Abstract
Previous studies have demonstrated that cyclooxygenase-2 (COX-2) inhibitor NS-398 decrease aromatase activity at the transcriptional level in breast cancer cells. However, N-Methyl NS-398, which does not have COX-2 inhibitory activity but has very similar structure to NS-398, decreases aromatase activity and transcription in MCF-7 and MDA-MB-231 breast cells to the same extent as NS-398. This suggests that NS-398 decrease aromatase expression in breast cancer cells via other mechanism(s). Further investigations find that both compounds only decrease aromatase activity stimulated by forskolin/phorbol ester at the transcriptional level in both breast cancer cell lines and in breast stromal cells from patients. They do not affect aromatase expression and activity stimulated by dexamethasone. Both compounds also suppress MCF-7 cell proliferation stimulated by testosterone. Aromatase inhibition studies using placental microsomes demonstrate that the compounds show only weak direct aromatase inhibition. These results suggest that NS-398 and its N-methyl analog suppress aromatase expression and activity with multiple mechanisms.
Keywords: Breast cancer, Aromatase, Prostaglandin E2, Cyclooxygenase-2, NS-398
1. Introduction
Breast cancer is the second leading cause of cancer death in women in the United States, and an estimated 212,930 women in the United States will be diagnosed with breast cancer in 2006 [1]. Approximately two-thirds of breast cancers are termed hormone-dependent breast cancers, contain estrogen receptors (ER), and require estrogen for tumor growth. Treatment of ER-positive breast cancer cases includes hormonal therapies such as aromatase inhibitors (AIs) or antiestrogens/selective estrogen modulators (SERMs) [2].
The most commonly used SERM is tamoxifen, which inhibits the growth of breast tumors by competitive antagonism of estrogen at the ER ligand binding site. However, development of drug resistance and increased uterine and endometrial cancer reduce its clinical application [3–5]. In contrast, AIs markedly suppress plasma estrogen levels by inhibiting aromatase, the cytochrome P450 enzyme responsible for the synthesis of estrogens from androgenic precursors. However, these compounds inhibit aromatase activity in a global fashion and thus could adversely impact sites where estrogen is required for normal function. The risk of important long-term reduction in bone density, including osteoporosis, may increase with the use of aromatase inhibitors [6–8]. Short-term use of letrozole has been associated with an increase in bone-resorption markers in plasma and urine, and adjuvant therapy with anastrozole appears to be associated with a higher incidence of fractures than adjuvant therapy with tamoxifen [9–11].
A new approach to reduce the risk of side effects is to develop agents that regulate aromatase expression in a tissue-specific manner. In postmenopausal women, estrogens are produced locally in the breast by aromatase. Higher levels of estrogens in breast cancer cells and breast adipose tissue stimulate tumor growth by binding to estrogen receptors in both an autocrine and a paracrine manner [12–14]. In humans, cytochrome P450 aromatase is encoded by the CYP19 gene, which contains nine coding exons (exons II to X). The expression of this gene is regulated in a tissue-specific manner by the alternative use of eight promoters, each one of them being associated with a specific 5′ untranslated exon I [15]. Furthermore, due to the unique organization of tissue-specific promoters, various promoters employ different signaling pathways and different transcription factors [16–20].
One of the major stimulators of aromatase expression in cancer breast is prostaglandin E2 (PGE2) derived from tumorous epithelium and/or infiltrating macrophages. PGE2 acts via EP1 and EP2 receptor subtypes to stimulate both the protein kinase A and protein kinase C pathways to increase aromatase expression through promoter II and promoter I.3 in breast stromal cells [21–24]. Moreover, expression of the CYP19 gene is correlated with COX-1 and COX-2 expression in human breast cancer [25]. This biochemical mechanism may explain epidemiological observations of the beneficial effect of nonsteroidal anti-inflammatory drugs (NSAIDs) on breast cancer. Recently, the COX-2 selective inhibitor, celecoxib, has shown strong chemopreventive activity against mammary carcinoma in rats [26].
COX-2 inhibitors can suppress aromatase activity in breast cancer cells by suppressing aromatase transcription [27]. However, the suppression varied significantly among COX-2 inhibitors. This observation suggests differences in the mechanisms by which these COX inhibitors modulate aromatase expression in breast cancer cells. Among all the tested COX-2 inhibitors, NS-398 showed the highest potency in suppressing aromatase transcription and activity compared to other COX-2 inhibitors studied. On the other hand, NS-398 is a weak inhibitor of COX-2. Thus, NS-398 may target aromatase gene regulation through other pathways independent of direct COX-2 inhibition. To investigate potential mechanisms for NS-398 suppression of aromatase, we synthesized N-Methyl NS-398 (Me-NS-398) (Figure 1), which does not have COX-2 inhibitory activity but has very similar chemical structure to NS-398 [28]. N-Methyl NS-398 suppresses aromatase expression and activity in SK-BR-3 breast cancer cells at similar extent with NS-398. In the present study, we investigated the effects of these two agents on the regulation of aromatase expression in other human breast cancer cells and in breast stromal cells from patients.
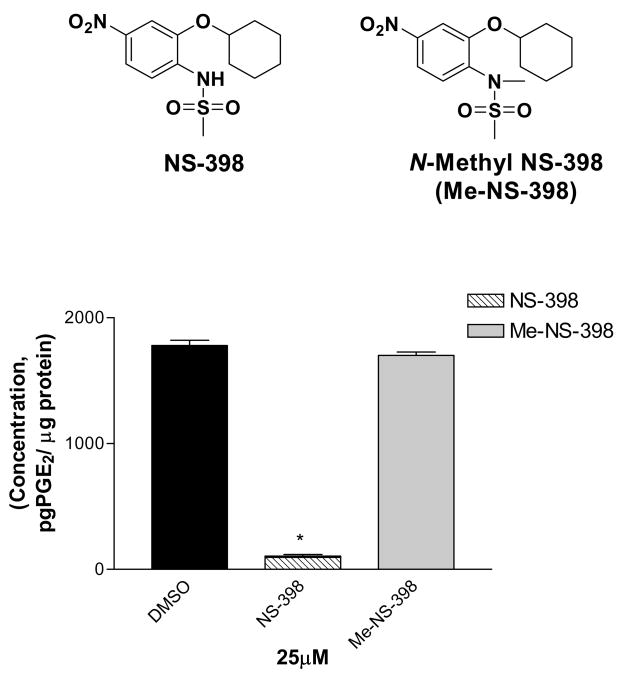
Figure 1.
Structure of NS-398 and N-Methyl NS-398 and their COX-2 inhibition results in MDA-MB-231 cell line [28].
2. Materials and Methods
2.1. Reagents
Radiolabeled [1β-3H]-androst-4-ene-3,17-dione was obtained from NEN Life Science Products (Boston, MA). NS-398 and Ru486 was purchased from Cayman Chemical (Ann Arbor, MI). Me-NS-398 was synthesized in our laboratory [28]. For in vitro experiments, these agents at various concentrations were dissolved in DMSO. Trypsin, TRIzol, and all enzymes were obtained from Invitrogen (Carlsbad, CA). Radioactive samples were counted on a LS6800 liquid scintillation counter (Beckman, Palo Alto, CA). Scintillation solution 3a70B was obtained from Research Prospect International Corp. (Mount Prospect, IL). Tetradecanoyl phorbol acetate (TPA), dexamethasone (DEX), forskolin (FSK), NADP+ and glucose-6-phosphate dehydrogenase were purchased from Sigma (St. Louis, MO). Human tissues (breast cancer, placenta) were obtained through OSUCCC Tissue Procurement under IRB-approved protocols OSU #2002H0104 and OSU #2002H0105.
2.2. Cell Culture
MCF-7 cells and MDA-MB-231 cells were obtained from ATCC (Rockville, MD). Cell cultures were maintained in phenol red-free custom media (MEM, Earle’s salts, 1.5x amino acids, 2x non-essential amino acids, L-glutamine, 1.5x vitamins, Gibco BRL) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 20 mg/L gentamycin. Adipose stromal cells were obtained as described [29] and maintained in DMEM/F12 media with 10% FBS, 2 mM L-glutamine and 20 mg/L gentamycin. Fetal bovine serum was heat inactivated for 30 min in a 56 °C water bath before use. Cell cultures were grown at 37 °C, in a humidified atmosphere of 5% CO2 in a Hereaus CO2 incubator. For all experiments, cells were plated in either T-25 flasks or 100 mM plates and grown to subconfluency. Before treatment, the media was changed to defined media consisting of DMEM/F12 media (Sigma) with 1.0 mg/mL human albumin (OSU Hospital Pharmacy), 5.0 mg/L human transferin and 5.0 mg/L bovine insulin.
2.3. Tritiated water-release assay in cells
Measurement of aromatase enzyme activity in cells was based on the tritium water release assay [24]. Cells in T-25 flasks or 100 mM plates were treated with DMSO (control), NS-398 and N-methyl NS-398 for 24 h. In the combination studies, cells were pre-treated with FSK/TPA or DEX for 24 h, then in combination with the indicated compounds for 24 h. After 24 hours, cells were then incubated for 6 h with fresh media containing drugs and 2 μCi [1β-3H]-androst-4-ene-3,17-dione (100nM). Subsequently, the reaction mixture was removed, and proteins were precipitated using 10% trichloroacetic acid at 42 °C for 20 min. After a brief centrifugation, the media was extracted three times with an equal amount of chloroform to extract unused substrate, and the aqueous layer subsequently treated with 1% dextran-treated charcoal. After centrifugation, a 250-μL aliquot containing the product was counted in 5 mL of liquid scintillation mixture. Each sample was run in triplicate and results were corrected for blanks and for the cell contents of culture flasks. Results were expressed as picomoles of 3H2O formed per hour incubation time per million cells (pmol/h/106 cells). To determine the amount of live cells in each flask, the cells were trypsinized and analyzed using the diphenylamine DNA assay adapted to a 96-well plate [24;30].
2.4. Preparation of Human Placental Microsomes
Human placentas were processed immediately after delivery from The Ohio State University Hospitals at 4 °C. The placenta was washed with normal saline, and connective and vascular tissue was removed. Microsomes were prepared from the remaining tissue using the method described by Kellis and Vickery [31]. Microsomal suspensions were stored at −80 °C until required.
2.5. Tritiated water-release assay with human placental microsomes
Inhibition of human placental aromatase was determined by monitoring the amount of tritium water released as the enzyme converts [1β-3H] androst-4-ene-3,17-dione to estrone [32]. Aromatase activity assays were carried in 0.1 M potassium phosphate buffer (pH 7.0) with 5% propylene glycol. All samples contained a NADPH regenerating system consisting of 2.85 mM glucose-6-phosphate, 1.8 mM NADP+ and 1.5 units of glucose-6-phosphate dehydrogenase. Samples contained 100 nM androst-4-ene-3, 17-dione (400,000 ~ 450,000 dpm). Reactions were initiated with the addition of 50 μg microsomal protein. The total incubation volume was 2.0 mL. Incubations were allowed to proceed for 15 min in a shaking water bath at 37 °C. Reactions were quenched by the addition of 2.0 mL of chloroform. Samples were then vortexed and centrifuged for 5 min and the aqueous layer was removed. The aqueous layer was subsequently extracted twice in the same manner with 2.0 mL chloroform. A 0.5 mL aliquot of the final aqueous layer was combined with 5 mL 3a70B scintillation cocktail and the amount of radioactivity was determined. Each sample was run in triplicate and background values were determined with microsomal protein inactivated by boiling.
2.6. RNA extraction
Total RNA was isolated using the TRIzol reagent according to the manufacturer’s protocol. Total RNA pellets were dissolved in DNase, RNase-free water and quantitated using a spectrophotometer. The quality of RNA samples was determined by electrophoresis through agarose gels and staining with ethidium bromide; the 18S and 28S rRNA bands were visualized under ultraviolet light.
2.7. cDNA synthesis
Isolated total RNA (2 μg) was treated with DNase I Amplification grade, according to the recommended protocol to eliminate any DNA before reverse transcription. Treated total RNA was denatured at 65 °C for 5 min in the presence of 2.5 ng/μL random hexamers and 0.5 mM dNTP mix. The samples were snap-cooled on ice and centrifuged briefly. Complementary DNA (cDNA) was synthesized using Superscript II reverse transcriptase according to the recommended protocol. Briefly, the reactions were conducted in the presence of 1X First-Strand Buffer and 20 mM DTT at 42 °C for 50 min and consequently inactivated at 70 °C for 15 min. The cDNA generated was used as a template in real-time PCR reactions.
2.8. Real-time PCR
Real-time PCR was performed using the Opticon™ 2 system from MJ Research (Waltham, MA). For the CYP19 total gene the PCR reaction mixture consisted of Taqman® Universal PCR Master Mix (Applied Biosystems), 600 nM of CYP19 primer (sense: 5′-TGT CTC TTT GTT CTT CAT GCT ATT TCT C-3′; antisense: 5′-TCA CCA ATA ACA GTC TGG ATT TCC-3′); 250 nM Taqman probe (6FAM 5′-TGC AAA GCA CCC TAA TGT TGA AGA GGC AAT-3′TAMRA)(Invitrogen), and 2.0 μL of each cDNA sample in a final volume of 20 μL. For the 18S house keeping total gene the PCR reaction mixture consisted of Taqman® Universal PCR Master Mix (Applied Biosystems), 500 nM of 18S primer (sense: 5′-CAG TTC ATA CAG CGG AAC ACT G-3′; antisense: 5′-TTT GCT GGA GAA CAG GGC TG-3′); 50 nM Taqman probe (6FAM 5′-TGC TGG CAC CAG ACT TGC CCT C-3′TAMRA) (Invitrogen), and 2.0 μL of each cDNA sample in a final volume of 20 μL. The Taqman probes for aromatase and 18S were designed to anneal to a specific sequence of the aromatase and 18S gene correspondingly between the forward and the reverse primers. Cycling conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 50 cycles at 95 °C for 15 s and 60 °C for 1 min.
2.9. Cell Proliferation Assay
MCF-7 cells were harvested, counted, and plated at a concentration of 1 × 104 cells/well in 400-μl total volume/well in 24 well plates. After 24 hours, the culture medium was removed and changed to DMEM/F12 media supplemented with 5% charcoal stripped FBS. After 24 hours, culture wells (n=6) were treated with the compounds combined with or without Testosterone (100nM) (400 μl volume) every two days for a total of six days. 24 hours after the last treatment, 3,(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2- (4-sulfophenyl)-2H-tetrazolium, inner salt and phenazine methosulfate were prepared in PBS at a final assay concentrations of 333 μg/ml and 25 μM respectively. These solutions were combined and 50 μl of this mixture were added to each well. After 1 hour of incubation at 37°C, absorbance at 490 nm was measured using a SPECTRAmax plate reader.
2.10. Statistical analysis
Statistical and graphical information was determined using GraphPad Prism software (GraphPad Software Incorporated) and Microsoft Excel (Microsoft Corporation). Determination of IC50 values were performed using nonlinear regression analysis. Statistically significant differences were calculated with the two-tailed unpaired Student’s t-test and P values reported at 95% confidence intervals.
3. Results
3.1. Decrease of Aromatase Enzymatic Activity in MCF-7 and MDA-MB-231 breast cancer cells
PGE2 produced in breast cancer tissue regulates aromatase expression through PKA/PKC driven pathway in an autocrine and paracrine manner in breast cells [23;24]. COX inhibitors were able to suppress aromatase of breast cancer cells of the transcriptional level. However, COX-2 inhibitors which have similar IC50 values for COX-2 inhibition differ significantly in their ability to suppress aromatase activity. NS-398 showed the highest potency in suppressing aromatase activity compared to other agents tested such as celecoxib [27]. We hypothesize that NS-398 may target regulation of aromatase gene transcription more effectively through other pathways than direct COX-2 inhibition. To test this hypothesis, we synthesized NS-398 derivative Me-NS-398(N-methyl-N-(2-cyclohexyloxy-4-nitrophenyl)methanesulfonamide), which does not inhibit PGE2 production (Figure 1) [28].
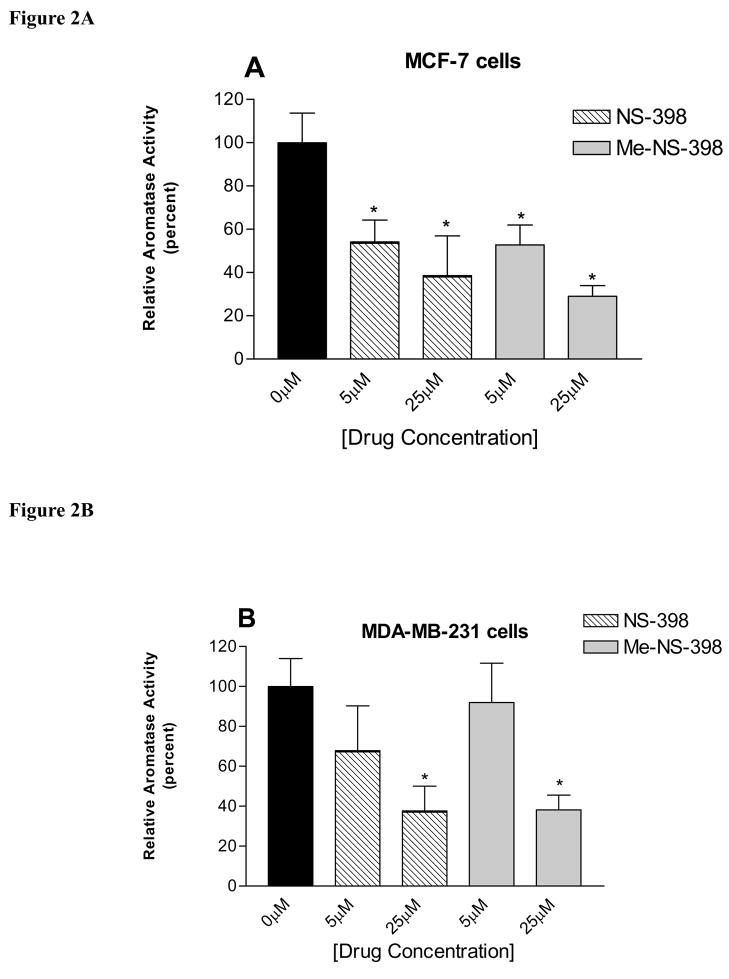
The effects of NS-398 and Me-NS-398 on aromatase activity are determined in MCF-7 and MDA-MB-231 breast cancer cells. At 25μM, both compounds significantly decreased aromatase activity in the two cell lines (Figure 2). At 5μM, both compounds only decreased aromatase activity in MCF-7 cells. Thus, Me-NS-398 and NS-398 exhibit similar effects on the suppression of aromatase activity in both breast cancer cell lines.
Figure 2. Suppression of aromatase activity in MCF-7 and MDA-MB-231 breast cancer cells.
Cells were treated with NS-398 and MNS-398 at indicated concentrations. The results were normalized against a control treatment with vehicle. Each data bar represents the mean results of three independent determinations. (A) MCF-7 cells, the value of 100% is equal to 0.0002 pmol/hr/106 cells. (B) MDA-MB-231 cells, the value of 100% is equal to 0.00007 pmol/hr/106 cells. *P <0.05 vs. control by unpaired t test.
3.2. Aromatase activity assay in human placental microsomes
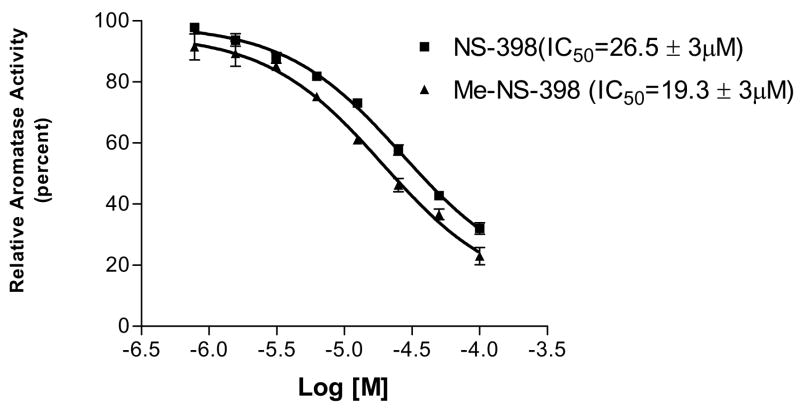
The compounds were tested for potential direct aromatase inhibition in the microsomal aromatase assay to determine if the decrease of aromatase activity is due to direct enzyme inhibition. These investigations find that NS-398 and Me-NS-398 produce only weak inhibition of aromatase with IC50 values of 26.5 μM and 19.3 μM, respectively (Figure 3). The results suggest that the suppression of aromatase activity by NS-398 and Me-NS-398 in MCF-7 and MDA-MB-231 cells by direct enzyme inhibition may only be observed at high concentrations.
Figure 3. Aromatase inhibition of microsomal activities of compounds NS-398 (■) and Me-NS-398 (▲).
Error bars represent standard error (n = 3), and the data were statistically analyzed by a nonlinear regression analysis method.
3.3. CYP19 mRNA Expression by Real-Time PCR in MCF-7 and MDA-MB-231 cells
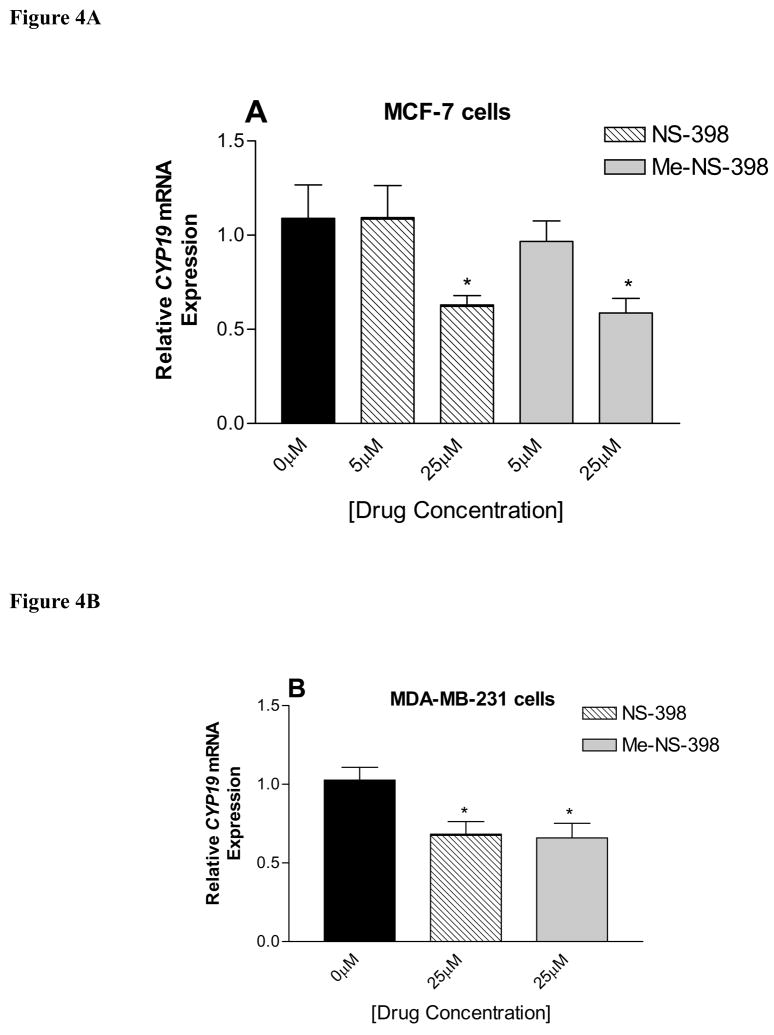
Analysis of total CYP19 mRNA transcripts was performed using real-time PCR in order to assess the extent of suppression in aromatase activity by NS-398 and Me-NS-398 through down-regulation of aromatase expression at transcriptional level. Cells were treated with NS-398 and Me-NS-398 for 24 h at 5 and 25μM. Total RNA was extracted after 24 h, and CYP19 transcript levels are compared to control (vehicle) treatment. Both compounds significantly decreased CYP19 gene expression in the two cell lines relative to the control at 25μM (Figure 4). No effect on the expression level of the housekeeping gene 18S rRNA was observed with any of the compounds. At 5μM, both compounds did not decrease CYP19 gene expression, but they suppressed aromatase activity in MCF-7 cells (Figure 3A). This suppression of enzyme activity is not likely to occur through direct enzyme inhibition, because the drug concentration in this experiment is much lower than the IC50 value from the microsomal aromatase inhibition study. This suggests that a non-genomic mechanism may also be involved in suppression of cellular aromatase activity. In all the experiments, Me-NS-398 decrease aromatase activity and expression to a similar extent; thus, NS-398 and Me-NS-398 apparently target other pathway(s) to suppress aromatase.
Figure 4. Real-time RT-PCR analysis of CYP19 mRNA expression in MCF-7 (A) and MDA-MB-231 cells (B).
Cells were treated for 24 h with the test agents at different concentrations, and total RNA was isolated. Results are expressed as means of CYP19 (normalized to 18S rRNA) ± SEM. *P <0.05 vs. control by unpaired t test (n = 9).
3.4. NS-398 and Me-NS-398 suppress aromatase stimulated by FSK/TPA in breast cancer and stromal cells
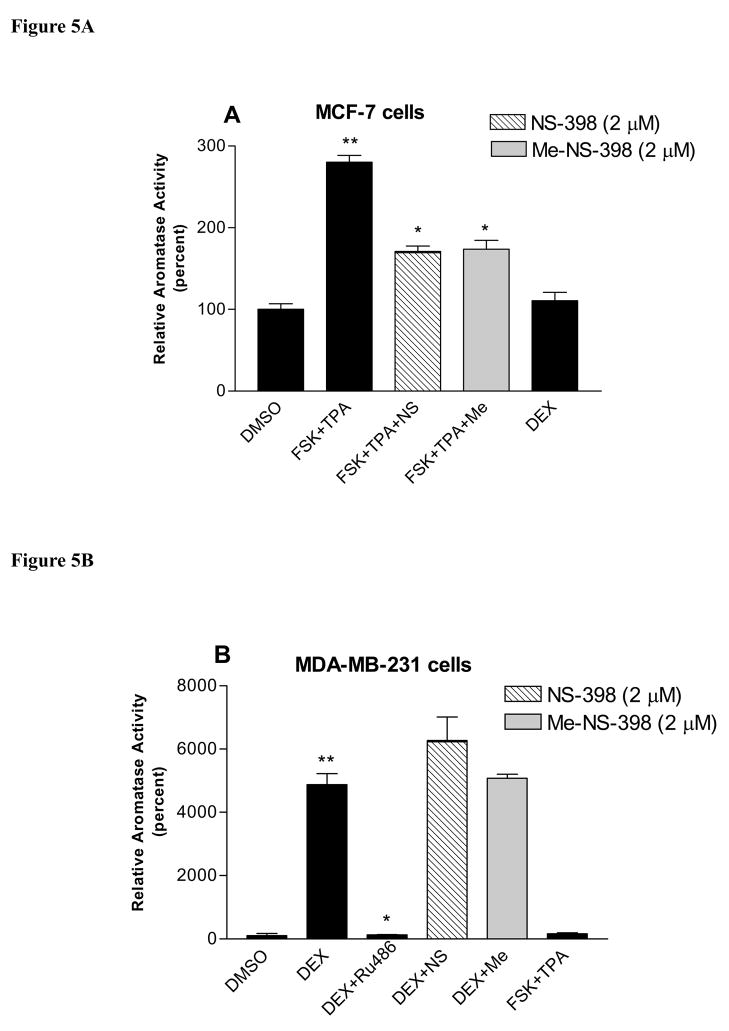
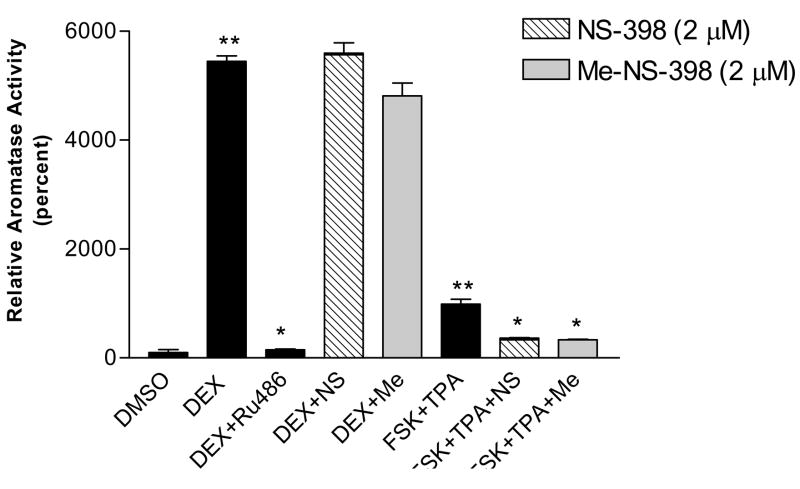
FSK/TPA can stimulate aromatase activity by activating PKA and PKC pathways, which are the major pathways to stimulate aromatase expression in breast cancer. DEX stimulates aromatase expression by binding to glucocorticoid receptor, which is the main pathway for aromatase expression in normal breast, skin and bone tissue [18;22;23]. The effect of NS-398 and Me-NS-398 on the aromatase activity stimulated by FSK (25μM)/TPA (10nM) or DEX (200nM) in MCF-7 and MDA-MB-231 cells was examined. NS-398 and Me-NS-398 at 2μM did not suppress aromatase activity stimulated by DEX, whereas DEX stimulation could be suppressed by RU486 (200nM), a glucocorticoid receptor antagonist, in MDA-MB-231 cells (Figure 5B). On the other hand, the two agents suppress aromatase activity stimulated by FSK/TPA in MCF-7 cells at 2μM (Figure 5A). Furthermore, we find that this suppression of aromatase activity occurs at the transcriptional level (Figure 6). This suggests that these agents affect PKA/PKC-driven transcription of aromatase. FSK/TPA does not stimulate aromatase in MDA-MB-231 cells, and DEX does not stimulate aromatase in MCF-7 cells (Figure 5B lane 5; Figure 5A lane 5). For MCF-7 cells, the aromatase transcription occurs primarily via promoter I.3 and II. Promoter I.4 is not detected in this cell line [17], which may explain why DEX can not stimulate promoter I.4 driven aromatase transcription.
Figure 5. Effect of NS-398 and Me-NS-398 on induced aromatase in breast cancer cells.
Cells were pre-treated with FSK(25μM)/TPA (10nM) or DEX(200nM) for 24 hr, then with the indicated compounds (2μM), Ru486(200nM) and FSK/TPA or DEX for 24 hr. Aromatase activity was subsequently determined during a 6 h assay as described. *p<0.05 vs FSK/TPA or DEX treatment for suppression of induced aromatase activity; **p<0.05 vs DMSO treatment for stimulation of aromatase activity by unpaired t test (n=3). (A) MCF-7 cells, the value of 100% is equal to 0.0002pmol/106 cells/hr. (B) MDA-MB-231 cells, the value of 100% is equal to 0.00007pmol/106 cells/hr. Similar results were obtained in at least two independent experiments.
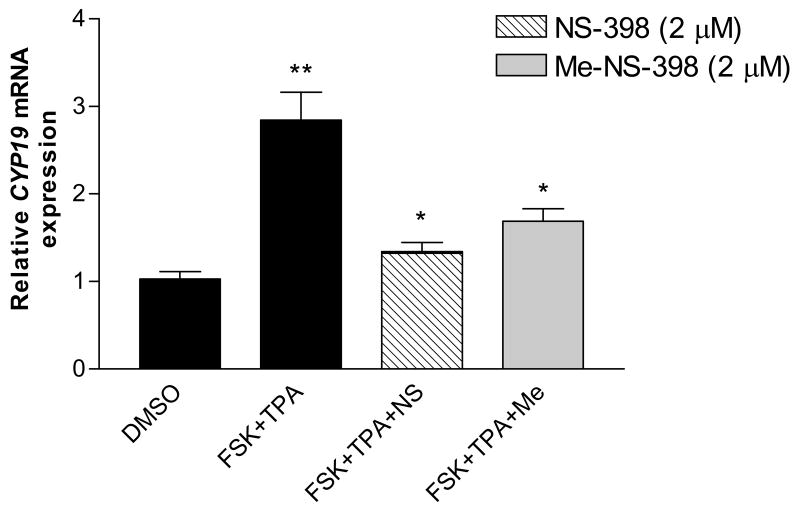
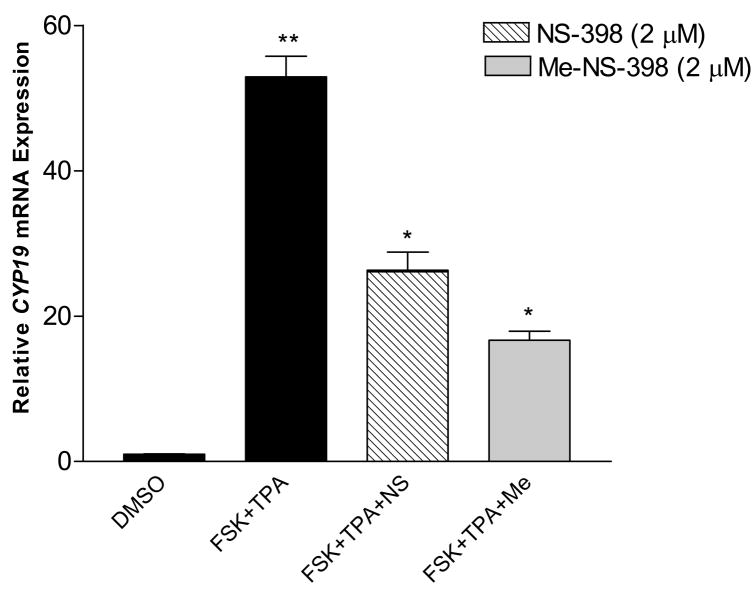
Figure 6. Effect of NS-398 and Me-NS-398 on induced CYP19 gene expression in MCF-7 breast cancer cells.
Cells were pre-treated with FSK(25μM)/TPA (10nM) for 24 hr, then with the indicated compounds (2μM) and FSK/TPA for 24 hr. Total RNA was isolated. Results are expressed as means of CYP19 (normalized to 18S rRNA) ± SEM., *p<0.05 vs FSK/TPA treatment for suppression of induced aromatase transcription; **p<0.05 vs DMSO treatment for stimulation of aromatase transcription by unpaired t test (n = 9). Similar results were obtained in at least two independent experiments.
In addition, NS-398 and Me-NS-398 at 2μM decreased the aromatase activity stimulated by FSK (10μM)/TPA (10nM) in breast stromal cells, and this suppression occurred at the transcriptional level (Figure 7, 8). NS-398 and Me-NS-398 at 2μM do not suppress aromatase activity stimulated by DEX (200nM).
Figure 7. Effect of NS-398 and Me-NS-398 on induced aromatase in breast stromal cells.
Cells were pre-treated with FSK(10μM)/TPA (10nM) or DEX(200nM) for 24 hr, then with the indicated compounds (2 μM), Ru486(200nM) and FSK/TPA or DEX for 24 hr. Aromatase activity was subsequently determined during a 6 h assay as described. *p<0.05 vs FSK/TPA or DEX treatment for suppression of induced aromatase activity; **p<0.05 vs DMSO treatment for stimulation of aromatase activity by unpaired t test (n = 3). The value of 100% is equal to 0.00012pmol/106 cells/hr. Similar results were obtained in at least two independent experiments.
Figure 8. Effect of NS-398 and Me-NS-398 on induced CYP19 gene expression in breast stromal cells.
Cells were pre-treated with FSK (10μM)/TPA (10nM) for 24 hr, then with the indicated compounds (2μM) and FSK/TPA for 24 hr. Total RNA was isolated. Results are expressed as means of CYP19 (normalized to 18S rRNA) ± SEM. *p<0.05 vs FSK/TPA treatment for suppression of induced aromatase transcription; **p<0.05 vs DMSO treatment for stimulation of aromatase transcription by unpaired t test (n=9). Similar results were obtained in at least two independent experiments.
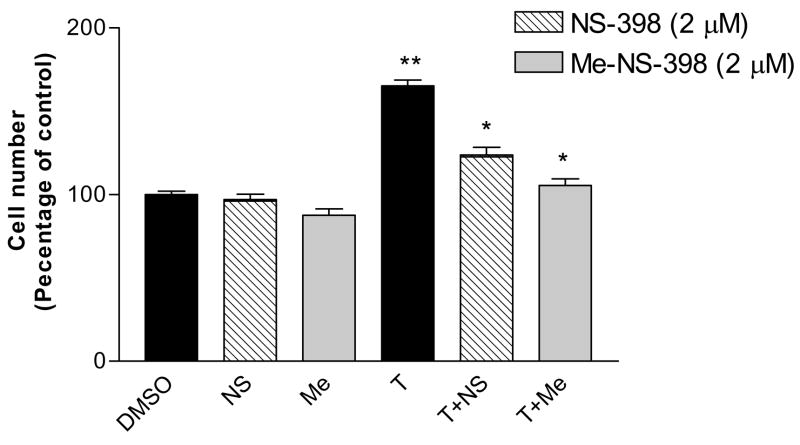
3.5. NS-398 and Me-NS-398 significantly suppressed testosterone induced MCF-7 cell proliferation
Treatment of MCF-7 cells with NS-398 or Me-NS-398 did not significantly affect normal MCF-7 cell proliferation at 2μM (Figure 9). Since the aromatase enzyme is present in MCF-7 cells, testosterone can be converted to estradiol and stimulate MCF-7 cell growth [33;34]. In this present study, testosterone (100nM) significantly increase MCF-7 cell proliferation. Both NS-398 and Me-NS-398 suppressed testosterone-induced cell proliferation in MCF-7 cells, presumable by suppression of aromatase expression and subsequently lack of conversion of testosterone to estradiol.
Figure 9. Antiproliferation effect of NS-398 and Me-NS-398 in MCF-7 cells.
MCF-7 cells were seeded into 24 wells plates at ~1×104/well and exposed to testosterone (T, 100nM) or the test agent (NS-398 and Me-NS-398at 2 μM) plus testosterone (100nM) in DMEM/F12 media supplemented 5% charcoal stripped fetal bovine serum. Cell viability was measured as described in the experimental section. The results were normalized against a control treatment with vehicle (DMSO). *p<0.05 vs testosterone treatment for suppression of induced proliferation; **p<0.05 vs control for stimulation of proliferation by unpaired t test (n=6). Similar results were obtained in at least two independent experiments.
4. Discussion
Increased aromatase expression in breast cancer tissue is associated with a switch in promoter usage from the normal adipose-specific promoter I.4 to the PGE2-responsive promoter II and promoter I.3. While promoter I.4 requires the synergistic actions of glucocorticoids and class I cytokines [19], promoters I.3 and II are both transactivated via PKA and PKC dependent signaling pathways [19;35]. This switch in the regulatory mechanism of aromatase expression from normal breast tissue to cancerous tissue has been extensively investigated [36]. Our hypothesis is that higher levels of COX-2 expression result in higher levels of PGE2, which in turn results in increased CYP19 expression. Potentially, COX-2 inhibitors could inhibit aromatase expression selectively in cancer breast tissue; therefore, COX-2 inhibitors may be classified as the first generation of selective aromatase expression regulators for ER positive breast cancer.
In our previous study [27], the COX-2 selective inhibitor NS-398 suppressed aromatase transcription and activity in breast cancer cells significantly better than other COX-2 inhibitors, even though NS-398 has weaker COX-2 inhibitory activity. These results suggested that other mechanisms may also be involved in regulation of aromatase expression by COX inhibitors. Various NSAIDs can influence the expression of certain transcription factors and other pathway mediators that could result in the suppression of aromatase.
To elucidate the phenomenon, Me-NS-398, which is devoid of COX-2 inhibitory activity, was synthesized and studied previously in SK-BR-3 breast cancer cells [28]. The biological activities of NS-398 and Me-NS-398 on aromatase were investigated here in MCF-7 and MDA-MB-231 breast cancer cells, normal breast stromal cells and also in microsomal aromatase assay. The results showed that Me-NS-398 and NS-398 suppressed aromatase activity and expression in these breast cancer cells. Microsomal aromatase inhibition studies further demonstrated that the compounds only inhibit aromatase enzyme at high concentrations. NS-398 and Me-NS-398 suppressed CYP19 gene expression in both MCF-7 and MDA-MB-231 cell lines as measured by real time PCR. These results suggest that the compounds suppress aromatase activity in breast cancer cells via multiply mechanisms.
To further investigate the possible mechanism(s) in suppression aromatase expression, the compounds were tested for their ability to suppress aromatase in breast cancer and stromal cells stimulated by FSK/TPA or DEX, which induce aromatase expression via different pathways and promoters. FSK/TPA stimulates aromatase via PKA and PKC pathways, and DEX induces aromatase via glucocorticoid receptor mediated transcription. The results demonstrated that NS-398 and Me-NS-398 suppressed aromatase activity by decreasing PKA/PKC driven transcription (FSK/TPA stimulated) and did not interfere with normal breast aromatase expression mediated through glucocorticoid receptor (DEX stimulated) in both breast epithelial and stromal cells. Additionally, the compounds inhibited testosterone stimulated MCF-7 cell proliferation, which also suggests that the compounds are able to decrease aromatase activity.
In summary, though N-methyl NS-398 does not show COX-2 inhibitory activity, it decreases aromatase activity and expression in breast cancer cells with similar extent as NS-398. This suggests NS-398 decrease aromatase expression in breast cells by other mechanism(s) other than COX-2 inhibition. These two compounds selectively affected aromatase stimulated from PKA/PKC pathway, which are the major pathways involved in aromatase expression in breast cancer. Consequently, our current research focuses on discerning the specific molecular target of the compounds and identifying a novel molecular basis for the development of selective aromatase expression regulators. In addition, investigations of structural requirements for aromatase suppression of NS-398 and analogs are on-going.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Grant R01 CA73698 (to R.W.B.), the NIH Chemistry and Biology Interface Training Program Grant T32 GM08512 (to E.S.D.-C.), and The Ohio State University Comprehensive Cancer Center Breast Cancer Research Fund.
ABBREVIATION LIST
- PGE2
Prostaglandin E2
- COX-2
cyclooxygenase-2
- NSAIDs
nonsteroidal anti-inflammatory drugs
- ER
estrogen receptors
- AIs
aromatase inhibitors
- SERMs
selective estrogen modulators
- TPA
tetradecanoyl phorbol acetate
- DEX
dexamethasone
- FSK
forskolin
- PKA
protein kinase A
- PKC
protein kinase C
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Cancer Society. Cancer Facts and Figures 2006. Atlanta: American Cancer Society; American Cancer Society. [Google Scholar]
- 2.Brueggemeier RW, Hackett JC, Diaz-Cruz ES. Aromatase inhibitors in the treatment of breast cancer. Endocr Rev. 2005;26:331–45. doi: 10.1210/er.2004-0015. [DOI] [PubMed] [Google Scholar]
- 3.Michalides R, Griekspoor A, Balkenende A, et al. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348:2431–42. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- 5.Bergman L, Beelen ML, Gallee MP, et al. Risk and prognosis of endometrial cancer after tamoxifen for breast cancer. Comprehensive Cancer Centres’ ALERT Group. Assessment of Liver and Endometrial cancer Risk following Tamoxifen. Lancet. 2000;356:881–7. doi: 10.1016/s0140-6736(00)02677-5. [DOI] [PubMed] [Google Scholar]
- 6.Bonneterre J, Thurlimann B, Robertson JF, et al. Anastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: results of the Tamoxifen or Arimidex Randomized Group Efficacy and Tolerability study. J Clin Oncol. 2000;18:3748–57. doi: 10.1200/JCO.2000.18.22.3748. [DOI] [PubMed] [Google Scholar]
- 7.Powles TJ, Hickish T, Kanis JA, et al. Effect of tamoxifen on bone mineral density measured by dual-energy x-ray absorptiometry in healthy premenopausal and postmenopausal women. J Clin Oncol. 1996;14:78–84. doi: 10.1200/JCO.1996.14.1.78. [DOI] [PubMed] [Google Scholar]
- 8.Love RR, Barden HS, Mazess RB, et al. Effect of tamoxifen on lumbar spine bone mineral density in postmenopausal women after 5 years. Arch Intern Med. 1994;154:2585–8. [PubMed] [Google Scholar]
- 9.Baum M, Budzar AU, Cuzick J, et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359:2131–9. doi: 10.1016/s0140-6736(02)09088-8. [DOI] [PubMed] [Google Scholar]
- 10.Harper-Wynne C, Ross G, Sacks N, et al. Effects of the aromatase inhibitor letrozole on normal breast epithelial cell proliferation and metabolic indices in postmenopausal women: a pilot study for breast cancer prevention. Cancer Epidemiol Biomarkers Prev. 2002;11:614–21. [PubMed] [Google Scholar]
- 11.Heshmati HM, Khosla S, Robins SP, et al. Role of low levels of endogenous estrogen in regulation of bone resorption in late postmenopausal women. J Bone Miner Res. 2002;17:172–8. doi: 10.1359/jbmr.2002.17.1.172. [DOI] [PubMed] [Google Scholar]
- 12.Santner SJ, Chen S, Zhou D, et al. Effect of androstenedione on growth of untransfected and aromatase-transfected MCF-7 cells in culture. J Steroid Biochem Mol Biol. 1993;44:611–6. doi: 10.1016/0960-0760(93)90267-z. [DOI] [PubMed] [Google Scholar]
- 13.Yue W, Zhou D, Chen S, Brodie A. A new nude mouse model for postmenopausal breast cancer using MCF-7 cells transfected with the human aromatase gene. Cancer Res. 1994;54:5092–5. [PubMed] [Google Scholar]
- 14.Sun XZ, Zhou D, Chen S. Autocrine and paracrine actions of breast tumor aromatase. A three-dimensional cell culture study involving aromatase transfected MCF-7 and T-47D cells. J Steroid Biochem Mol Biol. 1997;63:29–36. doi: 10.1016/s0960-0760(97)00068-x. [DOI] [PubMed] [Google Scholar]
- 15.Simpson ER, Mahendroo MS, Means GD, et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–55. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 16.Chen S. Aromatase and breast cancer. Front Biosci. 1998;3:d922–d933. doi: 10.2741/a333. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Zhou D, Esteban J, et al. Aromatase gene expression and its exon I usage in human breast tumors. Detection of aromatase messenger RNA by reverse transcription-polymerase chain reaction. J Steroid Biochem Mol Biol. 1996;59:163–71. doi: 10.1016/s0960-0760(96)00100-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Mendelson CR, Simpson ER. Characterization of the sequences of the human CYP19 (aromatase) gene that mediate regulation by glucocorticoids in adipose stromal cells and fetal hepatocytes. Mol Endocrinol. 1995;9:340–9. doi: 10.1210/mend.9.3.7776980. [DOI] [PubMed] [Google Scholar]
- 19.Zhou D, Clarke P, Wang J, Chen S. Identification of a promoter that controls aromatase expression in human breast cancer and adipose stromal cells. J Biol Chem. 1996;271:15194–202. doi: 10.1074/jbc.271.25.15194. [DOI] [PubMed] [Google Scholar]
- 20.Zhou D, Zhou C, Chen S. Gene regulation studies of aromatase expression in breast cancer and adipose stromal cells. J Steroid Biochem Mol Biol. 1997;61:273–80. [PubMed] [Google Scholar]
- 21.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Transcriptional regulation of CYP19 gene (aromatase) expression in adipose stromal cells in primary culture. J Steroid Biochem Mol Biol. 1997;61:203–10. doi: 10.1016/s0960-0760(97)80013-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhou D, Chen S. Identification and characterization of a cAMP-responsive element in the region upstream from promoter 1.3 of the human aromatase gene. Arch Biochem Biophys. 1999;371:179–90. doi: 10.1006/abbi.1999.1454. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Agarwal VR, Mendelson CR, Simpson ER. Estrogen biosynthesis proximal to a breast tumor is stimulated by PGE2 via cyclic AMP, leading to activation of promoter II of the CYP19 (aromatase) gene. Endocrinology. 1996;137:5739–42. doi: 10.1210/endo.137.12.8940410. [DOI] [PubMed] [Google Scholar]
- 24.Richards JA, Brueggemeier RW. Prostaglandin E2 regulates aromatase activity and expression in human adipose stromal cells via two distinct receptor subtypes. J Clin Endocrinol Metab. 2003;88:2810–6. doi: 10.1210/jc.2002-021475. [DOI] [PubMed] [Google Scholar]
- 25.Brueggemeier RW, Quinn AL, Parrett ML, et al. Correlation of aromatase and cyclooxygenase gene expression in human breast cancer specimens. Cancer Lett. 1999;140:27–35. doi: 10.1016/s0304-3835(99)00050-6. [DOI] [PubMed] [Google Scholar]
- 26.Harris RE, Alshafie GA, Abou-Issa H, Seibert K. Chemoprevention of breast cancer in rats by celecoxib, a cyclooxygenase 2 inhibitor. Cancer Res. 2000;60:2101–3. [PubMed] [Google Scholar]
- 27.Diaz-Cruz ES, Shapiro CL, Brueggemeier RW. Cyclooxygenase inhibitors suppress aromatase expression and activity in breast cancer cells. J Clin Endocrinol Metab. 2005;90:2563–70. doi: 10.1210/jc.2004-2029. [DOI] [PubMed] [Google Scholar]
- 28.Su B, Diaz-Cruz ES, Landini S, Brueggemeier RW. Novel sulfonanilide analogues suppress aromatase expression and activity in breast cancer cells independent of COX-2 inhibition. J Med Chem. 2006;49:1413–9. doi: 10.1021/jm051126f. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Nichols JE, Valdez R, et al. Tumor necrosis factor-alpha stimulates aromatase gene expression in human adipose stromal cells through use of an activating protein-1 binding site upstream of promoter 1.4. Mol Endocrinol. 1996;10:1350–7. doi: 10.1210/mend.10.11.8923461. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan N, Shambaugh GE, III, Elseth KM, et al. Adaptation of the diphenylamine (DPA) assay to a 96-well plate tissue culture format and comparison with the MTT assay. Biotechniques. 1994;17:166–71. [PubMed] [Google Scholar]
- 31.Kellis JT, Jr, Vickery LE. Purification and characterization of human placental aromatase cytochrome P-450. J Biol Chem. 1987;262:4413–20. [PubMed] [Google Scholar]
- 32.Su B, Hackett JC, Diaz-Cruz ES, et al. Lead optimization of 7-benzyloxy 2-(4′-pyridylmethyl)thio isoflavone aromatase inhibitors. Bioorg Med Chem. 2005;13:6571–7. doi: 10.1016/j.bmc.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 33.Sonne-Hansen K, Lykkesfeldt AE. Endogenous aromatization of testosterone results in growth stimulation of the human MCF-7 breast cancer cell line. J Steroid Biochem Mol Biol. 2005;93:25–34. doi: 10.1016/j.jsbmb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Cos S, Martinez-Campa C, Mediavilla MD, Sanchez-Barcelo EJ. Melatonin modulates aromatase activity in MCF-7 human breast cancer cells. J Pineal Res. 2005;38:136–42. doi: 10.1111/j.1600-079X.2004.00186.x. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal VR, Bulun SE, Leitch M, et al. Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J Clin Endocrinol Metab. 1996;81:3843–9. doi: 10.1210/jcem.81.11.8923826. [DOI] [PubMed] [Google Scholar]
- 36.Harada N. Aberrant expression of aromatase in breast cancer tissues. J Steroid Biochem Mol Biol. 1997;61:175–84. [PubMed] [Google Scholar]