The activation of Chk1 in response to stalled replication forks involves a pathway containing ATR, TopBP1, Rad17, and Claspin. We show that the Mre11-Rad50-Nbs1 (MRN) complex also has an important role in this pathway that is distinct from its role in response to double-stranded DNA breaks. These studies reveal a novel insight into the functions of the MRN complex.
Abstract
The activation of Chk1 in response to stalled replication forks in Xenopus egg extracts involves a complex pathway containing ATM and Rad3-related (ATR), topoisomerase IIβ-binding protein 1 (TopBP1), Rad17, the Rad9-Hus1-Rad1 (9-1-1) complex, and Claspin. We have observed that egg extracts lacking the Mre11-Rad50-Nbs1 (MRN) complex show greatly, although not completely, reduced activation of Chk1 in response to replication blockages. Depletion of both Rad17 and MRN leads to a further, essentially complete, reduction in the activation of Chk1. Thus, Rad17 and MRN act in at least a partially additive manner in promoting activation of Chk1. There was not an obvious change in the binding of RPA, ATR, Rad17, or the 9-1-1 complex to chromatin in aphidicolin (APH)-treated, MRN-depleted extracts. However, there was a substantial reduction in the binding of TopBP1. In structure–function studies of the MRN complex, we found that the Mre11 subunit is necessary for the APH-induced activation of Chk1. Moreover, a nuclease-deficient mutant of Mre11 cannot substitute for wild-type Mre11 in this process. These results indicate that the MRN complex, in particular the nuclease activity of Mre11, plays an important role in the activation of Chk1 in response to stalled replication forks. These studies reveal a previously unknown property of the MRN complex in genomic stability.
INTRODUCTION
The cell cycle must occur in a properly regulated manner for the continual production of normal cells throughout the lifetime of an organism (Morgan, 1997). A key part of the cell cycle involves duplication of the genomic DNA. Any aberrations in this process must be detected promptly and rectified accurately. Otherwise, genetic abnormalities may accumulate over time and provoke detrimental effects, such as the development of cancer (Sancar et al., 2004).
Key aspects of DNA replication involve separation of the duplex into single-stranded DNA (ssDNA) and duplication of the resulting templates by DNA polymerases (Méndez and Stillman, 2003; Sclafani and Holzen, 2007). Because ssDNA is a precarious and vulnerable structure, cells regulate its generation and metabolism very precisely. Initial unwinding of the DNA is confined to limited regions known as origins of replication. Moreover, a cascade of proteins ensures that these origins are utilized in a correctly regulated manner. The culmination of this process involves activation of the replicative helicase by S-phase kinases (Labib, 2010). This helicase, known as the CMG complex, consists of the Cdc45, MCM, and GINS proteins. On initial unwinding of the duplex, the ssDNA-binding protein RPA associates with the newly fired origin and thereby facilitates recognition of the DNA by various polymerases.
Cells possess elaborate checkpoint mechanisms that monitor the fidelity of DNA replication through a multistage, combinatorial process (Sancar et al., 2004; Cimprich and Cortez, 2008; Ciccia and Elledge, 2010; Nam and Cortez, 2011). These mechanisms appear to detect multiple structural features of replication forks that have experienced various difficulties. For example, a complex of ATR and ATR-interacting protein (ATRIP), a master checkpoint regulatory kinase, docks onto RPA-coated ssDNA (Cortez et al., 2001; Zou and Elledge, 2003). Likewise, a trimeric protein clamp composed of Rad9, Hus1, and Rad1 (known as the 9-1-1 complex) recognizes junctions between ssDNA and double-stranded DNA (Ellison and Stillman, 2003; Parrilla-Castellar et al., 2004). A clamp loader containing the Rad17 protein facilitates the deposition of the 9-1-1 complex onto these junctions. Moreover, the checkpoint mediator protein Claspin associates with chromatin immediately after Cdc45 and appears to monitor some aspect of DNA unwinding (Lee et al., 2003).
Another pivotal step in the activation of Chk1 involves a protein called topoisomerase IIβ-binding protein 1 (TopBP1). Under conditions of genotoxic stress, TopBP1 associates with and directly activates ATR-ATRIP (Kumagai et al., 2006; Mordes et al., 2008). This function involves a discrete ATR-activating domain of TopBP1. Significantly, TopBP1 also has an important role in DNA replication by promoting the incorporation of Cdc45 into the replicative helicase (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003). These dual roles of TopBP1 place it in an excellent position to respond to conditions of replicative stress. In vertebrates, docking of the N-terminal region of TopBP1 containing BRCA1 carboxy-terminal (BRCT) domains 1 and 2 with the 9-1-1 complex is critical for the activation of ATR-ATRIP (Delacroix et al., 2007; Lee et al., 2007). Thereupon activated ATR-ATRIP phosphorylates and activates the checkpoint effector kinase Chk1 with the assistance of Claspin (Guo et al., 2000; Hekmat-Nejad et al., 2000; Liu et al., 2000; Kumagai et al., 2004). Chk1 serves as the workhorse of checkpoint response by delaying the cell cycle and controlling the replication apparatus while cells cope with DNA lesions (Perry and Kornbluth, 2007; Ge and Blow, 2010).
Double-stranded DNA breaks (DSBs) represent another type of DNA damage that evokes a different class of checkpoint response. At the apex of this response, early-acting regulators, such as the Mre11-Rad50-Nbs1 (MRN) complex and master kinase ataxia-telangiectasia mutated (ATM), collaborate to initiate a cascade of events (Lee and Paull, 2007; Kanaar and Wyman, 2008; Stracker and Petrini, 2011). For example, processing of DSBs to yield resected 3′ overhangs of ssDNA is an important step in DSB checkpoint and repair pathways (Symington and Gautier, 2011). This processing leads to the recruitment of RPA to the ssDNA regions, which promotes the sequential activation of ATR-ATRIP and Chk1 (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Yoo et al., 2007).
The MRN complex is an intricate biochemical machine (Lee and Paull, 2007; Williams et al., 2010; Stracker and Petrini, 2011). The Mre11 protein possesses both ssDNA endonuclease and 3′-5′ exonuclease activities. These nucleolytic functions appear to promote the resection of damaged DNA under various circumstances, but the full physiological significance of these activities is enigmatic. Rad50 is a member of the ATP-binding cassette family of ATPases and contains coiled-coil domains characteristic of structural maintenance of chromosomes (SMC) proteins. Rad50 possesses both ATPase and adenylate kinase activities. Overall, Rad50 is important for the conformational dynamics of the MRN complex in response to various DNA structures. Finally, Nbs1 is important for docking of the complex with other regulators (Williams et al., 2010; Stracker and Petrini, 2011). The N-terminal region of Nbs1 contains both a forkhead-associated (FHA) domain and tandem BRCT domains. These modules mediate various phosphorylation-dependent protein–protein interactions. Nbs1 also contains a C-terminal ATM-binding domain that promotes MRN-mediated activation of ATM.
Interestingly, the MRN complex associates with replicating chromatin under apparently unperturbed conditions (Costanzo et al., 2001; Mirzoeva and Petrini, 2003). These findings have suggested that the MRN complex plays a preventive role against accumulation of DSBs during replication. Also, several lines of evidence have indicated that the MRN complex localizes to stalled replication forks (Mirzoeva and Petrini, 2003; Robison et al., 2004; Olson et al., 2007; Tittel-Elmer et al., 2009). The role of the MRN complex under conditions of replicative stress was suggested to involve structural support for the replisome or regulation of ATR-dependent phosphorylation (Stiff et al., 2005; Olson et al., 2007; Tittel-Elmer et al., 2009).
In previous studies, we reported that the MRN complex collaborates with TopBP1 in response to DSBs in Xenopus egg extracts (Yoo et al., 2009). The MRN complex associates specifically with TopBP1 in an Nbs1-dependent manner and thereby recruits ATM to TopBP1. Thereupon ATM phosphorylates TopBP1 on Ser-1131. This phosphorylation enhances the affinity of TopBP1 for ATR-ATRIP, boosts the ATR-activating capacity of TopBP1, and leads to further ATR-catalyzed phosphorylation of Ser-1131 in a positive-feedback loop. In pursuing these studies, we noted that depletion of the MRN complex also compromised the activation of Chk1 in response to stalled replication forks. We found that this effect involves the nuclease activity of the Mre11 subunit of the complex. These results indicate that the MRN complex also has an important role in checkpoint signaling from stalled replication forks. Moreover, these studies reveal a novel insight into the biological significance of the nuclease activity of Mre11.
RESULTS
The MRN complex is necessary for full activation of Chk1 in response to stalled replication forks
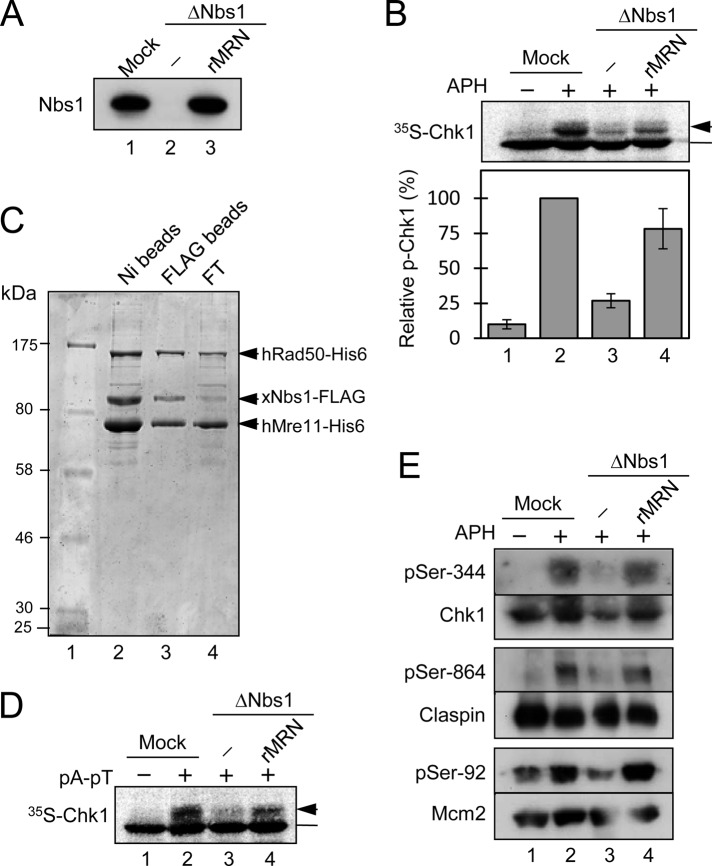
During our studies of Nbs1 in Xenopus egg extracts, we noted that depletion of this protein caused a very substantial, although not complete, reduction in the phosphorylation of Chk1 in response to stalled replication forks. As shown in Figure 1A, we prepared Nbs1-depleted egg extracts with affinity-purified anti-Nbs1 antibodies. This procedure effectively removed Nbs1 and codepleted essentially all of the Mre11 (see also Figure 5 later in the paper), which suggests that Xenopus MRN exists as a tight complex in egg extracts. Next we added to the extracts demembranated sperm chromatin and aphidicolin (APH), a DNA polymerase inhibitor that causes stalling of replication forks (Dasso and Newport, 1990; Kumagai et al., 1998). As a readout for checkpoint activation, we also included [35S]-radiolabeled Xenopus Chk1 and monitored its mobility in SDS gels (Figure 1B). In control mock-depleted extracts, APH elicited a characteristic reduction in the electrophoretic mobility of Chk1 that is indicative of phosphorylation (Kumagai et al., 1998). However, in Nbs1-depleted extracts, there was a large reduction in the phosphorylation of Chk1, typically around three-quarters of the total. Significantly, we could not ablate this phosphorylation completely, even through rigorous immunodepletion of Nbs1.
FIGURE 1:
The MRN complex is involved in the pathway that activates Chk1 in response to DNA replication blockages in Xenopus egg extracts. (A) Interphase egg extracts were subjected to an immunodepletion procedure with beads containing control antibodies (lane 1) or anti-Nbs1 antibodies (lanes 2 and 3). rMRN, prepared as described in (C), was added back to a level close to the endogenous amount (lane 3). Samples were immunoblotted for Nbs1. (B) Egg extracts from (A) were incubated with sperm chromatin and [35S]Chk1 in the absence (lane 1) or presence (lanes 2–4) of APH. Nuclear fractions from the extracts were processed for SDS–PAGE and phosphorimaging to assess phosphorylation of Chk1. Unshifted Chk1 was marked with a bar and slower-migrating band(s) with an arrow. Phosphorylation was expressed as a percentage of the level in mock-depleted extracts (lane 2). Results (mean ± SD) are from three independent experiments. (C) Expression and purification of rMRN from insect cells. Sf9 insect cells were coinfected with recombinant baculoviruses encoding human Mre11 (hMre11-His6), human Rad50 (hRad50-His6), and Xenopus Nbs1 (xNbs1-FLAG). Cell lysates were incubated with nickel (Ni) agarose, and bound proteins were eluted with imidazole buffer (lane 2). Next, this eluate was incubated with anti-FLAG antibody beads to achieve further purification. The flow-through fraction (FT) from this step is shown in lane 4. Finally, the bead-bound proteins were eluted with FLAG peptide (lane 3). Lane 1 contains molecular size markers. Samples were separated by SDS–PAGE and stained with Coomassie Brilliant Blue. (D) Egg extracts from (A) were incubated in the absence (lane 1) or presence (lanes 2–4) of pA-pT. [35S]-labeled Chk1 protein was included in extracts in order to monitor checkpoint activation. Samples were analyzed by SDS–PAGE and phosphorimaging. (E) Samples prepared as in (B) were analyzed for the phosphorylation status of various endogenous proteins by immunoblotting with antibodies that detect Ser-344 of Chk1, Ser-864 of Claspin, and Ser-92 of Mcm2. Samples were also immunoblotted for the corresponding protein antigens.
FIGURE 5:
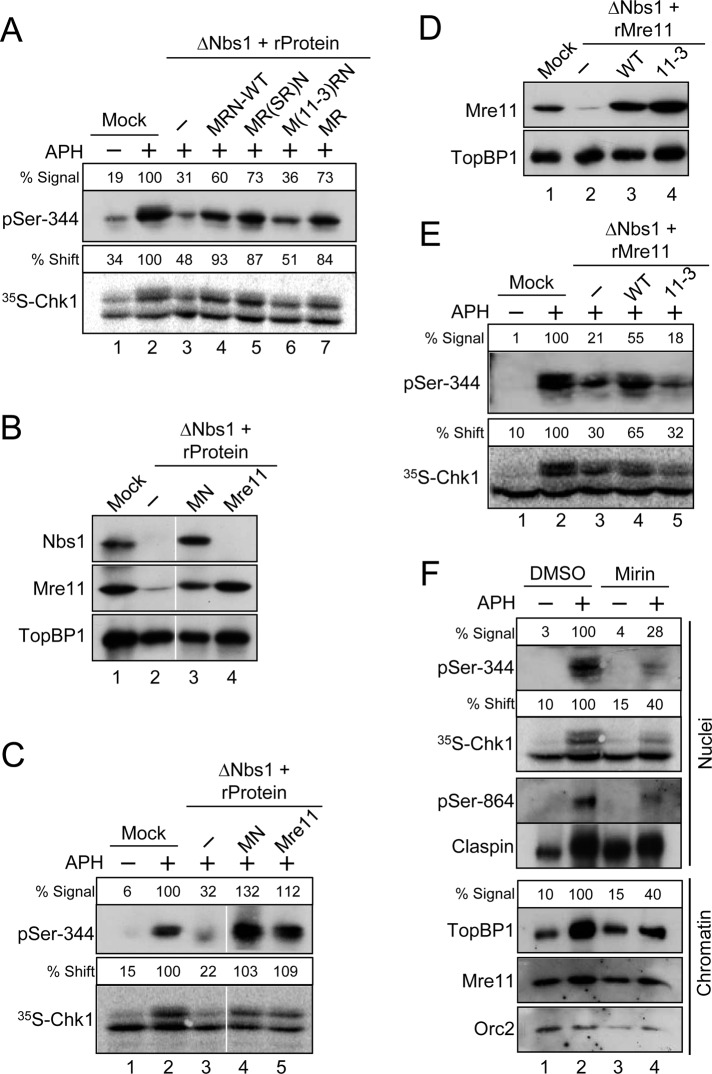
Mre11 is essential for the DNA replication checkpoint in egg extracts. (A) Mock-depleted (lanes 1 and 2) or Nbs1-depleted (lanes 3–7) extracts were supplemented with the following: control buffer (lanes 1–3), recombinant MRN complexes containing all wild-type subunits (lane 4), a Rad50-SR mutant subunit (lane 5), or an Mre11-3 mutant subunit (lane 6), and dimeric MR complex (lane 7). Extracts were incubated with [35S]Chk1 in the absence (lane 1) or presence (lanes 2–7) of APH. Nuclear fractions from the extracts were processed for immunoblotting with anti–pSer-344 Chk1 antibodies (top) or for phosphorimaging to detect radiolabeled Chk1 (bottom). Numbers above each lane denote quantitation of phosphorylation relative to mock-depleted, APH-treated extracts. (B) Mock-treated (lane 1) or Nbs1-depleted extracts (lanes 2–4) were supplemented with control buffer (lanes 1 and 2), recombinant Mre11-Nbs1 complex (MN, lane 3) or recombinant Mre11 protein (lane 4). Extracts were immunoblotted for the indicated proteins. (C) Extracts from (B) were incubated with [35S]Chk1 in the absence (lane 1) or presence (lanes 2–5) of APH. Nuclear fractions from the extracts were processed for immunoblotting with anti–pSer-344 Chk1 antibodies (top) or for phosphorimaging to detect radiolabeled Chk1 (bottom). (D) Mock-treated (lane 1) or Nbs1-depleted (lanes 2–4) extracts were supplemented with control buffer (lanes 1 and 2), recombinant wild-type Mre11 protein (lane 3), or the mutant Mre11-3 protein (lane 4). Extracts were immunoblotted for the indicated proteins. (E) Extracts from (D) were incubated with [35S]Chk1 in the absence (lane 1) or presence (lanes 2–5) of APH. Nuclear fractions from the extracts were processed for immunoblotting with anti–pSer-344 Chk1 antibodies (top) or for phosphorimaging to detect radiolabeled Chk1 (bottom). (F) Effects of mirin on the DNA replication checkpoint. Egg extracts were preincubated for 20 min in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 0.8 mM mirin. At this point, extracts were supplemented with sperm chromatin and [35S]Chk1 and incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of APH. Nuclear fractions (top four panels) were prepared and processed for phosphorimaging and immunoblotting with the indicated antibodies. Numbers above the lanes containing Chk1 denote phosphorylation relative to APH-containing extracts treated with DMSO. Chromatin fractions (bottom three panels) were prepared and immunoblotted for TopBP1, Mre11, and Orc2. The numbers above the lanes containing TopBP1 represent the binding of TopBP1 to chromatin relative to APH-containing extracts treated with DMSO.
Because the MRN complex is best known for its role in the response to DSBs, and APH does not induce DSBs in egg extracts under the conditions of our experiments (Li et al., 2004), we performed extensive studies to examine the specificity of this effect. For example, we asked whether a recombinant MRN complex could restore phosphorylation of Chk1 to Nbs1-depleted extracts. Toward this end, we coinfected Sf9 insect cells with baculoviruses encoding His-tagged human Mre11 (hMre11-His6), His-tagged human Rad50 (hRad50-His6), and FLAG-tagged Xenopus Nbs1 (xNbs1-FLAG). We purified the recombinant MRN complex (rMRN) by sequential chromatography on nickel agarose and anti-FLAG antibody beads. Coomassie Blue staining of such preparations showed a good stoichiometric ratio of the three subunits (Figure 1C).
To test the functionality of this complex, we added it to MRN-depleted extracts that were treated with the annealed double-stranded oligonucleotides poly(dA)70-poly(dT)70 (referred to hereafter as pA-pT). We have previously demonstrated that this DNA template is an effective inducer of the response to DSBs, a pathway that depends on the MRN complex (Kumagai and Dunphy, 2000; Yoo et al., 2009). As shown in Figure 1D, the rMRN complex was able to rescue the phosphorylation of Chk1 in response to pA-pT very effectively in MRN-depleted extracts.
Next we added the rMRN complex back to the APH-treated, MRN-depleted extracts (Figure 1, A and B). We observed that the phosphorylation of Chk1 was restored almost completely. We obtained similar results by monitoring the phosphorylation of Chk1 with anti-phosphopeptide antibodies that detect the ATR-catalyzed phosphorylation of Chk1 on Ser-344 (Figure 1E). These observations indicate that the reduced phosphorylation of Chk1 in the Nbs1-depleted extracts is specifically due to the absence of the MRN complex.
To probe these results further, we examined phosphorylation of other components in the ATR-dependent signaling pathway. We observed that the checkpoint-dependent phosphorylation of both Claspin on Ser-864 and Mcm2 on Ser-92 was significantly reduced in MRN-depleted extracts (Figure 1E). Both of these phosphorylations depend on ATR in APH-treated extracts (Kumagai and Dunphy, 2003; Yoo et al., 2004). Altogether, these results suggest that removal of the MRN complex substantially compromises the ATR-dependent response to stalled replication forks in egg extracts. Accordingly, inhibition of ATM with the small compound KU55933 did not compromise APH-induced activation of Chk1 in egg extracts under conditions in which it effectively blocked phosphorylation of Chk1 in response to DSBs created by the restriction enzyme PflMI (Supplemental Figure S1). These findings also raised the possibility that MRN might act at or near steps that regulate the activation of ATR.
The MRN complex associates with chromatin during the course of DNA replication
A number of proteins involved in the DNA replication checkpoint associate with the replication apparatus in a very specific manner and accumulate in elevated amounts at stalled replication forks (Lee et al., 2003; Petermann and Caldecott, 2006). Therefore, we sought to understand the chromatin-binding behavior of the MRN complex during normal and perturbed DNA replication in egg extracts. We incubated demembranated sperm chromatin in interphase egg extracts, removed aliquots of the extracts at various times, and examined the association of Nbs1 with chromatin (Figure 2A). Some Nbs1 initially associated quite rapidly with the chromatin (within 5 min). Thereafter its levels rose steadily for 30–40 min, by which time replication has typically undergone initiation. By 100 min, when replication usually nears completion, the amounts of Nbs1 on chromatin diminished to the low initial levels. In this system, nuclear envelope assembly, which is necessary for DNA replication, is normally complete by 30 min. Therefore these observations suggest that there are two modes for binding of Nbs1 to chromatin: an initial, membrane-independent binding and a slower binding that coincides with nuclear envelope assembly and DNA replication. When we added APH or the restriction enzyme PflMI to the extracts in order to induce the formation of stalled replication forks or DSBs, respectively, Nbs1 accumulated on chromatin at much higher levels than in the control extracts (Figure 2A; see also Figure 2B). Interestingly, Nbs1 underwent a signification modification in PflMI-treated extracts that was not evident in the APH-treated extracts.
FIGURE 2:
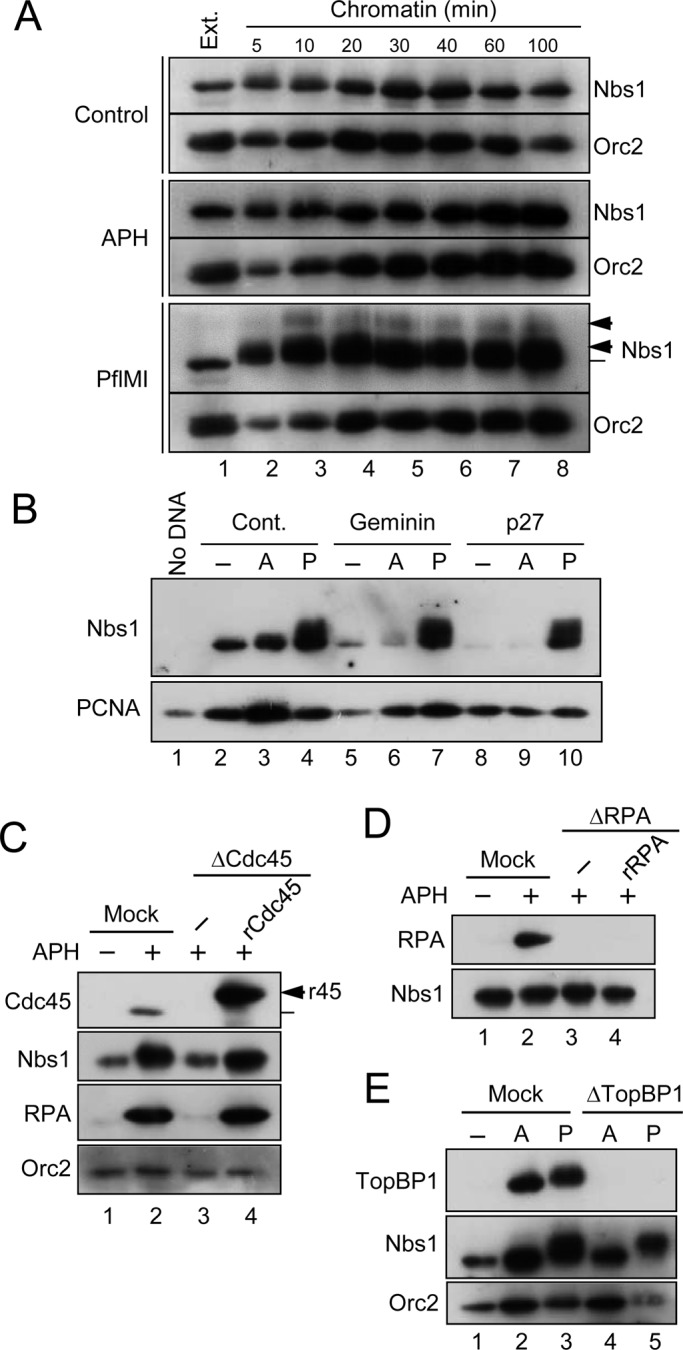
The MRN complex associates with chromatin in a replication-dependent manner. (A) Time course for binding of Nbs1 to chromatin. Interphase egg extracts treated with control buffer (top), APH (middle), or PflMI (bottom) were incubated with sperm chromatin (lanes 2–8). Chromatin fractions were prepared at the indicated times for immunoblotting with anti-Nbs1 and anti-Orc2 antibodies. Lane 1 depicts 1 μl of egg extract. (B) Extracts incubated with no checkpoint inducer (lanes 2, 5, and 8), APH (A; lanes 3, 6, and 9), or PflMI (P; lanes 4, 7, and 10) were treated with control buffer (lanes 2–4), Geminin (lanes 5–7), or p27 (lanes 8–10). Chromatin fractions were prepared and immunoblotted for Nbs1 (top) and PCNA (bottom). The sample for lane 1 lacked any added sperm chromatin. (C) Chromatin fractions were prepared from mock-treated (lanes 1 and 2) or Cdc45-depleted (lanes 3 and 4) extracts that had been incubated in the absence (lane 1) or presence (lanes 2–4) of APH. Recombinant Xenopus Cdc45 (r45) was added back in lane 4 (arrow). Samples were immunoblotted for the indicated proteins. (D) Chromatin fractions were prepared from mock-treated (lanes 1 and 2) or RPA-depleted (lanes 3 and 4) extracts that had been incubated in the absence (lane 1) or presence (lanes 2–4) of APH. Recombinant human RPA (rRPA) was added to the extract in lane 4. Samples were immunoblotted with antibodies against Xenopus Nbs1 and RPA70. Note that human RPA70 does not cross-react with the anti-Xenopus antibodies. (E) Chromatin fractions were prepared from mock-treated (lanes 1–3) or TopBP1-depleted (lanes 4–5) extracts that had been incubated in the absence (lane 1) or presence of either APH (A; lanes 2 and 4) or PflMI (P; lanes 3 and 5). Fractions were immunoblotted for the indicated proteins.
To understand more precisely how MRN associates with replicating chromatin, we blocked chromosomal DNA replication at various discrete stages. For example, we inhibited formation of the prereplicative complex (pre-RC) by treatment with geminin or by immunodepleting Cdc6 (Coleman et al., 1996; McGarry and Kirschner, 1998). We also blocked origin firing by adding the cyclin-dependent kinase inhibitor p27 (Walter and Newport, 2000). All of these treatments greatly reduced the binding of Nbs1 to chromatin in extracts that had been incubated in the absence or presence of APH (Figures 2B and S2). However, geminin and p27 did not affect the binding of Nbs1 to PflMI-treated chromatin, which suggests that the interactions of the MRN complex with replication forks and DSBs are distinct.
Because the key step triggered by Cdk2 during replication involves loading of Cdc45 onto the pre-RC complex (Mimura and Takisawa, 1998), we also examined the relationship between Cdc45 and Nbs1. For this purpose, we removed Cdc45 from the extracts with specific antibodies. As shown in Figure 2C, binding of Nbs1 to chromatin was substantially reduced in Cdc45-depleted extracts, and recombinant Cdc45 (rCdc45) could restore this binding to normal levels. The residual amount of Nbs1 on Cdc45-depleted chromatin likely reflects the nuclear membrane–independent mode of binding.
The formation of RPA-coated ssDNA at unwound replication origins depends on Cdc45 (Walter and Newport, 2000). Moreover, RPA is a key regulator in the ATR-dependent activation of Chk1 (Cimprich and Cortez, 2008; Nam and Cortez, 2011). Significantly, we observed that binding of Nbs1 to chromatin was unaffected in RPA-depleted extracts (Figure 2D). Finally, we examined the chromatin-binding properties of Nbs1 in the absence of TopBP1. The recruitment of Cdc45 to replication origins requires TopBP1 (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003). Consequently, the binding of Nbs1 to chromatin was greatly reduced in APH-treated extracts lacking TopBP1 (Figure 2E). Interestingly, depletion of TopBP1 also reduced the binding of Nbs1 to PflMI-digested chromatin, which suggests that TopBP1 may help to stabilize the presence of MRN at DSBs (Yoo et al., 2009).
Taken together, these results indicate that the MRN complex associates with the replication apparatus in a very precise manner. In particular, its binding depends on formation of the pre-RC and activation of the replicative helicase, but not on the formation of RPA-coated ssDNA.
Relationship between Rad17 and the MRN complex in the activation of Chk1
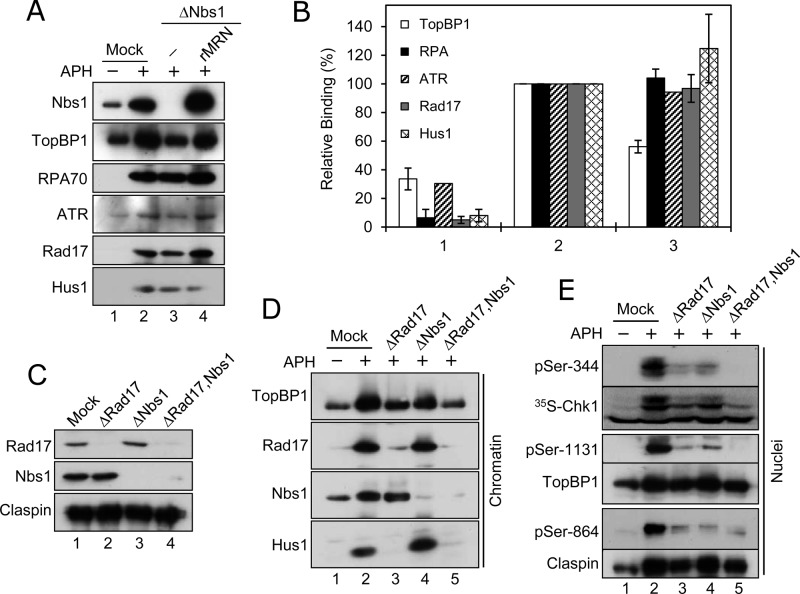
To probe the mechanism by which depletion of MRN compromises the checkpoint response to APH, we examined the binding of key checkpoint-relevant proteins to chromatin in Nbs1-depleted extracts. We could not detect an obvious change in the levels of RPA, ATR, Rad17, or Hus1 (a component of the 9-1-1 complex) on Nbs1-depleted chromatin in APH-treated extracts (Figure 3, A and B). Also, we could not observe any effect on the binding of Claspin to chromatin in the absence of Nbs1 (unpublished data). However, there was a significant reduction in the amount of TopBP1 on chromatin in Nbs1-depleted extracts (Figure 3, A and B). This effect appeared to be quite specific, because addition of recombinant MRN restored binding of TopBP1 efficiently.
FIGURE 3:
MRN and Rad17 regulate activation of Chk1 in an additive manner. (A) Mock-treated (lanes 1 and 2) or Nbs1-depleted (lanes 3 and 4) extracts were incubated in the absence (lane 1) or presence (lanes 2–4) of APH. Recombinant MRN was added back to the incubation shown in lane 4. Chromatin fractions were prepared and immunoblotted for the indicated proteins. (B) Quantitation of binding of proteins to chromatin in mock-treated extracts incubated in the absence (1) or presence (2) of APH and in Nbs1-depleted extracts incubated in the presence of APH (3). Values, compiled from two independent experiments, are expressed relative to mock-treated, APH-containing extracts. (C) Egg extracts were subjected to an immunodepletion procedure with control antibodies (lane 1), anti-Rad17 antibodies (lane 2), anti-Nbs1 antibodies (lane 3), or both anti-Rad17 and anti-Nbs1 antibodies (lane 4). Extracts were immunoblotted for the indicated proteins. (D) Extracts from (C) were incubated in the absence (lane 1) or presence (lanes 2–5) of APH. Chromatin fractions were immunoblotted for the indicated proteins. (E) Extracts from (C) were incubated with [35S]Chk1 in the absence (lane 1) or presence (lanes 2–5) of APH. Nuclear fractions from the extracts were processed for phosphorimaging to detect [35S]Chk1 (second panel from top) or for immunoblotting with antibodies that detect pSer-344 of Chk1, pSer-864 of Claspin, Claspin, pSer-1131 of TopBP1, and TopBP1, as indicated in the remaining panels.
As reported previously, depletion of Rad17 also reduces the binding of TopBP1 to APH-treated chromatin in egg extracts (Lee and Dunphy, 2010). Furthermore, the fact that Nbs1-depleted extracts still contain some residual phosphorylation of Chk1 is reminiscent of the behavior of Rad17-depleted extracts (Jones et al., 2003; Lee et al., 2003; Lee and Dunphy, 2010). We therefore examined the relationship between Rad17 and MRN in the checkpoint response to APH. For this purpose, we depleted egg extracts of Rad17 or Nbs1 or both (Figure 3C). As expected from previous studies, depletion of Rad17 abolished the binding of Hus1 to chromatin and substantially reduced the binding of TopBP1 (Figure 3D). Significantly, depletion of both Rad17 and Nbs1 resulted in less binding of TopBP1 to chromatin than depletion of Rad17 or Nbs1 alone. We also examined phosphorylation of Chk1 in these experiments. Depletion of Rad17 or Nbs1 alone reduced phosphorylation of Chk1 substantially but not totally (Figure 3E). Notably, however, depletion of both Rad17 and Nbs1 abolished phosphorylation almost completely.
We also examined the phosphorylation of other checkpoint proteins in extracts lacking either Rad17 or Nbs1 or both. For example, as shown in Figure 3E, we examined phosphorylation of TopBP1 on Ser-1131 (Hashimoto et al., 2006; Yoo et al., 2007). As was the case with phosphorylation of Chk1, phosphorylation of Ser-1131 on TopBP1 was greatly reduced in the absence of either Rad17 or Nbs1, but abolished completely in the absence of both proteins. We also assessed the phosphorylation of Claspin on Ser-864 (Figure 3E). This phosphorylation depends on ATR, but is carried out by a different kinase (Kumagai and Dunphy, 2003; Meng et al., 2011). We found that phosphorylation of Claspin on Ser-864, as detected with anti-phosphopeptide antibodies, was greatly reduced in the absence of either Rad17 or Nbs1. Although depletion of both Nbs1 and Rad17 did not appear to further reduce reactivity with the anti-phosphopeptide antibodies, the lack of both proteins did completely reverse the checkpoint-dependent mobility shift of Claspin. Taken together, these results indicate that both Rad17 and MRN contribute to the activation of Chk1 in an additive manner.
The Nbs1 subunit of the MRN complex is dispensable for the activation of Chk1 in response to stalled replication forks
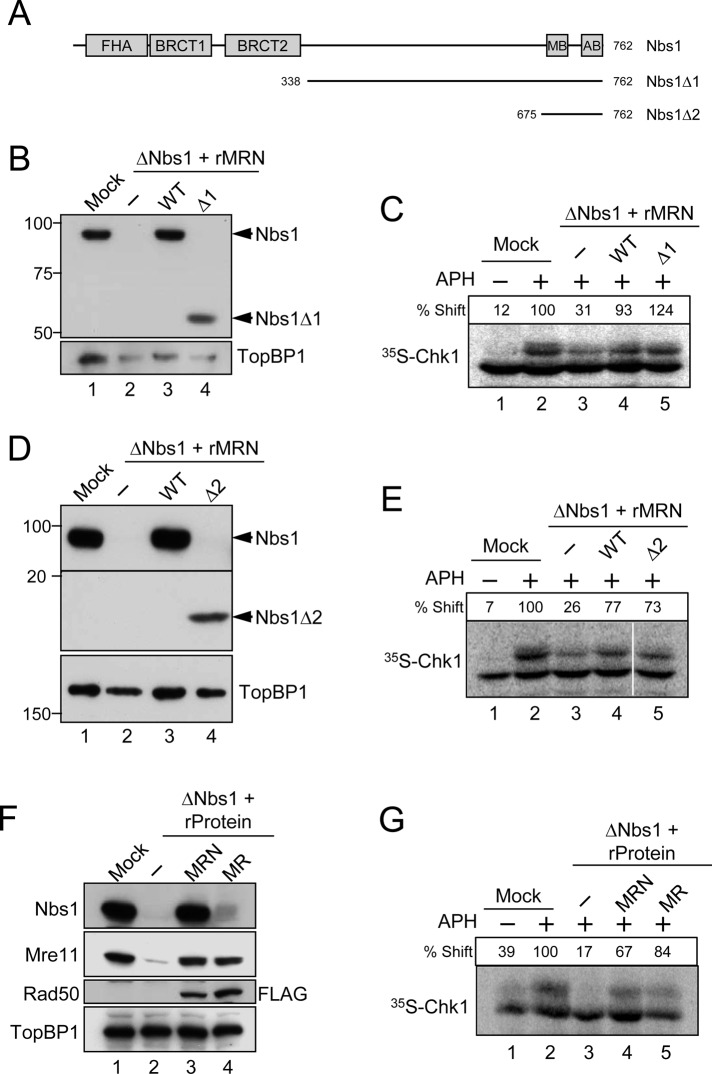
Next we sought to examine which subunit(s) of the MRN complex is necessary for the activation of Chk1 in response to APH. We initially focused on Nbs1, because we had previously studied the role of this protein in the TopBP1-dependent activation of Chk1 in response to DSBs (Yoo et al., 2009). As depicted in Figure 4A, Nbs1 contains a number of protein–protein interaction domains in its N-terminal region, including an FHA domain and tandem BRCT domains (Williams et al., 2010). The tandem BRCT domains are important for the association of Nbs1 with TopBP1 in egg extracts undergoing a checkpoint response to DSBs (Yoo et al., 2009). We prepared a deletion mutant of Nbs1 (designated as Δ1) lacking the entire N-terminal region that houses the FHA and BRCT domains. Next we expressed and purified a trimeric MRN complex that contains this mutated subunit (MRNΔ1; see Figure S3A). On addition to MRN-depleted extracts, we found that the mutant MRNΔ1 complex restored APH-induced phosphorylation of Chk1 as efficiently as the wild-type MRN complex (Figure 4, B and C).
FIGURE 4:
The Nbs1 component of the MRN complex is dispensable for the APH-induced activation of Chk1. (A) Domains of the Xenopus Nbs1 protein. The protein contains an FHA domain and two BRCT domains in its N-terminal region and an Mre11-binding domain (MB) and ATM-binding domain (AB) in its C-terminal region. Two deletion constructs of Nbs1 (∆1 and ∆2) are depicted. (B and D) Egg extracts were mock-treated with control antibodies (lane 1) or immunodepleted with anti-Nbs1 antibodies (lanes 2–4). Extracts were supplemented with control buffer (B and D, lanes 1 and 2), wild-type rMRN complex (B and D, lane 3), mutant rMRN∆1 complex (B, lane 4), or mutant rMRN∆2 complex (D, lane 4). Extracts were immunoblotted with anti-Nbs1 and anti-TopBP1 antibodies. (C and E) Extracts from (B) and (D), respectively, were incubated with [35S]Chk1 in the absence (lane 1) or presence (lanes 2–5) of APH. Nuclear fractions from the extracts were processed for phosphorimaging to assess phosphorylation-dependent shifting of Chk1. Numbers above each lane denote quantitation of phosphorylation relative to mock-depleted, APH-treated extracts. (F) Mock-treated (lane 1) or Nbs1-depleted extracts (lanes 2–4) were supplemented with control buffer (lanes 1 and 2), wild-type rMRN complex (lane 3), or recombinant MR complex lacking Nbs1 (lane 4). For these experiments, the Rad50 subunit in the trimeric MRN and dimeric MR complexes contained a C-terminal FLAG tag (see Materials and Methods). Extracts were immunoblotted for the indicated proteins. (G) Extracts in (F) were incubated with [35S]Chk1 in the absence (lane 1) or presence (lanes 2–5) of APH. Nuclear fractions from the extracts were processed for phosphorimaging.
To pursue these observations further, we made a more extreme deletion of Nbs1 that lacks most of the protein except for a C-terminal region containing the binding sites for Mre11 and ATM. We named this mutant Nbs1Δ2. We observed that a trimeric MRN complex containing this mutant (MRNΔ2) was also fully effective in rescuing phosphorylation of Chk1 in extracts lacking endogenous MRN (Figures 4, D and E, and S3B).
Finally, we prepared a dimeric Mre11-Rad50 (MR) complex that lacks Nbs1 completely (Figure S3C). Significantly, this complex was able to support phosphorylation of Chk1 as efficiently as the complete MRN complex (Figure 4, F and G). Thus Nbs1 appears to be dispensable for the APH-induced activation of Chk1 under these conditions.
Mre11 is necessary for the APH-dependent activation of Chk1
We proceeded to examine the other subunits of the complex (Mre11 and Rad50). In one approach, we analyzed MRN complexes in which these subunits contain mutations in key enzymatic activities. Rad50 contains an ATP-binding domain that possesses ATPase and adenylate kinase activities. Mre11 exhibits both ssDNA endonuclease and 3′-5′ exonuclease activities. We utilized a form of Rad50 in which a mutation of Ser-1202 to arginine in the signature motif of the ATP-binding domain impairs biological function (known as the Rad50-SR mutant). We also used a nuclease-deficient version of Mre11 in which His-129 and Asp-130 were changed to leucine and valine, respectively (known as the Mre11-3 mutant). Both the Rad50-SR and Mre11-3 mutants have been well characterized in biochemical studies (Arthur et al., 2004; Moncalian et al., 2004; Bhaskara et al., 2007).
We prepared trimeric MRN complexes containing either of these mutated subunits (see Figure S3D) and examined their capacities to rescue phosphorylation of Chk1 in APH-treated, Nbs1-depleted extracts. We observed that the complex containing the Rad50-SR protein rescued phosphorylation of Chk1 effectively, whereas the complex containing the Mre11-3 mutant was deficient (Figure 5A). Consistent with the results in Figure 4G, the dimeric MR complex also rescued phosphorylation well in these experiments. In contrast with the response to APH, the phosphorylation of Chk1 in response to pA-pT was not affected in Nbs1-depleted extracts that had been reconstituted with MRN complexes containing either the Rad50-SR or Mre11-3 proteins (Figure S4). However, phosphorylation of Chk1 in response to pA-pT was reduced in extracts containing the MR complex, which is consistent with the fact that Nbs1 plays a key role in the response to DSBs (You et al., 2005; Yoo et al., 2009). Finally, in order to address the possibility that some other function of Rad50 might be important for the response to APH, we also prepared a dimeric Mre11-Nbs1 complex (MN) that lacks Rad50 (Figure S3C). We observed that this complex also restored phosphorylation of Chk1 efficiently (Figure 5, B and C).
Next, we examined the isolated wild-type Mre11 and mutant Mre11-3 proteins (Figures 5, B–E, and S3E). Interestingly, the wild-type Mre11 protein itself could also rescue phosphorylation well. In comparison, the Mre11-3 protein was deficient. As another means to examine this issue, we utilized mirin, a chemical inhibitor of the nuclease activity of Mre11 (Dupré et al., 2008). We observed that addition of mirin to APH-treated extracts reduced phosphorylation of Chk1 to a similar extent as found in MRN-depleted extracts (Figure 5F). Similarly, mirin also diminished the phosphorylation of Claspin on Ser-864. Finally, we observed that treatment with mirin reduced the binding of TopBP1 to APH-treated chromatin in a manner similar to depletion of MRN (Figure 5F). Taken together, these results suggest that Mre11 is the key component of the MRN complex that promotes activation of Chk1 in response to APH. More specifically, the nuclease activity of Mre11 appears to be critical for this process.
DISCUSSION
In this study, we have uncovered a role for the MRN complex in the checkpoint response to stalled DNA replication forks in Xenopus egg extracts. In particular, we observed that depletion of the MRN complex from these extracts substantially reduced activation of the checkpoint effector kinase Chk1 in response to addition of the DNA polymerase inhibitor APH. This defect is specifically due to the absence of MRN because addition of a recombinant MRN complex to the depleted extracts effectively reversed the reduction. Nonetheless, the absence of MRN did not totally abolish the phosphorylation of Chk1. We and others have made a similar observation with Rad17-depleted extracts, which also display significant residual phosphorylation of Chk1 (Jones et al., 2003; Lee et al., 2003; Lee and Dunphy, 2010). Significantly, however, removal of both MRN and Rad17 does abolish the phosphorylation of Chk1. These observations suggest that both Rad17 and the MRN complex contribute to the activation of Chk1 in an additive manner. Because correct replication of the genome is extremely vital to the survival of the organism, this type of redundancy would be advantageous.
A specific role for MRN in a DNA replication checkpoint response is consistent with the fact that this complex associates in a highly specific manner with chromatin during S phase in egg extracts. Although we observe a small amount of replication-independent binding to chromatin, most of the binding coincides with DNA replication and depends on certain key replication proteins. In particular, this binding depends on Cdc45, a component of the replicative helicase. Interestingly, however, binding does not depend on RPA. Not only does RPA enable replication by the polymerases, it also serves as a critical regulator of checkpoint responses by recruiting ATR-ATRIP (Nam and Cortez, 2011). The chromatin-binding characteristics of MRN are generally similar to those of Claspin (Lee et al., 2003). Thus both of these important checkpoint regulators associate with the replication apparatus around the time of origin unwinding.
The absence of MRN also compromises a number of other ATR-dependent phosphorylations in egg extracts, including those of TopBP1, Claspin, and Mcm2. These phosphorylations occur upstream of Chk1. Moreover, inhibition of ATM in egg extracts does not affect APH-induced activation of Chk1. These observations suggest that there is some problem with the activation of ATR in MRN-depleted extracts. This defect most likely involves TopBP1 in some manner. Depletion of TopBP1 from egg extracts results in little, if any, phosphorylation of Chk1 in response to APH (Kumagai et al., 2006). For example, TopBP1-depleted extracts that have been reconstituted with a mutant of TopBP1 (W1138R) that is deficient for activation of ATR but proficient for DNA replication show minimal phosphorylation of Chk1. The implication is that both the MRN-dependent and Rad17-dependent pathways for activation of Chk1 ultimately funnel through TopBP1.
The lack of MRN also leads to a reduction in the amount of TopBP1 on APH-treated chromatin, which could help to explain the reduced activation of Chk1 under these circumstances. Although the pathway containing Rad17 and the 9-1-1 complex regulates the binding of TopBP1 to APH-stressed chromatin, the reduced binding of TopBP1 in the absence of MRN does not appear to involve the Rad17/9-1-1 pathway. As we have described, depletion of MRN from egg extracts does not affect binding of Rad17 or Hus1 to chromatin. Furthermore, depletion of Rad17 does not affect binding of MRN.
By performing structure–function studies on the MRN complex in egg extracts, we have observed that the nuclease activity of Mre11 is necessary for ATR-regulated activation of Chk1. Therefore an important mechanistic question is how this nuclease activity would contribute to the activation of Chk1 in response to incomplete replication. Stalled replication forks possess a number of features that would be absent from or less prominent in normal forks. These characteristics would include uncoupling of helicase from DNA polymerase activity, accumulation of ssDNA, repeated appearance of short primers on the DNA, and a buildup of numerous checkpoint and replication proteins on the chromatin (Lee et al., 2003; Sancar et al., 2004; Byun et al., 2005; Van et al., 2010; Nam and Cortez, 2011). Processing of primers by Mre11 may adjust the spacing between primers and/or length of primers for better activation of the checkpoint. Nonetheless, processing by Mre11 does not appear to result in extensive generation of RPA-coated ssDNA, since we do not observe a change in the total amount of RPA on chromatin in MRN-depleted extracts. Indeed, mechanisms exist to restrain excessive activity of Mre11 at stalled forks to avoid conditions that might lead to deterioration of forks (Hashimoto et al., 2010, 2012).
Previous studies have examined the role of the MRN complex in ATR-regulated pathways in mammalian cells. For example, Stiff et al. (2005) implicated Nbs1 in the phosphorylation of Chk1 upon treatment with hydroxyurea (HU). In addition, Zhong et al. (2005) observed that ablation of Rad50 compromises UV-induced phosphorylation of Chk1, but they did not examine replication inhibitors, such as HU and APH. Moreover, Olson et al. (2007) found that both Mre11 and Nbs1 are important for phosphorylation of Chk1 at low but not high doses of UV and HU. Although these studies are consistent with our general conclusion, there are some apparent inconsistencies about which subunit of MRN might be most critical. Ablation of one component of the MRN complex from living cells would be expected to affect the other components (Stewart et al., 1999; Olson et al., 2007). An advantage of the Xenopus egg–extract system is that one can remove proteins from extracts and examine immediate physiological consequences. In addition, the flexibility of this system has allowed us to add back either dimeric MR or monomeric Mre11 to MRN-depleted extracts and show that Mre11 is sufficient for APH-induced phosphorylation of Chk1. Our results do not necessarily rule out the possibility that Rad50 and Nbs1 could play some role that is not apparent under the conditions of our experiments.
Substitution of the Mre11 gene in mice with a mutant allele that lacks nuclease activity results in embryonic lethality (Buis et al., 2008). Mouse embryonic fibroblasts harboring this mutant could activate ATM normally but displayed more prolonged chromosome fragmentation following ionizing radiation (IR), sensitivity to both IR and APH, and defects in homologous recombination. These cells also exhibited reduced phosphorylation of Chk1 in response to UV. Although the authors did not test HU or APH, treatment with UV may elicit activation of Chk1, at least in part due to fork stalling in this context. If so, this observation would be consistent with our observations on APH-treated extracts containing nuclease-deficient Mre11.
The MRN complex and kinase ATM are two key regulatory components in checkpoint responses to DSBs (Lee and Paull, 2007; Stracker and Petrini, 2011). Cells with inherited hypomorphic alleles due to mutations in genes encoding Mre11, Rad50, or Nbs1 share a number of defects with cells from patients with ataxia-telangiectasia (A-T), which contain mutated ATM (Stracker and Petrini, 2011). For example, patients with Nijmegen breakage syndrome (mutated in Nbs1) or A-T like disorder (mutated in Mre11) show A-T abnormalities, such as hypersensitivity to IR, defects in IR-induced cell cycle checkpoints, and chromosome fragility (Shiloh, 2006; Stracker and Petrini, 2011). However, knockout of any one component of MRN in mice leads to embryonic lethality, whereas lack of ATM is not lethal (Stracker et al., 2004; Stracker and Petrini, 2011; Shiloh, 2006; Buis et al., 2008). The essentiality of the MRN complex during embryogenesis is reminiscent of the indispensability of ATR or Chk1 (Shiloh, 2006; Cimprich and Cortez, 2008). Collectively these findings have suggested that the MRN complex has essential protective roles in DNA metabolism besides activation of ATM. The observations in this study indicate that the MRN complex operates during a DNA replication checkpoint response without the involvement of ATM. Furthermore, the requirements for association of MRN with chromatin during a replication checkpoint response are clearly different from those during a checkpoint response to DSBs. Finally, unlike the indispensable involvement of Nbs1 in the response to DSBs, Nbs1 is not required for the replication checkpoint in egg extracts, according to our observations. Therefore the role of the MRN complex in the ATR-dependent activation of Chk1 appears to be largely unrelated to its role in the activation of ATM. This feature of the MRN complex is likely to be a significant aspect in its critical role during embryogenesis and maintenance of genomic integrity.
MATERIALS AND METHODS
Xenopus egg extracts and checkpoint assays
Cytostatic factor-arrested (M-phase) extracts from Xenopus eggs and demembranated sperm chromatin were prepared as described (Murray, 1991). Interphase egg extracts were prepared by addition of CaCl2 (0.4 mM) and cycloheximide (100 μg/ml). For induction of a checkpoint response with DNA oligonucleotides, annealed 70-mers of dA and dT, poly(dA)70-poly(dT)70, were incubated in extracts at a concentration of 50 μg/ml in the presence of 3 μM tautomycin for 60 min (Kumagai and Dunphy, 2000). For induction of the DNA replication checkpoint, sperm nuclei (3000/μl) were incubated in extracts with APH (50 μg/ml) for 100 min. For induction of chromosomal DSBs, sperm chromatin was incubated in extracts in the presence of the restriction endonuclease PflMI (0.02 U/μl) for 100 min. Total nuclear fractions and chromatin fractions were prepared as previously described (Lee et al., 2003). The Xenopus Chk1 protein was radiolabeled in vitro with the TnT system (Promega, Madison, WI) in the presence of TRAN35S-LABEL (MP Biomedicals, Santa Ana, CA).
Antibodies and immunodepletion
Affinity-purified rabbit polyclonal antibodies against Xenopus Nbs1, Chk1, Claspin, pSer-864 of Claspin, Orc2, Cdc6, Cdc45, RPA70, Rad17, TopBP1, pSer-1131 of TopBP1, pSer-92 of Mcm2, ATR, and Hus1 were described previously (Coleman et al., 1996; Kumagai and Dunphy, 2003; Kumagai et al., 2006; Lee et al., 2003; Lee and Dunphy, 2010; Yoo et al., 2007, 2009). For immunodepletion of Nbs1, anti-Nbs1 antibodies (50 μg) were coupled to protein A Dynabeads (Invitrogen, Irvine, CA). Half of the beads were incubated in interphase egg extracts (100 μl) for 40 min on ice. After removal of the beads, extracts were treated with the other half of the beads in the same manner. For mock depletions, control immunoglobulin G (Zymed Laboratories, South San Francisco, CA) was used in parallel. Immunodepletions of Cdc45, RPA, Rad17, and TopBP1 were also described before (Lee et al., 2003; Lee and Dunphy, 2010; Kumagai et al., 2006). Antisera and affinity-purified antibodies against Xenopus Mre11 were kindly provided by V. Costanzo (CRUK, United Kingdom) and H. Lindsay (Lancaster University, United Kingdom), respectively. Anti-Xenopus Mcm7 antibodies were the generous gift of J. Blow (University of Dundee, United Kingdom). We purchased antibodies against the following antigens from commercial sources: pSer-345 of human Chk1, equivalent to pSer-344 of Xenopus Chk1 (Cell Signaling Technology, Danvers, MA); and PCNA and Mcm2 from BD Biosciences PharMingen (San Diego, CA).
Preparation of recombinant proteins
Recombinant baculoviruses encoding His-tagged human Rad50 (hRad50-His6) and His-tagged human Mre11 (hMre11-His6) were kindly provided by T. Paull (University of Texas, Austin, TX; Lee and Paull, 2006). The baculovirus encoding recombinant Xenopus Nbs1 containing the FLAG epitope (xNbs1-FLAG) was previously described (Yoo et al., 2009). For some applications, pFastBac was engineered to contain sequences for the SV40 nuclear localization signal (NLS), an S-tag, and a 3X FLAG tag between the SphI and HindIII sites. This construct was designated as the pFastBac-NSF vector. The open reading frame of human Rad50 was amplified by PCR and subcloned between the RsrII and XhoI sites in this vector to generate pFB-hRad50-NSF. For preparation of the signature motif mutation of hRad50 (Moncalian et al., 2004; Bhaskara et al., 2007), Ser-1202 was substituted with arginine to make pFB-hRad50-SR-NSF. For preparation of the nuclease-deficient hMre11-3 mutant from pTP17 (Arthur et al., 2004), His-129 and Asp-130 were changed into leucine and valine, respectively, to make pFB-hMre11-3. The pFB-xNbs1-FLAG vector was used to generate the Δ1 and Δ2 mutants of Nbs1. The Δ2 version of xNbs1-FLAG also contained an ectopic SV40 NLS. Baculoviruses were generated with the Bac-to-Bac system (Invitrogen).
For preparation of recombinant proteins and protein complexes, (6–12) × 107 cells were infected with the appropriate combination of baculoviruses and incubated for 3 d. Cell pellets were briefly sonicated in 1 ml lysis buffer (10 mM HEPES-KOH, pH 7.6, 150 mM NaCl, 0.5% Triton X-100, and 5 mM ethylene glycol tetraacetic acid) containing 10 μg/ml each of pepstatin, chymostatin, and leupeptin. Cell lysates were clarified by centrifugation at 11,700 × g for 10 min. The supernatants were then incubated with nickel agarose beads for 1 h. After the beads were washed, proteins were eluted into 0.4 ml of elution buffer I (10 mM HEPES-KOH, pH 7.6, 150 mM NaCl, and 200 mM imidazole). The eluates were subsequently incubated with 15 μl of anti-FLAG M2 antibody beads (Sigma-Aldrich, St. Louis, MO) for 40 min. After washing, proteins were eluted into 50 μl of elution buffer II (10 mM HEPES-KOH, pH 7.6, 100 mM NaCl, and 10% glycerol) containing 100 μg/ml of 3X FLAG peptide (Sigma-Aldrich). Aliquots were frozen in liquid nitrogen for storage at −80°C. In the case of preparation of hMre11-His6, only a single purification with nickel beads was used. Preparation of recombinant Xenopus Cdc45-His6 and human RPA were described previously (Lee et al., 2003).
Quantitation
Radioactivity from [35S]Chk1 was detected with a PhosphorImager screen and quantitated with the ImageQuant program (Molecular Dynamics, Sunnyvale, CA). Immunoblot signals were detected on x-ray film using enhanced chemiluminescence (Thermo Fisher Scientific, Rockford, IL), and their scanned images were quantitated with the ImageJ program (National Institutes of Health [NIH], Bethesda, MD).
Miscellaneous
Preparation and use of Geminin and p27 were previously described (Lee et al., 2003). The ATM inhibitor KU55933 (Hickson et al., 2004) and the Mre11 inhibitor mirin were purchased from Tocris Biosciences (Bristol, United Kingdom) and Enzo Life Science (Farmingdale, NY), respectively. These chemicals were dissolved in dimethyl sulfoxide (DMSO). Typically, 0.25 mM KU55933 and 0.8 mM mirin were equilibrated in egg extracts for 20 min prior to the addition of sperm chromatin and checkpoint-inducing reagents.
Supplementary Material
Acknowledgments
We are grateful to laboratory members for helpful advice and comments on the manuscript. This work was supported by NIH grants GM070891 and GM043974 to W.G.D.
Abbreviations used:
- 9-1-1
Rad9-Hus1-Rad1
- APH
aphidicolin
- A-T
ataxia-telangiectasia
- ATM
ataxia-telangiectasia mutated
- ATR
ATM and Rad3-related
- ATRIP
ATR-interacting protein
- BRCT
BRCA1 carboxy-terminal
- DMSO
dimethyl sulfoxide
- DSB
double-stranded DNA break
- FHA
forkhead-associated
- HU
hydroxyurea
- IR
ionizing radiation
- MN
Mre11-Nbs1
- MR
Mre11-Rad50
- MRN
Mre11-Rad50-Nbs1
- NLS
nuclear localization signal
- pA-pT
poly(dA)70-poly(dT)70
- pre-RC
prereplicative complex
- rMRN
recombinant MRN
- ssDNA
single-stranded DNA
- TopBP1
topoisomerase IIβ-binding protein 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0025) on March 6, 2013.
REFERENCES
- Arthur LM, Gustausson K, Hopfner KP, Carson CT, Stracker TH, Karcher A, Felton D, Weitzman MD, Tainer J, Carney JP. Structural and functional analysis of Mre11-3. Nucleic Acids Res. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:231–243. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Blow JJ. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol. 2010;191:1285–1297. doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for ATR in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol. 2012;19:17–24. doi: 10.1038/nsmb.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You Z, Yee M, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Jones RE, Chapman JR, Puligilla C, Murray JM, Car AM, Ford CC, Lindsay HD. XRad17 is required for the activation of XChk1 but not XCds1 during checkpoint signaling in Xenopus. Mol Biol Cell. 2003;14:3898–3910. doi: 10.1091/mbc.E03-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Wyman C. DNA repair by the MRN complex: break it to make it. Cell. 2008;135:14–16. doi: 10.1016/j.cell.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat Cell Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Kim S-M, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J Biol Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells. Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–539. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Li W, Kim SM, Lee J, Dunphy WG. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J Cell Biol. 2004;165:801–812. doi: 10.1083/jcb.200402095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Méndez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- Meng Z, Capalbo L, Glover DM, Dunphy WG. Role for casein kinase 1 in the phosphorylation of Claspin on critical residues necessary for the activation of Chk1. Mol Biol Cell. 2011;22:2834–2847. doi: 10.1091/mbc.E11-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Lee AY, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–22952. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Perry JA, Kornbluth S. Cdc25 and Wee1: analogous opposites. Cell Div. 2007;2:12. doi: 10.1186/1747-1028-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Caldecott KW. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 2006;5:2203–2209. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P, Cobb JA. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 2009;28:1142–1156. doi: 10.1038/emboj.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van C, Yan S, Michael WM, Waga S, Cimprich KA. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol. 2010;189:233–246. doi: 10.1083/jcb.200909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Shevchenko A, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Bryson A, Eckersdorff M, Ferguson DO. Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum Mol Genet. 2005;14:2685–2693. doi: 10.1093/hmg/ddi302. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.