Abstract
Muscular dystrophies are a class of disorders that cause progressive muscle wasting. A major hurdle for discovering treatments for the muscular dystrophies is a lack of reliable assays to monitor disease progression in animal models. We have developed a novel mouse model to assess disease activity noninvasively in mice with muscular dystrophies. These mice express an inducible luciferase reporter gene in muscle stem cells. In dystrophic mice, muscle stem cells activate and proliferate in response to muscle degeneration, resulting in an increase in the level of luciferase expression, which can be monitored by noninvasive, bioluminescence imaging. We applied this noninvasive imaging to assess disease activity in a mouse model of the human disease limb girdle muscular dystrophy 2B (LGMD2B), caused by a mutation in the dysferlin gene. We monitored the natural history and disease progression in these dysferlin-deficient mice up to 18 months of age and were able to detect disease activity prior to the appearance of any overt disease manifestation by histopathological analyses. Disease activity was reflected by changes in luciferase activity over time, and disease burden was reflected by cumulative luciferase activity, which paralleled disease progression as determined by histopathological analysis. The ability to monitor disease activity noninvasively in mouse models of muscular dystrophy will be invaluable for the assessment of disease progression and the effectiveness of therapeutic interventions.
Introduction
Muscular dystrophies are a class of inherited muscle disorders that are characterized by progressive muscle weakness and wasting. These diseases often result from mutations of genes that are critical for muscle cell structure or function (1). Therapeutic strategies to treat muscular dystrophies, including gene therapies and small molecule therapies, are being investigated, but currently there are few treatments available and none that substantially alter disease progression (2). Many dystrophic animal models exist and provide valuable resources for understanding the disease pathogenesis and for testing therapeutic interventions (3). One of the major limitations to the study of therapeutic agents for the treatment of muscular dystrophies is the absence of reliable assays of disease activity in living animals.
The gold standard for monitoring disease progression or the response to treatments in animals is the analysis of muscle histopathology. This approach is labor intensive, difficult to quantify, and usually terminal for the experimental animal. As such, investigators have sought methods to assess disease activity or progression using noninvasive or minimally invasive methods. In animal models, levels of serum biomarkers, strength measurements, and MRI evaluations have been used to assess disease activity and progressive deterioration of dystrophic muscle (4–6). However, these techniques are either highly variable (especially serum biomarkers), nonspecific (especially strength measurements), expensive (especially MRI), or some combination of the three. As such, there remains a critical need for a method to provide quantitative and reliable assessment of ongoing and cumulative disease activity that closely reflects the histopathological changes occurring in the muscle in dystrophic animal models.
In this report, we describe a novel mouse model in which muscle regeneration, reflecting the response to degeneration that occurs in the muscular dystrophies, can be measured noninvasively and quantitatively in living mice over time. This mouse expresses an estrogen-responsive Cre-recombinase under the control of the Pax7 locus and a luciferase reporter gene that is Cre dependent. Following tamoxifen treatment, luciferase is expressed only in muscle satellite cells, since these are the only cells in the adult (other than cells in small regions in the brain) that express Pax7 (7, 8). Therefore, each time the muscle undergoes degeneration and regeneration, luciferase-expressing satellite cells give rise to progeny that also express the reporter gene as they proliferate and differentiate to repair the muscle, and that luciferase activity can be measured noninvasively in a highly quantitative manner (9–12). We applied this model to the study of a mouse model of a form of limb girdle muscular dystrophy (LGMD) and found a remarkable correlation between the results of noninvasive imaging and disease activity and progression as determined histopathologically over the course of 18 months. This technology, which is applicable to all murine models of muscular dystrophy, will dramatically improve characterizations of the natural history and progression of muscle diseases and will be an invaluable tool for measuring the effectiveness of experimental therapeutics.
Results
Characterization of the “regeneration reporter” strain.
With the goal of developing a mouse model to monitor muscle regeneration as a surrogate for ongoing disease activity in mice with muscular dystrophies, we used Pax7CreER/LuSEAP mice in which an estrogen-responsive Cre-recombinase is induced to permanently activate a luciferase gene in muscle satellite cells (10). To characterize this “regeneration reporter” strain, mice were first imaged prior to administration of tamoxifen in order to determine the baseline level of the luciferase signal (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI68458DS1). Tamoxifen was then administered for 5 days when the mice were 2 months of age. Mice were sacrificed 7 days after the final dose of tamoxifen, and muscles were assessed for the presence of luciferase-positive satellite cells. Indeed, luciferase-positive satellite cells were observed in every muscle examined (Figure 1A), and at the dose of tamoxifen used, an average of 65% of the satellite cells expressed luciferase (Supplemental Figure 2). No other cells were observed to be luciferase positive, demonstrating the specificity of the Pax7CreER driver for the satellite cell lineage.
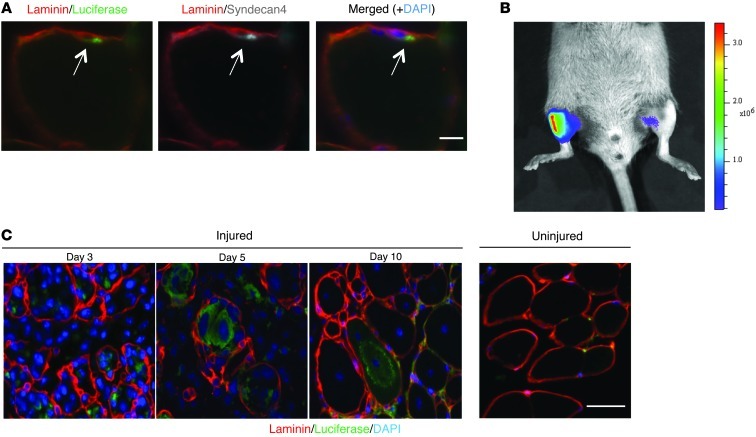
Figure 1. The expression of luciferase in resting and injured muscle in Pax7CreER/LuSEAP mice.
(A) A luciferase-expressing cell (arrows) residing under the basal lamina (laminin staining) and expressing the satellite cell marker syndecan-4 1 week after tamoxifen administration to a Pax7CreER/LuSEAP mouse. Scale bar: 10 μm. (B) Three days after an acute injury to the right TA muscle of a Pax7CreER/LuSEAP mouse, luciferase signals were detectable only in the injured limb. Scale to the right of the image represents photon emission from the tissue surface and is expressed as p/s/cm2/sr (or radiance). (C) Luciferase-expressing cells contributed to regenerative myotubes and nascent myofibers during the regenerative process (days 3–10), but were absent in the uninjured muscle. Scale bar: 50 μm.
To demonstrate that the proliferative amplification of these cells could be detected as an increase in luciferase signal, we injured one of the tibialis anterior (TA) muscles, leaving the contralateral TA muscle uninjured, in a cohort of mice. Three days after the injury, the luciferase signals from the injured muscles were dramatically increased, whereas the signals in the uninjured muscles remained at baseline levels (Figure 1B). There is always a very low level of luciferase activity even in uninjured muscles of Pax7CreER/LuSEAP mice following tamoxifen injection, and this signal is presumably related to the quiescent satellite cell pool that then expresses luciferase. Histological examination of the injured muscle showed luciferase-positive myotubes and nascent myofibers during the process of muscle regeneration (Figure 1C). As myofibers continued to mature, the intensity of the luciferase signal declined in association with the increase in cytoplasmic volume and corresponded to a plateau of the luciferase signal determined by bioluminescence imaging (Supplemental Figure 3).
These findings in wild-type mice clearly demonstrated the value of this reporter strain to reflect the proliferative activity of satellite cells and their progeny, providing a noninvasive readout of the regenerative response in vivo. Furthermore, the specificity of the luciferase signal for the satellite cell lineage was highly encouraging for the application of this strain to reflect disease activity in dystrophic muscle. As such, we crossed this reporter strain with a mouse strain, the SJL strain that is an animal model for the human disease LGMD2B. LGMD2B is caused by mutations in the dysferlin gene and preferentially affects the proximal limb muscles (13). There are several mouse models of LGMD2B (14, 15), including the naturally occurring SJL model (16, 17). As typical of a progressive muscular dystrophy, histopathological analysis in the SJL model and in other genetic dysferlinopathy models reveals ongoing degeneration and regeneration of muscle fibers and cumulative fibrosis and adiposis (15, 16, 18, 19).
Monitoring the natural history and progression of disease activity over time.
A hallmark of dystrophic muscle is the degeneration of mature muscle fibers followed by the regeneration of new fibers mediated by muscle satellite cell progeny (20). We expected that the expression of luciferase in dystrophic muscle would increase over time, reflecting the regenerative response of the muscle to the ongoing degeneration (Figure 2A). Cohorts of 2-month-old wild-type (Dysf+/+) and dysferlin-deficient (Dysf–/–) Pax7CreER/LuSEAP mice were injected with tamoxifen to induce luciferase expression in satellite cells. As additional controls, we injected Dysf–/–/Pax7CreER/LuSEAP mice with vehicle alone (corn oil) and found no evidence of luciferase expression (Supplemental Figure 4A). In tamoxifen-treated mice, the luciferase signal increased dramatically over time in hind limb muscles of the dysferlin-deficient strain, but not in the wild-type strain (Figure 2, B and C, and Supplemental Figure 4B). This enhanced regenerative activity in dysferlin-deficient mice was apparent as early as 3 months of age and increased continuously through 18 months of age (Figure 2C and Supplemental Figure 4C). Comparisons between male and female mice showed no significant sex differences with regard to disease progression (Figure 2D).
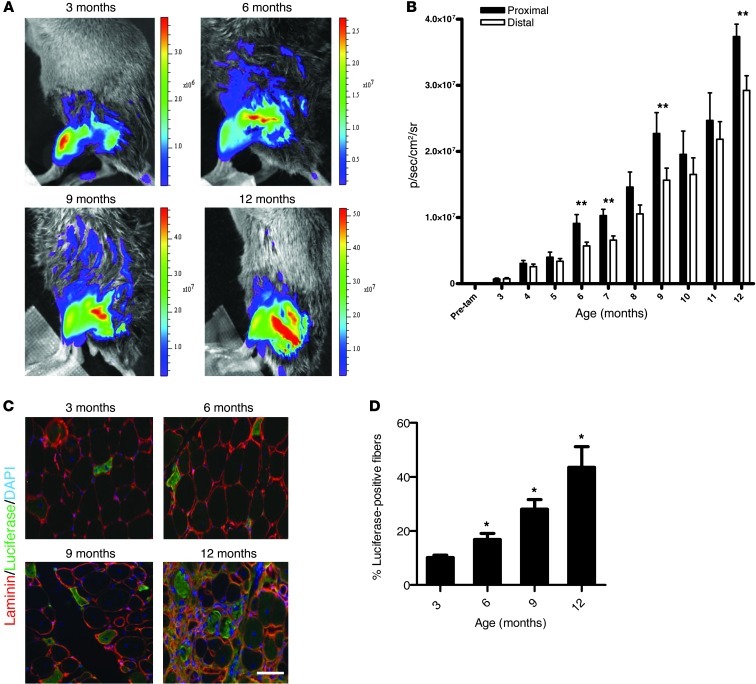
Figure 2. Monitoring disease activity in dysferlin-deficient mice by noninvasive bioluminescence imaging.
(A) Diagrammatic representation of the temporal pattern of luciferase expression in dystrophic Pax7CreER/LuSEAP mice. After tamoxifen administration, a subset of satellite cells express luciferase. In response to injury or muscle degeneration, these luciferase-positive satellite cells activate and proliferate, giving rise to luciferase-positive myoblasts that ultimately differentiate and fuse to form new myofibers, which will then be luciferase positive. (B) Wild-type (left) and dysferlin-deficient (right) Pax7CreER/LuSEAP mice, injected with tamoxifen at 2 months of age, imaged together at 7 months of age. Scale to the right of the image represents photon emission from the tissue surface and is expressed as p/s/cm2/sr (or radiance). (C) The luciferase signals from distal hind limb muscles of both wild-type and dysferlin-deficient mice were measured before tamoxifen administration (Pre-tam), at 3 months of age, and then monthly up to 18 months of age. P < 0.05 for each measurement between 3 and 18 months of age in the dysferlin-deficient compared with the control strain; n = 12 Dysf+/+ compared with Dysf–/– at each time point. (D) Luciferase signals from the distal hind limb muscles of male and female dysferlin-deficient mice (n = 12; same mice as in C).
Luciferase activity in proximal versus distal limb muscles.
LGMD2B manifests initially and primarily in the proximal limb muscles, with less involvement of distal limb muscles (21). We assessed disease activity by bioluminescent signals according to anatomical locations (Figure 3A). Measurements of the luciferase activity indicated that the proximal limb muscles had significantly more regeneration than the distal muscles beginning at 6 months of age (Figure 3B). The luciferase activity in both proximal and distal muscle groups increased continuously over time, with differences between them persisting up to 12 months of age (Figure 3B).
Figure 3. Regenerative activity in hind limb muscles of Dysf–/–/Pax7CreER/LuSEAP mice as a function of age.
(A) Hind limb muscles of Dysf–/–/Pax7CreER/LuSEAP mice imaged at 3, 6, 9, and 12 months of age. The luciferase signals increase in the hind limb muscles over time, with more signals emanating from the proximal muscles after 6 months of age. Scale to the right of the image represents photon emission from the tissue surface and is expressed as p/s/cm2/sr (or radiance). (B) Luciferase signals were quantified in proximal or distal hind limb muscles of Dysf–/–/Pax7CreER/LuSEAP mice. **P < 0.05; n = 12. (C) Representative images demonstrating the accrual of luciferase-positive myofibers in quadriceps muscles of Dysf–/–/Pax7CreER/LuSEAP mice at different ages. Scale bar: 50 μm. (D) Quantitation of the percentage of luciferase-positive quadriceps fibers from mice sacrificed at 3, 6, 9, and 12 months of age. *P < 0.05; n ≥ 3; value at each time point compared with that at 3 months of age.
Measurement of luciferase expression by histological and biochemical analyses over time.
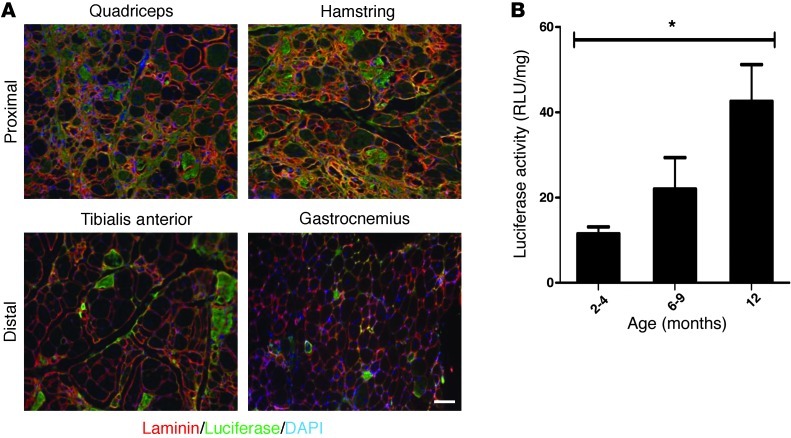
As a correlate of the changes in luciferase activity detected by noninvasive imaging and as further validation of the model, we sacrificed mice at different ages and isolated muscles for both biochemical and histological studies of luciferase expression. Based on the temporal progression of luciferase activity determined by noninvasive imaging (Figure 2C), we expected to see an increase in the number of luciferase-positive myofibers over time as a result of the regenerative activity in the muscles. Indeed, we found a significant increase in the number of luciferase-positive fibers over time (Figure 3, C and D), with the proximal limb muscles showing a greater accretion of luciferase-positive fibers with age than the distal muscles (Figure 4A). Surprisingly, there were relatively few luciferase-positive fibers in the gastrocnemii (Figure 4A). Measurements of luciferase activity in whole-muscle extracts also showed an increase with age (Figure 4B), consistent with the results of noninvasive imaging. Luciferase-positive fibers were not detected in nondystrophic animals, and only background levels of luciferase activity could be detected in muscles of 12-month-old dysferlin-deficient mice that were injected with vehicle (Supplemental Figure 5).
Figure 4. Regenerative activity in proximal and distal hind limb muscles of Dysf–/–/Pax7CreER/LuSEAP mice.
(A) Representative images of luciferase-positive fibers detected in proximal and distal hind limb muscles of 12-month-old Dysf–/–/Pax7CreER/LuSEAP mice. Scale bar: 50 μm. (B) Quadriceps muscles of dysferlin-deficient mice have increasing luciferase enzyme activity over time. *P < 0.05; n = 3. Luciferase activities were measured biochemically in muscles of mice from specific age groups, as indicated.
Noninvasive luciferase imaging in the Dysf–/–/Pax7CreER/LuSEAP mouse correlates with conventional histopathological manifestations of disease activity.
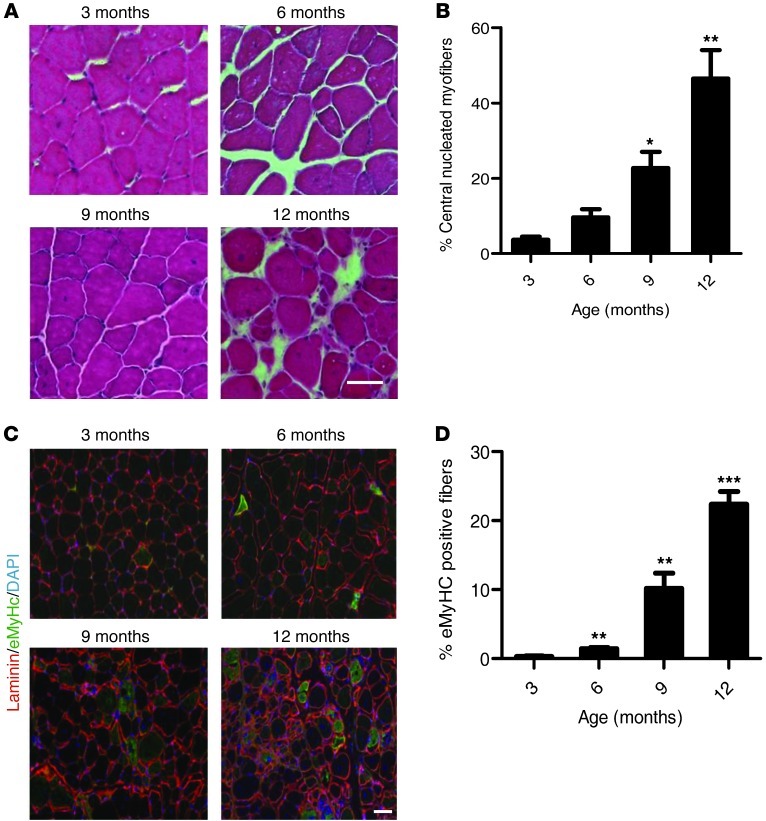
Classic markers of muscle regeneration in mice include the appearance of centrally localized nuclei within the skeletal muscle fibers as well as the appearance of fibers that express developmental proteins such as embryonic myosin heavy chain (eMyHc) (22). To validate further the noninvasive imaging of luciferase activity in Dysf–/–/Pax7CreER/LuSEAP mice as a reflection of disease activity, we examined these conventional markers (Figure 5, A–D). Centrally nucleated myofibers began to appear at 3 months of age and continuously accrued as the animals aged (Figure 5, A and B). By 12 months of age, nearly 50% of the fibers were centrally nucleated in the dysferlin-deficient strain (Figure 5B). Newly regenerated myofibers that express eMyHc could also be found in increasing numbers as the mice aged (Figure 5, C and D). In wild-type mice, fewer than 1% of fibers were centrally nucleated, and no eMyHc-positive fibers were detected. Therefore, both in terms of time course and magnitude of change, noninvasive imaging faithfully mirrored disease activity and progression by conventional analysis.
Figure 5. Histological analyses of muscles of Dysf–/–/Pax7CreER/LuSEAP mice over time.
(A) Cryosections of quadriceps muscles of 3-, 6-, 9-, and 12-month-old mice were stained with H&E to assess for histopathologic evidence of disease progression. (B) Quantitative analysis of the percentages of centrally nucleated myofibers in quadriceps of Dysf–/–/Pax7CreER/LuSEAP mice as a function of age. *P < 0.05, **P < 0.005; n = 3; value at each time point compared with that at 3 months of age. (C) Analysis of serial sections from muscles of mice as in panel A were stained with an antibody against eMyHc to assay for regenerating myofibers. (D) Quantitative analysis of the percentages of eMyHc-positive fibers in the quadriceps muscles of Dysf–/–/Pax7CreER/LuSEAP mice as a function of age. **P < 0.05, ***P < 0.001; n = 3; value at each time point compared with that at 3 months of age. Scale bars: 50 μm.
Discussion
Here, we present a novel mouse model in which muscle regeneration, a feature of disease activity in muscular dystrophies, can be followed noninvasively using bioluminescence imaging. By activation of a satellite cell–specific Cre-recombinase, we marked a majority of muscle satellite cells with the genetic lineage tracer luciferase, which enabled us to follow muscle regeneration over the course of 18 months in dysferlin-deficient mice. The Pax7CreER strain has been used previously for lineage tracing of muscle satellite cells in wild-type mice (9, 10, 12, 23). In one study, postnatal muscle growth, as assessed by measurement of the incorporation of Pax7-expressing progenitors into growing myofibers, was monitored for 12 weeks after tamoxifen administration to wild-type mice at 4 weeks of age (10). We performed our studies after this postnatal growth period, at a time when the hind limb muscles have fully matured (24, 25), to monitor disease activity as a function of regeneration in dystrophic muscle without any confounding aspects of muscle growth.
We validated the Pax7CreER/LuSEAP mice as a “regeneration reporter” strain by demonstrating that an acute muscle injury resulted in a luciferase signal localized only to the injured region of the hind limb. We observed luciferase-positive myotubes and nascent myofibers with central nuclei, indicating that these newly regenerated fibers were formed (at least in part) from the progeny of luciferase-expressing satellite cells (Figure 1C). In the absence of any injury, luciferase-expressing myofibers were not found (Figure 1C). After establishing that this mouse model was capable of reporting regeneration activity sensitively and reliably, we began to monitor the Dysf–/–/Pax7CreER/LuSEAP mice to assess the changes in luciferase signals over time. We found that it is possible to monitor disease activity noninvasively, corroborating our results with histopathological analysis and discovering aspects of the natural history of the SJL mouse model in the process. The reliability and reproducibility of this model was extremely high and comparable to the gold standard of histological measures of disease activity. Furthermore, individual mice showed patterns of increases in luciferase activity over time that paralleled the changes seen in the whole cohort (Supplemental Figure 4B), providing further support for this imaging system to be used for monitoring disease activity in different kinds of therapeutic trials in which individual mice can be used as their own controls in crossover type studies. However, this technology is not optimal for imaging of muscles, such as the diaphragm, within the body cavity, as the light signal is attenuated through layers of tissues and cannot be distinguished from any signal arising from surface musculature. Finally, it should be noted that the LuSEAP strain was designed as a dual reporter strain, expressing not only luciferase but also secreted alkaline phosphatase following Cre-mediated recombination. However, and as previously noted (10), we did not find assays of serum alkaline phosphatase levels to provide a reliable measure of cell number. We thus focused exclusively on the luciferase reporter for the purpose of these studies.
We specifically chose to use the SJL mouse model for several reasons, several of which are discussed in more detail below. First, it is a model that exhibits progressive and steady changes over the lifetime of the mouse as opposed to other models of human dystrophy, such as the mdx mouse, which shows an early burst of degeneration and regeneration followed by a much more stable course after several months (26–28). Second, there is a longer “preclinical” and even “prepathological” phase during which we would be able to assess the sensitivity of our noninvasive imaging methodology for subtle indications of disease onset as noted above, a finding that will be critically important in the assessment of therapeutic interventions. Third, there is an interesting anatomical localization of disease severity in the mouse, mimicking that in humans, which could also be assessed by our noninvasive imaging modality. Finally, several reports suggest that different pharmacological interventions may be of benefit in the progressive changes seen in the SJL mouse (29–31), making it particularly amenable for noninvasive imaging to be combined with experimental therapeutics.
Clinical manifestations of LGMD2B usually appear during the second or third decade of life. The rate of progression of this disease is highly variable among patients, with no correlation between the age of onset of symptoms and the rate at which the muscles deteriorate (32, 33). The SJL mouse has previously been described as having a slowly progressive muscular dystrophy by 6 months of age followed by rapid disease progression in subsequent months (16, 17). Likewise, the A/J mouse, another naturally occurring mouse model of LGMD2B, had virtually no histological evidence of disease activity until after 5 months of age (14). Our imaging studies revealed that enhanced regenerative activity began much earlier in the SJL mouse than had been concluded by the use of conventional markers of disease activity, demonstrating the enhanced sensitivity of this imaging technology. By noninvasive imaging, and corroborated by the detection of luciferase-positive fibers in situ as well as biochemical analyses, we found evidence of disease onset by 3 months of age (Figure 2C, Figure 3, C and D, and Figure 4B). Although the cumulative luciferase signal was low compared with later time points, the differences between the SJL and control strains were highly significant even at these early time points (Supplemental Figure 4C). This regenerative response increased continuously over time, which is reflective of the progressive nature of LGMD2B (Figure 2C).
Dysferlin deficiency causes a wide range of clinical phenotypes in humans (13). LGMD2B is generally characterized by the onset of disease symptoms in the proximal lower limb muscles irrespective of sex, whereas Miyoshi myopathy (MM), a disease also caused by mutations in the dysferlin gene, has been described as initially affecting posterior, distal limb muscles (17, 21). Mouse models of LGMD2B show that proximal limb muscles are primarily affected with little disease activity detected in distal muscles (16). In agreement with these findings, we show that the proximal limb muscles have more regenerative activity than distal muscles beginning at 6 months of age (Figure 3B). Although luciferase signals were greater in the proximal muscle groups, disease activity was also detected in the distal limb muscles at 3 months of age (Figure 3B). We found no significant sex differences in the onset (or progression) of disease activity in the mice (Figure 2D). Additionally, a greater number of luciferase-positive myofibers were detected in the TA muscles than in the gastrocnemii (Figure 4A), a feature that distinguishes between the 2 diseases, LGMD2B and MM, resulting from mutations in the dysferlin gene, supporting the conclusion that these mice recapitulate the anatomical distribution of disease activity of LGMD2B.
Previously, it was reported that disease activity in the SJL mouse model could be imaged by MRI using the enhanced contrasting agent Gadofluorine-M (34). While it was possible to detect increased permeability of muscle fibers to this agent, and this correlated with advanced disease progression, it was not sensitive enough to detect early pathologic changes. Additionally, the detection limit was dependent upon the amount of contrasting agent administered to the mice, which may have been limiting in the analysis (34). Although MRI does not require breeding of dystrophic mice with reporter mice, noninvasive imaging of disease activity using luciferase bioluminescence is less expensive, more widely available, and much more versatile for laboratory studies to monitor disease progression noninvasively for natural history studies or studies of response to experimental therapeutics.
The goal of this study was to generate a mouse model in which muscle regenerative activity could be measured over time to monitor the progression of disease activity or response to therapy for muscular dystrophies. Traditionally, the efficacy of drug treatments for the muscular dystrophies has been assayed using, alone or in combination, strength measurements, serum-biomarker concentrations, and changes in histopathology. As an alternative, the Pax7CreER/LuSEAP system will be invaluable for testing pharmacological, cellular, and genetic treatments for muscular dystrophies. Increased sensitivity, reliable quantitation, the ability to use each animal as its own control, and obviating the need to do labor- and time-intensive histopathology all will allow for more high-throughput analysis of treatments in vivo with this noninvasive method to monitor disease progression.
Methods
Animals.
The Pax7CreER and LuSEAP mice were a gift of Charles Keller, Oregon Health and Science University, Portland, Oregon, USA. SJL mice were purchased from Jackson Laboratories.
Bioluminescence imaging.
Bioluminescent imaging was performed using the Xenogen IVIS-Spectrum System (Caliper Life Sciences). Mice were anesthetized using 2% isoflurane and 100% oxygen at a flow rate of 2.5 l/min. Then 300 μl of a 50 mg/ml sterile D-Luciferin firefly substrate (Biosynth International Inc.) dissolved in PBS was administered by i.p. injection, and 23 minutes after substrate injection, the mice were imaged for 30 seconds at the maximal light collection (f-stop 1) at the highest resolution (small binning). Each image was saved for subsequent analysis. The scales to the right of images in Figure 1B, Figure 2B, and Figure 3A represent the photon emission from the tissue surfaces and are expressed as photons/second/centimeter squared/steradian (p/s/cm2/sr) (or radiance).
Image analysis.
Analysis of each image was performed using Living Image Software, version 4.0 (Caliper Life Sciences). Briefly, a manually-generated circle was placed on top of the region of interest (ROI) and resized to completely surround the limb or the specified region on the wild-type mouse. This ROI outline was then duplicated and placed over the limb of the dysferlin-deficient mouse. In cases where a control wild-type mouse was not imaged simultaneously, the ROI was encircled only for the experimental mouse.
Skeletal muscle injury.
Mice were anesthetized using 2% isoflurane and 100% oxygen at a flow rate of 2.5 l/min. The hind limbs were shaved, and 25 μl of 1.2% BaCl2 was injected into TA muscles using 10-cc insulin syringes (9).
Histology and immunohistochemistry.
At the specified time points, mice were euthanized using CO2 asphyxiation followed by cervical dislocation. Hind limb muscles were submerged in 0.5% PFA for 5 hours and then dehydrated in 20% sucrose overnight at 4°C. The muscles were then submerged in OCT (Sakura Finetek USA Inc.) and frozen for approximately 30 seconds in liquid nitrogen chilled in isopentane. Muscle cryosections (8-μm-thick) were placed on coated glass slides, which were then processed for histological or immunohistological staining. Antibodies used in this study were as follows: chicken anti–syndecan-4 (1:1,000; a gift from Bradley Olwin (University of Colorado, Boulder, Colorado, USA), rabbit anti-laminin (1:1,000; Sigma-Aldrich L9393), mouse anti-luciferase (1:1,000; Sigma-Aldrich L2164), and mouse anti-eMyHc (1:100; DSHB clone F1.652-s).
Luciferase biochemical assay.
Mice were euthanized as described above, and muscles were weighed immediately following isolation. The muscles were then placed into 5 ml of a 2-mg/ml solution of collagenase II for 1 hour at 37°C with agitation. The muscles were then triturated, and the samples were centrifuged. Supernatants were aspirated, and muscle tissue was lysed as per the manufacturer’s instructions for the Luciferase Assay System (Promega Inc.).
Isolation of satellite cells.
Mice were euthanized and hind limb muscles were collected. Muscles were resuspended in 0.2% collagenase II and digested for 1 hour and 45 minutes, after which the muscle suspension was triturated until fully dissociated. The muscles were then subjected to a second digestion in 0.5% collagenase II and 1% dispase for an additional half hour. Satellite cells were dissociated from the fibers by passing the digested muscle through a 20-gauge needle. Cells were then collected by centrifugation at 183.7 g and resuspended in 500 μl of Ham’s F-10 medium (Mediatech Inc.). The cell suspension was incubated with the following primary antibodies: biotin-labeled VCAM (CD106), FITC-CD45, FITC-CD31, and FITC-Sca1 (1/100; BD Biosciences — Pharmingen) for 45 minutes at 4°C on a shaking platform. Cells were then washed thoroughly and incubated with anti-FITC microbeads (1/10; MACS Miltenyi Biotec) for 15 minutes. Cells were then placed in an AutoMACS cell separation machine according to the manufacturer’s instructions (MACS Miltenyi Biotec). Cells that were not FITC positive were then incubated with anti-biotin microbeads for 15 minutes (1/10; MACS Miltenyi Biotec) and again subjected to MACS separation according to the manufacturer’s instructions. The cells were counted and plated on ECM-coated (1/1,000; Sigma-Aldrich) microscope chamber slides (Lab Tek II; Fisher Scientific) and incubated at 37°C until fixed.
Statistics.
All quantitative data are represented as means, and error bars indicate the SEM. Data were subjected to either 2-tailed Mann-Whitney test (for disease progression analysis comparing wild-type to dysferlin-deficient mice) or 2-tailed Student’s t tests (when comparing male and female imaging data, proximal and distal hind limb, and quantitations from histological analyses) to determine the significance of calculated differences. These differences were considered statistically significant at P < 0.05.
Study approval.
Animals were handled and housed according to the guidelines set forth by the Veterinary Medical Unit of the VA Palo Alto Health Care System, and all procedures were preapproved by the IACUC prior to being performed.
Supplementary Material
Acknowledgments
We would like to acknowledge Charles Keller for his seminal contributions to the field in terms of the generation of the transgenic mouse strains that were used in these studies and that we and others have found to be so valuable in studies of muscle stem cell biology and muscle disease studies. This work was supported by grants from the Jain Foundation and the NIH (Director’s Pioneer Award DP1 OD000392) to T.A. Rando.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2013;123(5):2298–2305. doi:10.1172/JCI68458.
See the related Commentary beginning on page 1931.
References
- 1.Cohn RD, Campbell KP. Molecular basis of muscular dystrophies. Muscle Nerve. 2000;23(10):1456–1471. doi: 10.1002/1097-4598(200010)23:10<1456::AID-MUS2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 2.Goyenvalle A, Seto JT, Davies KE, Chamberlain J. Therapeutic approaches to muscular dystrophy. Hum Mol Genet. 2011;20(R1):R69–R78. doi: 10.1093/hmg/ddr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vainzof M, et al. Animal models for genetic neuromuscular diseases. J Mol Neurosci. 2008;34(3):241–248. doi: 10.1007/s12031-007-9023-9. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman JS, Taylor RG, Fowler WM. Serum creatine phosphokinase variations in dystrophic mice. Exp Neurol. 1981;73(3):716–724. doi: 10.1016/0014-4886(81)90207-7. [DOI] [PubMed] [Google Scholar]
- 5.Dellorusso C, Crawford RW, Chamberlain JS, Brooks SV. Tibialis anterior muscles in mdx mice are highly susceptible to contraction-induced injury. J Muscle Res Cell Motil. 2001;22(5):467–475. doi: 10.1023/A:1014587918367. [DOI] [PubMed] [Google Scholar]
- 6.McIntosh LM, Baker RE, Anderson JE. Magnetic resonance imaging of regenerating and dystrophic mouse muscle. Biochem Cell Biol. 1998;76(2–3):532–541. doi: 10.1139/o98-033. [DOI] [PubMed] [Google Scholar]
- 7.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi: 10.1016/S0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 8.Buckingham M, et al. Myogenic progenitor cells in the mouse embryo are marked by the expression of Pax3/7 genes that regulate their survival and myogenic potential. Anat Embryol (Berl). 2006;211(suppl 1):51–56. doi: 10.1007/s00429-006-0122-0. [DOI] [PubMed] [Google Scholar]
- 9.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317(5839):807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 10.Nishijo K, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23(8):2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quach NL, Biressi S, Reichardt LF, Keller C, Rando TA. Focal adhesion kinase signaling regulates the expression of caveolin 3 and beta1 integrin, genes essential for normal myoblast fusion. Mol Biol Cell. 2009;20(14):3422–3435. doi: 10.1091/mbc.E09-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30(2):232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amato AA, Brown RH. Dysferlinopathies. Handb Clin Neurol. 2011;101:111–118. doi: 10.1016/B978-0-08-045031-5.00007-4. [DOI] [PubMed] [Google Scholar]
- 14.Ho M, et al. Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency. Hum Mol Genet. 2004;13(18):1999–2010. doi: 10.1093/hmg/ddh212. [DOI] [PubMed] [Google Scholar]
- 15.Bansal D, et al. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423(6936):168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 16.Bittner RE, et al. Dysferlin deletion in SJL mice (SJL-Dysf) defines a natural model for limb girdle muscular dystrophy 2B. Nat Genet. 1999;23(2):141–142. doi: 10.1038/13770. [DOI] [PubMed] [Google Scholar]
- 17.Weller AH, Magliato SA, Bell KP, Rosenberg NL. Spontaneous myopathy in the SJL/J mouse: pathology and strength loss. Muscle Nerve. 1997;20(1):72–82. doi: 10.1002/(SICI)1097-4598(199701)20:1<72::AID-MUS10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Nemoto H, Konno S, Nakazora H, Miura H, Kurihara T. Histological and immunohistological changes of the skeletal muscles in older SJL/J mice. Eur Neurol. 2007;57(1):19–25. doi: 10.1159/000097005. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi K, Izawa T, Kuwamura M, Yamate J. The distribution and characterization of skeletal muscle lesions in dysferlin-deficient SJL and A/J mice. Exp Toxicol Pathol. 2010;62(5):509–517. doi: 10.1016/j.etp.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol. 2009;71:37–57. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- 21.Bushby KM. Dysferlin and muscular dystrophy. Acta Neurol Belg. 2000;100(3):142–145. [PubMed] [Google Scholar]
- 22.Reznik M, Engel WK. Ultrastructural and histochemical correlations of experimental muscle regeneration. J Neurol Sci. 1970;11(2):167–185. doi: 10.1016/0022-510X(70)90126-7. [DOI] [PubMed] [Google Scholar]
- 23.Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48(7):424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz E. Satellite cell proliferative compartments in growing skeletal muscles. Dev Biol. 1996;175(1):84–94. doi: 10.1006/dbio.1996.0097. [DOI] [PubMed] [Google Scholar]
- 25.White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A. 1984;81(4):1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dangain J, Vrbova G. Muscle development in mdx mutant mice. Muscle Nerve. 1984;7(9):700–704. doi: 10.1002/mus.880070903. [DOI] [PubMed] [Google Scholar]
- 28.DiMario JX, Uzman A, Strohman RC. Fiber regeneration is not persistent in dystrophic (MDX) mouse skeletal muscle. Dev Biol. 1991;148(1):314–321. doi: 10.1016/0012-1606(91)90340-9. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki N, et al. Continuous administration of poloxamer 188 reduces overload-induced muscular atrophy in dysferlin-deficient SJL mice. Neurosci Res. 2012;72(2):181–186. doi: 10.1016/j.neures.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Nemoto H, et al. Anti-TNF therapy using etanercept suppresses degenerative and inflammatory changes in skeletal muscle of older SJL/J mice. Exp Mol Pathol. 2011;90(3):264–270. doi: 10.1016/j.yexmp.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Rayavarapu S, Van der Meulen JH, Gordish-Dressman H, Hoffman EP, Nagaraju K, Knoblach SM. Characterization of dysferlin deficient SJL/J mice to assess preclinical drug efficacy: fasudil exacerbates muscle disease phenotype. PLoS One. 2010;5(9):e12981. doi: 10.1371/journal.pone.0012981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paradas C, et al. Redefining dysferlinopathy phenotypes based on clinical findings and muscle imaging studies. Neurology. 2010;75(4):316–323. doi: 10.1212/WNL.0b013e3181ea1564. [DOI] [PubMed] [Google Scholar]
- 33.Gayathri N, et al. Dysferlinopathy: spectrum of pathological changes in skeletal muscle tissue. Indian J Pathol Microbiol. 2011;54(2):350–354. doi: 10.4103/0377-4929.81636. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt S, et al. Dysferlin-deficient muscular dystrophy: gadofluorine M suitability at MR imaging in a mouse model. Radiology. 2009;250(1):87–94. doi: 10.1148/radiol.2501080180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.