Abstract
Aromatase expression and enzyme activity in breast cancer patients is greater in or near the tumor tissue compared with the normal breast tissue. Complex regulation of aromatase expression in human tissues involves alternative promoter sites that provide tissue-specific control. Previous studies in our laboratories suggested a strong association between aromatase (CYP19) gene expression and the expression of cyclooxygenase (COX) genes. Additionally, nonsteroidal anti-inflammatory drugs (NSAIDs) and COX selective inhibitors can suppress CYP19 gene expression and decrease aromatase activity. Our current hypothesis is that pharmacological regulation of aromatase and/or cyclooxygenases can act locally to decrease the biosynthesis of estrogen and may provide additional therapy options for patients with hormone-dependent breast cancer. Two pharmacological approaches are being developed, one involving mRNA silencing by selective siRNA molecules and the second utilizing small molecule drug design. In the first approach, short interfering RNAs (siRNA) were designed against either human aromatase mRNA or human COX-2 mRNA. Treatment of breast cancer cells with siAROMs completely masked the aromatase enzyme activity. Treatment with COX-2 siRNAs decreased the expression of COX-2 mRNA; furthermore, the siCOX-2-mediated decrease also resulted in suppression of CYP19 mRNA. The small molecule drug design approach focuses on the synthesis and biological evaluation of a novel series of sulfonanilide analogs derived from the COX-2 selective inhibitors. The compounds suppress aromatase enzyme activity in SK-BR-3 breast cancer cells in a dose and time dependent manner, and structure activity analysis does not find a correlation between aromatase suppression and COX inhibition. Real-time PCR analysis demonstrates that the sulfonanilide analogs decrease aromatase gene transcription in breast cells. Thus, these results suggest that the siRNAs and novel sulfonanilides targeting aromatase expression may be valuable tools for selective regulation of aromatase in breast cancer.
Keywords: aromatase, cyclooxygenase-2, prostaglandin E2, NSAIDs, COX-2 inhibitors, siRNA, sulfonanilides
Introduction
Estradiol is the most potent endogenous estrogen and is biosynthesized from androgens by the cytochrome P450 enzyme complex called aromatase [1]. The highest levels of enzyme are present in the ovaries of premenopausal women, in the placenta of pregnant women, and in the peripheral adipose tissues of postmenopausal women and of men. Aromatase activity has also been demonstrated in breast tissue in vitro [2–4]. Furthermore, expression of aromatase is highest in or near breast tumor sites [3, 5], and the importance of intratumoral aromatase and local estrogen production is being unraveled [3, 6, 7]. Aromatase has been measured in the stromal cell component of normal breast and breast tumors, but the enzyme has also been detected in the breast epithelial cells in vitro [2, 5 ,7–9]. Furthermore, expression of aromatase is highest in or near breast tumor sites [5, 7]. The exact cellular location(s) of aromatase must await more rigorous analysis by several labs with a new monoclonal antibody now being developed and evaluated [10].
Regulation of aromatase in various tissues is complex, and several tissue-specific promoter regions have been identified upstream from the CYP19 gene [11–13]. These tissue-specific promoters include promoter PI.1, PI.3, PI.4, PI.6, PI.7, and PII. Promoter PI.1 is the major promoter used in placental tissues and is the farthest upstream. The PII promoter is utilized in the ovary and in breast cancer tissues, and it contains a cAMP response element. Promoters PI.3, PI.4, PI.6, and PI.7 are the promoters used in extraglandular sites. Promoter PI.4 is the primary promoter used in normal adipose tissue and is responsive to glucocorticoids and cytokines such as IL-1β, IL-6 and TNFα.
The increased expression of aromatase cytochrome P450 observed in breast cancer tissues was associated with a switch in the major promoter region utilized in gene expression. In the normal breast cells, aromatase expression is primarily derived by the tissue-specific promoter I.4 for transcription, whereas expression from breast cancer patients switches from promoter I.4 to promoter I.3 and promoter II [14]. As a result of the use of the alternate promoter, the regulation of estrogen biosynthesis switches from one controlled primarily by glucocorticoids and cytokines to a promoter regulated through cAMP-mediated pathways [14]. The prostaglandin PGE2 increases intracellular cAMP levels and stimulates estrogen biosynthesis [14], whereas other autocrine factors such as IL-1β do not appear to act via PGE2 [15].
Aromatase and Cyclooxygenases in Breast Cancer
Prostaglandin G/H endoperoxide synthase, also referred to as cyclooxygenase (COX), is a key enzyme which catalyzes the conversion of arachidonic acid to prostaglandins. Local production of PGE2 via the cyclooxgenase isozymes (constitutive COX-1 isozyme and inducible COX-2 isozyme) can influence estrogen biosynthesis and estrogen-dependent breast cancer. This biochemical mechanism may explain epidemiological observations of the beneficial effects of NSAIDs on breast cancer [16–19]. Previous studies in our laboratory suggest a relationship between CYP19 gene expression and the expression of COX genes [20]. Gene expressions of CYP19, COX-1, and COX-2 were performed in 20 human breast cancer specimens and in 5 normal control breast tissue samples. A positive correlation was observed between CYP19 expression and the greater extent of breast cancer cellularity, in agreement with literature reports showing that aromatase levels were higher in tumors than in normal tissue. Furthermore, a positive linear correlation was observed between COX-2 expression breast cancer cellularity in each sample. Linear regression analysis using a bivariate model shows a strong linear association between CYP19 expression and the sum of COX-1 and COX-2 expression. Similar correlations between CYP19 expression and COX-2 expression in breast cancer patient specimens have been confirmed in other laboratories [21].
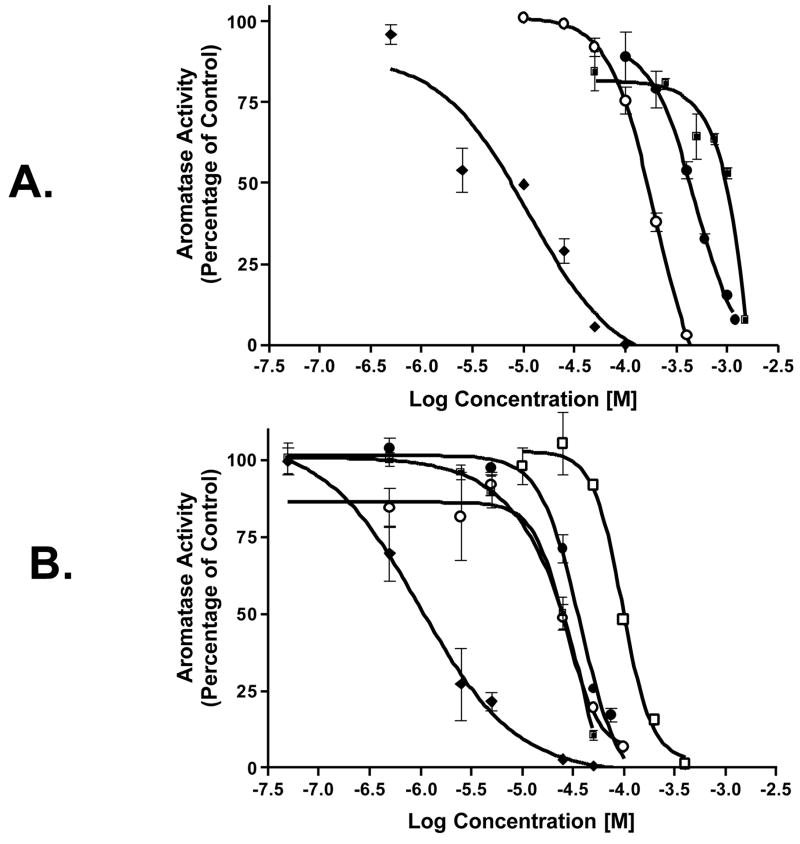
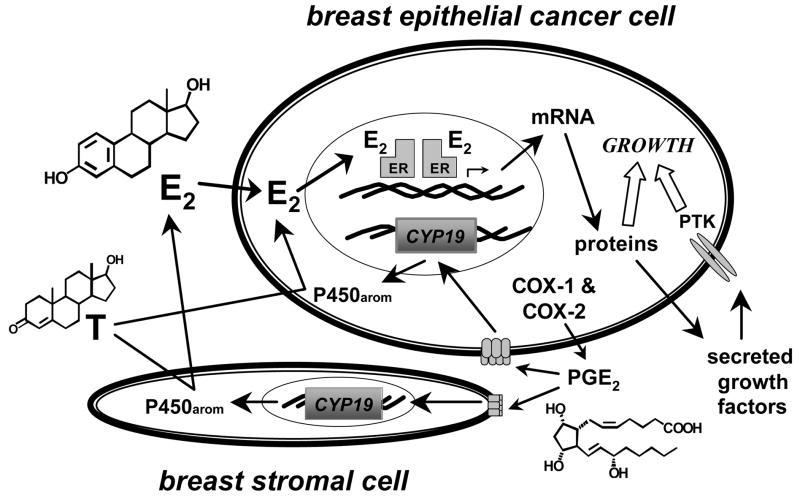
This significant relationship between the aromatase and cyclooxygenase enzyme systems suggests that autocrine and paracrine mechanisms may be involved in hormone-dependent breast cancer development via growth stimulation from local estrogen biosynthesis. In human breast stromal cells, PGE2 acts via two G-protein coupled receptors, EP1 and EP2 receptors, to stimulate aromatase gene expression via protein kinase A and protein kinase C signaling pathways [22]. NSAIDs, COX-1 and COX-2 selective inhibitors produce dose-dependent decreases in aromatase activity in breast cancer tissues (Figure 1) [23, 24]. Real time PCR analysis of aromatase gene expression showed a significant decrease in mRNA levels by these agents, and the effect of COX inhibitors on aromatase expression occurs through suppression at the tissue specific promoters PI.3, PI.4, and PII. This significant relationship between the aromatase and cyclooxygenase enzyme systems suggests that autocrine and paracrine mechanisms may be involved in hormone-dependent breast cancer development via growth stimulation from local estrogen biosynthesis (Figure 2).
Figure 1. Effect of NSAIDs and COX-specific Inhibitors on Aromatase Enzyme Activity.
(A)SK-BR-3 cells were treated with indomethacin (❍), piroxicam (●), ibuprofen (■), or SC-560 (◆), and aromatase activity was measured using the tritiated water release assay. (B) SK-BR-3 cells were treated with NS-398 (◆), nimesulide (❍), SC-58125 (■), celecoxib (●), or niflumic acid (□), and aromatase activity was measured using the tritiated water release assay.
Figure 2.
Model of Autocrine and Paracrine Pathways of Aromatase and Cyclooxygenases in Hormone-dependent Breast Cancer.
Our current research focuses on pharmacological regulation of aromatase and/or cyclooxygenases by agents that can act locally to decrease the biosynthesis of estrogen and may provide additional therapy options for patients with hormone-dependent breast cancer. Two pharmacological approaches are being developed, one involving mRNA silencing by selective siRNA molecules and the second utilizing small molecule drug design.
MATERIALS and METHODS
Reagents
The synthesis of the sulfonanilide analogs are described in our recent publication [25].
Cell Culture
The MCF-7 and SK-BR-3 cell lines were obtained from ATCC (Rockville, MD). Cell cultures were maintained in phenol red-free custom media (MEM, Earle’s salts, 1.5x amino acids, 2x non-essential amino acids, L-glutamine, 1.5x vitamins, Gibco BRL) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 20 mg/l gentamycin. Fetal calf serum was heat inactivated for 30 min in a 56 C water bath before use. Cell cultures were grown at 37 C, in a humidified atmosphere of 5% CO2 in a Hereaus CO2 incubator. For all experiments, cells were plated in either T-25 flasks or 100 mM plates and grown to subconfluency. Before treatment, the media was changed to a defined one containing DMEM/F12 media (Sigma) with 1.0 mg/ml human albumin (OSU Hospital Pharmacy), 5.0 mg/l human transferin and 5.0 mg/l bovine insulin.
Tritiated water-release assay
Measurement of aromatase enzyme activity was based on the tritium water release assay [27]. Cells in T-25 flasks or 100 mM plates were treated with 0.1% DMSO (control), NSAIDs (ibuprofen, piroxicam, and indomethacin), COX-1 selective inhibitor SC-560, and COX-2 selective inhibitors (SC-58125, NS-398, celecoxib, niflumic acid and nimesulide) at the indicated concentrations. After 24 h, the cells were incubated for 6 hours with fresh media along with 50 nM androstenedione including 2 μCi [1β-3H]-androst-4-ene-3,17-dione. Subsequently, the reaction mixture was removed, and proteins were precipitated using 10% trichloroacetic acid at 42 C for 20 min. After a brief centrifugation, the media was extracted three times with an equal amount of chloroform to extract unused substrate and further dextran-treated charcoal. After centrifugation, a 250-μl aliquot containing the product was counted in 5 ml of liquid scintillation mixture. Results were corrected for blanks and for the cell contents of culture flasks, and results were expressed as picomoles of 3H2O formed per hour incubation time per million live cells (pmol/h/106 cells). To determine the amount of live cells in each flask, the cells were trypsinized and analyzed using the diphenylamine DNA assay.
RNA extraction
Total RNA was isolated using the TRIzol reagent according to the manufacturer’s protocol. Total RNA pellets were dissolved in DNase, RNase-free water and quantitated using a spectrophotometer. The quality of RNA samples was determined by electrophoresis through agarose gels and staining with ethidium bromide; the 18S and 28S rRNA bands were visualized under ultraviolet light.
cDNA synthesis
Isolated total RNA (2 μg) was treated with DNase I, Amplification grade, according to the recommended protocol to eliminate any DNA before reverse transcription. Treated total RNA was denatured at 65 C for 5 min in the presence of 2.5 ng/μl random hexamers and 0.5 mM dNTP mix. The samples were snap-cooled on ice and centrifuged briefly. Complementary DNA (cDNA) was synthesized using Superscript II reverse transcriptase according to the recommended protocol. Briefly, the reactions were conducted in the presence of 1X First-Strand Buffer and 20 mM DTT at 42 C for 50 min and consequently inactivated at 70 C for 15 min. The cDNA generated was used as a template in real-time PCR reactions.
Real-time PCR
Real-time PCR was performed using the Opticon™ 2 system from MJ Research (Waltham, MA). For the CYP19 total gene the PCR reaction mixture consisted of Taqman® Universal PCR Master Mix (Applied Biosystems), 600 nM of each primer (Invitrogen), 250 nM Taqman probe, 18S rRNA (Applied Biosystems, Foster City, CA), and 2.5 μl of each RT sample in a final volume of 25 μl. Cycling conditions were 50 C for 2 min and 95 C for 10 min, followed by 50 cycles at 95 C for 15 s and 60 C for 1 min. For the specific exon I promoter regions and TATA-box-binding protein (TBP), the PCR reaction mixture consisted of DyNAmo Hot Start SYBR Green qPCR kit (MJ Research), 600 nM of each primer (Table 1), and 2.5 μl of each RT sample in a final volume of 20 μl. SYBR Green uses a dye that will bind to double stranded DNA. In this methodology, the primers are carefully designed to each of the promoter regions of aromatase exon I. Cycling conditions were 95 C for 15 min, followed by 50 cycles at 94 C for 10 s and 60 C for 25 s and 72 C for 30 s.
Statistical analysis
Statistical and graphical information was determined using GraphPad Prism software (GraphPad Software Incorporated) and Microsoft Excel (Microsoft Corporation). Determination of IC50 values were performed using nonlinear regression analysis. Statistically significant differences were calculated with the two-tailed unpaired Student’s t-test and P values reported at 95% confidence intervals.
RESULTS
RNA Interference of Aromatase and Cyclooxygenase-2
Newly developed RNA interference (RNAi) technology was utilized to further probe the interactions between aromatase and cyclooxygenases in breast cancer. RNAi technology permits transient suppression of the levels of endogenous proteins in mammalian cells by enhancing the degradation of target mRNA [28]. One RNAi approach involves transfection of 21–23 nucleotide double-stranded, short interfering RNAs (siRNA) into mammalian cells. These siRNAs are then incorporated into the RNA-inducing silencing complex (RISC), and RISC unwinds the siRNA duplex using ATP. The unwound, single-stranded antisense strand guides RISC to mRNA that has a complementary sequence, and the complex results in the endonucleolytic cleavage of the target mRNA.
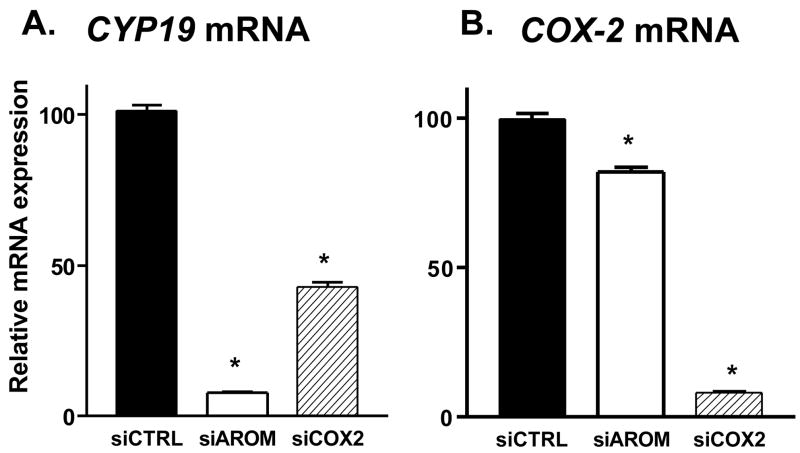
Short interfering RNAs (siRNA) were designed to target either human CYP19 mRNA or human COX-2 mRNA. The designed siRNAs were transiently transfected in SK-BR-3 cells, and several were effective in suppressing CYP19 mRNA and in decreasing COX-2mRNA [23]. Two siRNA molecules, termed siAROM and siCOX2, were chosen for further investigations in SK-BR-3 cells. The transfection of siAROM resulted in suppression of CYP19 mRNA levels by greater than 90% compared with cells transfected with a nonspecific control siRNA, termed siCTRL (Figure 3A). The siAROM produced little suppression of COX-2 mRNA expression (Figure 3B). Also shown in Figure 3B, transfection of siCOX2 suppressed COX-2 mRNA levels by greater than 90% compared with cells transfected with siCTRL. This siCOX2 also resulted in suppression of CYP19 mRNA levels by approximately 60% (Figure 3A). Thus, the suppression of COX-2 mRNA, which lowered levels of COX-2 enzyme and decreased prostaglandin production, results in suppression of CYP19 mRNA levels.
Figure 3. Suppression of CYP19 mRNA (A) and COX-2 mRNA (B) by siRNA Molecules Transfected into SK-BR-3 Breast Cancer Cells.
Each data bar represents the mean results of three independent determinations. *P <0.05 vs. control by unpaired t test.
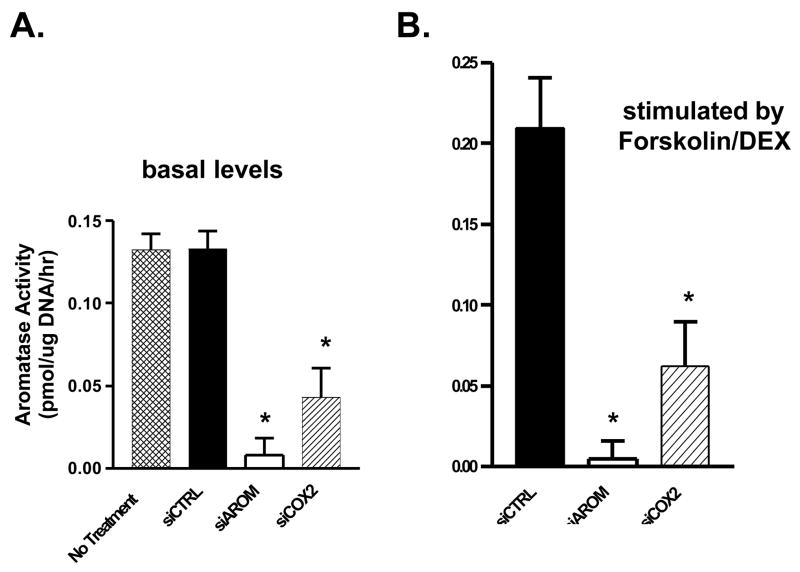
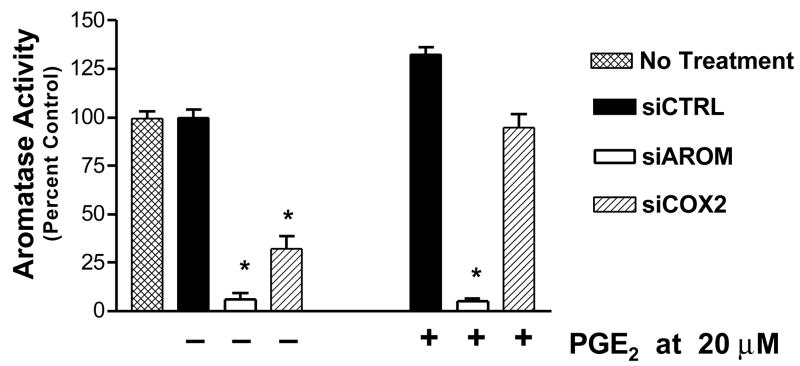
The effects of siAROM and siCOX2 on aromatase enzyme activity in SK-BR-3 cells were also examined. The transfection of siAROM resulted in suppression of basal levels of aromatase activity by greater than 90% compared with cells transfected with a nonspecific control siRNA or untreated cells (Figure 4A). Also shown in this figure, the siCOX2 also resulted in suppression of aromatase activity by approximately 67%. Furthermore, treatment of SK-BR-3 cells with the combination of forskolin and dexamethasone results in stimulation of aromatase activity, and the siAROM and siCOX2 both significantly suppressed the induced aromatase activity in these cells (Figure 4B). Finally, the administration of PGE2 to cells treated with the siRNAs results in antagonism of only the siCOX2 and restores aromatase activity to untreated levels (Figure 5).
Figure 4. Suppression of Basal Aromatase Activity (A) and stimulated Aromatase Activity (B) by siRNA Molecules Transfected into SK-BR-3 Breast Cancer Cells.
Each data bar represents the mean results of three independent determinations. *P <0.05 vs. control by unpaired t test.
Figure 5.
Novel Sulfonanilide Analogs for Suppression of Aromatase Expression and Activity
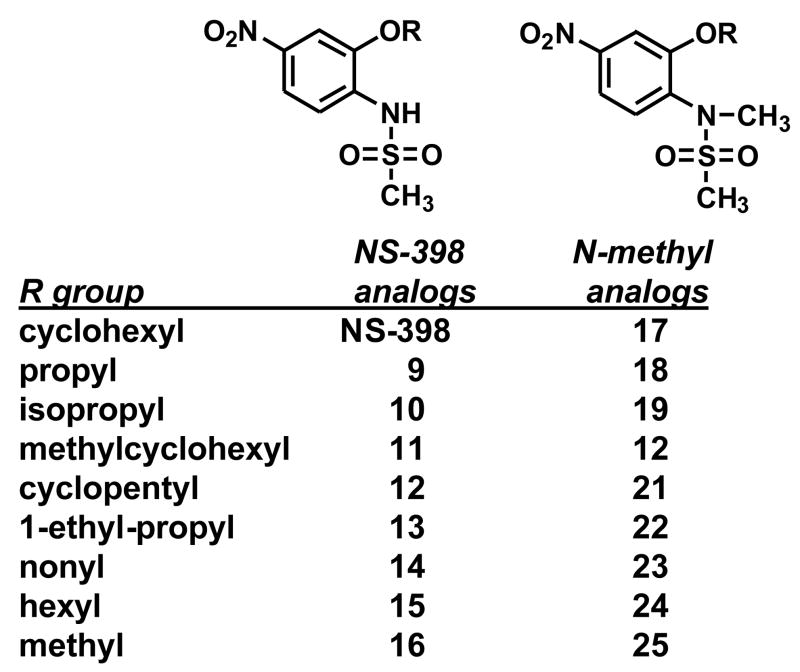
As shown in Figure 1, different COX-2 inhibitors with similar IC50 values (concentration for 50% inhibition) for COX-2 inhibition differ significantly in their ability to suppress aromatase activity. This observation suggests differences in the mechanisms by which these COX inhibitors modulate aromatase expression in SK-BR-3 cells. It is noteworthy that the effect of aromatase suppression by the COX-2 selective inhibitor NS-398 was greater than other COX-2 inhibitors, even though NS-398 has weak COX-2 inhibitory activity. To determine whether the modulation of aromatase expression by NS-398 required the inhibition of COX-2 enzyme activity, we designed and synthesized NS-398 analogs with no COX-2 inhibitory activity (Figure 6) [25]. Introduction of a methyl group at the N atom of the sulfonamide group to the COX-2 inhibitor nimesulide resulted in no COX-2 inhibitory activity [26]. This structural modification was utilized in our drug design. The nitrate group at the 4 position of NS-398 was retained and modifications of the sulfonamide and of the 2 position alkyl group were made to generate the new compounds [25].
Figure 6.
Synthetic Novel Sulfonanilides.
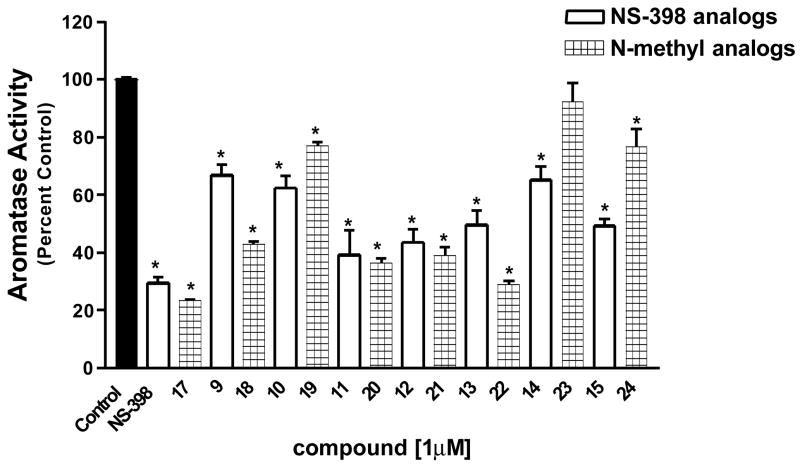
To investigate whether these compounds decrease aromatase activity in breast cancer cells, we performed a 1 μM bioassay in SK-BR-3 breast cancer cells, and most compounds significantly decreased aromatase activity (Figure 7). In an effort to discriminate among compounds in this library, dose response studies of the active compounds were performed, and the resulting IC50 values of the compounds ranged from approximately 0.20 to 6.00 μM. Our results suggest that the length of the group on position 2 of the compounds is important for the suppression of aromatase activity. Compounds containing a methoxy (16 and 25) or an isopropyloxy (10 and 19), which are relatively short, have low ability to suppress aromatase activity. Extremely long chain substituents (15, 24, 14 and 23) have reduced activity as well, which may also be due to the poor solubility of the compounds. All the N-methyl compounds exhibited better activity than their corresponding unsubstituted compounds with the exception of compounds 23 and 24. One possible explanation is the pKa value of the reagents.
Figure 7. Suppression of Aromatase Activity in SK-BR-3 Breast Cancer Cells.
SK-BR-3 cells were treated with indicated compounds (1 μM). Aromatase activity was measured as described in the experimental section. The results were normalized against a control treatment with vehicle. The value of 100% is equal to 0.03 pmol/hr/106 cells. Each data bar represents the mean results of three independent determinations. *P <0.05 vs. control by unpaired t test.
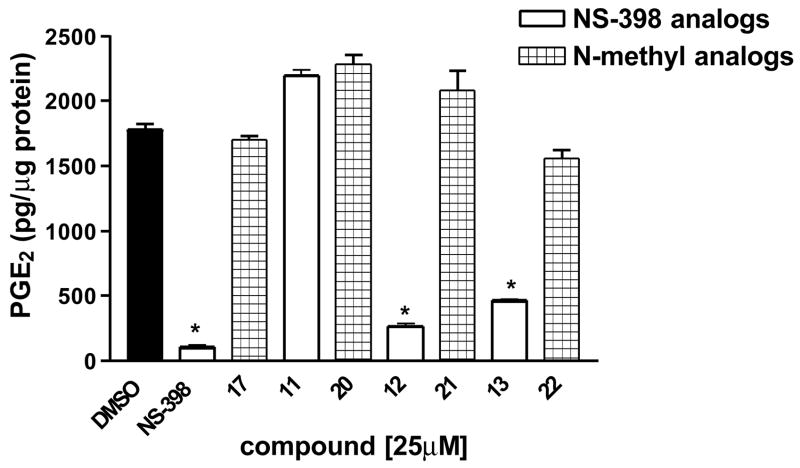
The production of PGE2 was measured in cells treated with NS-398 and the novel sulfonanilide derivatives. NS-398, compound 12 and 13 resulted in a significantly decrease in PGE2 production, whereas compounds 17, 20, 21 and 22 did not show any inhibitory activity (Figure 8) [25]. This is consistent with our design approach that the introduction of a methyl group in to the N atom of the sulfonamide group results in analogs that cannot be deprotonated and thus loses COX-2 inhibitory activity. In addition, compound 11 did not show any COX-2 inhibitory activity, and compound 11 has one carbon longer side chain comparing with NS-398. This result suggests that the size of the side chain is very important for the COX-2 inhibitory activity and that this extension affects the binding of the compound with COX-2 and results in no COX-2 inhibitory activity.
Figure 8. Effect of NS-398 derivatives on PGE2 production of MDA-MB-231 cells.
Cells were treated for 24 h with the indicated agents at 25 μM. Results are expressed as means of the concentration of PGE2 produced per microgram protein ± SEM. , *P < 0.05 vs. control by unpaired t test (n = 6).
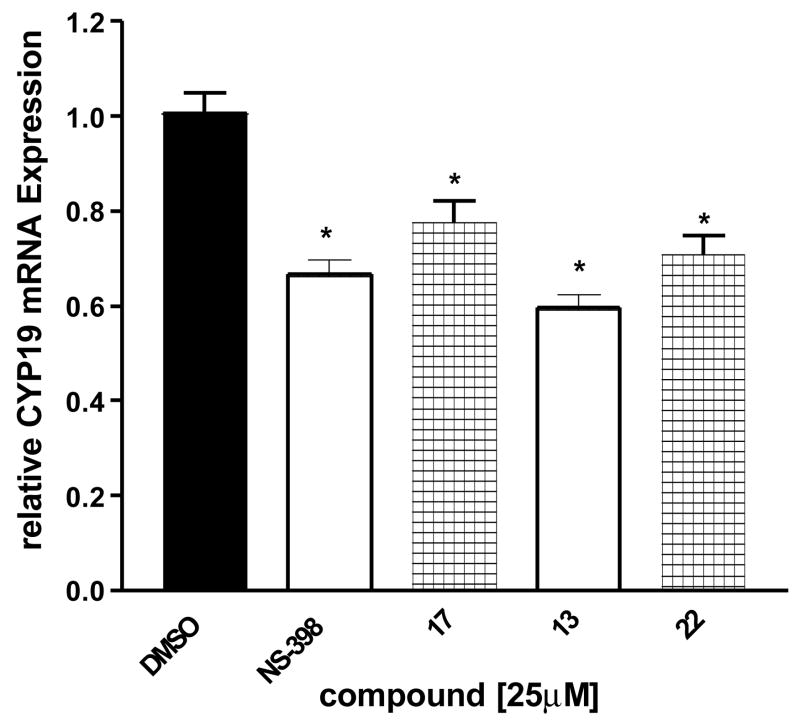
Analysis of total CYP19 mRNA transcripts was performed using real-time PCR in order to determine whether the decrease in aromatase activity by NS-398 in SK-BR-3 cells was due to a down-regulation of aromatase expression at transcriptional level. SK-BR-3 cells were treated with NS-398, compound 17, 13 and 22 for 24 h at concentrations at 25 μM. Total RNA was extracted at 24 h, and CYP19 transcript levels were compared to control (vehicle) treatment. All four compounds significantly decreased CYP19 gene expression in SK-BR-3 cells relative to the control (Figure 9). No effect on the expression level of the housekeeping 18S rRNA was observed with any of the compounds. Compounds 17 and 22, which do not show COX-2 inhibitory activity, decreased aromatase expression at similar levels. This suggests that the compounds interfere with pathways affecting aromatase expression in breast cancer cells that do not involve prostaglandins and COX enzyme activities. Similar results on CYP19 gene expression and aromatase activity have been observed in MCF-7 human breast cancer cell cultures and in primary cultures of isolated breast fibroblasts.
Figure 9. Real-time RT-PCR analysis of CYP19 mRNA expression in SK-BR-3 cells.
Cells were treated for 24 h with the indicated agents at 25 μM, and total RNA was isolated. Results are expressed as means of CYP19 (normalized to 18S rRNA) ± SEM., *P <0.05 vs. control by unpaired t test (n = 9).
CONCLUSIONS
Local regulation of aromatase by both endogenous factors as well as exogenous medicinal agents will influence the levels of estrogen available for breast cancer growth. The prostaglandin PGE2 increases intracellular cAMP levels and stimulates estrogen biosynthesis, and previous studies in our laboratories have shown a strong linear association between aromatase (CYP19) expression and expression of the cyclooxygenases (COX-1 and COX-2) in breast cancer specimens. Using selective pharmacological agents, dose-dependent decreases in aromatase activity were observed following treatment with NSAIDs, COX-1 selective inhibitor, and COX-2 selective inhibitors. Real time PCR analysis of aromatase gene expression showed a significant decrease in CYP19 mRNA levels in treated cells when compared to vehicle control. These results suggest that the effect of COX inhibitors on aromatase occurs at the transcriptional level.
Investigations using RNA interference (RNAi) technology confirmed the interactions between aromatase and cyclooxygenases in breast cancer. Short interfering RNAs (siRNA) were designed against either human CYP19 mRNA or human COX-2 mRNA. Treatment of breast cancer cells with siAROM suppressed CYP19 mRNA and aromatase enzyme activity. Treatment with stCOX2 downregulated the expression of COX-2 mRNA; furthermore, the siCOX-2-mediated suppression of COX-2 also resulted in suppression of aromatase mRNA. Finally, the administration of PGE2 to cells treated with the siRNAs results in antagonism of only the siCOX2 and restores aromatase activity to untreated levels.
The small molecule drug design approach focused on the synthesis and biological evaluation of a novel series of sulfonanilide analogs derived from the COX-2 selective inhibitors. The compounds suppressed aromatase enzyme activity in SK-BR-3 breast cancer cells in a dose dependent manner, and structure activity analysis did not find a correlation between aromatase suppression and COX inhibition. Real-time PCR analysis demonstrated that the sulfonanilide analogs decrease aromatase gene transcription in breast cells. Thus, these results suggest that the novel sulfonanilides targeting aromatase expression may be valuable tools for selective regulation of aromatase in breast cancer.
Thus, the regulation of aromatase and cyclooxygenases in breast cancer involves complex autocrine and paracrine interactions, resulting in significant consequences on the pathogenesis of hormone-dependent breast cancer via growth stimulation from local estrogen biosynthesis (Figure 2). Higher levels of COX-2 expression and COX-2 enzyme activity could result in higher levels of PGE2, which in turn could increase CYP19 expression through increases in intracellular cAMP levels and activation of promoter 1.3 and promoter II. Thus, the breast cancer tissue microenvironment can influence the extent of estrogen biosynthesis and metabolism, resulting in altered levels of hormonally active estrogens and therefore influencing breast tumor development and growth. Furthermore, siRNAs and novel sulfonanilides targeting aromatase expression may be valuable tools for selective regulation of aromatase in breast cancer.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Grant R01 CA73698 (R.W.B.), the NIH Chemistry and Biology Interface Training Program Grant T32 GM08512 (E.S.D.-C.), and The Ohio State University Comprehensive Cancer Center Breast Cancer Research Fund.
Footnotes
Presented at AROMATASE 2006: VIII International Aromatase Conference, Baltimore, Maryland, September 17-20, 2006.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 2.James VH, McNeill JM, Lai LC, Newton CJ, Ghilchik MW, Reed MJ. Aromatase activity in normal breast and breast tumor tissues: in vivo and in vitro studies. Steroids. 1987;50:269–279. doi: 10.1016/0039-128x(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 3.Miller WR, O’Neill J. The importance of local synthesis of estrogen within the breast. Steroids. 1987;50:537–548. doi: 10.1016/0039-128x(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 4.Reed MJ, Owen AM, Lai LC, Coldham NG, Ghilchik MW, Shaikh NA, James VH. In situ oestrone synthesis in normal breast and breast tumour tissues: effect of treatment with 4-hydroxyandrostenedione. Int J Cancer. 1989;44:233–237. doi: 10.1002/ijc.2910440208. [DOI] [PubMed] [Google Scholar]
- 5.Bulun SE, Price TM, Aitken J, Mahendroo MS, Simpson ER. A link between breast cancer and local estrogen biosynthesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J Clin Endocrinol Metab. 1993;77:1622–1628. doi: 10.1210/jcem.77.6.8117355. [DOI] [PubMed] [Google Scholar]
- 6.Dowsett M, Macaulay V, Gledhill J, Ryde C, Nicholls J, Ashworth A, McKinna JA, Smith IE. Control of aromatase in breast cancer cells and its importance for tumor growth. J Steroid Biochem Mol Biol. 1993;44:605–609. doi: 10.1016/0960-0760(93)90266-y. [DOI] [PubMed] [Google Scholar]
- 7.Miller WR, Mullen P, Sourdaine P, Watson C, Dixon JM, Telford J. Regulation of aromatase activity within the breast. J Steroid Biochem Mol Biol. 1997;61:193–202. [PubMed] [Google Scholar]
- 8.Reed MJ, Topping L, Coldham NG, Purohit A, Ghilchik MW, James VH. Control of aromatase activity in breast cancer cells: the role of cytokines and growth factors. J Steroid Biochem Mol Biol. 1993;44:589–596. doi: 10.1016/0960-0760(93)90264-w. [DOI] [PubMed] [Google Scholar]
- 9.Quinn AL, Burak WE, Brueggemeier RW. Effects of matrix components on aromatase activity in breast stromal cells in culture. J Steroid Biochem Mol Biol. 1999;70:249–256. doi: 10.1016/s0960-0760(99)00113-2. [DOI] [PubMed] [Google Scholar]
- 10.Sasano H, Edwards DP, Anderson TJ, Silverberg SG, Evans DB, Santen RJ, Ramage P, Simpson ER, Bhatnagar AS, Miller WR. Validation of new aromatase monoclonal antibodies for immunohistochemistry: progress report. J Steroid Biochem Mol Biol. 2003;86:239–244. doi: 10.1016/s0960-0760(03)00363-7. [DOI] [PubMed] [Google Scholar]
- 11.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Corbin CJ, Mendelson CR. Tissue-specific promoters regulate aromatase cytochrome P450 expression. J Steroid Biochem Mol Biol. 1993;44:321–330. doi: 10.1016/0960-0760(93)90235-o. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Transcriptional regulation of CYP19 gene (aromatase) expression in adipose stromal cells in primary culture. J Steroid Biochem Mol Biol. 1997;61:203–210. doi: 10.1016/s0960-0760(97)80013-1. [DOI] [PubMed] [Google Scholar]
- 13.Simpson ER, Clyne C, Rubin G, Boon WC, Robertson K, Britt K, Speed C, Jones M. Aromatase--a brief overview. Annu Rev Physiol. 2002;64:93–127. doi: 10.1146/annurev.physiol.64.081601.142703. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Estrogen biosynthesis proximal to a breast tumor is stimulated by PGE2 via cyclic AMP, leading to activation of promoter II of the CYP19 (aromatase) gene. Endocrinology. 1996;137:5739–5742. doi: 10.1210/endo.137.12.8940410. [DOI] [PubMed] [Google Scholar]
- 15.Hughes R, Timmermans P, Schrey MP. Regulation of arachidonic acid metabolism, aromatase activity and growth in human breast cancer cells by interleukin-1beta and phorbol ester: dissociation of a mediatory role for prostaglandin E2 in the autocrine control of cell function. Int J Cancer. 1996;67:684–689. doi: 10.1002/(SICI)1097-0215(19960904)67:5<684::AID-IJC16>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Harris RE, Namboodiri KK, Farrar WB. Nonsteroidal antiinflammatory drugs and breast cancer. Epidemiology. 1996;7:203–205. doi: 10.1097/00001648-199603000-00017. [DOI] [PubMed] [Google Scholar]
- 17.Harris RE, Robertson FM, Abou-Issa HM, Farrar WB, Brueggemeier R. Genetic induction and upregulation of cyclooxygenase (COX) and aromatase (CYP19): an extension of the dietary fat hypothesis of breast cancer. Med Hypotheses. 1999;52:291–292. doi: 10.1054/mehy.1998.0009. [DOI] [PubMed] [Google Scholar]
- 18.Arun B, Goss P. The role of COX-2 inhibition in breast cancer treatment and prevention. Semin Oncol. 2004;31:22–29. doi: 10.1053/j.seminoncol.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 19.Terry MB, Gammon MD, Zhang FF, Tawfik H, Teitelbaum SL, Britton JA, Subbaramaiah K, Dannenberg AJ, Neugut AI. Association of frequency and duration of aspirin use and hormone receptor status with breast cancer risk. JAMA. 2004;291:2433–2440. doi: 10.1001/jama.291.20.2433. [DOI] [PubMed] [Google Scholar]
- 20.Brueggemeier RW, Quinn AL, Parrett ML, Joarder FS, Harris RE, Robertson FM. Correlation of aromatase and cyclooxygenase gene expression in human breast cancer specimens. Cancer Lett. 1999;140:27–35. doi: 10.1016/s0304-3835(99)00050-6. [DOI] [PubMed] [Google Scholar]
- 21.Brodie AM, Lu Q, Long BJ, Fulton A, Chen T, Macpherson N, DeJong PC, Blankenstein MA, Nortier JW, Slee PH, van dV, van Gorp JM, Elbers JR, Schipper ME, Blijham GH, Thijssen JH. Aromatase and COX-2 expression in human breast cancers. J Steroid Biochem Mol Biol. 2001;79:41–47. doi: 10.1016/s0960-0760(01)00131-5. [DOI] [PubMed] [Google Scholar]
- 22.Richards JA, Brueggemeier RW. Prostaglandin E2 regulates aromatase activity and expression in human adipose stromal cells via two distinct receptor subtypes. J Clin Endocrinol Metab. 2003;88:2810–2816. doi: 10.1210/jc.2002-021475. [DOI] [PubMed] [Google Scholar]
- 23.Brueggemeier RW, Diaz-Cruz ES, Li PK, Sugimoto Y, Lin YC, Shapiro CL. Translational studies on aromatase, cyclooxygenases, and enzyme inhibitors in breast cancer. J Steroid Biochem Mol Biol. 2005;95:129–136. doi: 10.1016/j.jsbmb.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Cruz ES, Shapiro CL, Brueggemeier RW. Cyclooxygenase inhibitors suppress aromatase expression and activity in breast cancer cells. J Clin Endocrinol Metab. 2005;90:2563–2570. doi: 10.1210/jc.2004-2029. [DOI] [PubMed] [Google Scholar]
- 25.Su B, Diaz-Cruz ES, Landini S, Brueggemeier RW. Novel sulfonanilide analogues suppress aromatase expression and activity in breast cancer cells independent of COX-2 inhibition. J Med Chem. 2006;49:1413–1419. doi: 10.1021/jm051126f. [DOI] [PubMed] [Google Scholar]
- 26.Julemont F, de L, Michaux XC, Damas J, Charlier C, Durant F, Pirotte B, Dogne JM. Spectral and crystallographic study of pyridinic analogues of nimesulide: determination of the active form of methanesulfonamides as COX-2 selective inhibitors. J Med Chem. 2002;45:5182–5185. doi: 10.1021/jm020920n. [DOI] [PubMed] [Google Scholar]