Abstract
Establishment of primary and immortalized cultures of many cell types has facilitated efforts to understand the signals involved in proliferation and differentiation and yielded tools to rapidly assay new molecules targeting specific receptor pathways. Taste cells are specialized sensory epithelial cells which reside within taste buds on the lingual epithelium. Only recently have successful culturing protocols been developed which maintain essential molecular and functional characteristics. These protocols provide a tractable tool to examine the molecular, regenerative, and functional properties of these unique sensory cells within a controlled environment. The method involves an enzymatic isolation procedure and standardized culture conditions, and may be applied to either dissected rodent tissue or human fungiform papillae obtained by biopsy. Human fungiform cells can be maintained in culture for more than seven passages, without loss of viability and with retention of the molecular and biochemical properties of acutely isolated taste cells. Cultured primary human fungiform papillae cells also exhibit functional responses to taste stimuli indicating the presence of taste receptors and at least some relevant signaling pathways. While the loss of the three-dimensional structure of the intact taste bud must be taken into consideration in interpreting results obtained with these cells, this culture protocol provides a useful model for molecular studies of the proliferation, differentiation, and physiological function of mammalian taste receptor cells.
Keywords: Fungiform, Culture, Taste, Gustducin, Taste receptor, Regeneration, Sweet
1. Introduction
Taste receptor cells are specialized epithelial cells with unique histological, molecular, and physiological characteristics that permit detection of a wide range of both simple and structurally complex molecules. In mammals, taste buds are associated with fungiform papillae on the anterior two-thirds of the tongue, and circumvallate and foliate papillae are on the posterior third of the tongue. Taste buds are also present in the epithelium of the soft palate and pharynx (1). Taste receptor molecules and their downstream signaling components have also been found in enteroendocrine cells throughout the lining of the gastrointestinal tract (2). Human fungiform papillae contain from zero to over 25 taste buds, with over half having no taste buds and the rest having an average of three or four buds (3). Each taste bud contains 50–100 cells of four morphologically and functionally distinct types, which exhibit properties of both neuronal and epithelial cells (4). About half of the cells in the taste bud are spindle-shaped type I (dark) cells, which appear to have a glial like function because they surround other cell types and express molecules involved in neu-rotransmitter inactivation (5). An additional 25% of the taste bud cells are type II (light) cells, which express several proteins including the G-protein alpha-gustducin, phospholipase C-β2 (PLC-β2), inositol 1,4,5-trisphosphate receptor type 3 (IP3R3), and transient receptor potential channel M5 (TRPM5), which have been implicated in transduction of sweet, umami, and bitter taste responses (6, 7). Current evidence indicates that G-protein-coupled receptors (GPCRs) implicated in sweet and umami taste (T1Rs) and bitter taste (T2Rs) are expressed in nonoverlapping subsets of type II cells (8, 9). Cells mediating sour (acid) taste are likely to be a subset of type III cells (10), which comprise an additional 15% of the taste cells. The cells mediating salty taste have not yet been identified. Importantly, taste buds are one of the four very few truly regenerative organs in the human body, with taste cells having an average life span of about 12 days (11). A small number of type IV or basal cells in each bud are generally considered to be the stem cells giving rise to the other cell types (12, 13). Properties of taste cells often vary among species, and it is presently not known how maturational stage or lineage relates to the stimulus–response properties of these cells (12).
To date, no immortalized taste cell lines or long-term (i.e., months) taste culture methods have been published. Because of the unavailability of a human taste cell culture model, investigators are now dependent on freshly isolated primary cells, explant cultures, or nonhuman species to study taste cell development and physiological properties and require the use of a large number of experimental animals (14, 15). Importantly, these methods are not conducive to screening large number of chemicals for taste activity. Although heterologous systems expressing known specific taste receptors are used routinely to screen the activity of putative taste compounds, results may fail to reflect the actual more complicated taste detection processes. Functional studies of taste cells have been done using freshly isolated cells in primary culture, explant cultures from rodents, or semi-intact taste buds in tissue slices (14, 15). While each of these preparations has advantages, the development of long-term cultures would have provided significant benefits, particularly for studies of taste cell proliferation and differentiation. Most attempts to culture taste cells have reported limited viability, with cells typically not lasting beyond 3–5 days (16–19). We recently reported on a method for the extended culture of rodent taste cells (20). Here, we describe the establishment of long-term primary cultures of cells from human fungiform taste papillae that have molecular and physiological properties consistent with both developing and mature taste cells. In addition, these cultures were amenable to the use of moderate throughput screening (MTS) to examine responses to individual or combinations of taste stimuli. The establishment of this human fungiform cell culture protocol provides an important in vitro model to study intracellular signaling mechanisms, the impact of trophic or toxic agents on taste cell growth and function, and the assessment of potential taste stimuli and taste modifiers.
2. Materials
2.1. Human Fungiform Taste Papillae Biopsy
Biopsy instruments: Small fine-tip forceps and extra fine spring scissors (Fine Science Tools), surgical razor.
Taste cell isolation solution: 26 mM NaHCO3, 2.5 mM NaH2PO4, 20 mM glucose, 65 mM NaCl, 20 mM KCl, and 1 mM EDTA dissolved in nuclease-free water and filter sterilize.
Enzyme mixture: Mix collagenase type 1 (550 U/mL, Worthington), elastase (10 U/mL, Worthington), and soy bean trypsin inhibitor (0.9 mg/mL, Worthington) in calcium-free Ringer solution just before use.
Enzyme mixture (for alternative route): Mix pronase E (1.5 mg/mL, Sigma-Aldrich®), elastase (1 mg/mL, Sigma-Aldrich®) in calcium-free Ringer solution just before use.
2.2. Cell Culture
Tissue culture plastics: T25 (25 cm2), T75 (75 cm2) tissue culture flasks and other tissue culture plastics (i.e., conical tubes, 96-well plates).
Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco®/BRL) supplemented with 10% fetal bovine serum (FBS, HyClone).
MCDB 153 medium (Sigma-Aldrich®): Dissolve a package of MCDB 153 (17.6 g) completely in 1 L of tissue culture grade water, supplemented with 1.18 g/L sodium bicarbonate according to the manufacturer’s instructions. pH 7.0–7.1 (see Note 1).
Taste cell culture medium: IMDM containing 10% FBS, 1:5 ratio of MCDB 153, and a triple cocktail of antibiotics (100 U/mL/100 μg/mL, penicillin/streptomycin, 2.5 μg/mL gentamicin, and 0.5 μg/mL fungizone) (see Note 1).
Preparation of coverslips: Prior to use, treat coverslips with 2 M NaOH for 1 h and leave overnight in 70% nitric acid (HNO3). Wash coverslips with 9 M HCl acid for 1 h, autoclave in water, rinse with 70% ethanol and 100% ethanol, and then air-dry.
Coating coverslips with rat tail collagen type-1: Dilute rat tail collagen type-1 (3.96 mg/mL, BD Sciences) with sterile nuclease-free water at 1:4 ratio. Add 0.5–1 mL of rat tail collagen type-1 onto coverslips into a 12-well tissue culture plate for 15 min at room temperature. Remove rat tail collagen type-1 and let coverslip air-dry for 10–20 min.
Cloning cylinder 10–12 mm in diameter, sterilized sealed onto coverslip by using autoclaved grease coated on glass piece.
0.25% (w/v) trypsin/EDTA (Life Technologies™).
PBS with calcium and magnesium.
Cloning cylinder, 10–12 mm in diameter.
2.3. Cryopreservation of Cultured Cells
Freezing medium: Add DMSO into FBS to make final volume 5%.
Sterile cryovials, 2 mL.
Freezing container (e.g., Mr. Frosty, NUNC).
Isopropanol.
15 mL conical tubes.
2.4. Immunocyto-chemistry
4% Paraformaldehyde in PBS.
Deperoxidase solution: 4 mL PBS, 0.5 mL 100% methanol (4°C), 0.5 mL 30% H2O2 (4°C).
Blocking and antibody dilution solution: 3% Normal goat serum, 3% bovine serum albumin, and 0.3% Triton X-100 in PBS.
Primary antibodies (see Table 1).
Secondary antibodies (see Table 1).
Mounting medium and nuclear stain: Vecta® Shield with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories).
Leica TCS SP2 Spectral Confocal Microscope (Leica Microsystems Inc.).
Table 1.
Antibodies used for detecting expression of specific molecules
| Primary antibody | Source | Host | Dilution | Secondary antibody | Source | Host | Dilution |
|---|---|---|---|---|---|---|---|
| Gustducin | SantaCruz | Rabbit | 1:500 | Anti-rabbit IgG Alexa 633 | Molecular Probes | Goat | 1:500 |
| PLC-β2 | SantaCruz | Rabbit | 1:500 | Anti-rabbit IgG Alexa 633 | Molecular Probes | Goat | 1:500 |
2.5. Reverse Transcription-Polymerase Chain Reaction
Total RNA from cultured human fungiform cells.
#x02022; 2. Superscript First Strand Synthesis System for RT-PCR® (Life Technologies™).
1× AmpliTaq® Gold PCR buffer (Applied Biosystems™).
2.5 mM MgCl2: Add 0.0050 g MgCl2·6H2O into 10 mL ddH2O. Autoclave solution.
1 mM deoxynucleoside triphosphates: From 100 mM stock of dTTP, dATP, dGTP, dCTP, add 10 μL of each to 60 μL of ddH2O to make a 100 μL mix of dNTPs.
0.4 μM of each primer: For an amount of 38.5 nM of lyophilized primer provided by the primer supplier, 385 μL of PCR grade sterile water were added to get 100 μM/L stock solution. From this stock solution, to prepare 10 μM/L (or 10 picom/μL) as a PCR working solution, dilute stock solution 1:100 to give a concentration of 10 μM.
AmpliTaq® Gold polymerases (Applied Biosystems™).
1× TBE buffer: Dissolve 108 g Tris base, 55 g boric acid, 9.3 g EDTA in 800 mL H2O and adjust volume to 1 L with additional distilled dH2O.
2% Agarose gels in 1× TBE buffer.
0.2 μg/mL of ethidium bromide (10 mg/mL): Dissolve 0.2 g ethidium bromide to 20 mL water. Mix well and store at 4°C in the dark.
2.6. Moderate Throughput Screening
Modified Ringer solution (pH 7.1–7.2, 300–310 mOsmol): Dissolve 4.67 g NaCl, 0.373 g KCl, 0.203 g MgCl2, 0.147 g CaCl2, 0.110 g Na-pyruvate, and 4.76 g HEPES-Na in 1 L water and adjust pH to 7.1–7.2 and osmolarity to 300–310 mOsm/L by 5 M NaCl. Filter sterilize (0.2 μm).
Cell loading solution: 1 mM Fura-2 AM (Molecular Probes Inc.) in 10 mg/mL Pluronic F127 (Molecular Probes Inc.) in modified Ringer solution.
Tastant dissolved in modified Ringer solution: 1 mM denatonium; 0.446 g of denatonium benzoate dissolved in 50 mL of modified Ringer solution. 1 mM sucralose; 0.019 g of sucralose dissolved in 50 mL of modified Ringer solution. 250 ppm AceK; 25 mg of AceK was dissolved in 50 mL modified Ringer solution. 3 mM mono potassium glutamate (MPG); 0.025 g of MPG dissolved in 50 mL of modified Ringer solution. All solutions are filter sterilized.
3. Methods
3.1. Isolation, Culture, and Maintenance of Human Fungiform Cells
Remove 4–8 human fungiform taste papillae from the dorsal surface of the anterior portion of the tongue using curved spring microscissors.
Place immediately taste papillae into the taste isolation solution (see Subheading 2.1, step 2).
Digest fungiform papillae with collagenase, elastase, and soy bean trypsin inhibitor (see Subheading 2.1, step 3) under O2 bubbling in water bath with circulation for 30 min (see Note 2).
For alternative digestion protocol, please refer to Note 2.
Remove taste isolation solution and add 1 mL of taste cell culture medium (see Subheading 2.1, step 4).
Transfer digested fungiform papillae into glass dish.
Dissect fungiform papillae with surgical razor. Dissect gently to dissociate tissue pieces.
Add 250 μL of dissected papillae into cloning cylinder onto rat tail collagen type-1-coated coverslip (see Note 3).
Add 1 mL of taste cell culture medium into each well.
Incubate plate at 36°C (see Note 4) in a humidified incubator containing 5% CO2.
Place in an incubator undisturbed for 2 days prior to the first change of complete medium. Remove cloning cylinder from plate and aspirate medium completely. Add 1 mL of taste cell medium into each well (see Fig. 1a).
Replace 1/3 of medium every 6–7 days.
Fig. 1.
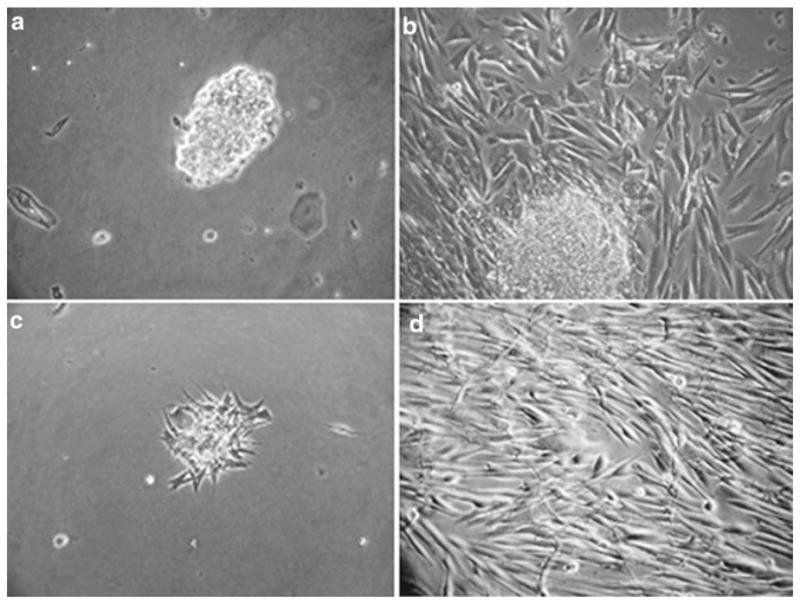
Attachment and morphology of cultured human fungiform taste cells. (a) Primary cell cultures grown on collagen type-1-coated plates were imaged after 2 days. (b) Cells from human fungiform papillae grew for up to 2–4 weeks under attached cell clusters. (c and d) Represent day 2 and 4 weeks after harvesting, respectively. During this period we did not observe growth of cells with the appearance of non-taste epithelial cells.
3.2. Passage of Cultured Human Fungiform Cells
Once 40–50% of the site of cloning cylinder is covered in expanding taste cells (see Fig. 1b), trypsinize cells using 0.25% (w/v) trypsin/EDTA for 2–3 min at 36°C.
Remove cells from well(s) and add into a 15 mL conical tube. Add 3 volumes of taste cell culture medium followed by centrifugation at 1,500 × g for 5 min at room temperature.
Remove supernatant and resuspend cells with 1 mL of taste cell medium.
3.3. Propagation of Human Fungiform Taste Cells
Transfer cells into T25 flask and add 4 mL of taste cell medium. Maintain cells at 36°C (see Note 5) in a humidified incubator containing 5% CO2 (see Fig. 1c).
Replace 1/3 of medium every 6–7 days until cultured taste cells have reached 100% confluence (see Fig. 1d).
To passage taste cells, wash cells once with sterile PBS and then trypsinize cells using 0.25% (w/v) trypsin/EDTA for 2–3 min at 36°C.
After centrifugation as described above, resuspend in complete taste cell medium and transfer cells to fresh T-75 flasks (passage 1).
Replace 1/3 of medium every 6–7 days.
Repeat steps 3 and 4 when cells have reached close to 100% confluence.
Cells are now ready to be processed for immunohistochemis-try, RNA isolation, calcium imaging, or other applications.
Split cells at a highest 1:4 dilution in a T75 flask for maintaining adequate growth of the cells over time. We then choose whether to proceed with freezing numerous vials of passage-1 cells for archival purposes.
3.4. Freezing and Thawing Cultured Human Fungiform Cells
To freeze stocks of primary taste cells, after trypsinization (see Subheading 3.2, step 1), add complete taste cell medium and transfer cells to sterile 15 mL conical centrifuge tubes. Centrifuge at 1,000 × g for 5 min at room temperature.
Carefully discard the supernatant and gently resuspend cells with appropriate volume of freezing medium (see Subheading 2.3, step 1).
Transfer cells to labeled, sterile cryovials, cap tightly, and place in a freezing container containing isopropanol (see Subheading 2.3). Place into a −80°C freezer for at least 1 day prior to transferring indefinitely to liquid nitrogen.
To thaw a cryovial, transfer a vial of cells directly to 37°C water bath and swirl vial to thaw as quickly as possible.
Spray vial with ethanol. Transfer cells into a sterile T25 or T75 cell culture flask having taste cell culture medium.
Continue to culture cells according to Subheadings 3.1–3.6 (see Note 5).
3.5. Confocal Immunofluorescence for Taste Cell Markers
Human fungiform cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature.
After washing coverslips in PBS, the cells were treated with 3% deperoxidase solution (see Subheading 2.4, step 2) to remove endogenous peroxidase activity.
Cells were blocked with blocking solution (see Subheading 2.4, step 3) for 30–60 min and then incubated with primary antibodies (see Table 1) overnight at 4°C.
After washing coverslips with PBS, cells were then incubated with secondary antibodies (see Table 1) diluted in blocking buffer for 30 h at room temperature.
After washing in PBS (3 × 15 min) and water (3 × 20 min), cov-erslips were mounted with Vectashield® mounting solution with DAPI (see Note 6).
Fluorescent images were captured with a Leica TCS SP2 Spectral Confocal Microscope (see Fig. 2).
Fig. 2.
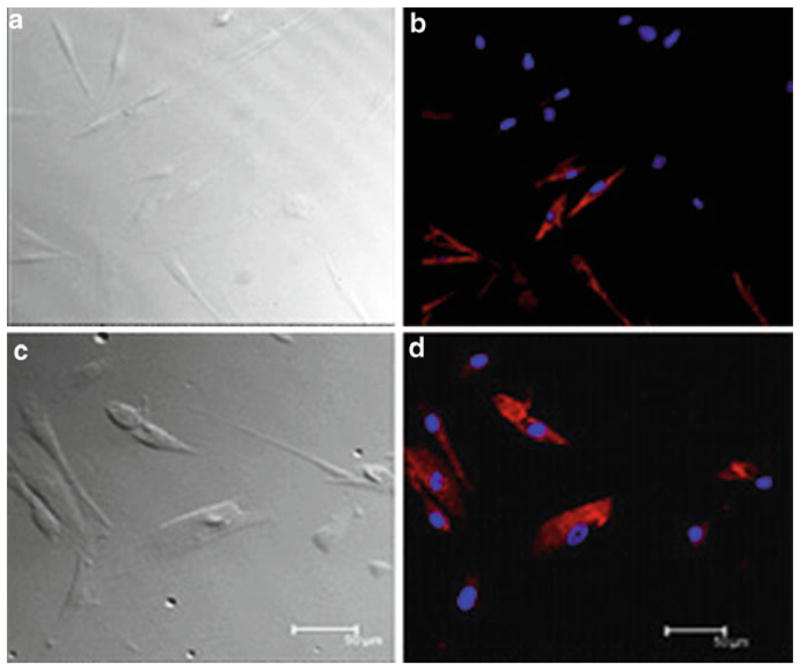
Immunostaining of cultured human fungiform papillae cells showed the presence of taste cell-speci fic biomarkers. Images were acquired with a Leica TCS-SP2 confocal laser scanning microscope. (a and c) Transmission images of corresponding fields are shown on the left. (b) Immunoreactivity was observed for gustducin in about 60% of cultured cells. (d) Immunoreactivity for PLC-β2 was also observed in about 25% of cultured cells (h). Nuclei of cells were stained as blue with DAPI. For controls, immunostaining with antibody-specific immunoglobulin demonstrated the absence of nonspecific immunoreactivity (data not shown). Scale bars = 50 μm.
3.6. Reverse Transcription-Polymerase Chain Reaction for Determining Taste Cell Markers
Total RNA (0.5 μg) was reverse transcribed for 50 min at 42°C using the Superscript® First Strand Synthesis System for RT-PCR (see Note 7).
PCR amplification of cDNA for each RT reaction was performed in a final volume of 25 μL containing 1 μL of cDNA, 1× AmpliTaq Gold PCR buffer, 2.5 mM MgCl2, 1 mM deoxy-nucleoside triphosphates, 0.4 μM of each primer (see Table 2), and 0.25 U/μL of AmpliTaq Gold polymerases.
PCR products were separated on 2% agarose gels and stained with 0.2 μg/mL of ethidium bromide (see Subheading 2.5, step 10) to verify their expected size (see Fig. 3).
Table 2.
Primers and conditions used for detecting taste-specific molecules
| Gene | Sequence | PCR | condition | Expected size (bp) | Reference |
|---|---|---|---|---|---|
| β-Actin | GGACTTCGAGCAAGAGATGG | 95 | 7 min | 234 | NM_001101.3 |
| AGCACTGTGTTGGCGTACAG | 94 | 45 s | |||
| 53 | 45 s | ||||
| 72 | 45 s | ||||
| 72 | 7 min | ||||
|
| |||||
| Gustducin | TCTGGGTATGTGCCAAATGA | 95 | 7 min | 386 | NM_001102386 |
| GGCCCAGTGTATTCTGGAAA | 94 | 45 s | |||
| 53 | 45 s | ||||
| 72 | 45 s | ||||
| 72 | 7 min | ||||
|
| |||||
| PLC-β2 | GTCACCTGAAGGCATGGTCT | 95 | 3 min | 333 | NM_004573 |
| TTAAAGGCGCTTTCTGCAAT | 94 | 30 s | |||
| 53 | 30 s | ||||
| 72 | 60 s | ||||
| 72 | 7 min | ||||
|
| |||||
| T2R5 | TATGGTTTGCCACCTTCCTC | 95 | 7 min | 394 | NM_01898 |
| AAGGACTTCAGCGCAGTGAT | 94 | 45 s | |||
| 53 | 45 s | ||||
| 72 | 45 s | ||||
| 72 | 7 min | ||||
|
| |||||
| T1R3 | CTTTTGTGGCCAGGATGAGT | 95 | 4 min | 345 | NM_152228 |
| TGCAGGAAGAGTGTGCTCAG | 94 | 45 s | |||
| 56 | 45 s | ||||
| 72 | 50 s | ||||
| 72 | 10 min | ||||
|
| |||||
| TRPM5 | TGGTAGAGCGCATGATGAAG | 95 | 10 min | 301 | NM_001101 |
| ACCAACAGGAAGGTGACCAG | 94 | 20 s | |||
| 63 | 30 s | ||||
| 72 | 45 s | ||||
| 72 | 7 min | ||||
Fig. 3.
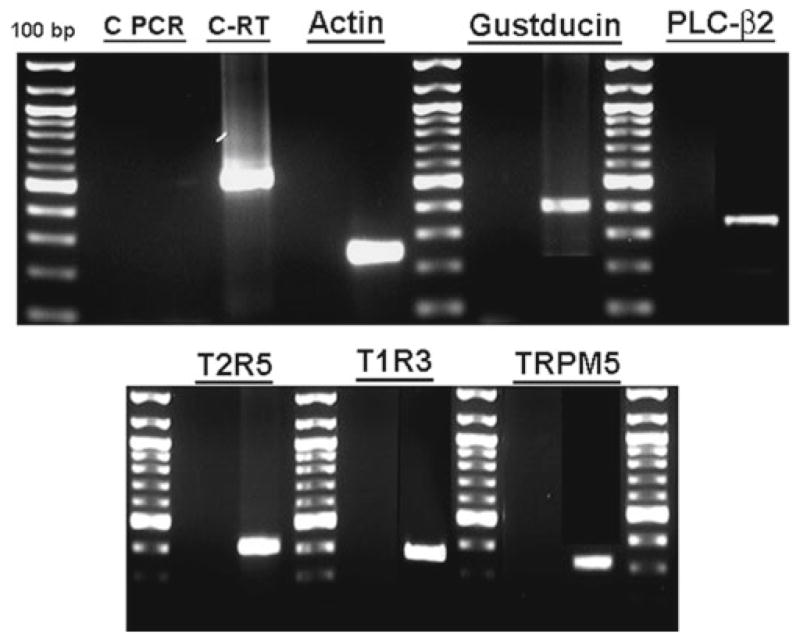
The expression taste cell marker mRNAs were demonstrated by RT-PCR. The expression taste cell markers of β-actin, gustducin, PLC β2, T1R3, T2R5, and TRPM5 mRNA were shown in cultured human fungiform taste cells. The cDNA transcribed from total RNA was amplified with intron spanning-specific primers (Table 2). PCR products were found at expected size and confirmed by sequencing. Specific mRNA was not detected in control experiments without reverse transcriptase indicating no genomic DNA contamination. M = marker (100-bp division).
3.7. Moderate Throughput Screening
Cultured human fungiform cells were grown for 2–4 days in 96-well plates.
Cells were loaded with loading solution in modified Ringer solution (see Subheading 2.6) for 1 h at 36°C (see Note 8).
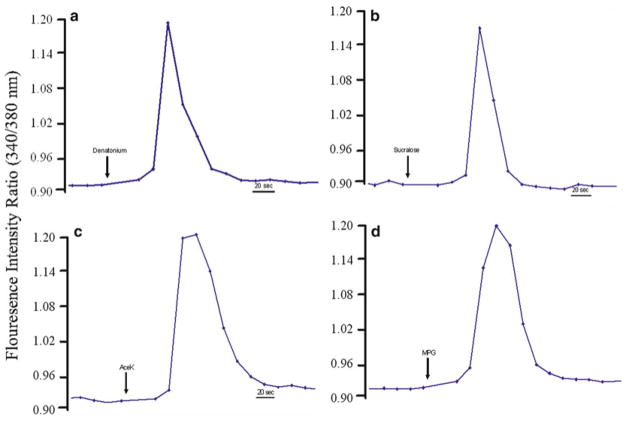
Cells were first superfused with bath solution, then exposed to taste stimuli (see Table 3) via superfusion, with each stimulus applied for 30–60 s, and washed with modified Ringer for at least 2 min (see Fig. 4).
Table 3.
Response frequency of cultured human fungiform taste cells to different chemical stimuli
| Stimuli | Response frequency (%) |
|---|---|
| Denatonium 2 mM | 15 |
| Sucralose 1 mM | 2 |
| AceK 250 ppm | 2 |
| MPG 3 mM | 1 |
Fig. 4.
Cultured taste cells responded to sweet, umami, and bitter stimuli. Changes in intracellular calcium levels ([Ca2+]i) in cultured human fungiform taste cell were measured in 96-well plates using fura-2, a medium-throughput system. Stimuli were dissolved in modified Ringer solution and adjusted for pH and osmolarity. Graphs illustrate representative changes in [Ca2+]i levels in individual cells during exposure to (a) denatonium benzoate (2 mM), (b) sucralose (1 mM), (c) Acesulfame-K (AceK, 250 ppm), (d) mono potassium glutamate (MPG, 3 mM).
Acknowledgments
We acknowledge the technical assistance of Esi Quayson, Aimee Myers, Linda Wysocki, and Valerie Audige. This work was supported in part by P50-DC006760, National Science Foundation (NSF) 0216310, and Givaudan Inc. grant.
Footnotes
Store at 4°C and protect from light by covering medium bottle with aluminum foil. MCDB 153 should be colorless after preparation; any change of color is indication of expiration. MCDB 153 medium expires in 3 months.
Alternative protocol: Digest fungiform papillae with pronase and elastase mixed in isolation solution at room temperature for 30–45 min and then follow step 5 of Subheading 3.1. These two protocol steps give the same result.
Taste cells will eventually attach to the coated coverslip, though it may not be clearly visible in 1–3 days, and you may observe some cubical cells which may be distinct from majority of cells which do not show any sign of proliferation. These cells will be eliminated after the first passaging.
The exact temperature (36°C) of incubator was found to be critical for growth of cultured taste papillae cells.
Experiments are typically performed using cells at passage-2 through -7; however, cells may be used beyond that passage number.
Controls for immunofluorescence consisted of omitting the primary antibody or substituting the primary antibody with the host IgG from which the antibody was generated. In all cases these controls revealed no artifactual labeling. Immunoreactive cells were counted in at least three sampling fields.
As a control to check genomic DNA contamination and nonspecific amplification, samples of RNA were treated in parallel in the presence and absence of reverse transcriptase and used for PCR by amplifying with primers designed for detection of β-actin, gustducin, PLC-β2, T2R5, T1R3, and Trpm5 (see Table 2). Primers were chosen to span one or more introns to exclude confusion with amplified fragments from genomic DNA.
Changes in intracellular calcium levels ([Ca2+]i) in response to taste stimuli were measured in a 96-well format using a Discovery-1 imaging station and Metamorph software® (Molecular Devices). The cells in each well were visualized with an inverted fluorescence microscope using excitation wavelengths of 340 and 380 nm and an emission wavelength set by a band-pass filter centered at 510 nm. A series of image pairs was acquired for each well at 20× with a CCD camera (Photometrics®) as follows: four baseline, 12 poststimulus, and four baseline, for each of the three stimuli. A 2-min washout period followed each poststimulus block of images. Stimulus selection and delivery, focusing, and image acquisition, as well as plate movement were controlled by custom-designed Discovery-1 software (Molecular Devices).
References
- 1.Arvidson K. Location and variation in number of taste buds in human fungiform papillae. Scand J Dent Res. 1979;87:435–442. doi: 10.1111/j.1600-0722.1979.tb00705.x. [DOI] [PubMed] [Google Scholar]
- 2.Barlow LA, Northcutt RG. Analysis of the embryonic lineage of vertebrate taste buds. Chem Senses. 1994;19:715–724. doi: 10.1093/chemse/19.6.715. [DOI] [PubMed] [Google Scholar]
- 3.Beidler LM, Smallman RL. Renewal of cells within taste buds. J Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezencon C, Le Coutre J, Damak S. Taste-signaling proteins are coexpressed in solitary intestinal epithelial cells. Chem Senses. 2007;32:41–49. doi: 10.1093/chemse/bjl034. [DOI] [PubMed] [Google Scholar]
- 5.Caicedo A, Jafri MS, Roper SD. In situ Ca2+ imaging reveals neurotransmitter receptors for glutamate in taste receptor cells. J Neurosci. 2000;20:7978–7985. doi: 10.1523/JNEUROSCI.20-21-07978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 7.Finger TE. Cell types and lineages in taste buds. Chem Senses. 2005;30(Suppl 1):i54–i55. doi: 10.1093/chemse/bjh110. [DOI] [PubMed] [Google Scholar]
- 8.Gilbertson TA, Damak S, Margolskee RF. The molecular physiology of taste transduction. Curr Opin Neurobiol. 2000;10:519–527. doi: 10.1016/s0959-4388(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 9.Huang YA, Maruyama Y, Stimac R, Roper SD. Presynaptic (Type III) cells in mouse taste buds sense sour (acid) taste. J Physiol. 2008;586:2903–2912. doi: 10.1113/jphysiol.2008.151233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishi M, Emori Y, Tsukamoto Y, Abe K. Primary culture of rat taste bud cells that retain molecular markers for taste buds and permit functional expression of foreign genes. Neuroscience. 2001;106:217–225. doi: 10.1016/s0306-4522(01)00184-1. [DOI] [PubMed] [Google Scholar]
- 11.Mbiene JP, Maccallum DK, Mistretta CM. Organ cultures of embryonic rat tongue support tongue and gustatory papilla morphogenesis in vitro without intact sensory ganglia. J Comp Neurol. 1997;377:324–340. doi: 10.1002/(sici)1096-9861(19970120)377:3<324::aid-cne2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Ozdener H, Yee KK, Cao J, Brand JG, Teeter JH, Rawson NE. Characterization and long-term maintenance of rat taste cells in culture. Chem Senses. 2006;31:279–290. doi: 10.1093/chemse/bjj030. [DOI] [PubMed] [Google Scholar]
- 13.Paran N, Mattern CF, Henkin RI. Ultrastructure of the taste bud of the human fungiform papilla. Cell Tissue Res. 1975;161:1–10. doi: 10.1007/BF00222109. [DOI] [PubMed] [Google Scholar]
- 14.Perez CA, Huang L, Rong M, Kozak JA, Preuss AK, Zhang H, Max M, Margolskee RF. A transient receptor potential channel expressed in taste receptor cells. Nat Neurosci. 2002;5:1169–1176. doi: 10.1038/nn952. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz CJ, Stone LM, McPheeters M, Ogura T, Bottger B, Lasher RS, Finger TE, Kinnamon SC. Maintenance of rat taste buds in primary culture. Chem Senses. 2001;26:861–873. doi: 10.1093/chemse/26.7.861. [DOI] [PubMed] [Google Scholar]
- 16.Spielman AI, Mody I, Brand JG, Whitney G, MacDonald JF, Salter MW. A method for isolating and patch-clamping single mammalian taste receptor cells. Brain Res. 1989;503:326–329. doi: 10.1016/0006-8993(89)91684-3. [DOI] [PubMed] [Google Scholar]
- 17.Stone LM, Wilcox CL, Kinnamon SC. Virus-mediated transfer of foreign DNA into taste receptor cells. Chem Senses. 2002;27:779–787. doi: 10.1093/chemse/27.9.779. [DOI] [PubMed] [Google Scholar]
- 18.Stone LM, Tan SS, Tam PP, Finger TE. Analysis of cell lineage relationships in taste buds. J Neurosci. 2002;22:4522–4529. doi: 10.1523/JNEUROSCI.22-11-04522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 20.Travers SP, Nicklas K. Taste bud distribution in the rat pharynx and larynx. Anat Rec. 1990;227:373–379. doi: 10.1002/ar.1092270313. [DOI] [PubMed] [Google Scholar]