The Polo-like kinase homologue in Trypanosoma brucei (TbPLK) regulates the assembly of a series of organelles necessary for positioning the parasite's flagellum. An analogue-sensitive strategy is used to acutely and specifically inhibit the kinase with a small molecule, making it possible to identify novel TbPLK functions.
Abstract
Polo-like kinases are important regulators of cell division, playing diverse roles in mitosis and cytoskeletal inheritance. In the parasite Trypanosoma brucei, the single PLK homologue TbPLK is necessary for the assembly of a series of essential organelles that position and adhere the flagellum to the cell surface. Previous work relied on RNA interference or inhibitors of undefined specificity to inhibit TbPLK, both of which have significant experimental limitations. Here we use an analogue-sensitive approach to selectively and acutely inhibit TbPLK. T. brucei cells expressing only analogue-sensitive TbPLK (TbPLKas) grow normally, but upon treatment with inhibitor develop defects in flagellar attachment and cytokinesis. TbPLK cannot migrate effectively when inhibited and remains trapped in the posterior of the cell throughout the cell cycle. Using synchronized cells, we show that active TbPLK is a direct requirement for the assembly and extension of the flagellum attachment zone, which adheres the flagellum to the cell surface, and for the rotation of the duplicated basal bodies, which positions the new flagellum so that it can extend without impinging on the old flagellum. This approach should be applicable to the many kinases found in the T. brucei genome that lack an ascribed function.
INTRODUCTION
Trypanosoma brucei causes severe illnesses in humans and animals that create substantial health and economic problems in sub-Saharan Africa. The few viable treatments for trypanosomiasis are extremely toxic, and parasite resistance to available drugs is a worsening problem (Bouteille et al., 2003). T. brucei is an obligate extracellular parasite that confines all of its exocytosis and endocytosis to a single compartment in the posterior of the cell (Gull, 2003; Field and Carrington, 2009). This compartment, known as the flagellar pocket, also contains the trypanosome's single flagellum, which is nucleated by the basal body docked at the base of the pocket (Lacomble et al., 2009). The flagellum traverses the pocket and then passes through the top of the compartment, which is cinched tightly against the flagellum. The top of the pocket is marked by two cytoskeletal structures—the flagellar pocket collar (FPC) and the bilobe—which encircle the flagellum and may be responsible for ensuring the tight connection between the plasma membrane and flagellum, which delineates the pocket as a distinct membrane compartment (Bonhivers et al., 2008; Gadelha et al., 2009; Esson et al., 2012).
Once outside the cell, the flagellum is adhered to the cell surface and extends toward the anterior of the cell. The attachment of the flagellum is mediated by a structure found within the cell body known as the flagellum attachment zone (FAZ). The FAZ comprises an electron-dense filament and a unique set of four microtubules termed the quartet (MtQ). The filament links together a series of junctions that contact the flagellar membrane, which keep the flagellum firmly attached to the cell body (Vickerman, 1969; Gull, 1999). The MtQ nucleates next to the basal body, wraps around the flagellar pocket, and then extends beside the FAZ filament along the whole length of the cell body (Lacomble et al., 2009). Depletion of bilobe, FPC, or FAZ components causes defects in flagellar attachment and in cytokinesis, usually leading to growth arrest (LaCount et al., 2002; Selvapandiyan et al., 2007; Bonhivers et al., 2008; Shi et al., 2008; Vaughan et al., 2008).
The genome sequence of T. brucei shows that the parasite has undergone substantial selection by its environment. The trypanosome cytoskeleton has reduced the role of the acto-myosin network to the point that actin appears to be dispensable in one life stage of the parasite, whereas the role of tubulin has been enhanced (García-Salcedo et al., 2004; Berriman et al., 2005). T. brucei possesses a large complement of protein kinases, comprising almost 2% of its genome (Parsons et al., 2005). The kinome includes members of most eukaryotic kinase families, along with many unique kinases that may be important for the unique features of the trypanosome life cycle and for pathogenicity.
Among the T. brucei kinases that have been studied is the single Polo-like kinase homologue TbPLK (Kumar and Wang, 2006; Hammarton et al., 2007; de Graffenried et al., 2008). In mammalian cells the four PLK homologues play important roles in mitosis, centriole biogenesis, and the DNA damage response (Archambault and Glover, 2009). The function of TbPLK has been specialized toward the inheritance of the flagellum, which is duplicated during cell division (Ikeda and de Graffenried, 2012). TbPLK is expressed early in the cell cycle, initially localizing to the new MtQ as it nucleates. The kinase is then found on the basal body as it duplicates and forms a new flagellum, which then rotates around the old flagellum to assume a position in the posterior of the cell (Lacomble et al., 2010). This rotation event also coincides with the formation of a new flagellar pocket (Gadelha et al., 2009). TbPLK then migrates to the bilobe as the structure duplicates. After bilobe duplication, TbPLK localizes to the tip of the new FAZ as it extends toward the anterior of the cell, disappearing just before cytokinesis (Ikeda and de Graffenried, 2012). Depletion of TbPLK causes defects in bilobe, FPC, and FAZ duplication (de Graffenried et al., 2008; Ikeda and de Graffenried, 2012). Basal body duplication does occur, but the replicated structures cannot separate.
Small-molecule inhibitors have played an essential role in elucidating the function of important mitotic kinases such as CDK1 and PLK1. These molecules, which are usually modeled after the universal phosphate donor ATP (Peters et al., 2006; Vassilev et al., 2006; Lénárt et al., 2007), rapidly block the activity of kinases within cells and allow the consequences of this acute inhibition to be assessed. Unfortunately, the utility of many inhibitors is limited by their lack of specificity. Early kinase inhibitors used ATP as a starting point for inhibitor design because its binding site is highly conserved, but this made it difficult to develop drugs that targeted a single kinase (Cohen, 1999). The large number of kinase crystal structures has allowed subtle differences in the ATP-binding site and conformational effects to be exploited to generate specific inhibitors, but this process is synthetically laborious and requires extensive screening to ensure that the compounds are truly specific (Tong et al., 1997; Gray et al., 1998; Schindler et al., 2000). The advent of RNA interference (RNAi) provided a new method for inhibition by depleting the kinase of interest, which frequently can be done without targeting related proteins. However, inhibition by RNAi requires protein turnover, which takes much longer than small-molecule inhibition and may be more prone to the up-regulation of other kinases that compensate for the loss of activity (Weiss et al., 2007). Kinase depletion may also lead to additional phenotypes if the protein is part of a complex or has other functions.
Considering its importance in cell division, a specific inhibitor of TbPLK would be extremely useful to study the acute effects of kinase inhibition. An inhibitor of mammalian PLK1 has been shown to inhibit TbPLK in vitro and causes cytokinesis defects in vivo, but the specificity of this drug is not well established (Li et al., 2010). Parasite-specific inhibitors are under development (Urbaniak et al., 2012), but it is unlikely that inhibitors against the many different kinases present in T. brucei will be available in the near future. The ability to clearly identify the function of individual kinases would also facilitate the discovery of potential drug targets.
A general method for kinase inhibition has been established that takes advantage of the conservation within the ATP-binding site (Bishop et al., 1998, 2000; Garske et al., 2011). A conserved large hydrophobic residue (known as the gatekeeper) that contacts the N6 group of ATP is mutated to alanine or glycine, generating a novel pocket within the active site. A potent but nonspecific ATP-competitive kinase inhibitor is then modified with a bulky substituent that docks into this additional pocket, which renders the modified compound incapable of inhibiting kinases that lack the accommodating mutation. When introduced into cells the mutated, termed analogue-sensitive, kinase should be the only kinase whose activity is blocked by addition of the modified inhibitor. The specificity of the modified inhibitor can be further tested by adding it to wild-type cells, which should not show any response. By generating orthogonal kinase-inhibitor pairs, it is now possible to use modified inhibitors to acutely inhibit many different protein kinases with limited concerns about off-target effects.
In this work we generated procyclic T. brucei cells that exclusively express the analogue-sensitive variant of TbPLK (TbPLKas). Using acute inhibition and cell cycle synchronization achieved by elutriation, we were able to dissect key roles played by TbPLK in new FAZ formation and basal body rotation during cell division. Our results show the utility of the analogue-sensitive method in T. brucei, which will allow the acute and specific inhibition of many of the kinases present in the parasite without the burden of identifying new inhibitors.
RESULTS
The PLK family belongs to a small subset of kinases that lose the bulk of their activity upon mutation of the gatekeeper residue within the ATP-binding site that is necessary for the analogue-sensitive strategy. A second site suppressor screen identified a compensatory mutation that restores sufficient activity to PLK to support growth (Zhang et al., 2005). We were able to identify both the gatekeeper residue (L118) and the second site suppressor (C57) in TbPLK due to the high degree of conservation with other PLK family members within the kinase domain (Burkard et al., 2007). We generated a TbPLKas mutant by mutating L118 to glycine and C57 to valine. Recombinant baculoviruses containing the hexahistidine-tagged wild-type and TbPLKas sequences were generated and used to infect insect cells to produce the proteins. Affinity-purified kinases were tested for their sensitivity to the bulky purine analogue 3MB-PP1, which was previously used to inhibit PLK1as (Burkard et al., 2007). TbCentrin2, a small calcium-binding protein phosphorylated by TbPLK in vitro, was expressed in Escherichia coli and used as a substrate (de Graffenried et al., 2008; Yu et al., 2012). Wild-type TbPLK was not inhibited by 3MB-PP1 within the concentration range tested (50 nM to 10 μM), whereas TbPLKas showed an IC50 value of ∼250 nM (Supplemental Figure S1). TbPLKas was less active than the wild-type kinase, requiring a longer exposure. This result shows that TbPLK tolerates the gatekeeper and second-site suppressor mutations and is sensitive to 3MB-PP1. Of importance, the wild-type kinase was not inhibited by the drug.
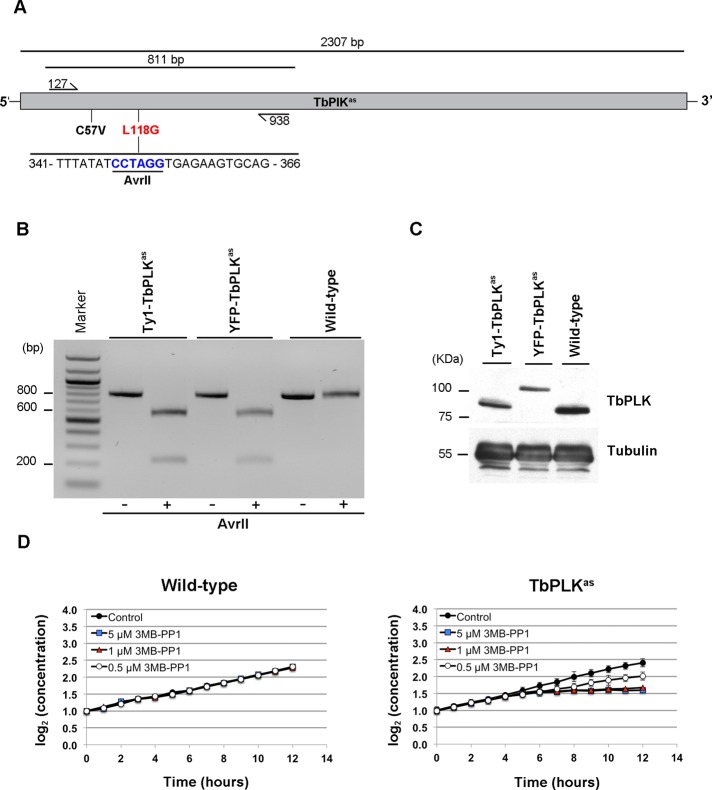
Once the in vitro activity and 3MB-PP1 sensitivity of TbPLKas were established, we made a procyclic T. brucei cell line that exclusively expressed the mutant kinase. Modifying the endogenous TbPLK loci using homologous recombination was necessary because the kinase is mitotically regulated, and constitutive overexpression can cause premature cytokinesis (Kumar and Wang, 2006). We generated a cell line in which one TbPLK allele was replaced with a puromycin resistance gene and the second one with a construct containing both analogue-sensitive mutations and a blasticidin resistance cassette. To allow us to identify clones in which both mutations were incorporated into the TbPLK loci, the nucleotides that introduced the L118G mutation also included a unique AvrII restriction site (Figure 1A). Two versions of the targeting construct were used, with either an N-terminal yellow fluorescent protein (YFP) or a Ty1 tag, so that the TbPLKas allele could be identified directly. Amplified genomic DNA from doubly resistant clones was screened by PCR and digestion with AvrII to identify clones in which both mutations had been incorporated (Figure 1B). Lysates from the Ty1-TbPLKas, YFP-TbPLKas, and wild-type cells were blotted and probed with a monoclonal antibody against TbPLK. The Ty1 and YFP-TbPLKas cell lines both lack untagged TbPLK and express slightly lower levels of kinase than the wild-type cell line (Figure 1C).
FIGURE 1:
Incorporation of analogue-sensitive mutations into TbPLK in procyclic cells. (A) A schematic of the TbPLK loci and the strategy used to introduce analogue-sensitive mutations. Primers flanking the positions of the C57V (black) and L11G (red) mutations were used to amplify an 811–base pair fragment of the TbPLK gene. If the L11G mutant is present, a unique AvrII restriction site is present in the fragment. (B) PCR using the primers described in A from genomic DNA isolated from cells expressing only Ty1-TbPLKas, YFP-TbPLKas, and wild-type cells. The fragment was digested with AvrII to test for incorporation of the analogue-sensitive mutations. The fragment and the product of the AvrII digest for each isolated genomic DNA were run on an agarose gel. Only the fragments isolated from the TbPLKas cell lines were sensitive to AvrII, showing that the mutations were present. (C) Cell lysates from Ty1-TbPLKas, YFP-TbPLKas, and wild-type cells were fractionated by SDS–PAGE, transferred to nitrocellulose, and incubated with an antibody against TbPLK and tubulin as a loading control. (D) Wild-type and TbPLKas cells were treated with different concentrations of 3MB-PP1 or a vehicle control, and their growth was monitored with a cell counter for 12 h. Error bars, SD of three biological replicates.
The effect of 3MB-PP1 on growth of wild-type and TbPLKas cells was tested to ensure that the introduced mutations had made the kinase sensitive to the drug in vivo. Wild-type and TbPLKas cells were seeded in culture, and different concentrations of 3MB-PP1, ranging from 5 μM to 500 nM, or a vehicle control (dimethyl sulfoxide [DMSO]) were added. The growth of the treated cultures was monitored every hour for 12 h using a cell counter. A full round of cell division in procyclic T. brucei takes ∼8.5 h (Sherwin and Gull, 1989). In the absence of drug, the growth of the wild-type and TbPLKas cell lines was almost identical, showing that the mutations in the TbPLKas allele were well tolerated (Figure 1D). The growth of wild-type cells was not affected by the drug at any concentration tested. The intermediate cell line lacking one TbPLK allele that was used to construct the TbPLKas cell line was also insensitive to the drug at all concentrations (Supplemental Figure S2). The growth of the TbPLKas cells was strongly inhibited at 1 and 5 μM, with a clear growth defect appearing 6 h after the addition of drug. At this point the cells ceased to divide for the duration of the experiment. This result shows that TbPLKas cells treated with at least 1 μM 3MB-PP1 do not undergo cytokinesis within the first cell cycle. TbPLKas cells treated with 500 nM drug grew at ∼50% the rate of control cells.
The cell cycle phenotypes produced by TbPLKas inhibition were identified to determine whether they were similar to previously published results using other methods for inhibiting or depleting TbPLK. Early in the cell cycle trypanosomes contain one nucleus and one kinetoplast (1N1K). The kinetoplast duplicates before the nucleus (1N2K); subsequent nuclear division results in a 2N2K cell, which then undergoes cytokinesis. An exponentially growing culture contains ∼80% 1N1K cells; the remaining 20% of the culture is near evenly split between 1N2K and 2N2K cells. TbPLK activity has been depleted from cells using RNAi and inhibited using a drug that inhibits human PLK1 (Kumar and Wang, 2006; Hammarton et al., 2007; de Graffenried et al., 2008; Li et al., 2010). Cells lacking TbPLK activity have fewer 1N1K cells, an increased number of 2N2K cells, and aberrant 2N1K cells where the nucleus has duplicated prior to the kinetoplast.
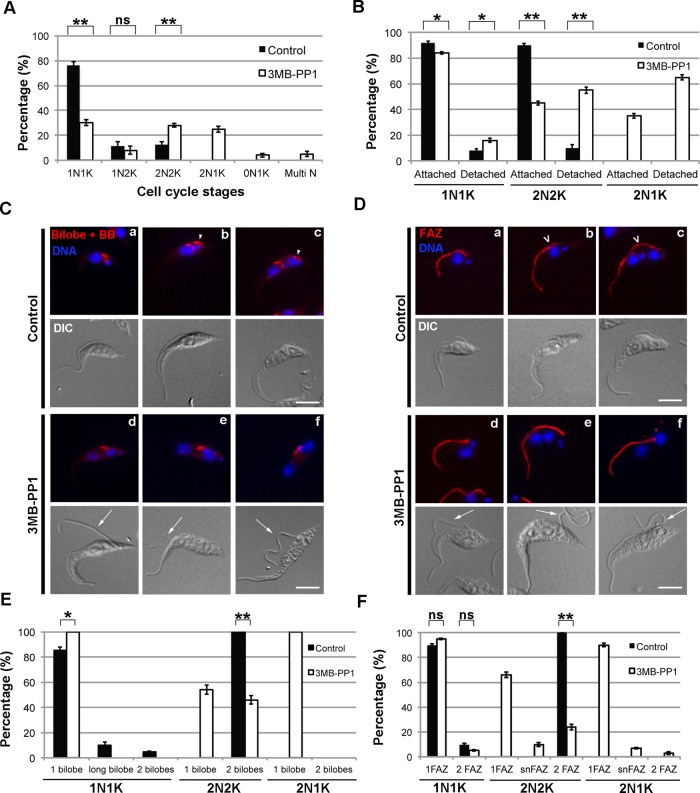
These 2N1K cells are a hallmark of TbPLK inhibition and arise due to a lack of segregation of the duplicated basal bodies (Ikeda and de Graffenried, 2012). To compare these results with TbPLKas inhibition, we treated TbPLKas cells with 3MB-PP1 or vehicle control for 9 h and then fixed and stained them with 4′,6-diamidino-2-phenylindole (DAPI) to assess their DNA state. Vehicle control–treated TbPLKas cells had a DNA distribution identical to that of wild-type cells (Figure 2A). Cells treated with 3MB-PP1 showed a significant change in DNA state, with the 1N1K population dropping to only 30% of total. The number of 1N2K cells decreased slightly, whereas the number of 2N2K cells doubled. 2N1K cells also appeared, comprising >20% of the population. These results show that TbPLKas inhibition has a very similar effect on the cell cycle as depletion of the kinase by RNAi.
FIGURE 2:
Cells expressing TbPLKas and treated with 3MB-PP1 have DNA, bilobe, and FAZ defects. (A) TbPLKas cells were treated with 3MB-PP1 or vehicle control for 9 h and then fixed and stained with DAPI to label DNA. The DNA content of the cells was determined by fluorescence microscopy. Cells treated with 3MB-PP1 showed an increase in 2N2K cells and the appearance of aberrant 2N1K cells. (B) TbPLKas cells treated as in A were fixed and evaluated for flagellar attachment by differential interference contrast microscopy. Cells treated with 3MB-PP1 showed high levels of flagellar detachment. (C) TbPLKas cells were treated as in A and then fixed and labeled with anti-TbCentrin4 antibody (Bilobe + BB; red) and DAPI (DNA; blue) to label DNA. The bilobe is present between the kinetoplast and nucleus (a). The new bilobe forms toward the posterior of the cell (arrowhead) in a subset of 1N1K cells (b) and moves away from the old bilobe as the cell cycle progresses (c). Cells treated with 3MB-PP1 had difficulty duplicating the bilobe (d–f) and have detached new flagella (arrows). (D) TbPLKas cells were treated as in A and then fixed and labeled with anti-FAZ1 antibody (FAZ; red) and DAPI (DNA; blue) to label DNA. The FAZ underlies the flagellum and runs from the posterior to the anterior end of the cell (a). The new FAZ (empty arrowhead) forms toward the posterior of the cell (b), then extends until just before cytokinesis (c). Cells treated with 3MB-PP1 were not able to assemble a new FAZ and had detached flagella (d–f; arrows). (E) Quantitation of the cells in C. (F) Quantitation of the cells in D. Scale bars, 5 μm. Error bars, SD of three biological replicates with 300 cells counted per condition. *p < 0.05, **p < 0.01, ns, not significant.
The cell cycle defects observed upon TbPLK depletion are caused by difficulties in assembling and separating cytoskeletal elements that are necessary for kinetoplast segregation and cytokinesis (Ikeda and de Graffenried, 2012). In TbPLK-depleted cells the basal bodies are able to undergo the normal maturation and duplication cycle, producing a new flagellum, but the replicated structures are not able to separate. The bilobe structure and the FAZ, which are involved in positioning and adhering the flagellum to the cell surface, do not duplicate, generating a new flagellum that is detached from the cell body. In TbPLKas cells treated with 3MB-PP1 for 9 h, cells at all cell cycle stages showed an increase in detached new flagella compared with vehicle control-treated samples (Figure 2B). In 2N cells, more than half of the population had detached new flagella. The small number of detached flagella present in the vehicle control samples is most likely due to a fixation artifact, as detached flagella were never observed in live cells in this sample.
To test whether bilobe duplication was perturbed, TbPLKas cells were treated with 3MB-PP1 or vehicle control for 9 h, fixed, and then stained for the bilobe and basal body marker TbCentrin4 (Figure 2C; Selvapandiyan et al., 2007; Shi et al., 2008). In wild-type cells the bilobe duplicates at the 1N1K state (Figure 2C, b, arrowhead). The first evidence of duplication is the elongation of the bilobe to 2.2 μm or longer. This elongated structure then is separated into two distinct bilobes, which are moved further apart as the cell goes through the 1N2K and 2N2K states (Figure 2C, c, arrowhead). In vehicle control-treated cells, ∼18% of 1N1K cells contained either elongated or duplicated bilobes, whereas all 2N2K cells contained two bilobes (Figure 2E). In 3MB-PP1-treated cells, there was no evidence of elongated (>2.2 μm) or duplicated bilobes in 1N1K cells (Figure 2E). Greater than 50% of 2N2K cells had only one bilobe (Figure 2, E and C, d and e), whereas all the aberrant 2N1K cells had a single bilobe (Figure 2, E and C, f). As noted previously, the 3MB-PP1–treated cells frequently had detached flagella (Figure 2C, d–f, arrows).
Considering the extent of new flagellum detachment and the defects in bilobe assembly, it was likely that FAZ assembly was also impeded by the absence of TbPLK activity. To evaluate the new FAZ, we treated TbPLKas cells with 3MB-PP1 or vehicle control for 9 h and then fixed and stained them with an antibody against FAZ1, a FAZ component (Figure 2D; Kohl et al., 1999; Vaughan et al., 2008). In vehicle control–treated cells new FAZ assembly begins in 1N1K cells and continues through the 1N2K and 2N2K stages, reaching completion just before cytokinesis (Figure 2D, b and c, open arrowheads). In vehicle control–treated cells, FAZ duplication proceeds as in wild-type cells, with ∼10% of 1N1K cells containing two FAZ, whereas all 1N2K and 2N2K cells had two FAZ (Figure 2F). In 3MB-PP1–treated cells the number of 1N1K cells with two FAZ decreased (Figure 2, D, d, and F). In the 2N2K population, 23% had only a single FAZ, whereas 10% contained a short new FAZ (snFAZ), <5 μm, which is the shortest-length new FAZ that was observed in wild-type 2N2K cells (Figure 2, D, e, and F). We hypothesize either that cells with snFAZ prematurely aborted the assembly of the new FAZ or that the assembly process was somehow delayed. FAZ defects were very evident in 2N1K cells, with 90% of these cells containing only a single FAZ (Figure 2, D, f, and F). Cells that lacked a new FAZ or had an snFAZ almost always had detached new flagella (Figure 2D, d–f, arrows).
To further confirm that the defects observed in drug-treated TbPLKas cells were specific, we treated wild-type cells with 3MB-PP1 or a vehicle control for 9 h and evaluated their DNA state and ability to assemble a new bilobe and FAZ. Wild-type cells treated with drug or the vehicle control had nearly identical distributions of 1N1K, 1N2K, and 2N2K states and lacked any cells with aberrant DNA content, such as 2N1K (Supplemental Figure S3A). Wild-type cells were able to duplicate their bilobe and FAZ in the presence of 3MB-PP1, showing that the drug only has an effect on cells carrying the TbPLKas allele (Supplemental Figure S3, B and C).
Inhibition of TbPLKas is phenotypically similar to depletion of TbPLK by RNAi. Both methods produce cells that contain aberrant DNA states, detached flagella, and defects in bilobe and FAZ duplication. The onset of the phenotype appears to be more rapid and severe when the TbPLKas system is used, which is expected because the small molecule can directly inhibit the kinase, whereas RNAi relies on protein turnover for its effects. One important difference is that small-molecule inhibition leaves its target intact, allowing changes in the localization pattern of the enzyme to be established. Because TbPLK is highly motile and PLK homologues rely on their activity to generate some of their own binding sites, identifying what happens to the kinase upon inhibition may provide some insight into how TbPLK localizes to different positions within the cell (Neef et al., 2003).
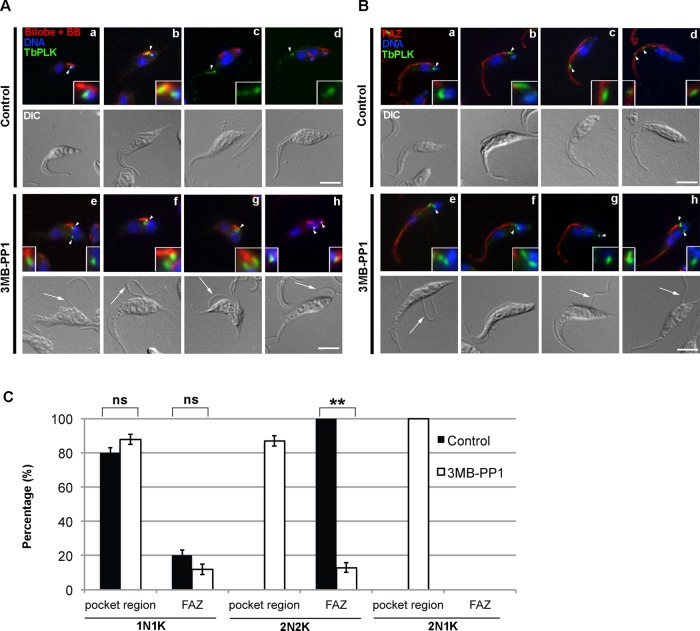
TbPLKas was localized in 3MB-PP1– and vehicle-control–treated cells to determine whether inhibition of the kinase affected its localization. TbPLKas cells were treated for 9 h with 3MB-PP1 or vehicle control, then fixed and labeled with antibodies against TbPLK and either TbCentrin4 or FAZ1 antibody to mark the bilobe or FAZ, respectively. In control cells, TbPLK was first present on a structure next to the kinetoplast, which corresponds to the new MtQ (Figure 3, A, a, and B, a; arrowheads; Ikeda and de Graffenried, 2012). The kinase then moved to the basal body and bilobe (Figure 3, A, b, and B, b; arrowheads), followed by accumulation on the tip of the new FAZ (Figure 3, A, c and d, and B, c and d; arrowheads). The location of the kinase in 3MB-PP1–treated and vehicle control cells was quantitated and placed in two categories: 1) TbPLK localization to the MtQ, basal body, and bilobe (pocket region), and 2) TbPLK presence on the new FAZ. Sixty percent of cells expressed TbPLK in both conditions, arguing that inhibition does not lead to changes in kinase expression (data not shown). In vehicle control–treated cells, 80% of 1N1K cells expressing TbPLK had labeling in the pocket region, whereas in 20% the kinase had already migrated onto the growing tip of the new FAZ (Figure 3C). In 2N2K cells, all cells expressing TbPLKas had labeling exclusively on the FAZ. In the 3MB-PP1–treated samples 1N1K cells showed slightly elevated levels of pocket region localization. There was a striking change in localization in 2N2K cells. The kinase, which localized exclusively to the FAZ in untreated cells, was now found within the pocket region >80% of the time (Figure 3, A, e–h, and B, e–h, arrowheads, and C). 2N2K cells with pocket region TbPLKas frequently had defects in bilobe and FAZ duplication, along with detached new flagella (Figure 3, A, e–f, and B, e–f; arrows). In 2N1K cells TbPLKas was found exclusively in the pocket region (Figure 3, A, g–h, and B, g–h; arrows).
FIGURE 3:
Inhibition of TbPLK causes the kinase to remain in the pocket region. TbPLKas cells were treated with 3MB-PP1 or vehicle control for 9 h, then fixed and stained with anti-TbPLK antibodies and organelle markers to identify the location of TbPLK. Insets in A and B are threefold magnifications of the area around the TbPLK signal. (A) Cells were stained with anti-TbCentrin4 (Bilobe + BB; red), anti-TbPLK (TbPLK; green), and DAPI (DNA; blue). In control cells TbPLK is initially present in the pocket region, which consists of the basal body, MtQ, and bilobe (a, b; arrowheads), then migrates out toward the anterior of the cell (c, d; arrowheads) once kinetoplast and nuclear duplication occur. In 3MB-PP1–treated cells, the kinase remains in the pocket region even after nuclear duplication had occurred (e–h; arrowheads). (B) Cells were stained with anti-FAZ1 (FAZ; red), anti-TbPLK (TbPLK; green), and DAPI (DNA; blue). In the control cells the TbPLK signal shows the same migration pattern as in A (a–d; arrowheads). In 3MB-PP1–treated cells, the kinase does not migrate toward the posterior of the cell (e–h; arrowheads) and remains near the flagellar pocket. (C) Quantitation of the data in A and B. Scale bars, 5 μm. Error bars, SD of three biological replicates with 300 cells counted per condition. **p < 0.01, ns, not significant.
Because TbPLKas plays multiple roles at different time points and locations during cell division, a cell cycle synchronization approach would allow a much more precise assignment of kinase function. The ability to couple the rapid inhibition afforded by the TbPLKas system with cell synchronization would allow us to dissect the roles of the kinase at specific points of the cell cycle. Synchronization techniques for trypanosomes have been established that rely on either treatment with hydroxyurea (HU) or cell starvation (Gale and Parsons, 1993; Chowdhury et al., 2008). Treatment with HU produces a synchronous population of cells on the 1N1K/1N2K boundary, which proceeds through the cell cycle upon washout. However, the point of synchronization is after many of the critical events that require TbPLK, such as bilobe duplication and the initiation of the new FAZ. Cell starvation can be used to produce synchronized cells at an earlier point in the cell cycle corresponding to nuclear G1, but the cells take 10 h to progress into S phase, suggesting that the cells may be in a quiescent G0-like state due to starvation (Li et al., 2010). This makes it uncertain whether any phenotypes identified using this method are due to conventional cell cycle progression.
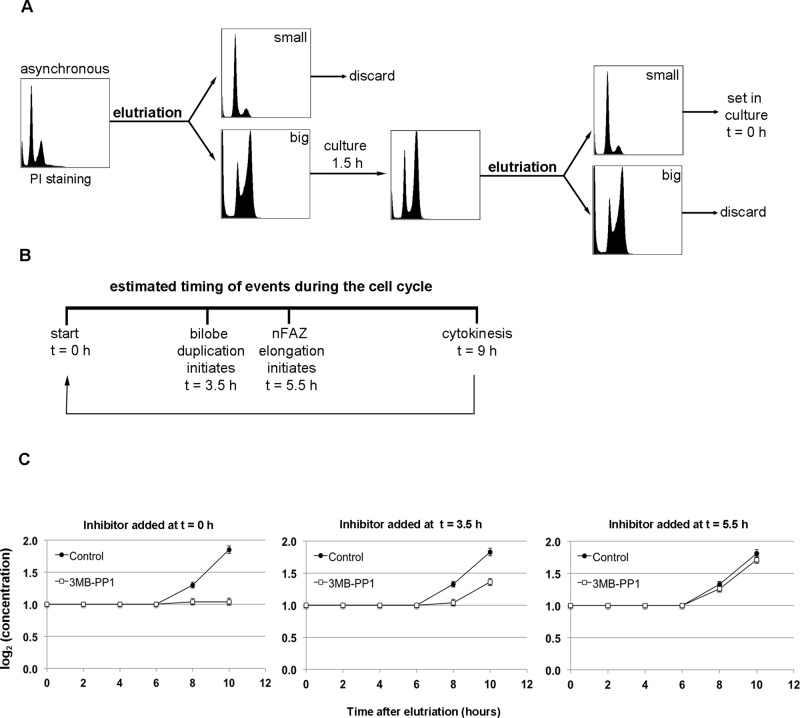
A promising double-cut elutriation strategy has been described that synchronizes cells at a very early point in the cell cycle, allowing all subsequent events to be monitored (Archer et al., 2011). Elutriation discriminates between cells based on their size and density, which correlates well with their cell cycle stage (Banfalvi, 2008). In the double-cut protocol, the largest cells isolated during elutriation are put back into culture for 1.5 h (Figure 4A). Because the largest cells are most likely ones that are about to undergo cytokinesis, this incubation time produces a large number of cells that have just finished dividing. This population of recently divided cells is then isolated by a second round of elutriation, this time selecting for the smallest cells in the culture.
FIGURE 4:
Double-cut elutriation of TbPLKas cells produces synchronous cultures. (A) The protocol for double-cut elutriation. An asynchronous culture is elutriated, selecting for the largest cells, which are set in culture for 1.5 h so that they can undergo division. The population is elutriated again, and the smallest cells, which have just finished dividing, are selected and used for further experiments. (B) A simplified view of the cell cycle in T. brucei, highlighting bilobe duplication (t = 3.5 h) and new FAZ elongation (t = 5.5 h). The cell cycle completes with cytokinesis at 9 h. (C) TbPLKas cells were synchronized by double-cut elutriation, and then 3MB-PP1 or vehicle control was added to the cells at different points in the cell cycle. Addition of drug just after elutriation (t = 0) totally blocked cell division, whereas addition at t = 3.5 caused a 50% block. Addition at t = 5.5 had essentially no effect on division. Error bars, SD of three biological replicates.
We used the double-cut elutriation protocol to synchronize TbPLKas cells. Elutriated cells were fixed and stained at different time points with propidium iodide to monitor DNA synthesis (Supplemental Figure S4). Cells fixed just after the elutriation procedure (t = 0 h) showed a substantial increase in G1 cells, very few cells in S phase, and a suppressed G2 population compared with an asynchronous culture. At t = 3 h, the population shifted into S phase, followed by most of the cells progressing to G2 at t = 5 h. The cells began to return to G1 at t = 7 h, indicating that cytokinesis was beginning. The cells then started another round of division, shifting into S phase at t = 10 h. Labeling elutriated cells with DAPI and TbPLK antibodies to look at individual cells by fluorescence microscopy showed that the culture was 99% 1N1K cells at t = 0 h, with only 10% of the population expressing TbPLK, compared with 60% of the 1N1K population in an asynchronous culture (data not shown). By following the TbPLK expression profile of the synchronized cells over time, we showed that all the cells in our synchronized cultures were within a 2.5-h portion of the cell cycle (data not shown).
Once the efficacy of the double-cut elutriation strategy in the TbPLKas background had been established, we synchronized cells and treated them with 3MB-PP1 or vehicle control at specific points of the cell cycle. Cells were treated at four points in the cell cycle: immediately after completing the elutriation (t = 0 h), just before TbPLK expression (t = 1.5 h), at the point when TbPLK is on the bilobe and the structure begins to duplicate (t = 3.5 h), and when TbPLK is present on the new FAZ (t = 5.5 h; Figure 4B). These points were chosen based on previous work that established the timing of these cell cycle events and the position of TbPLK (Ikeda and de Graffenried, 2012). Elutriated TbPLKas cells treated with vehicle control began to divide after 7 h in culture and completed a full round of division within 10 h (Figure 4C, control). Treatment with drug at t = 0 or 1.5 h caused total growth arrest (only t = 0 h is shown), whereas treatment at t = 3.5 h caused a 50% decline in growth. Cells treated at t = 5.5 h showed little or no growth defects compared with vehicle control within the time frame of the experiment.
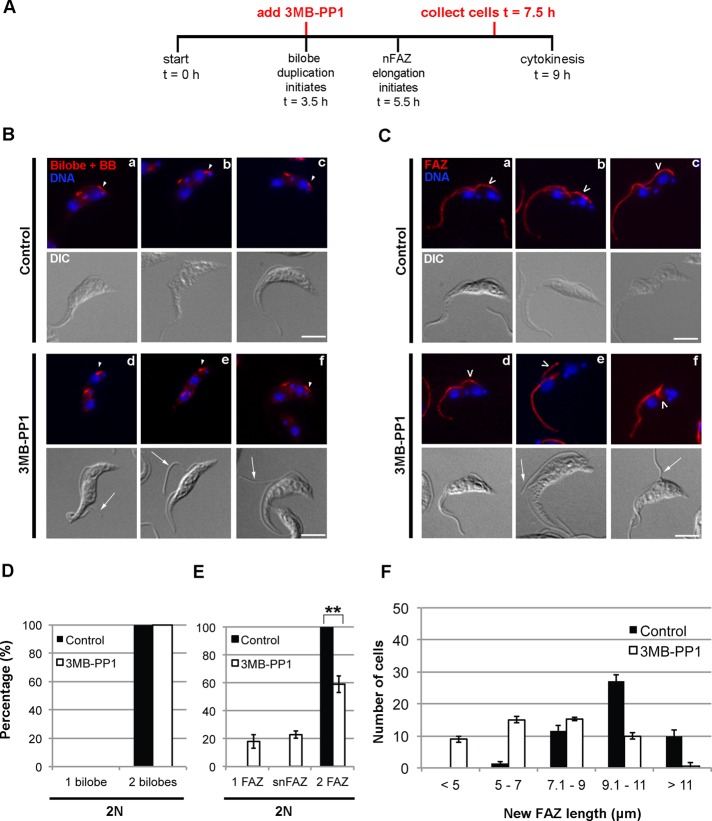
The elutriated TbPLKas cells were treated with 3MB-PP1 or vehicle control at t = 0, 1.5, 3.5, or 5.5 h and then harvested at t = 7.5 h to ensure that most of the cells had not undergone cytokinesis. The cells were then fixed and stained with DAPI and antibodies against bilobe and FAZ markers. Treatment with drug at t = 0 or 1.5 h led to decreased numbers of 1N2K cells and the appearance of 2N1K cells, but adding the drug at later points had little effect on DNA state (Supplemental Figure S5A). We chose to concentrate on 2N cells for bilobe and FAZ analysis to be certain that we were analyzing cells that had not divided since the drug had been added. The duplication of the bilobe was impaired by the addition of 3MB-PP1 at t = 0 or 1.5 h, leading to 2N cells with a single bilobe (Supplemental Figure S5B). Treatment at later time points had no effect on bilobe duplication. FAZ duplication was completely blocked by 3MB-PP1 treatment at t = 0 or 1.5 h, whereas treatment at t = 5.5 h had no effect (Supplemental Figure S5C). Treatment at 3.5 h produced an intermediate phenotype, where 40% of 2N cells had either a single FAZ or an snFAZ. A summary of the effects of TbPLKas inhibition at different time points on DNA content, bilobe, and FAZ is presented in Table 1. We chose to study the t = 3.5 h time point to establish the precise bilobe and FAZ phenotype in more detail.
TABLE 1:
Summary of the DNA, bilobe, and FAZ states of synchronized cells upon treatment with 3MB-PP1 at different cell cycle stages.
| Cells harvested at t = 7.5 h | |||
|---|---|---|---|
| 3MB-PP1 added at t (h) | Abnormal DNA states? | Bilobe duplication? | FAZ duplication? |
| 0 | Yes | Impaired | Impaired |
| 3.5 | No | Normal | Partial |
| 5.5 | No | Normal | Normal |
Cells synchronized by elutriation were treated with 3MB-PP1 after 0, 3.5, or 5 h in culture. The cells were then fixed and stained with DAPI to evaluate their DNA state, along with antibodies against the FAZ or bilobe to assess the status of these structures.
Considering the close spatial association of the bilobe and FAZ and how the structures duplicate within a narrow time window, it has been difficult to say with certainty whether one requires the other for assembly (Esson et al., 2012; Ikeda and de Graffenried, 2012). If bilobe duplication is necessary for new FAZ assembly, then the absence of new FAZ in cells lacking TbPLK activity could be due to defects in bilobe duplication. To address this issue, we treated TbPLKas cells with 3MB-PP1 or a vehicle control 3.5 h after synchronization, then harvested them at 7.5 h and fixed and stained them with TbCentrin4 or FAZ1 antibodies (Figure 5A). Cells treated with 3MB-PP1 were able to assemble new bilobes but frequently had detached flagella, suggesting a FAZ defect (Figure 5B, d–f; arrowheads depict new bilobes, arrows show detached flagella; Figure 5D). Forty percent of 2N cells had either no new FAZ or an snFAZ (Figure 5C, d–f, empty arrowheads, and E). This result shows that TbPLK activity is directly necessary for FAZ elongation. We measured the length of the new FAZ in 2N cells in vehicle control– and drug-treated samples to further establish the FAZ phenotype. In vehicle control–treated samples, new FAZ length ranged from 5 to >11 μm, with a median length of 10.1 μm (Figure 5F). There were no FAZ <5 μm. In 3MB-PP1–treated samples, cells with aberrant snFAZ of <5 μm were observed, but the median length of the new FAZ also dropped to 7.1 μm. TbPLK activity is not only necessary for the initiation of a new FAZ, but it also is required for the structure to reach its full length.
FIGURE 5:
Treatment of synchronized TbPLKas cells with 3MB-PP1 at t = 3.5 h produces cells that duplicate the bilobe but have new FAZ defects. (A) TbPLKas cells were synchronized and treated with 3MB-PP1 or vehicle control at t = 3.5, followed by fixation at t = 7.5. (B) Cells treated as in A were labeled with anti-TbCentrin4 antibody (Bilobe+BB; red) and DAPI (DNA; blue). Cells that had reached the 2N state after treatment with 3MB-PP1 at t = 3.5 were able to duplicate their bilobe (arrowheads, d–f) but frequently had detached flagella (arrows). (C) Cells treated as in A were labeled with anti-FAZ1 antibody (FAZ; red) and DAPI (DNA; blue). Cells that had reached the 2N state after treatment with 3MB-PP1 at t = 3.5 frequently had FAZ that were too short (d–f; empty arrowheads) and had detached flagella (arrows). (D) Quantification of the bilobe state of the 2N cells in B. (E) Quantification of the FAZ state of the 2N cells in C. (F) Quantification of new FAZ length in 2N cells in C. Cells treated with 3MB-PP1 at t = 3.5 have shorter new FAZ than cells treated with vehicle control. Scale bars, 5 μm. Error bars, SD of three biological replicates. For D and E, 80 2N cells were counted per condition. **p < 0.01. For F, 50 cells were counted per condition. A Student's t-test was performed to compare the means of the two distributions, giving p < 0.05.
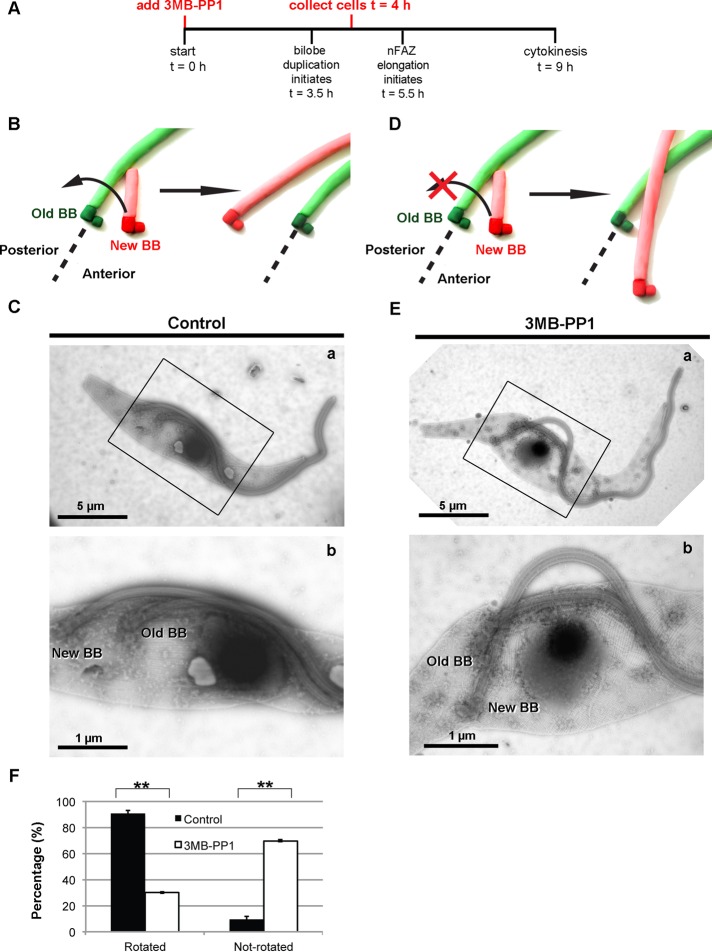
Having shown that cell synchronization along with TbPLKas inhibition allows us to study cell cycle events with high temporal resolution, we decided to revisit the function of TbPLK in the duplication and inheritance of the basal body. Recent work shows that the newly assembled basal body, which nucleates the new flagellum, rotates around the old flagellum in a counterclockwise motion before the new flagellum emerges from the flagellar pocket (Lacomble et al., 2010). This rotation may facilitate the production of a new flagellar pocket and situates the new flagellum in the posterior of the cell, where it can extend properly. This rotation occurs early in the cell cycle when TbPLK is still in the pocket region; if this process does not occur, it could lead to basal body segregation defects due to steric clashes between the assembled organelles.
To test for defects in basal body rotation, we synchronized TbPLKas cells, treated them with 3MB-PP1 or vehicle control at t = 0 h, and then harvested them at t = 4 h (Figure 6A). This endpoint was chosen because almost all cells should have undergone rotation by this time. The cells were placed on grids, and whole-mount cytoskeletons were prepared by extraction with detergent, followed by labeling with aurothioglucose for negative-stain electron microscopy (Höög et al., 2010). At early points in the cell cycle, the probasal body is found on the anterior side of the mature basal body. When the probasal body matures, it docks with the flagellar pocket membrane and nucleates a new flagellum, which contacts the old flagellum (Figure 6B). Before the new flagellum emerges from the flagellar pocket, the new basal body rotates in a counterclockwise direction around the old flagellum, assuming a position in the posterior of the cell (Figure 6B). In vehicle control–treated cells the new basal body was positioned on the posterior side of the old basal body in 90% of cells, indicating that rotation had occurred (Figure 6, C, a–b, and F). If basal body rotation failed to occur, the new basal body would remain on the anterior side of the old basal body (Figure 6D). In 3MB-PP1–treated cells the new flagellum was frequently detached from the cell body, indicating a defect in new FAZ formation (Figure 6E, a–b). The new basal body had not rotated in 70% of the cells (Figure 6, E, a–b, and F). The flagella appeared to cross one another at the point where they emerged from the flagellar pocket. Additional images of 3MB-PP1–treated cells where basal body rotation has been inhibited are shown in Supplemental Figure S6.
FIGURE 6:
Treatment of synchronized TbPLKas cells with 3MB-PP1 at t = 0 h blocks rotation of the duplicated basal body. (A) TbPLKas cells were synchronized and treated with 3MB-PP1 or vehicle control at t = 0 and then collected at t = 4 and processed for negative-stain electron microscopy. (B) A schematic of the progression of basal body rotation. The old basal body (Old BB) and flagellum are shown in green, and the new basal body (New BB) and flagellum are shown in red. The new basal body rotates around the old basal body before the new flagellum emerges from the flagellar pocket. (C) A 1N cell from a vehicle control–treated (Control) sample. The new flagellum is present in the posterior of the cell, having rotated from the anterior side of the old flagellum during its maturation process. (D) A schematic of the aberrant case in which basal body rotation does not occur. (E) In the 3MB-PP1–treated cell, the new flagellum is detached from the cell body and has not rotated around the old flagellum (a, b). (F) Quantification of the data in B. Scale bar size is noted in the individual images. Error bars, SD of three biological replicates with between 55 and 65 cells counted per condition. **p < 0.01.
DISCUSSION
TbPLK tolerates the analogue-sensitive mutations and responds to the modified inhibitor 3MB-PP1 in vitro and in vivo. TbPLKas cells responded to 3MB-PP1 treatment while treatment of wild-type cells with drug had no effect on growth. This clearly shows that the phenotypes observed in the TbPLKas cell line upon drug treatment are due to inhibition of TbPLKas. The recombinant analogue-sensitive kinase was not as active as wild-type TbPLK. However, the activity of the sensitized kinase was sufficient to support near-wild-type growth in procyclic cells, even at slightly lower expression levels. Previous work showed that a human PLK1 inhibitor blocks the activity of TbPLK in vitro and causes defects in cytokinesis in vivo, but the specificity of the inhibitor in trypanosomes was not established (Li et al., 2010). The drug inhibits other human kinases such as Aurora and CDK2 in vitro with IC50 values that fall within the concentration range used for in vivo experiments with trypanosomes (5 μM), raising the possibility that the trypanosome homologues of these kinases were also affected and contributed to the observed phenotypes.
In asynchronous cells, TbPLKas inhibition and TbPLK depletion by RNAi lead to similar phenotypes. The fact that two distinct methods give very similar phenotypes argues strongly that TbPLK plays an important role in FAZ and bilobe duplication. The similarity between the RNAi and small-molecule inhibition phenotypes suggests that the essential functions of TbPLK are mediated by its enzymatic activity. TbPLK is the sole PLK family member in trypanosomes, making up-regulation of a compensatory enzyme unlikely. TbPLK, like other polo homologues, appears to be degraded at the end of each cell cycle, which improves the effectiveness of RNAi (Lindon and Pines, 2004). Previously we were able to observe near-quantitative depletion of TbPLK from cultures using RNAi within 8 h of induction (de Graffenried et al., 2008; Ikeda and de Graffenried, 2012). Even with this rapid effect, the TbPLK-depleted cultures did not show any growth defects at this point, whereas TbPLKas inhibition yields growth defects within 6 h of treatment.
Inhibition of TbPLK causes the kinase to remain in the flagellar pocket region of the cell throughout the cell cycle. The inability of the kinase to migrate may be due to the absence of the new FAZ, which could recruit the kinase and function to transport it toward the anterior of the cell. At early points in the cell cycle TbPLK localizes to the MtQ (Ikeda and de Graffenried, 2012). If these microtubules are not initiated or if their extension is cut short, the kinase may be trapped within the flagellar pocket region. PLKs are frequently recruited by upstream kinases such as Cdk1 that generate phosphosites recognized by the polo-box domain of PLK (Preisinger et al., 2005; Crasta et al., 2008). The upstream kinases that control TbPLK localization may continue to generate binding sites for TbPLK within the pocket region even though its activity is blocked. Migration of TbPLK may require the kinase to generate its own binding sites on the FAZ, which would explain its inability to localize to this structure upon inhibition. PLK1 creates a subset of its own binding sites during anaphase (Neef et al., 2007). Previous work showed that kinase-dead versions of TbPLK, the use of which is analogous to the inhibition strategy used here, remain associated with the FAZ (Sun and Wang, 2011; Yu et al., 2012). However, the kinase-dead TbPLK was overexpressed using a very strong promoter, which can lead to mislocalization of the protein. It is also possible that the kinase-dead mutants produce a different phenotype from small-molecule inhibition, perhaps allowing the new FAZ to form to some extent.
Combining analogue-sensitive inhibition with cell synchronization established the precise role of TbPLK at different points of the cell cycle. Previous work with TbPLK RNAi on asynchronous cultures was hampered by the fact that TbPLK plays many different roles in cell division, and loss of activity at different points in the cell cycle most likely has different outcomes. Cells that are affected by TbPLK depletion early in the experiment may change their appearance over time due to the progression of cell cycle events that are not governed by TbPLK, such as karyokinesis. Using synchronous cells, we were able to show that inhibition at early points of the cell cycle blocks cell division and causes severe defects in bilobe and FAZ duplication. Treatment at late time points had little or no effect on growth, which suggests that early phosphorylation events that trigger cytoskeletal duplication are essential events in the T. brucei cell cycle.
Using synchronous cells, we showed that TbPLKas inhibition led directly to FAZ defects. Previous work suggested that FAZ assembly requires TbPLK, but this result relied on RNAi in asynchronous cells (Ikeda and de Graffenried, 2012). This limitation meant that although TbPLK depletion led to cells with FAZ defects, these cells almost always had defects in bilobe duplication or basal body segregation. Because depletion of bilobe and basal body components blocks formation of the new FAZ, it seemed possible that the FAZ defects in TbPLK RNAi cells could be secondary effects (Selvapandiyan et al., 2007; Shi et al., 2008). By using synchronous cells and acute inhibition, we can inhibit TbPLK activity at the point when FAZ duplication occurs but after bilobe duplication. This allows us to clearly show that FAZ duplication and extension rely on TbPLK activity and are not just blocked due to bilobe defects. Inhibition of TbPLK also shortened the overall length of the new FAZ. Considering that treatment of cells with drug at t = 3.5 h had a partial effect on growth, it is likely that some of the shorter new FAZ were still functional. This result argues that TbPLK is involved in FAZ extension late in the cell cycle but that once the new FAZ elongates beyond a critical length, which is at least 5 μm, cytokinesis can proceed.
Inhibition of TbPLKas early in the cell cycle leads to defects in basal body rotation. The rotation of the new basal body resolves a series of steric clashes between the new flagellum and MtQ that would arise if it remained on the anterior side of the old basal body and may allow the new flagellar pocket to form by pulling membrane along the old MtQ, which delineates the new pocket (Gadelha et al., 2009; Lacomble et al., 2010). TbPLK is present on the basal bodies early in the cell cycle, just before bilobe duplication, which is consistent with the timing of the rotation (Ikeda and de Graffenried, 2012). The forces responsible for basal body rotation, which includes movement of the new MtQ and probasal body, are not known. PLK1 plays an important role in centriole separation in mammalian cells by acting on the kinesin Eg5 (Le Guellec et al., 1991; Hagan and Yanagida, 1992; Smith et al., 2011). It is possible that the rotation is mediated by a related kinesin. It is important to note that the basal bodies in trypanosomes contact the flagellar pocket membrane and are linked to the kinetoplast, which puts additional constraints on their movement (Robinson and Gull, 1991; Lacomble et al., 2009). Basal body rotation usually occurs before emergence of the new flagellum and may be essential for the formation of a new flagellar pocket (Lacomble et al., 2010). In cells that have not managed to undergo rotation, the new flagellum may emerge from the same flagellar pocket collar, as is the case with TbPLK RNAi cells (Ikeda and de Graffenried, 2012). It appears likely that once the two flagella are threaded through the same FPC, the cell will not be able to generate a new pocket and the two flagella may be confined within the same membrane compartment, making their separation unlikely.
To our knowledge, our modification of TbPLK is the first time that the analogue-sensitive method has been used in the kinetoplastids. Kinases comprise approximately 2% of the sequenced kinetoplastid genomes, which argues that protein phosphorylation plays important and diverse roles in these organisms (Parsons et al., 2005). In novel kinases in which the gatekeeper residue can be easily identified and its mutation tolerated, the analogue-sensitive approach should provide a viable alternative to depletion by RNAi. Many of the kinetoplastid kinases are members of kinase families that are amenable to the analogue-sensitive strategy, such as the CDKs and MAPKs (Naula et al., 2005; Parsons et al., 2005; Morand et al., 2012). It is likely that many of these kinases could be studied using this method, allowing acute inhibition without concerns for off-target effects.
MATERIALS AND METHODS
Cell culture
All experiments were performed in the procyclic T. brucei brucei 427 strain. Cells were cultured at 27°C in SDM-79 medium supplemented with 7.5 μg/ml hemin and 20% heat-inactivated fetal calf serum (Sigma-Aldrich, St. Louis, MO). Cell growth was monitored using a particle counter (Z2 Coulter Counter; Beckmann Coulter, Brea, CA).
Sf9 and High Five insect cells were cultured according to the manufacturer's instructions (Growth and Maintenance of Insect Cell Lines; Invitrogen, Carlsbad CA).
TbPLK inhibitor
3MB-PP1 was originally obtained from Kevan Shokat (University of California, San Francisco, San Francisco, CA) and later purchased from Calbiochem (La Jolla, CA).
Antibodies
The anti-FAZ1 (L3B2) was provided by Keith Gull (University of Oxford, Oxford, United Kingdom). The mouse anti-TbCentrin4 monoclonal and rabbit anti-TbPLK polyclonal antibodies were reported previously (de Graffenried et al., 2008; Ikeda and de Graffenried, 2012). The rat monoclonal anti-TbPLK will be described in a subsequent publication (de Graffenried and Warren, unpublished data).
TbPLKas cell line construction
A knockout construct comprising the puromycin resistance gene flanked by 500 base pairs of TbPLK 5′ and 3′ untranslated regions (UTRs) was transfected into 427 cells, which were cloned and put under puromycin selection. Puromycin-resistant clones were screened by PCR to assure proper targeting of the puromycin cassette. One positive clone was chosen and transfected with an endogenous tagging construct that appends either Ty1 or YFP to the N-terminus of proteins under blasticidin resistance (Morriswood et al., 2009). This construct was directed against the TbPLK genomic loci using 500 base pairs of the TbPLK 5′ UTR and first 456 base pairs of the open reading frame. The portion of the open reading frame used for targeting contained the two point mutations (L118G and C57V) necessary to produce TbPLKas. The sequence around the L118G mutation included silent mutations that introduced an AvrII restriction site (nt 341-TTTATATCCTAGGTGAGAAGTGCAG-nt 366). After transfection, cells were cloned and selected with puromycin and blasticidin. Genomic DNA was isolated from doubly resistant clones, and PCR was performed to amplify an ∼800–base pair fragment of TbPLK using the primers 5′-TTCCGTTGCGGAAGGATGCTCG-3′ and 5′-TGTAGAAGAGTGTGGTAGGTGCTGTCGTCG-3′. This fragment contains both mutation sites. The PCR product was subjected to digestion with AvrII to determine whether both mutations were present. The fragment was then submitted for DNA sequencing.
Western blotting
Cells were washed once in phosphate-buffered saline (PBS) and then lysed directly in SDS–PAGE loading buffer. Then 3 × 106 cell equivalents were loaded per sample and fractionated by SDS–PAGE. Proteins were then transferred to nitrocellulose and probed with rat anti-TbPLK, followed by horseradish peroxidase–labeled secondary antibodies. Enhanced chemiluminescence and film were used for detection.
In vitro TbPLKas expression
The gatekeeper mutation (L118G) and the compensatory mutation (C57V) were introduced into N-terminal hexahistidine-tagged TbPLK (de Graffenried and Warren, unpublished data) using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) according to the manufacturer's instructions. This construct was cloned into pFastBac HT, and recombinant baculovirus was generated using the Bac-to-Bac Expression System (Invitrogen). Sf9 cells were transfected with the baculovirus DNA to produce virus, which was then used to infect additional Sf9 cells to produce high-titer virus. The obtained high-titer virus was then used to infect High Five cells, which were then grown for 60 h before lysis in lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 1% Triton X-100, 1% glycerol, 300 mM NaCl, and 15 mM imidazole). The recombinant TbPLK was purified using TALON metal affinity resin (Clontech, Mountain View, CA) according to the manufacturer's conditions.
Kinase assays
The kinase assay protocol was adapted from a previously published protocol (Hammarton et al., 2007). Purified TbPLK or TbPLKas were mixed with 10 μg of recombinant TbCentrin2 and different concentrations of 3MB-PP1 or vehicle control (DMSO) in kinase assay buffer (50 mM 3-(N-morpholino)propanesulfonic acid, pH 7.2, 20 mM MgCl2, 10 mM ethylene glycol tetraacetic acid, and 2 mM dithiothreitol). ATP, 20 μM, containing 0.5 μCi of [γ-32P]ATP was then added to the reactions, which were then incubated for 30 min at 30°C. SDS–PAGE loading buffer was then added, followed by boiling. Thirty percent of each reaction was fractionated using SDS–PAGE, then stained with GelCode (ThermoScientific, Waltham, MA) to visualize proteins. The gel was dried under vacuum and exposed to a phosphorimaging plate, followed by visualization with a Typhoon scanner (GE Healthcare, Piscataway, NJ). The resultant image file was quantified in ImageJ (National Institutes of Health, Bethesda, MD; 1.47h) using the “plot lanes” tool.
Statistics
All quantifications were generated from three biological replicates. For comparisons of groups where the outcome variable is categorical the chi-squared test was used. For comparisons with data that fit a normal distribution, the Student's t test (two tailed, equal variance, unpaired) was used. The p value and number of cells counted are given in the figure legends.
Immunofluorescence microscopy
Cells were taken from culture and washed once with PBS and then adhered onto coverslips before fixation in −20°C methanol for 15 min. Samples were rehydrated with PBS and blocked overnight at 4°C in 3% bovine serum albumin in PBS.
Primary antibody incubations (anti-FAZ1, diluted 1:200; anti-TbCentrin4, 1:400; rabbit anti-TbPLK, 1:200) were done in blocking buffer for 1 h at room temperature. Coverslips were then washed with PBS, blocked for 20 min, and incubated with the appropriate Alexa dye–conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. Cells were then washed three times in PBS and mounted using DAPI Fluoromount G (Southern Biotech, Birmingham, AL).
Images were acquired using an inverted microscope (Axio Observer Z1; Carl Zeiss MicroImaging, Jena, Germany) equipped with a PCO 1600 camera. Image processing was done using ImageJ and Photoshop (Adobe, San Jose, CA). For each condition and time point, 300 cells were analyzed in asynchronous cultures and 100 in synchronous ones.
Bilobe and FAZ length measurements
Bilobe and FAZ length measurements were done using the Freehand Line and Measure tools in ImageJ.
Electron microscopy
Negative-stain electron microscopy was performed as described previously (Höög et al., 2010; Ikeda and de Graffenried, 2012).
Double-cut elutriation
Cells were cultured in MEM (Life Technologies, Carlsbad, CA) supplemented with MEM nonessential amino acids (Life Technologies) and with 20% heat-inactivated fetal calf serum. Approximately 4 × 109 cells were collected for double-cut elutriation, which was performed as previously described (Archer et al., 2011), except for the interval between the first and second cuts, which was extended to 90 min. Synchronous cells were set back in culture in supplemented MEM, and growth was monitored for the next 10 h.
Flow cytometry analysis
Approximately 2 × 106 cells were harvested by centrifugation, washed once with PBS, and fixed by dropwise addition of ice-cold 70% ethanol/30% PBS while vortexing. Cells were kept at 4°C overnight, then collected by centrifugation and resuspended in 500 μl of PBS containing RNase A (10 μg/ml) and propidium iodide (10 μg/ml) and incubated at 37°C for 30 min. Cells were analyzed using a FACSCalibur cytometer (BD Biosciences, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Graham Warren for support and critical comments and the members of the Warren laboratory for comments on the manuscript. We also thank Stuart Archer (Zentrum für Molekulare Biologie, Heidelberg, Germany) and Cristina Purificato (Istituto Superiore di Sanità, Rome, Italy) for valuable advice on elutriation, Marisi Castañón for help with baculovirus expression, and Andras Aszodi for help with statistics. This work was supported by the Austrian Science Fund (FWF) by Grant P21550-B12 to Graham Warren and C.L.dG. and via the doctoral program Molecular Mechanisms of Cell Signaling (W1220-B09).
Abbreviations used:
- BB
basal body
- CDK1
cyclin-dependent kinase 1
- CDK2
cyclin-dependent kinase 2
- DMSO
dimethyl sulfoxide
- EM
electron microscopy
- FAZ
flagellum attachment zone
- FPC
flagellar pocket collar
- HU
hydroxyurea
- MtQ
microtubule quartet
- PLK
Polo-like kinase
- RNAi
RNA interference
- snFAZ
short new flagellum attachment zone
- TbPLK
Trypanosoma brucei Polo-like kinase
- TbPLKas
analogue-sensitive Trypanosoma brucei Polo-like kinase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0846) on February 27, 2013.
REFERENCES
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Archer SK, Inchaustegui D, Queiroz R, Clayton C. The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One. 2011;6:e18425. doi: 10.1371/journal.pone.0018425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfalvi G. Cell cycle synchronization of animal cells and nuclei by centrifugal elutriation. Nat Protoc. 2008;3:663–673. doi: 10.1038/nprot.2008.34. [DOI] [PubMed] [Google Scholar]
- Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Bonhivers M, Nowacki S, Landrein N, Robinson DR. Biogenesis of the trypanosome endo-exocytotic organelle is cytoskeleton mediated. PLoS Biol. 2008;6:e105. doi: 10.1371/journal.pbio.0060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouteille B, Oukem O, Bisser S, Dumas M. Treatment perspectives for human African trypanosomiasis. Fundam Clin Pharmacol. 2003;17:171–181. doi: 10.1046/j.1472-8206.2003.00167.x. [DOI] [PubMed] [Google Scholar]
- Burkard ME, Randall CL, Larochelle S, Zhang C, Shokat KM, Fisher RP, Jallepalli PV. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci USA. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury AR, Zhao Z, Englund PT. Effect of hydroxyurea on procyclic Trypanosoma brucei: an unconventional mechanism for achieving synchronous growth. Eukaryotic Cell. 2008;7:425–428. doi: 10.1128/EC.00369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The development and therapeutic potential of protein kinase inhibitors. Curr Opin Chem Biol. 1999;3:459–465. doi: 10.1016/S1367-5931(99)80067-2. [DOI] [PubMed] [Google Scholar]
- Crasta K, Lim HH, Giddings TH, Winey M, Surana U. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat Cell Biol. 2008;10:665–675. doi: 10.1038/ncb1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graffenried CL, Ho HH, Warren G. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J Cell Biol. 2008;181:431–438. doi: 10.1083/jcb.200708082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esson HJ, Morriswood B, Yavuz S, Vidilaseris K, Dong G, Warren G. Morphology of the trypanosome bilobe, a novel cytoskeletal structure. Eukaryotic Cell. 2012;11:761–772. doi: 10.1128/EC.05287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- Gadelha C, Rothery S, Morphew M, McIntosh JR, Severs NJ, Gull K. Membrane domains and flagellar pocket boundaries are influenced by the cytoskeleton in African trypanosomes. Proc Natl Acad Sci USA. 2009;106:17425–17430. doi: 10.1073/pnas.0909289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M, Parsons M. A Trypanosoma brucei gene family encoding protein kinases with catalytic domains structurally related to Nek1 and NIMA. Mol Biochem Parasitol. 1993;59:111–121. doi: 10.1016/0166-6851(93)90012-m. [DOI] [PubMed] [Google Scholar]
- García-Salcedo JA, Pérez-Morga D, Gijón P, Dilbeck V, Pays E, Nolan DP. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garske AL, Peters U, Cortesi AT, Perez JL, Shokat KM. Chemical genetic strategy for targeting protein kinases based on covalent complementarity. Proc Natl Acad Sci USA. 2011;108:15046–15052. doi: 10.1073/pnas.1111239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NS, et al. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- Gull K. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol. 1999;53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- Gull K. Host-parasite interactions and trypanosome morphogenesis: a flagellar pocketful of goodies. Curr Opin Microbiol. 2003;6:365–370. doi: 10.1016/s1369-5274(03)00092-4. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hammarton TC, Kramer S, Tetley L, Boshart M, Mottram JC. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol Microbiol. 2007;65:1229–1248. doi: 10.1111/j.1365-2958.2007.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höög JL, Gluenz E, Vaughan S, Gull K. Ultrastructural investigation methods for Trypanosoma brucei. Methods Cell Biol. 2010;96:175–196. doi: 10.1016/S0091-679X(10)96008-1. [DOI] [PubMed] [Google Scholar]
- Ikeda KN, de Graffenried CL. Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J Cell Sci. 2012;125:3173–3184. doi: 10.1242/jcs.101162. [DOI] [PubMed] [Google Scholar]
- Kohl L, Sherwin T, Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- Kumar P, Wang CC. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryotic Cell. 2006;5:92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J Cell Sci. 2009;122:1081–1090. doi: 10.1242/jcs.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei. J Cell Sci. 2010;123:2884–2891. doi: 10.1242/jcs.074161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount DJ, Barrett B, Donelson JE. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J Biol Chem. 2002;277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- Le Guellec R, Paris J, Couturier A, Roghi C, Philippe M. Cloning by differential screening of a Xenopus cDNA that encodes a kinesin-related protein. Mol Cell Biol. 1991;11:3395–3398. doi: 10.1128/mcb.11.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters J-M. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Li Z, Umeyama T, Li Z, Wang CC. Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryotic Cell. 2010;9:705–716. doi: 10.1128/EC.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand S, Renggli CK, Roditi I, Vassella E. MAP kinase kinase 1 (MKK1) is essential for transmission of Trypanosoma brucei by Glossina morsitans. Mol Biochem Parasitol. 2012;186:73–76. doi: 10.1016/j.molbiopara.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Morriswood B, He CY, Sealey-Cardona M, Yelinek J, Pypaert M, Warren G. The bilobe structure of Trypanosoma brucei contains a MORN-repeat protein. Mol Biochem Parasitol. 2009;167:95–103. doi: 10.1016/j.molbiopara.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Naula C, Parsons M, Mottram JC. Protein kinases as drug targets in trypanosomes and Leishmania. Biochim Biophys Acta. 2005;1754:151–159. doi: 10.1016/j.bbapap.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters U, Cherian J, Kim JH, Kwok BH, Kapoor TM. Probing cell-division phenotype space and Polo-like kinase function using small molecules. Nat Chem Biol. 2006;2:618–626. doi: 10.1038/nchembio826. [DOI] [PubMed] [Google Scholar]
- Preisinger C, Körner R, Wind M, Lehmann WD, Kopajtich R, Barr FA. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Selvapandiyan A, Kumar P, Morris JC, Salisbury JL, Wang CC, Nakhasi HL. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol Biol Cell. 2007;18:3290–3301. doi: 10.1091/mbc.E07-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- Shi J, Franklin JB, Yelinek JT, Ebersberger I, Warren G, He CY. Centrin4 coordinates cell and nuclear division in T. brucei. J Cell Sci. 2008;121:3062–3070. doi: 10.1242/jcs.030643. [DOI] [PubMed] [Google Scholar]
- Smith E, et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011;30:2233–2245. doi: 10.1038/emboj.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang CC. The structural basis of localizing polo-like kinase to the flagellum attachment zone in Trypanosoma brucei. PLoS One. 2011;6:e27303. doi: 10.1371/journal.pone.0027303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Pav S, White DM, Rogers S, Crane KM, Cywin CL, Brown ML, Pargellis CA. A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket. Nat Struct Biol. 1997;4:311–316. doi: 10.1038/nsb0497-311. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Mathieson T, Bantscheff M, Eberhard D, Grimaldi R, Miranda-Saavedra D, Wyatt P, Ferguson MAJ, Frearson J, Drewes G. Chemical proteomic analysis reveals the drugability of the kinome of Trypanosoma brucei. ACS Chem Biol. 2012;7:1858–1865. doi: 10.1021/cb300326z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S, Kohl L, Ngai I, Wheeler RJ, Gull K. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist. 2008;159:127–136. doi: 10.1016/j.protis.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Vickerman K. On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci. 1969;5:163–193. doi: 10.1242/jcs.5.1.163. [DOI] [PubMed] [Google Scholar]
- Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Liu Y, Li Z. Structure-function relationship of the Polo-like kinase in Trypanosoma brucei. J Cell Sci. 2012;125:1519–1530. doi: 10.1242/jcs.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kenski DM, Paulson JL, Bonshtien A, Sessa G, Cross JV, Templeton DJ, Shokat KM. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.