j.cbpa.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Li C, Li B, Umekawa M, Yamamoto K, Zhang X, Wang LX. J. Am. Chem. Soc. 2009;131:2214. doi: 10.1021/ja8074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, Yang Q, Umekawa M, Yamamoto K, Wang LX. ChemBioChem. 2010;11:1350. doi: 10.1002/cbic.201000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Zhang X, Ju T, Cummings RD, Wang LX. Org. Biomol. Chem. 2010;8:5224. doi: 10.1039/c0ob00341g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz F, Huang W, Li C, Schulz BL, Lizak C, Palumbo A, Numao S, Neri D, Aebi M, Wang LX. Nat. Chem. Biol. 2010;6:264. doi: 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Li J, Wang LX. ChemBioChem. 2011;12:932. doi: 10.1002/cbic.201000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou G, Ochiai H, Huang W, Yang Q, Li C, Wang LX. J. Am. Chem. Soc. 2011;133:18975. doi: 10.1021/ja208390n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amin MN, Huang W, Mizanur RM, Wang LX. J. Am. Chem. Soc. 2011;133:14404. doi: 10.1021/ja204831z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan SQ, Huang W, Wang LX. J. Biol. Chem. 2012;287:11272. doi: 10.1074/jbc.M112.340497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. J. Am. Chem. Soc. 2012;134:12308. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons TB, Patel MK, Boraston AB, Vocadlo DJ, Fairbanks AJ. Org. Biomol. Chem. 2010;8:1861. doi: 10.1039/b926078a. [DOI] [PubMed] [Google Scholar]
- 27.Parsons TB, Moir JW, Fairbanks AJ. Org. Biomol. Chem. 2009;7:3128. [Google Scholar]
- 28.Rising TW, Heidecke CD, Moir JW, Ling Z, Fairbanks AJ. Chem. Eur. J. 2008;14:6444. doi: 10.1002/chem.200800365. [DOI] [PubMed] [Google Scholar]
- 29.Heidecke CD, Ling Z, Bruce NC, Moir JW, Parsons TB, Fairbanks AJ. ChemBioChem. 2008;9:2045. doi: 10.1002/cbic.200800214. [DOI] [PubMed] [Google Scholar]
- 30.Rising TW, Claridge TD, Moir JW, Fairbanks AJ. ChemBioChem. 2006;7:1177. doi: 10.1002/cbic.200600183. [DOI] [PubMed] [Google Scholar]
- 31.Wang LX. Trends Glycosci. Glycotechnol. 2011;23:33. doi: 10.4052/tigg.23.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grass S, Buscher AZ, Swords WE, Apicella MA, Barenkamp SJ, Ozchlewski N, St. Geme JW., III Mol. Microbiol. 2003;48:737. doi: 10.1046/j.1365-2958.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- 33.Choi KJ, Grass S, Paek S, St Geme JW, III, Yeo HJ. PLoS One. 2010;5:e15888. doi: 10.1371/journal.pone.0015888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grass S, Lichti CF, Townsend RR, Gross J, St. Geme JW., III PLoS Pathog. 2010;6:e1000919. doi: 10.1371/journal.ppat.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawai F, Grass S, Kim Y, Choi KJ, St. Geme JW, III, Yeo HJ. J. Biol. Chem. 2011;286:38546. doi: 10.1074/jbc.M111.237602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz F, Fan YY, Schubert M, Aebi M. J. Biol. Chem. 2011;286:35267. doi: 10.1074/jbc.M111.277160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad Z, Huang KP. J. Biol. Chem. 1981;256:757. [PubMed] [Google Scholar]
- 38.Chan DC, Chutkowski CT, Kim PS. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15613. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan DC, Kim PS. Cell. 1998;93:681. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 40.Wang LX, Song H, Liu S, Lu H, Jiang S, Ni J, Li H. ChemBioChem. 2005;6:1068. doi: 10.1002/cbic.200400440. [DOI] [PubMed] [Google Scholar]
- 41.Huang W, Ochiai H, Zhang X, Wang LX. Carbohydr. Res. 2008;343:2903. doi: 10.1016/j.carres.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deras IL, Takegawa K, Kondo A, Kato I, Lee YC. Bioorg. Med. Chem. Lett. 1998;8:1763. doi: 10.1016/s0960-894x(98)00306-0. [DOI] [PubMed] [Google Scholar]
- 43.Umekawa M, Huang W, Li B, Fujita K, Ashida H, Wang LX, Yamamoto K. J. Biol. Chem. 2008;283:4469. doi: 10.1074/jbc.M707137200. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi M, Tanaka T, Gyakushi H, Kobayashi A, Shoda SI. J. Org. Chem. 2009;74:2210. doi: 10.1021/jo8024708. [DOI] [PubMed] [Google Scholar]
- 45.Fan JQ, Lee YC. J. Biol. Chem. 1997;272:27058. doi: 10.1074/jbc.272.43.27058. [DOI] [PubMed] [Google Scholar]
- 46.Fujita K, Tanaka N, Sano M, Kato I, Asada Y, Takegawa K. Biochem. Biophys. Res. Commun. 2000;267:134. doi: 10.1006/bbrc.1999.1963. [DOI] [PubMed] [Google Scholar]