Abstract
Uropathogenic Escherichia coli (UPEC) is the leading cause of urinary tract infections in women, causing significant morbidity and mortality in this population. Adherence to host epithelial cells is a pivotal step in the pathogenesis of UPEC. One of the most important virulence factors involved in mediating this attachment is the type 1 pilus (type 1 fimbria) encoded by a set of fim genes arranged in an operon. The expression of type 1 pili is controlled by a phenomenon known as phase variation, which reversibly switches between the expression of type 1 pili (Phase-ON) and loss of expression (Phase-OFF). Phase-ON cells have the promoter for the fimA structural gene on an invertible DNA element called fimS, which lines up to allow transcription, whereas transcription of the structural gene is silenced in Phase-OFF cells. The orientation of the fimS invertible element is controlled by two site-specific recombinases, FimB and FimE. Environmental conditions cause transcriptional and post-transcriptional changes in UPEC cells that affect the level of regulatory proteins, which in turn play vital roles in modulating this phase switching ability. The role of fim gene regulation in UPEC pathogenesis will be discussed.
Keywords: Type 1 fimbriae, Type 1 pili, Gene regulation, Uropathogenic Escherichia coli, Urinary tract
ROLE OF TYPE 1 PILI IN UROPATHOGENIC ESCHERICHIA COLI PATHOGENESIS
Uropathogenic Escherichia coli (UPEC) is the number one cause of urinary tract infections in the United States[1,2]. Approximately 6-7 million people are afflicted with a urinary tract infection each year in the United States at a cost of $2.5 billion per year. Urinary tract infections are modeled as ascending infections. In women, the UPEC bacteria move from the rectum to the vaginal surface to the urinary tract. Although UPEC can express several different varieties of pili, type 1 pili may be the most important in the human lower urinary tract. Agglutination of guinea pig erythrocytes in the absence of mannose is an important characteristic of type 1 pili[3,4]. Besides Escherichia coli (E. coli), type 1 pili are found on several other species within the Enterobacteriaceae family[5]. The role of type 1 piliated UPEC cells in the pathogenesis of human urinary tract infections was first demonstrated in the early 1980s and has continued in more recent studies[6-12]. Moreover, these human patient studies have been supported by several murine urinary tract infection model studies that have shown the importance of type 1 pili in UPEC pathogenesis[11,13-15]. This culminated in a study by Connell et al[16], who compared a fimA mutant strain to the wild-type parent to show the critical role of type 1 pili in UPEC colonization of the lower urinary tract.
GENETIC ORGANIZATION OF THE UPEC fim OPERON
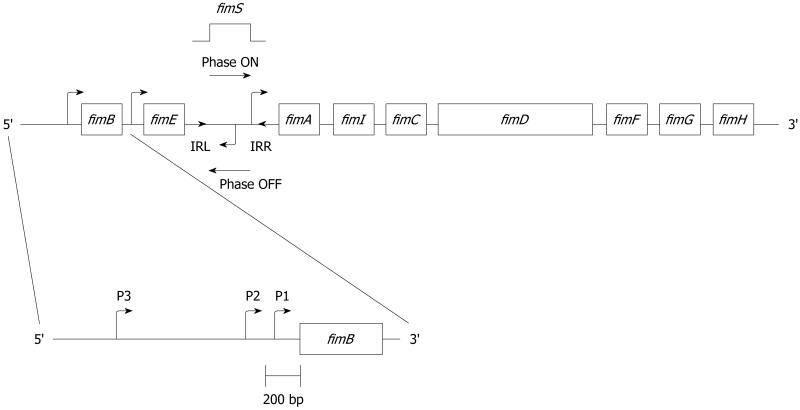
Type 1 pili are produced from a contiguous DNA segment, labeled the fim operon, which encodes the genes necessary for their synthesis, assembly, and regulation. The fim cluster was mapped to the 98 min on the E. coli chromosome[17]. Nine genes have now been identified within the gene cluster (Figure 1).
Figure 1. Schematic of the fim operon, including the characterized promoter sites.
The pilin structural gene, fimA, encodes a 158-159 amino acid polypeptide with an approximate molecular weight of 17 kDa[18,19]. Immediately upstream of the fimA gene is a 314-bp invertible DNA element called fimS, which contains the promoter for fimA with 9 bp inverted repeats (IRs) flanking this segment of DNA (5′ TTGGGGCCA), labeled IRL and IRR (Figure 1)[20,21]. The fimA promoter sequence undergoes site-specific recombination, positioning the invertible element in either the Phase-ON (piliated phenotype) or Phase-OFF (nonpiliated phenotype) orientation. This switching phenomenon is known as phase variation. Two genes upstream of the fimS invertible element, fimB and fimE, encode proteins thought to be involved in positioning the fimS DNA and will be discussed further below.
The fimI gene was the last gene within the fim operon to be characterized[22]. FimI’s function is not known. Within the fim gene cluster, there are two additional genes involved in transport and assembly of type 1 pili: fimC and fimD. FimC is a periplasmic chaperone protein[23-25] that helps translocate the fimbrial proteins through the periplasm until the FimC-Fim protein complex reaches the FimD usher. FimD is an integral outer membrane protein that serves as an usher, allowing surface localization of the nascently forming type 1 pilus[26-28].
Although the FimA monomers comprise the bulk of the type 1 pilus structure, FimA does not mediate binding to the mannose containing receptor. An adhesin, encoded by the fimH gene, is responsible for this binding[29-33]. The two remaining genes in the fim operon are fimF and fimG. FimF and FimG are associated with FimH adhesin, forming a fibrillum structure that anchors the adhesin to the pilus shaft and controls the length of the type 1 pilus[29,30,34-37].
PHASE VARIATION’S ROLE IN TYPE 1 PILUS EXPRESSION
Phase variation is a reversible process, which, in the case of UPEC, leads to an oscillation between Phase-ON piliated cells and Phase-OFF nonpiliated cells. Using fimA-lacZ operon fusions, rates of 10−3 to 10−4/cell/generation were originally calculated for type 1 pilus expression[38,39]. Phase variation results in agar and, particularly, broth cultures of UPEC to comprise a mixture of piliated and nonpiliated cells.
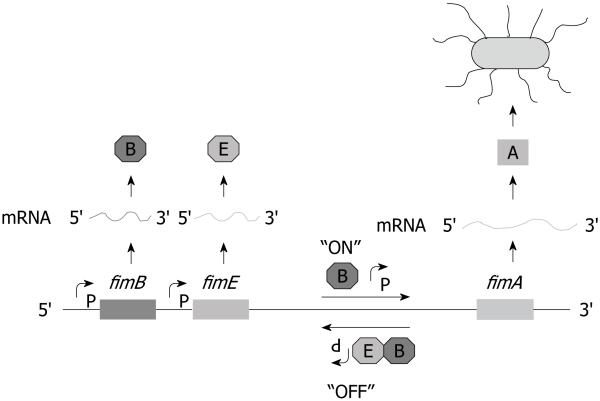
The site-specific recombination that allows phase variation to occur requires two trans-acting factors located proximally upstream of fimS, encoded by fimB and fimE[40]. Sequence analysis of fimB and fimE indicated that the predicted proteins were highly basic, a property of many DNA-binding proteins[41]. The predicted amino acid sequences show homology with the DNA binding domain of integrase[42] and contain a tetrad of conserved amino acids required for the recombinase activity[43-45]. Furthermore, FimB and FimE have 48% amino acid homology with each other[40]. Klemm[40] originally suggested that FimB and FimE might act independently to switch the fimS element unidirectionally, either Phase-ON to Phase-OFF or vice versa, via the two 9 bp invertible repeat elements, IRL and IRR. FimB can bind to the fimS element to either switch from Phase-ON to Phase-OFF or vice versa, with a slight bias towards the Phase-OFF over the Phase-ON orientation (Figure 2)[46-56]. By contrast, FimE binds to switch fimS from Phase-ON to Phase-OFF. In rare cases, FimE has been shown to initiate a Phase-OFF to Phase-ON switch[57] or when specific amino acid substitutions are made[45]. Orientation of the fimS element in the Phase-OFF position leads to the production of antisense transcripts from the fimA promoter[49,58].
Figure 2. A schematic showing how the FimB and FimE proteins orient the fimS element.
FimB-mediated recombination occurs at the rate of 10−3 to 10−4 per cell per generation that was originally described; however, FimE-mediated switching occurs more often at a frequency of 0.3 per cell per generation[52,59]. Base substitutions within fimS demonstrated that FimB and FimE used the same DNA cleavage and religation sites within IRL and IRR, allowing more DNA base variations for FimB than FimE[60]. When fimB and fimE were provided in trans on plasmids, they affected pilin expression, suggesting that the ratio of FimB and FimE is important.
The promoters for both fimB and fimE have been mapped[61-63]. For the fimB gene, the number of promoters varies between one and three. Promoters P1 and P2, which were mapped by Schwan et al[63] in two UPEC strains (Figure 1), were confirmed by another group[61]. A potential third fimB promoter was also identified by Schwan et al[63], approximately 650 bp upstream of the fimB P2 promoter, and around 840 bp upstream of the translational start site of fimB. This third fimB promoter has not been confirmed by other groups and could be an anomaly. It could also be a third fimB promoter connected to sialic acid regulation of fimB (see below). Certainly, strain differences could explain the different numbers of fimB promoters. Only one promoter has been identified for the fimE gene[62].
OTHER CO-FACTOR PROTEINS THAT AFFECT PHASE SWITCHING
Besides the fim gene cluster, other genes and their gene products contribute to the expression of type 1 pili. Early work mapped a gene, pilG, at 27 min on the E. coli chromosome that affected inversion of the fimS region[21]. A mutation of the pilG gene increased the inversion of the fimS region by up to 100-fold as measured with a fimA-lac fusion[21]. The pilG locus was shown to be allelic to bglY[64], drdX[65], and osmZ[66]. Later, it was determined that the pilG and osmZ genes were in fact alleles of the hns gene[66-68]. The hns gene encodes the H-NS global regulatory protein[69].
H-NS possibly controls the phase variation of the fimS region both directly and indirectly[61,62,70-74]. For a potential direct effect, H-NS binds to sequences adjacent to the fimS invertible element[72,75].
Indirectly, H-NS represses the transcription of both fimB and fimE[62,71,74]. H-NS binds, with a high degree of specificity, to both the P1 and P2 promoter sites for fimB[71,72]. The DNA-binding regulatory protein also binds to the fimE promoter[71]. Moreover, H-NS also represses lrp transcription[76], which would in turn affect the phase switching of the fimS element, as described below. Thus, transcriptional repression of the fimB and fimE site-specific recombinase genes would indirectly influence the position of the fimS element, which would indirectly affect phase variation.
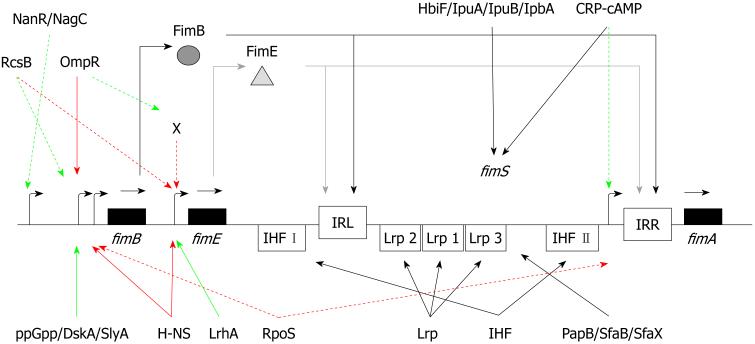
Besides H-NS, integration host factor (IHF) and leucine-responsive protein (Lrp) are additional co-factors that affect type 1 pilus phase variation. Both proteins cause sharp bends in the DNA structure, introducing hairpin loops that facilitate recombination events within UPEC. IHF is a two-component protein consisting of IHF encoded by ihfA[77] and IHF encoded by ihfB[78]. Both Eisenstein et al[42] and Dorman et al[43] showed that IHF plays a role in type 1 pilus switching. Mutations in either ihfA or ihfB locked the fimS region in either the Phase-OFF or Phase-ON orientation[79]. In both studies, an IHF binding site (IHF II) proximal to IRR was identified (Figure 3). In addition, an IHF binding site was also identified between IRL and the 3′ end of fimE (IHF I)[80]. A mutational analysis of this IHF I site demonstrated that FimB-mediated recombination was more adversely affected, suggesting a directional bias for FimB recombination[73,75,79,81,82].
Figure 3. Schematic model of the actions of 20 auxiliary proteins on the regulation of type 1 pili.
The inverted repeat left and right (IRL and IRR) are shown as open boxes. Binding sites for integration host factor (IHF I and II) and leucine-responsive protein (Lrp1, 2, and 3) are also represented as open boxes. Genes are displayed as black boxes and the promoters are shown as bent black arrows. The dark gray arrows correspond to FimB and the light gray arrows are for FimE. Black arrows signify an effect on the fimS element. Solid green arrows indicate confirmed binding associated with stimulatory effects, whereas dashed green arrows indicate presumed stimulatory effects. Solid red arrows indicate confirmed binding associated with repressing effects, whereas dashed red arrows indicate presumed repressing effects.
The leucine-responsive regulatory protein (Lrp) is another protein that has been shown to affect the fimS region. Lrp is a global regulator of genes involved in metabolic functions within E. coli, including pili synthesis[83]. Mutations of the lrp gene cause a lower frequency of recombination of the fimS element[80,84]. Lrp binds to three distinct sites within the fimS element that are closer to the IRL site. When the high affinity sites 1 and 2 are mutated, the recombination frequency declines[79,85]. Lrp binding to the low affinity site 3 inhibits recombination[86,87]. Lrp and IHF can bend the fimS DNA; therefore, they would allow the proper positioning of IRL and IRR that facilitates recombination[80,87]. The levels of specific amino acids will also affect Lrp binding to the fimS element and subsequently phase variation[86]. Lrp binding causes an orientational bias to the fimS element. When neither Lrp nor IHF are present at sufficient levels, H-NS will bind and maintain the Phase-OFF orientation[88]. Although Lrp binds to multiple sites within the fimS element, Lrp directly regulates neither fimB nor fimE.
Another protein that regulates type 1 pilus expression is the LysR-type regulator, LrhA[89]. LrhA was first identified to be associated with RpoS degradation[90]. Microarray analysis of mRNA populations from an lrhA mutant vs wild-type bacteria revealed increased expression of the fimAICDFGH operon. Purified LrhA protein bound to the promoter regions of both fimB and fimE; however, there was higher affinity for the fimE promoter. The use of fimB- or fimE-lacZ translational fusions indicated there was a greater effect with the fimE-lacZ fusion. Thus, LrhA appears to activate fimE, which would repress type 1 pilus expression.
Three other proteins have unexplained effects on type 1 pilus expression in E. coli: OmpX, IbeA, and IbeT. Inactivation of ompX, encoding an outer membrane protein OmpX, caused an increased production of FimA[91]. A disruption caused by the loss of OmpX would change the cell surface, which would affect cell-surface interactions. It is likely that OmpX acts indirectly to regulate type 1 pilus expression. A deletion of the ibeA gene caused diminished type 1 pilus expression, as well as lower transcription of fimB and fimE, whereas an ibeT mutant was shown to have the fimS element preferentially in the Phase-OFF orientation[92]. How each of these proteins works to regulate the fim genes has not been determined.
The regulatory alarmone, ppGpp, has been connected to the regulation of multiple genes in E. coli, including the fim operon. ppGpp-deficient strains exhibited diminished type 1 pili expression compared to the wild-type strain[93]. Furthermore, primer-extension analysis indicated that ppGpp activated the fimB P2 promoter. A follow through study demonstrated that DskA, a cofactor required for ppGpp-mediated positive regulation of several amino acid biosynthesis promoters[94], also activated transcription from the fimB P2 promoter[95].
Besides FimB and FimE, there are four other site-specific recombinases that could affect phase switching of the fimS element: HbiF, IpuA, IpuB, and IpbA. The HbiF-mediated inversion of the fimS element occurs primarily from Phase-OFF to Phase-ON[96]. Constitutive expression of HbiF locked the fimS DNA in the Phase-ON position. The three other site-specific recombinases (IpuA, IpuB, and IpbA) were discovered by sequence analysis of the UPEC strain CFT073 genome because of their high homology with the fimB and fimE genes[97]. Both IpuA and IbpA bind to the fimS element and mediate phase switching. IpuA functions like FimB, allowing a Phase-OFF to Phase-ON switch as well as Phase-ON to Phase-OFF switching, whereas IpbA can switch fimS from Phase-OFF to Phase-ON. It is not clear under what environmental growth conditions these alternative site-specific recombinases affect the fimS element positioning.
Also linked to the fimS genetic switch are Rho and LeuX. Transcriptional termination of fimE was determined to be Rho-dependent, based on the use of a rho mutant or by treatment with bicyclomycin, an antibiotic that interferes with Rho[98,99]. Thus, when the phase switch is in the Phase-OFF position, there is a Rho-dependent termination of the fimE sense transcript, leading to a truncated, unstable mRNA that is readily degraded. Less FimE site-specific recombinase would allow FimB to bind and switch the fimS element to the Phase-ON position. The minor leucyl tRNA, LeuX, affects the fimS element switching from Phase-OFF to Phase-ON[100,101]. Placing the leuX gene on a multicopy plasmid caused greater expression from the fimAICDFGH operon[102].
All of the studies examining fimB regulation described above have concentrated on the P1 and P2 promoter regions. However, several other studies have shown that the intergenic region between the yjhATS operon and the fimB gene also plays a role in genetic regulation of fimB[103-105]. Sialic acid and N-acetylglucosamine inhibit the FimB recombinase. Two proteins, NagC (a N-acetylglucosamine-6P-responsive protein) and NanR (a sialic acid-responsive protein), linked to sialic acid and N-acetylglucosamine catabolism[106,107], bind to two deoxyadenosine methylation sites within the intergenic region[103-105] that align with P3 fimB promoter described earlier[58]. In addition, NagC also binds to an operator site 212 bp closer to the fimB translational start site[105]. Both proteins are thought to act as antirepressors that allow fimB transcription to occur[103]. However, a urinary tract infection caused by type 1 piliated UPEC will elicit an inflammatory response[108], leading to increased levels of both sialic acid and N-acetylglucosamine that will, in turn, activate some cis-active regulatory protein that shuts off fimB transcription.
Regulatory proteins for other pilus systems can also regulate type 1 pilus expression through a cross-talk mechanism. PapB, which affects the phase variation of the pyelonephritis associated pilus (pap) operon[109,110], also regulates the orientation of the fimS element[111-113]. In contrast to FimB, PapB inhibits the Phase-OFF to Phase-ON switching. Two proteins associated with S pili, SfaB and SfaX, also have a negative effect on Phase-OFF to Phase-ON switching[111,114]. Thus, there appears to be an expression competition between the different pilus operons. These regulatory proteins that allow expression of other types of pili in other environments counter the need for type 1 pili under growth conditions where type 1 pili are not needed.
In stationary phase-grown E. coli cells, type 1 pilus expression is diminished compared to logarithmic grown cells. The alternative sigma factor, RpoS, which is activated during stationary phase, represses fimB transcription[115]. Another regulatory signal active in a logarithmic phase culture may be provided by glucose acting as a catabolite repressor by increasing internal cAMP concentrations, which allow for greater interactions with its receptor protein, CRP[116]. For type 1 pilus expression, the role of cAMP and glucose is opaque. Early studies indicated that cAMP affected pilus expression in some strains of E. coli[117] and in cya (adenyl cyclase) mutants of Salmonella enterica serovar Typhimurium[118]. However, in a later study, glucose had no effect on pilus expression, even when added with exogenous cAMP or when tested in adenlyate cyclase mutants[119]. Unfortunately, some of the early work was done with the CSH50 strain of E. coli, which has a fimE::IS1 mutation[52], so the role of catabolite repression remained unclear, until recently. Using a more defined system, Müller et al[120] have shown that CRP-cAMP directly represses the fimA promoter and indirectly affects phase variation by limiting the switch from Phase-OFF to Phase-ON in a logarithmic stage population.
Two other proteins that activate fimB transcription are RcsB and SlyA. RcsB is part of the RcsC/RcsB two-component phosphorelay regulatory system[121]. Using an rcsB mutant, it was shown that under neutral pH/low osmolality growth conditions, RcsB appears to activate fimB[122]. Growth in an acidic environment did not affect fimB expression in the rcsB strain compared to wild-type cells. Recently, the SlyA global regulator was implicated in fimB gene activation[123], but the growth conditions that would favor slyA expression were not determined.
The last accessory protein with relevance to fim gene regulation is OmpR. OmpR is part of the EnvZ/OmpR two-component regulatory system that regulates genes under an osmotic stress[124]. A study by Schwan et al[74] found that an ompR mutant strain had de-repressed transcription of fimB and fimE compared to wild-type cells. More recently, they found that unphosphorylated OmpR bound to the P2 promoter of fimB to repress fimB transcription[125] (Rentchler, Lovrich, and Schwan, manuscript submitted). However, through DNase I footprinting analysis, neither unphosphorylated nor phosphorylated OmpR bound directly to the fimE promoter, suggesting another regulatory element that is regulated by OmpR-P would directly affect fimE transcription.
Thus, in addition to FimB and FimE, approximately 20 different auxiliary proteins have a role to play in the regulation of one or more fim genes or positioning the fimS element. These 20 proteins are represented in a schematic model shown in Figure 3. Some of the proteins repress fim gene expression (e.g. H-NS, OmpR, RpoS), whereas others appear to activate fim gene expression (e.g. DskA, LrhA, NagC, NanR, RcsB, SlyA). How some of these proteins may affect UPEC type 1 pilus expression during the course of a human or murine urinary tract infection is described below.
ENVIRONMENTAL SIGNALS WITHIN THE URINARY TRACT AFFECTING UPEC TYPE 1 PILUS EXPRESSION
The human or murine urinary tract is a dynamic environment. In the lower urinary tract, there are ample mannose receptors for FimH-mediated attachment of type 1 piliated UPEC cells[126]. The temperature in the urinary tract is around 37°C. Although one group showed Phase-OFF to Phase-ON switching increased at lower temperatures, others have demonstrated that the fimA promoter element is biased in its switch from the Phase-ON to the Phase-OFF orientation in broth cultures grown at 20°C, but the switch favors FimB recombination at 37°C[59,71,127]. More recently, Kuwahara et al[128] demonstrated that FimB-mediated recombination could be linked to a controlled downregulation of the Phase-ON to Phase-OFF switching rate based on a temperature-dependent suppression of the interplay of the FimE recombinase.
When the UPEC cells move from the vaginal surface, which has only a slightly acidic pH/low osmolality environment, to the urethra or ascend to the bladder, there is a switch to a moderate acidic pH/moderate to high osmolality environment[129,130]. Under the slightly acidic pH/low salt growth conditions found on the vaginal surface, proteins such as SlyA or RcsB may activate fimB and prevent H-NS from binding, allowing type 1 pili to be created and presented on the surface of the UPEC cells for attachment. When the bacteria move from the exterior opening of the urinary tract and ascend the urethra to the bladder, an acidic pH/moderate osmolality environment is encountered in the bladder[129,130]. A preliminary study implied that an acid tolerance system-induced protein is involved in the regulation of several fim genes (Schwan WR, unpublished results), which may begin to turn off the fim operon. Furthermore, a change in the osmolality would activate the EnvZ/OmpR two-component regulatory system, allowing OmpR to repress fimB transcription[74,125].
UPEC infections are ascending infections[13,131]; therefore, the presence of flagella on the UPEC cells would allow the bacteria to ascend to the kidneys. Expression of the flagella may coordinately turn off expression of the type 1 pili[132,133]. As the bacteria ascend to the kidneys, the pH would drop further and the osmolality would increase. OmpR becomes phosphorylated and activates an unknown gene whose gene product in turn potentially shuts down not only fimB, but also fimE expression. Moreover, H-NS may bind and repress both fimB and fimE at this time. This would lock the fimS element in the Phase-OFF position, creating nonpiliated UPEC cells. Furthermore, as the young E. coli population matures and moves into stationary phase, they trigger transcriptional activation of the rpoS gene. The acidic/high osmolality environment would cause greater translation of the rpoS transcripts[134], leading to more RpoS protein for repression of fimB transcription.
CONCLUSION
Several strains of UPEC have been shown to become nonpiliated in the murine kidney over time[13,135]. There are very few mannose receptors in human or murine kidneys[136,137] and the innate immune system is more apt to target type 1 piliated bacteria[138]; therefore, the regulatory loss of type 1 pili on UPEC cells in the human kidney would be an evolutionary advantage for these bacteria. Thus, the ability to phase vary their type 1 pilus expression offers several advantages to the UPEC. On vaginal surfaces, the outer rim of the urinary tract, and within the urethra and bladder, type 1-piliated cells benefit the bacteria because there are ample mannose receptors. When the bacteria ascend into the kidneys, the growth environment may turn off expression of an unneeded external surface structure that may target the bacteria for elimination by the host’s innate defenses.
ACKNOWLEDGMENTS
I would like to thank the University of Wisconsin-La Crosse for grant support for my laboratory and also thank all of the undergraduate and graduate students whom I have mentored.
Supported by National Institutes of Health Grant, No. 1R15AI-065432-01A2
Footnotes
Author contributions: Schwan WR wrote the review and prepared the figures.
REFERENCES
- 1.Foxman B, Brown P. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect Dis Clin North Am. 2003;17:227–241. doi: 10.1016/s0891-5520(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 2.Litwin MS, Saigal CS, Yano EM, Avila C, Geschwind SA, Hanley JM, Joyce GF, Madison R, Pace J, Polich SM, Wang M. Urologic diseases in America Project: analytical methods and principal findings. J Urol. 2005;173:933–937. doi: 10.1097/01.ju.0000152365.43125.3b. [DOI] [PubMed] [Google Scholar]
- 3.Duguid JP, Gillies RR. Fimbriæ and adhesive properties in dysentery bacilli. J Pathol Bacteriol. 1957;74:397–411. [Google Scholar]
- 4.Salit IE, Gotschlich EC. Hemagglutination by purified type I Escherichia coli pili. J Exp Med. 1977;146:1169–1181. doi: 10.1084/jem.146.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clegg S, Gerlach GF. Enterobacterial fimbriae. J Bacteriol. 1987;169:934–938. doi: 10.1128/jb.169.3.934-938.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ofek I, Mosek A, Sharon N. Mannose-specific adherence of Escherichia coli freshly excreted in the urine of patients with urinary tract infections, and of isolates subcultured from the infected urine. Infect Immun. 1981;34:708–711. doi: 10.1128/iai.34.3.708-711.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pere A, Nowicki B, Saxén H, Siitonen A, Korhonen TK. Expression of P, type-1, and type-1C fimbriae of Escherichia coli in the urine of patients with acute urinary tract infection. J Infect Dis. 1987;156:567–574. doi: 10.1093/infdis/156.4.567. [DOI] [PubMed] [Google Scholar]
- 8.Mobley HL, Chippendale GR, Tenney JH, Hull RA, Warren JW. Expression of type 1 fimbriae may be required for persistence of Escherichia coli in the catheterized urinary tract. J Clin Microbiol. 1987;25:2253–2257. doi: 10.1128/jcm.25.12.2253-2257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith BR, Maurer L, Spears PA, Orndorff PE. Receptor-binding function of type 1 pili effects bladder colonization by a clinical isolate of Escherichia coli. Infect Immun. 1986;53:693–696. doi: 10.1128/iai.53.3.693-696.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kisielius PV, Schwan WR, Amundsen SK, Duncan JL, Schaeffer AJ. In vivo expression and variation of Escherichia coli type 1 and P pili in the urine of adults with acute urinary tract infections. Infect Immun. 1989;57:1656–1662. doi: 10.1128/iai.57.6.1656-1662.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim JK, Gunther NW, Zhao H, Johnson DE, Keay SK, Mobley HL. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect Immun. 1998;66:3303–3310. doi: 10.1128/iai.66.7.3303-3310.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HL. Role of phase variation of type 1 fimbriae in a uropathogenic Escherichia coli cystitis isolate during urinary tract infection. Infect Immun. 2006;74:1387–1393. doi: 10.1128/IAI.74.2.1387-1393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaeffer AJ, Schwan WR, Hultgren SJ, Duncan JL. Relationship of type 1 pilus expression in Escherichia coli to ascending urinary tract infections in mice. Infect Immun. 1987;55:373–380. doi: 10.1128/iai.55.2.373-380.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Struve C, Krogfelt KA. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology. 1999;145(Pt 10):2683–2690. doi: 10.1099/00221287-145-10-2683. [DOI] [PubMed] [Google Scholar]
- 15.Gunther NW, Lockatell V, Johnson DE, Mobley HL. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect Immun. 2001;69:2838–2846. doi: 10.1128/IAI.69.5.2838-2846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinton CC, Jr, Gemski P, Jr, Falkow S, Baron LS. Location of the piliation factor on the chromosome of Escherichia coli. Biochem Biophys Res Commun. 1961;5:293–298. [Google Scholar]
- 18.Orndorff PE, Falkow S. Nucleotide sequence of pilA, the gene encoding the structural component of type 1 pili in Escherichia coli. J Bacteriol. 1985;162:454–457. doi: 10.1128/jb.162.1.454-457.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemm P. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Nucleotide sequence and primary structure of the protein. Eur J Biochem. 1984;143:395–399. doi: 10.1111/j.1432-1033.1984.tb08386.x. [DOI] [PubMed] [Google Scholar]
- 20.Abraham JM, Freitag CS, Clements JR, Eisenstein BI. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spears PA, Schauer D, Orndorff PE. Metastable regulation of type 1 piliation in Escherichia coli and isolation and charac-terization of a phenotypically stable mutant. J Bacteriol. 1986;168:179–185. doi: 10.1128/jb.168.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valenski ML, Harris SL, Spears PA, Horton JR, Orndorff PE. The Product of the fimI gene is necessary for Escherichia coli type 1 pilus biosynthesis. J Bacteriol. 2003;185:5007–5011. doi: 10.1128/JB.185.16.5007-5011.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klemm P, Jørgensen BJ, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol Gen Genet. 1985;199:410–414. doi: 10.1007/BF00330751. [DOI] [PubMed] [Google Scholar]
- 24.Orndorff PE, Falkow S. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J Bacteriol. 1984;159:736–744. doi: 10.1128/jb.159.2.736-744.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CH, Pinkner JS, Nicholes AV, Slonim LN, Abraham SN, Hultgren SJ. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freitag CS, Eisenstein BI. Genetic mapping and transcriptional orientation of the fimD gene. J Bacteriol. 1983;156:1052–1058. doi: 10.1128/jb.156.3.1052-1058.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurer L, Orndorff PE. Identification and characterization of genes determining receptor binding and pilus length of Escherichia coli type 1 pili. J Bacteriol. 1987;169:640–645. doi: 10.1128/jb.169.2.640-645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klemm P, Christiansen G. The fimD gene required for cell surface localization of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1990;220:334–338. doi: 10.1007/BF00260505. [DOI] [PubMed] [Google Scholar]
- 29.Minion FC, Abraham SN, Beachey EH, Goguen JD. The genetic determinant of adhesive function in type 1 fimbriae of Escherichia coli is distinct from the gene encoding the fimbrial subunit. J Bacteriol. 1986;165:1033–1036. doi: 10.1128/jb.165.3.1033-1036.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham SN, Goguen JD, Sun D, Klemm P, Beachey EH. Identification of two ancillary subunits of Escherichia coli type 1 fimbriae by using antibodies against synthetic oligopeptides of fim gene products. J Bacteriol. 1987;169:5530–5536. doi: 10.1128/jb.169.12.5530-5536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham SN, Goguen JD, Beachey EH. Hyperadhesive mutant of type 1-fimbriated Escherichia coli associated with formation of FimH organelles (fimbriosomes) Infect Immun. 1988;56:1023–1029. doi: 10.1128/iai.56.5.1023-1029.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson MS, Brinton CC. Identification and characterization of E. coli type-1 pilus tip adhesion protein. Nature. 1988;332:265–268. doi: 10.1038/332265a0. [DOI] [PubMed] [Google Scholar]
- 33.Krogfelt KA, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 35.Krogfelt KA, Klemm P. Investigation of minor components of Escherichia coli type 1 fimbriae: protein chemical and immunological aspects. Microb Pathog. 1988;4:231–238. doi: 10.1016/0882-4010(88)90073-3. [DOI] [PubMed] [Google Scholar]
- 36.Jones CH, Pinkner JS, Roth R, Heuser J, Nicholes AV, Abraham SN, Hultgren SJ. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell PW, Orndorff PE. Lesions in two Escherichia coli type 1 pilus genes alter pilus number and length without affecting receptor binding. J Bacteriol. 1992;174:5923–5935. doi: 10.1128/jb.174.18.5923-5935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenstein BI. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science. 1981;214:337–339. doi: 10.1126/science.6116279. [DOI] [PubMed] [Google Scholar]
- 39.Orndorff PE, Spears PA, Schauer D, Falkow S. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J Bacteriol. 1985;164:321–330. doi: 10.1128/jb.164.1.321-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabo CO, Sauer RT. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 42.Eisenstein BI, Sweet DS, Vaughn V, Friedman DI. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorman CJ, Higgins CF. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J Bacteriol. 1987;169:3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith SG, Dorman CJ. Functional analysis of the FimE integrase of Escherichia coli K-12: isolation of mutant derivatives with altered DNA inversion preferences. Mol Microbiol. 1999;34:965–979. doi: 10.1046/j.1365-2958.1999.01657.x. [DOI] [PubMed] [Google Scholar]
- 45.Burns LS, Smith SG, Dorman CJ. Interaction of the FimB integrase with the fimS invertible DNA element in Escherichia coli in vivo and in vitro. J Bacteriol. 2000;182:2953–2959. doi: 10.1128/jb.182.10.2953-2959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulasekara HD, Blomfield IC. The molecular basis for the specificity of fimE in the phase variation of type 1 fimbriae of Escherichia coli K-12. Mol Microbiol. 1999;31:1171–1181. doi: 10.1046/j.1365-2958.1999.01257.x. [DOI] [PubMed] [Google Scholar]
- 47.McClain MS, Blomfield IC, Eisenstein BI. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holden N, Blomfield IC, Uhlin BE, Totsika M, Kulasekara DH, Gally DL. Comparative analysis of FimB and FimE recombinase activity. Microbiology. 2007;153:4138–4149. doi: 10.1099/mic.0.2007/010363-0. [DOI] [PubMed] [Google Scholar]
- 49.Pallesen L, Madsen O, Klemm P. Regulation of the phase switch controlling expression of type 1 fimbriae in Escherichia coli. Mol Microbiol. 1989;3:925–931. doi: 10.1111/j.1365-2958.1989.tb00242.x. [DOI] [PubMed] [Google Scholar]
- 50.Sohanpal BK, Kulasekara HD, Bonnen A, Blomfield IC. Orientational control of fimE expression in Escherichia coli. Mol Microbiol. 2001;42:483–494. doi: 10.1046/j.1365-2958.2001.02655.x. [DOI] [PubMed] [Google Scholar]
- 51.Freitag CS, Abraham JM, Clements JR, Eisenstein BI. Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J Bacteriol. 1985;162:668–675. doi: 10.1128/jb.162.2.668-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blomfield IC, McClain MS, Princ JA, Calie PJ, Eisenstein BI. Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J Bacteriol. 1991;173:5298–5307. doi: 10.1128/jb.173.17.5298-5307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orndorff PE, Falkow S. Identification and characterization of a gene product that regulates type 1 piliation in Escherichia coli. J Bacteriol. 1984;160:61–66. doi: 10.1128/jb.160.1.61-66.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClain MS, Blomfield IC, Eberhardt KJ, Eisenstein BI. Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1993;175:4335–4344. doi: 10.1128/jb.175.14.4335-4344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gunther NW, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HL. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect Immun. 2002;70:3344–3354. doi: 10.1128/IAI.70.7.3344-3354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gally DL, Leathart J, Blomfield IC. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 57.Stentebjerg-Olesen B, Chakraborty T, Klemm P. FimE-catalyzed off-to-on inversion of the type 1 fimbrial phase switch and insertion sequence recruitment in an Escherichia coli K-12 fimB strain. FEMS Microbiol Lett. 2000;182:319–325. doi: 10.1111/j.1574-6968.2000.tb08915.x. [DOI] [PubMed] [Google Scholar]
- 58.Schwan WR, Seifert HS, Duncan JL. Growth conditions mediate differential transcription of fim genes involved in phase variation of type 1 pili. J Bacteriol. 1992;174:2367–2375. doi: 10.1128/jb.174.7.2367-2375.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gally DL, Bogan JA, Eisenstein BI, Blomfield IC. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCusker MP, Turner EC, Dorman CJ. DNA sequence heterogeneity in Fim tyrosine-integrase recombinase-binding elements and functional motif asymmetries determine the directionality of the fim genetic switch in Escherichia coli K-12. Mol Microbiol. 2008;67:171–187. doi: 10.1111/j.1365-2958.2007.06037.x. [DOI] [PubMed] [Google Scholar]
- 61.Donato GM, Lelivelt MJ, Kawula TH. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol. 1997;179:6618–6625. doi: 10.1128/jb.179.21.6618-6625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen PB, Klemm P. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol Lett. 1994;116:95–100. doi: 10.1111/j.1574-6968.1994.tb06681.x. [DOI] [PubMed] [Google Scholar]
- 63.Schwan WR, Seifert HS, Duncan JL. Analysis of the fimB promoter region involved in type 1 pilus phase variation in Escherichia coli. Mol Gen Genet. 1994;242:623–630. doi: 10.1007/BF00285286. [DOI] [PubMed] [Google Scholar]
- 64.Lejeune P, Danchin A. Mutations in the bglY gene increase the frequency of spontaneous deletions in Escherichia coli K-12. Proc Natl Acad Sci USA. 1990;87:360–363. doi: 10.1073/pnas.87.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Göransson M, Sondén B, Nilsson P, Dagberg B, Forsman K, Emanuelsson K, Uhlin BE. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 66.Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 67.Hulton CS, Seirafi A, Hinton JC, Sidebotham JM, Waddell L, Pavitt GD, Owen-Hughes T, Spassky A, Buc H, Higgins CF. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 68.Kawula TH, Orndorff PE. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histonelike protein H-NS. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 70.Kawula TH, Lelivelt MJ. Mutations in a gene encoding a new Hsp70 suppress rapid DNA inversion and bgl activation, but not proU derepression, in hns-1 mutant Escherichia coli. J Bacteriol. 1994;176:610–619. doi: 10.1128/jb.176.3.610-619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olsen PB, Schembri MA, Gally DL, Klemm P. Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol Lett. 1998;162:17–23. doi: 10.1111/j.1574-6968.1998.tb12973.x. [DOI] [PubMed] [Google Scholar]
- 72.Donato GM, Kawula TH. Phenotypic analysis of random hns mutations differentiate DNA-binding activity from properties of fimA promoter inversion modulation and bacterial motility. J Bacteriol. 1999;181:941–948. doi: 10.1128/jb.181.3.941-948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Gara JP, Dorman CJ. Effects of local transcription and H-NS on inversion of the fim switch of Escherichia coli. Mol Microbiol. 2000;36:457–466. doi: 10.1046/j.1365-2958.2000.01864.x. [DOI] [PubMed] [Google Scholar]
- 74.Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT. Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect Immun. 2002;70:1391–1402. doi: 10.1128/IAI.70.3.1391-1402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schembri MA, Olsen PB, Klemm P. Orientation-dependent enhancement by H-NS of the activity of the type 1 fimbrial phase switch promoter in Escherichia coli. Mol Gen Genet. 1998;259:336–344. doi: 10.1007/s004380050820. [DOI] [PubMed] [Google Scholar]
- 76.Oshima T, Ito K, Kabayama H, Nakamura Y. Regulation of lrp gene expression by H-NS and Lrp proteins in Escherichia coli: dominant negative mutations in lrp. Mol Gen Genet. 1995;247:521–528. doi: 10.1007/BF00290342. [DOI] [PubMed] [Google Scholar]
- 77.Miller HI, Friedman DI. An E. coli gene product required for lambda site-specific recombination. Cell. 1980;20:711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- 78.Flamm EL, Weisberg RA. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985;183:117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- 79.Blomfield IC, Kulasekara DH, Eisenstein BI. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol Microbiol. 1997;23:705–717. doi: 10.1046/j.1365-2958.1997.2241615.x. [DOI] [PubMed] [Google Scholar]
- 80.Blomfield IC, Calie PJ, Eberhardt KJ, McClain MS, Eisenstein BI. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leathart JB, Gally DL. Regulation of type 1 fimbrial expression in uropathogenic Escherichia coli: heterogeneity of expression through sequence changes in the fim switch region. Mol Microbiol. 1998;28:371–381. doi: 10.1046/j.1365-2958.1998.00802.x. [DOI] [PubMed] [Google Scholar]
- 82.Dove SL, Dorman CJ. Multicopy fimB gene expression in Escherichia coli: binding to inverted repeats in vivo, effect on fimA gene transcription and DNA inversion. Mol Microbiol. 1996;21:1161–1173. doi: 10.1046/j.1365-2958.1996.681434.x. [DOI] [PubMed] [Google Scholar]
- 83.Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Mol Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 84.Kelly A, Conway C, O Cróinín T, Smith SG, Dorman CJ. DNA supercoiling and the Lrp protein determine the directionality of fim switch DNA inversion in Escherichia coli K-12. J Bacteriol. 2006;188:5356–5363. doi: 10.1128/JB.00344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gally DL, Rucker TJ, Blomfield IC. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J Bacteriol. 1994;176:5665–5672. doi: 10.1128/jb.176.18.5665-5672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roesch PL, Blomfield IC. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol Microbiol. 1998;27:751–761. doi: 10.1046/j.1365-2958.1998.00720.x. [DOI] [PubMed] [Google Scholar]
- 87.Corcoran CP, Dorman CJ. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol. 2009;74:1071–1082. doi: 10.1111/j.1365-2958.2009.06919.x. [DOI] [PubMed] [Google Scholar]
- 88.Lahooti M, Roesch PL, Blomfield IC. Modulation of the sensitivity of FimB recombination to branched-chain amino acids and alanine in Escherichia coli K-12. J Bacteriol. 2005;187:6273–6280. doi: 10.1128/JB.187.18.6273-6280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blumer C, Kleefeld A, Lehnen D, Heintz M, Dobrindt U, Nagy G, Michaelis K, Emödy L, Polen T, Rachel R, Wendisch VF, Unden G. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology. 2005;151:3287–3298. doi: 10.1099/mic.0.28098-0. [DOI] [PubMed] [Google Scholar]
- 90.Gibson KE, Silhavy TJ. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J Bacteriol. 1999;181:563–571. doi: 10.1128/jb.181.2.563-571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Otto K, Hermansson M. Inactivation of ompX causes increased interactions of type 1 fimbriated Escherichia coli with abiotic surfaces. J Bacteriol. 2004;186:226–234. doi: 10.1128/JB.186.1.226-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cortes MA, Gibon J, Chanteloup NK, Moulin-Schouleur M, Gilot P, Germon P. Inactivation of ibeA and ibeT results in decreased expression of type 1 fimbriae in extraintestinal pathogenic Escherichia coli strain BEN2908. Infect Immun. 2008;76:4129–4136. doi: 10.1128/IAI.00334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aberg A, Shingler V, Balsalobre C. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol Microbiol. 2006;60:1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- 94.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol Microbiol. 2008;67:1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- 96.Xie Y, Yao Y, Kolisnychenko V, Teng CH, Kim KS. HbiF regulates type 1 fimbriation independently of FimB and FimE. Infect Immun. 2006;74:4039–4047. doi: 10.1128/IAI.02058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bryan A, Roesch P, Davis L, Moritz R, Pellett S, Welch RA. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect Immun. 2006;74:1072–1083. doi: 10.1128/IAI.74.2.1072-1083.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hinde P, Deighan P, Dorman CJ. Characterization of the detachable Rho-dependent transcription terminator of the fimE gene in Escherichia coli K-12. J Bacteriol. 2005;187:8256–8266. doi: 10.1128/JB.187.24.8256-8266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Joyce SA, Dorman CJ. A Rho-dependent phase-variable transcription terminator controls expression of the FimE recombinase in Escherichia coli. Mol Microbiol. 2002;45:1107–1117. doi: 10.1046/j.1365-2958.2002.03081.x. [DOI] [PubMed] [Google Scholar]
- 100.Ritter A, Blum G, Emödy L, Kerenyi M, Böck A, Neuhierl B, Rabsch W, Scheutz F, Hacker J. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 101.Ritter A, Gally DL, Olsen PB, Dobrindt U, Friedrich A, Klemm P, Hacker J. The Pai-associated leuX specific tRNA5(Leu) affects type 1 fimbriation in pathogenic Escherichia coli by control of FimB recombinase expression. Mol Microbiol. 1997;25:871–882. doi: 10.1111/j.1365-2958.1997.mmi517.x. [DOI] [PubMed] [Google Scholar]
- 102.Newman JV, Burghoff RL, Pallesen L, Krogfelt KA, Kristensen CS, Laux DC, Cohen PS. Stimulation of Escherichia coli F-18Col-type-1 fimbriae synthesis by leuX. FEMS Microbiol Lett. 1994;122:281–287. doi: 10.1016/0378-1097(94)00337-8. [DOI] [PubMed] [Google Scholar]
- 103.El-Labany S, Sohanpal BK, Lahooti M, Akerman R, Blomfield IC. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol Microbiol. 2003;49:1109–1118. doi: 10.1046/j.1365-2958.2003.03624.x. [DOI] [PubMed] [Google Scholar]
- 104.Sohanpal BK, El-Labany S, Lahooti M, Plumbridge JA, Blomfield IC. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc Natl Acad Sci USA. 2004;101:16322–16327. doi: 10.1073/pnas.0405821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sohanpal BK, Friar S, Roobol J, Plumbridge JA, Blomfield IC. Multiple co-regulatory elements and IHF are necessary for the control of fimB expression in response to sialic acid and N-acetylglucosamine in Escherichia coli K-12. Mol Microbiol. 2007;63:1223–1236. doi: 10.1111/j.1365-2958.2006.05583.x. [DOI] [PubMed] [Google Scholar]
- 106.Plumbridge J, Kolb A. CAP and Nag repressor binding to the regulatory regions of the nagE-B and manX genes of Escherichia coli. J Mol Biol. 1991;217:661–679. doi: 10.1016/0022-2836(91)90524-a. [DOI] [PubMed] [Google Scholar]
- 107.Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylman-nosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381:77–80. doi: 10.1038/381077a0. [DOI] [PubMed] [Google Scholar]
- 109.Göransson M, Uhlin BE. Environmental temperature regulates transcription of a virulence pili operon in E. coli. EMBO J. 1984;3:2885–2888. doi: 10.1002/j.1460-2075.1984.tb02225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Båga M, Göransson M, Normark S, Uhlin BE. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J. 1985;4:3887–3893. doi: 10.1002/j.1460-2075.1985.tb04162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holden NJ, Uhlin BE, Gally DL. PapB paralogues and their effect on the phase variation of type 1 fimbriae in Escherichia coli. Mol Microbiol. 2001;42:319–330. doi: 10.1046/j.1365-2958.2001.02656.x. [DOI] [PubMed] [Google Scholar]
- 112.Holden NJ, Totsika M, Mahler E, Roe AJ, Catherwood K, Lindner K, Dobrindt U, Gally DL. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology. 2006;152:1143–1153. doi: 10.1099/mic.0.28677-0. [DOI] [PubMed] [Google Scholar]
- 113.Xia Y, Gally D, Forsman-Semb K, Uhlin BE. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 2000;19:1450–1457. doi: 10.1093/emboj/19.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sjöström AE, Balsalobre C, Emödy L, Westerlund-Wikström B, Hacker J, Uhlin BE. The SfaXII protein from newborn meningitis E. coli is involved in regulation of motility and type 1 fimbriae expression. Microb Pathog. 2009;46:243–252. doi: 10.1016/j.micpath.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 115.Dove SL, Smith SG, Dorman CJ. Control of Escherichia coli type 1 fimbrial gene expression in stationary phase: a negative role for RpoS. Mol Gen Genet. 1997;254:13–20. doi: 10.1007/s004380050385. [DOI] [PubMed] [Google Scholar]
- 116.Göransson M, Forsman K, Uhlin BE. Regulatory genes in the thermoregulation of Escherichia coli pili gene transcription. Genes Dev. 1989;3:123–130. doi: 10.1101/gad.3.1.123. [DOI] [PubMed] [Google Scholar]
- 117.Eisenstein BI, Beachey EH, Solomon SS. Divergent effects of cyclic adenosine 3′,5′-monophosphate on formation of type 1 fimbriae in different K-12 strains of Escherichia coli. J Bacteriol. 1981;145:620–623. doi: 10.1128/jb.145.1.620-623.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saier MH, Schmidt MR, Leibowitz M. Cyclic AMP-dependent synthesis of fimbriae in Salmonella typhimurium: effects of cya and pts mutations. J Bacteriol. 1978;134:356–358. doi: 10.1128/jb.134.1.356-358.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eisenstein BI, Dodd DC. Pseudocatabolite repression of type 1 fimbriae of Escherichia coli. J Bacteriol. 1982;151:1560–1567. doi: 10.1128/jb.151.3.1560-1567.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Müller CM, Aberg A, Straseviçiene J, Emody L, Uhlin BE, Balsalobre C. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 2009;5:e1000303. doi: 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 122.Schwan WR, Shibata S, Aizawa S, Wolfe AJ. The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J Bacteriol. 2007;189:7159–7163. doi: 10.1128/JB.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McVicker G, Sun L, Sohanpal BK, Gashi K, Williamson RA, Plumbridge J, Blomfield IC. SlyA protein activates fimB gene expression and type 1 fimbriation in Escherichia coli K-12. J Biol Chem. 2011;286:32026–32035. doi: 10.1074/jbc.M111.266619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 125.Rentschler AE. In vitro analysis of OmpR regulation of the fimB and fimE genes of uropathogenic Escherichia coli. University of Wisconsin-La Crosse; La Crosse, WI: 2010. [Google Scholar]
- 126.Virkola R, Westerlund B, Holthöfer H, Parkkinen J, Kekomäki M, Korhonen TK. Binding characteristics of Escherichia coli adhesins in human urinary bladder. Infect Immun. 1988;56:2615–2622. doi: 10.1128/iai.56.10.2615-2622.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dorman CJ, Ní Bhriain N. Thermal regulation of fimA, the Escherichia coli gene coding for the type 1 fimbrial subunit protein. FEMS Microbiol Lett. 1992;78:125–130. doi: 10.1016/0378-1097(92)90013-e. [DOI] [PubMed] [Google Scholar]
- 128.Kuwahara H, Myers CJ, Samoilov MS. Temperature control of fimbriation circuit switch in uropathogenic Escherichia coli: quantitative analysis via automated model abstraction. PLoS Comput Biol. 2010;6:e1000723. doi: 10.1371/journal.pcbi.1000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ross DL, Neely AE. Textbook of Urinalysis and Body Fluids. Appleton Century Crofts; Norwalk: 1983. [Google Scholar]
- 130.Loeb WF, Quimby FW. The clinical chemistry of laboratory animals. Pergamon Press; New York: 1989. [Google Scholar]
- 131.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Edén C. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun. 1983;40:273–283. doi: 10.1128/iai.40.1.273-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lane MC, Simms AN, Mobley HL. complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J Bacteriol. 2007;189:5523–5533. doi: 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Snyder JA, Haugen BJ, Lockatell CV, Maroncle N, Hagan EC, Johnson DE, Welch RA, Mobley HL. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect Immun. 2005;73:7588–7596. doi: 10.1128/IAI.73.11.7588-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hultgren SJ, Porter TN, Schaeffer AJ, Duncan JL. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect Immun. 1985;50:370–377. doi: 10.1128/iai.50.2.370-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Väisänen-Rhen V, Rhen M, Linder E, Korhonen TK. Adhesion of Escherichia coli to human kidney cryostat sections. FEMS Microbiol Lett. 1985;27:179–182. [Google Scholar]
- 137.Virkola R. Binding characteristics of Escherichia coli type 1 fimbriae in the human kidney. FEMS Microbiol Lett. 1987;40:257–262. [Google Scholar]
- 138.Silverblatt FJ, Dreyer JS, Schauer S. Effect of pili on susceptibility of Escherichia coli to phagocytosis. Infect Immun. 1979;24:218–223. doi: 10.1128/iai.24.1.218-223.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]