Talin-mediated integrin activation drives integrin-based adhesions. A simple binary switch—vinculin competitively displacing RIAM from talin—is found to play a central role in the maturation and evolving functions of integrin-based adhesions.
Abstract
Talin-mediated integrin activation drives integrin-based adhesions. Here we examine the roles of two proteins that induce talin–integrin interactions—vinculin and Rap1-GTP-interacting adaptor molecule (RIAM)—in the formation and maturation of integrin-based adhesions. RIAM-containing adhesions are primarily in the lamellipodium; RIAM is subsequently reduced in mature focal adhesions due to direct competition with vinculin for talin-binding sites. We show that vinculin binding to talin induces Rap1-independent association of talin with integrins and resulting integrin activation, in sharp contrast to Rap1-dependent RIAM-induced activation. Vinculin stabilizes adhesions, increasing their ability to transmit force, whereas RIAM played a critical role in lamellipodial protrusion. Thus displacement of RIAM by vinculin acts as a molecular switch that mediates the transition of integrin-based adhesions from drivers of lamellipodial protrusion to stable, force-bearing adhesions. Consequently changes in the abundance of two multiprotein modules within maturing adhesions, one regulated by Rap1 and one by tension, result in the temporal evolution of adhesion functions.
INTRODUCTION
Matrix adhesions, comprising a constellation of integrin-driven assemblies of structural and signaling components (Gardel et al., 2010; Hanein and Horwitz, 2012), play central roles in the biology of metazoans, controlling cell interactions with the extracellular matrix (ECM), migration, proliferation, and differentiation. In migrating cells, nascent adhesions (Choi et al., 2008) form at the cell anterior after ECM engagement of integrins and are associated with polymerizing actin. Some of the nascent adhesions disassemble within minutes (Choi et al., 2008), and the remainder grow and mature into focal complexes (∼0.5 μm) and then focal adhesions (FAs; 1–5 μm); this evolution is driven by force (Geiger et al., 2009). Adhesions are heterogeneous, exhibiting marked variability in composition, structure, and function (Gardel et al., 2010; Hanein and Horwitz, 2012), although they may have a modular nanostructure (Patla et al., 2010), and each module may play a distinct role in adhesion function.
Adhesions are initiated and maintained by integrin engagement of the ECM; however, integrins are expressed in a low-affinity state and require activation to bind ECM with high affinity (Shattil et al., 2010). The binding of talins to the integrin β-subunit cytoplasmic tail is a final common step in integrin activation in vitro, in vivo (Shattil et al., 2010), and in purified systems (Ye et al., 2010), and much has been learned about the details of talin-induced activation (Shattil et al., 2010). The entire spectrum of integrin-based adhesions contains talin (Gardel et al., 2010), which is required for adhesion formation (Brown et al., 2002); however, talin is autoinhibited, and most talin molecules are cytosolic (Beckerle et al., 1989) because integrin- and membrane-binding sites are masked by intramolecular interactions (Yan et al., 2001; Goksoy et al., 2008; Banno et al., 2012). Recent work has begun to decipher how the talin–integrin association is regulated to control integrin activation.
Rap GTPases are important regulators of integrin activation (Bos, 2005). Rap1-GTP-interacting adaptor molecule (RIAM) is a Rap1 effector implicated in activation (Lafuente et al., 2004). Lamellipodin is a RIAM paralogue that is also present in many cells (Krause et al., 2004), and MIG-10, the Caenorhabditis elegans orthologue, is involved in axonal path finding (Colo et al., 2012). RIAM overexpression induces β1 and β2 integrin–mediated cell adhesion, and RIAM depletion inhibits Rap1-dependent activation (Lafuente et al., 2004; Han et al., 2006), indicating that RIAM is downstream of Rap1. GTP-bound Rap1 induces formation of a complex containing RIAM and talin, resulting in talin recruitment to integrins and consequent activation (Han et al., 2006; Watanabe et al., 2008). Thus RIAM functions as a scaffold that in effect connects the membrane-targeting sequences in Ras GTPases to talin, thereby bringing talin to the plasma membrane, where it activates integrins.
Here we examine the relationships between two talin-binding proteins, vinculin and RIAM, in the formation and maturation of integrin-based adhesions. We report that vinculin and RIAM localize differentially: RIAM is most abundant at the plasma membrane in the lamellipodium rather than in mature FAs, sites where vinculin is most enriched. Furthermore, in maturing adhesions, RIAM initially precedes vinculin accumulation and is then subsequently reduced in mature adhesions due to direct competition between the two proteins for binding sites on talin. Vinculin deposition is known to stabilize adhesions and to increase their ability to transmit force (Ziegler et al., 2006; Humphries et al., 2007), whereas we report that RIAM promotes lamellipodial protrusion. These data delineate a simple biochemical molecular switch, the displacement of RIAM by vinculin, which leads to the transition of integrin-based adhesions from drivers of lamellipodial protrusion to stable FAs. In addition, we propose that these two proteins that induce talin binding to integrins lead to assembly of distinct multiprotein modules, one regulated by Rap1 and one by tension, and that change in the abundance of these modules within maturing adhesions contributes to the temporal evolution of their functions.
RESULTS
Vinculin and RIAM exhibit different localizations within adhesions
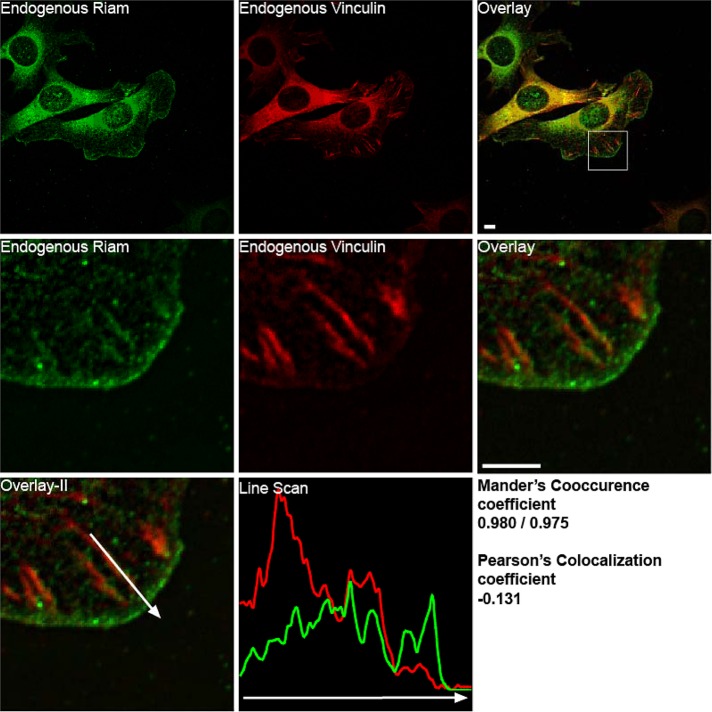
Previous work reported that RIAM is partially colocalized with talin at adhesion structures (Han et al., 2006) in the lamellipodium. We directly compared the localization of RIAM and vinculin in murine fibroblasts plated on fibrinogen or fibronectin. RIAM and vinculin were both present in the same focal complexes in the lamellipodium and in focal adhesion (Manders co-occurrence coefficient, 0.98/0.975; Manders et al., 1993). However within these structures, their abundance was not well correlated (Pearson r = –0.13; Adler and Parmryd, 2010). In particular, RIAM staining intensity was highest in the membrane and focal complexes in protrusions and was markedly reduced at the mature adhesions located nearer to the cell center. In contrast, vinculin staining was most concentrated in more centrally located mature FAs and notably less in the RIAM-enriched focal complexes in the protrusions (Figure 1 and Supplemental Figure S1).
FIGURE 1:
Localization of RIAM and vinculin in adhesion structures. Immunofluorescence localization of endogenous RIAM (green) and endogenous vinculin (red) in NIH 3T3 fibroblasts plated for 1 h on coverslips coated with fibrinogen (10 μg/ml) imaged with Olympus IX81 microscope with a spinning-disk head. The square inset shows the magnified region. A line scan, performed over an adhesion in the enlarged region, is also depicted (vinculin in red and RIAM in green). The Manders coefficient (for co-occurrence) and Pearson's coefficient were calculated for the enlarged region. Scale bar, 5 μm.
RIAM accumulation precedes vinculin within adhesions
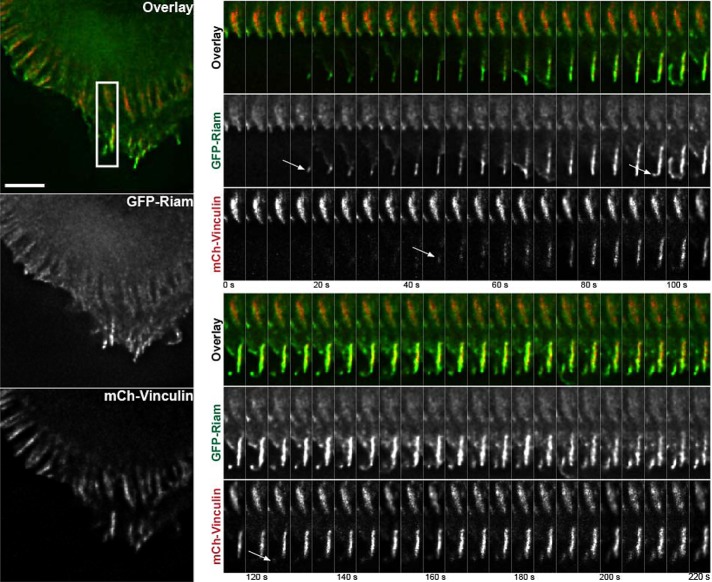
To assess the spatiotemporal patterns of localization of RIAM and vinculin in forming adhesions, we transiently transfected murine fibroblasts with plasmids encoding green fluorescent protein (GFP)–RIAM and mCherry-vinculin and examined the dynamics of each of these proteins by total internal reflectance microscopy (TIRFM). RIAM was abundant in the earliest detectable adhesions at the protruding membrane edge and in newly formed focal complexes near the edge. In contrast, vinculin was much less evident in the most membrane-proximal complexes but increased in the same adhesions as they matured (Figure 2 and Supplemental Video S1). To test whether the kinetics was integrin or cell type specific, we performed a similar experiment with CHO cells expressing α4β1 integrins (CHOα4) plated on fibronectin, as this integrin drives markedly increased protrusive activity (Liu et al., 1999), and a similar pattern of initial high RIAM abundance, followed by increasing vinculin abundance, was observed (Supplementa1 Figure S2 and Supplemental Video S2). As an alternative approach, these cells were adhered to the α4 integrin–binding CS1 fragment of fibronectin (Humphries et al., 1988) and stimulated by treatment with phorbol 12-myristate 13-acetate (Supplemental Figure S3 and Supplemental Video S3). Under these conditions, the adhesions matured by expanding retrograde from the membrane with little leading-edge retraction. RIAM preceded vinculin in these adhesions and was enriched in the membrane-proximal region, whereas vinculin was enriched in the membrane-distal region of the same adhesion (Supplemental Figure S3).
FIGURE 2:
RIAM precedes vinculin at newly formed adhesions. 3t3 cells were transiently transfected with cDNA encoding GFP-RIAM (green) and mCherry vinculin (red) and spread for 45 min on coverslips coated with 10 μg/ml fibrinogen before time-lapse imaging by TIRFM (Supplemental Video S1). The boxed region with active adhesion formation is enlarged and depicted as a montage of 5-s intervals for 220 s. The arrows point to the earliest observed recruitment of RIAM or vinculin to an adhesion. Scale bar, 5 μm.
RIAM drives lamellipodial membrane protrusions
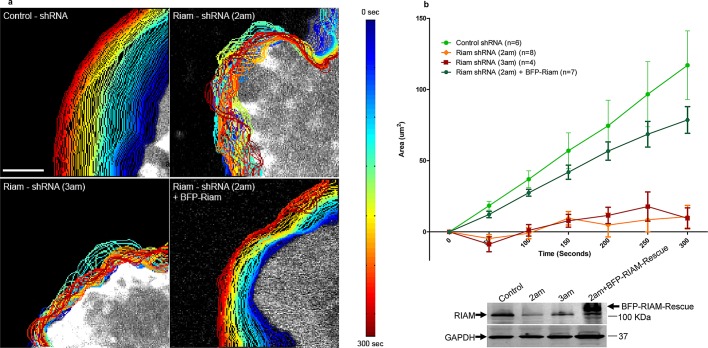
Vinculin causes the formation of stable adhesions that transmit force (Ziegler et al., 2006; Hanein and Horwitz, 2012); however, the functional role of RIAM-driven adhesions is not clear. We used vinculin-null fibroblasts to examine the role of RIAM in cellular behavior in the absence of vinculin-mediated events. The vinculin-null cells exhibited large lamellipodia, with multiple RIAM-containing adhesions throughout the lamellipodium (Supplemental Figure S4). These lamellipodia exhibited relatively uniform lamellipodial protrusions (Figure 3, a and b) and Supplemental Video S4). Silencing RIAM with two different shRNAs collapsed the lamellipodium; reexpression of siRNA-resistant RIAM restored lamellipodial protrusion (Figure 3, a and b). Similar effects were observed when RIAM was silenced in vinculin-replete murine fibroblasts (Supplemental Figure S5). Thus RIAM-driven adhesions are enriched in the lamellipodium, where they mediate protrusion.
FIGURE 3:
RIAM-based adhesions drive lamellipodial protrusion. (a) Vinculin-null MEFs were infected with lentiviruses encoding control shRNA, RIAM shRNA (2am), or RIAM shRNA (3am). These cells were then transfected with the membrane marker mCherry Ras-Caax either alone or in combination with cDNA encoding shRNA-resistant BFP-RIAM. The transfected cells were imaged at 5-s intervals for 5 min. An actively protruding part of the cell and cell edge was computationally detected as described in Materials and Methods, and the images of the edge at different times were superimposed. The time of imaging was indicated by the edge pseudocolor as depicted in the heat map to the right of the image. (b) The change in protrusion area over time in the four conditions depicted in (a). Each point depicts the mean ± SEM for the indicated number of cells. There was a statistically significant difference in control vs. 2am shRNA (p = 0.0062), control vs. 3am shRNA (p = 0.0079), and 2am shRNA + BFP-RIAM vs. 2am shRNA alone (p = 0.0067).
Vinculin competes with RIAM for binding to talin and displaces RIAM from adhesions
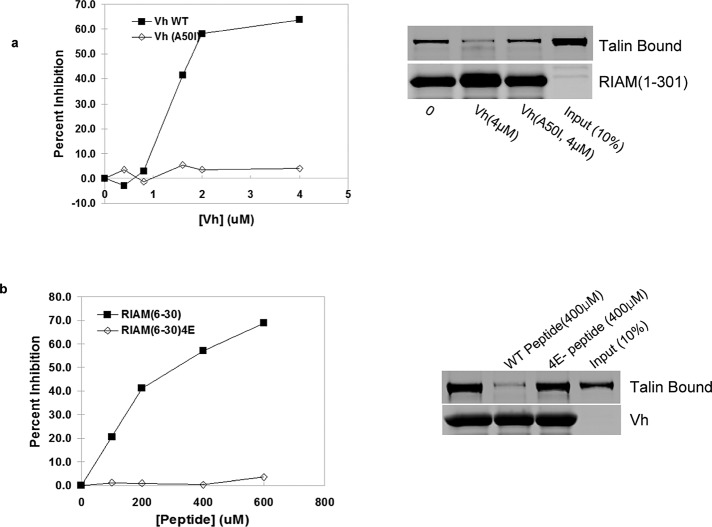
The N-terminal 30 residues of RIAM and the vinculin D1 domain (Vh; Gingras et al., 2010) bind directly to talin (Lee et al., 2009). The differential distribution of vinculin and RIAM described in the foregoing raised the possibility that the two proteins might compete with each other for binding to talin. Purified recombinant Vh inhibited the binding of talin to an N-terminal fragment of RIAM, whereas the Vh(A50I) mutant, which has a reduced affinity for talin (Bakolitsa et al., 2004), failed to do so (Figure 4a). Conversely, a synthetic RIAM peptide that contains the minimal talin-binding site (Lee et al., 2009) inhibited the binding of Vh to talin, whereas the talin binding–deficient RIAM-4E(6-30) mutant peptide (Lee et al., 2009) did not (Figure 4b). Thus RIAM and vinculin bind to talin in a mutually exclusive manner. Of importance, the IC50 of Vh was ∼100-fold lower than that of the RIAM peptide, suggesting that the favored reaction in cells would be displacement of RIAM by vinculin.
FIGURE 4:
Vinculin and RIAM compete for binding to talin. (a) Vh displaces RIAM (1–301) from talin. Increasing amounts of recombinant Vh or talin binding–defective His6-Vh (A50I) were incubated with the mixture of recombinant talin and GSH matrix-immobilized GST-RIAM (1–301). After washing, bound proteins were fractionated by SDS–PAGE, stained with Coomassie blue, and quantified by infrared scanning densitometry. (b) RIAM (6–30), comprising the minimal talin-binding fragment of RIAM, competes with vinculin for binding to talin. Increasing amounts of RIAM (6–30) peptide or the nonbinding RIAM-4E (6–30) peptide were added to the mixture of recombinant talin and GSH-bound GST-Vh. Bound proteins were separated by SDS–PAGE, stained by Coomassie blue, and quantified by infrared scanning densitometry. Mean values of duplicates are shown in (a) and (b).
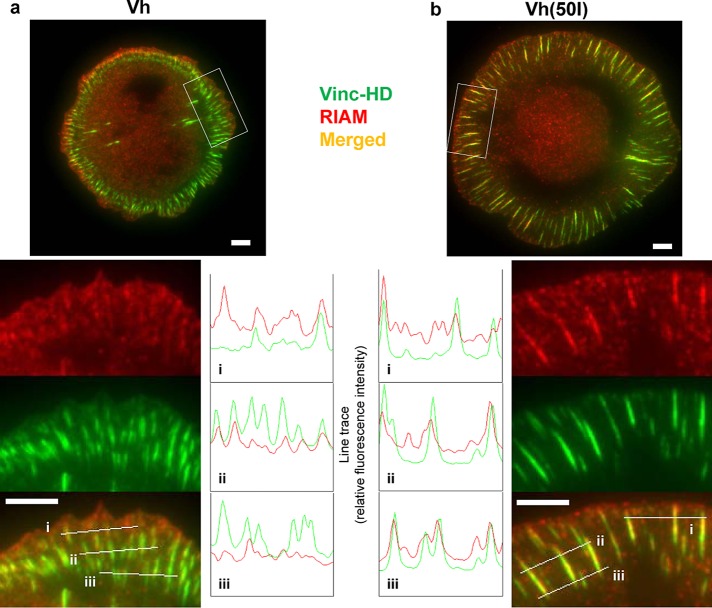
To determine whether vinculin could displace RIAM from adhesions in cells, we expressed mCherry Vh or Vh(A50I) in vinculin-null mouse embryo fibroblasts. mCherry-Vh was predominantly present in focal adhesions that contained little RIAM (Figure 5a), whereas RIAM was extensively colocalized in Vh(A50I)-containing adhesions (Figure 5b). The RIAM was present in small, vinculin-negative clusters at the cell edge in both Vh- and Vh(A50I)-transfected cells, indicating that a fraction of membrane-associated RIAM is not displaced by Vh. The failure of Vh to displace RIAM from the membrane edge is consistent with the ability of Rap1-RIAM to localize to membranes in the absence of talin (Han et al., 2006).
FIGURE 5:
Expression of Vh, a talin-binding fragment of vinculin, displaces RIAM from adhesion structures. Vin−/− MEF cells were transiently transfected with cDNA encoding (a) GFP-Vh (vinculin (2–258), D1 domain ) or (b) GFP-Vh(A50I) and adhered for 60 min to coverslips coated with 10 μg/ml fibronectin before fixation and staining for endogenous RIAM (red). Cells were imaged by TIRFM illumination set at 80-nm penetration depth. Line tracings parallel and at various distances to the cell edge indicate abundance of Vh relative to endogenous RIAM in adhesion structures. Scale bars, 5 μm.
Vinculin binding induces Rap1-independent talin–integrin association and integrin activation
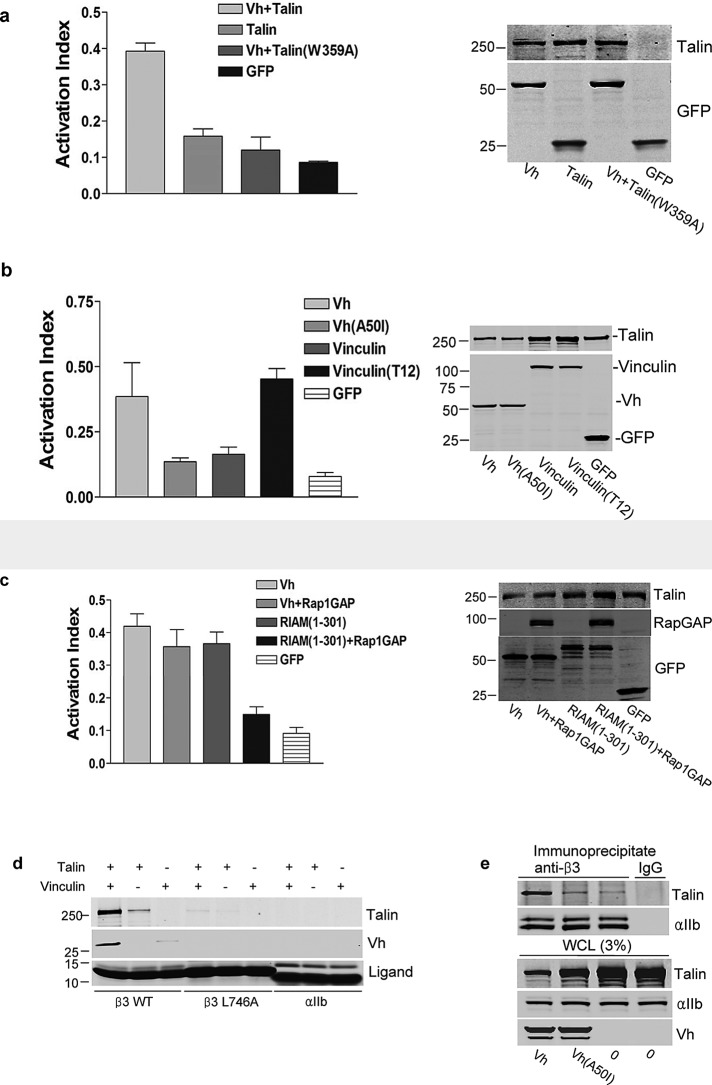
Rap1-RIAM can drive talin-mediated integrin activation (Lafuente et al., 2004; Han et al., 2006); it is unclear how integrin activation might be regulated if vinculin displaced RIAM during the processes of FA maturation. We tested whether vinculin has the ability to promote integrin activation by expressing Vh in combination with talin or talin(W359A), a mutant with reduced affinity for integrin β3 (Garcia-Alvarez et al., 2003), in cells expressing integrin αIIbβ3. Vh increased binding of PAC1, an activation-specific antibody, confirming (Humphries et al., 2007) that vinculin is able to activate integrins (Figure 6a). Activation depended on talin–integrin interaction, since Vh coexpression with talin(W359A) failed to enhance PAC1 binding (Figure 6a). Vinculin is autoinhibited via head–tail intramolecular interactions, which control talin binding (Ziegler et al., 2006). To test whether vinculin-mediated activation is regulated by its interaction with talin, we used two vinculin mutants to measure their ability to induce integrin activation: Vh(A50I), which has reduced capacity to bind talin, and vinculin T12, which is in an open conformation and constitutively binds talin (Cohen et al., 2005). When coexpressed with talin, Vh induced Pac1 binding and integrin activation, whereas Vh(A50I) did not (Figure 6b). Furthermore, Vinculin T12 greatly enhanced activation relative to full-length vinculin. These results indicate that the talin-binding capacity of vinculin, which is regulated by a vinculin head-to-tail interaction, is required for vinculin-induced integrin activation.
FIGURE 6:
Vinculin binding to talin induces integrin activation via a Rap1-independent mechanism. (a) Vh activates integrin αIIbβ3. CHO-A5 cells were cotransfected with cDNAs encoding HA-talin, integrin binding-defective HA-talin (W359A), or empty vector in combination with either GFP-Vh or GFP as indicated. After 24 h, cells were harvested, and the binding of activation-specific antibody PAC1 was measured by flow cytometry. Activation index (mean ± SE; n ≥ 3) was calculated as described in Materials and Methods. The difference between talin and talin(W359A) was statistically significant (p = 0.003). Expression of recombinant proteins was assessed by immunoblotting (right). (b) Vinculin-induced activation of integrins is dependent on vinculin–talin interaction. CHO-A5 cells were cotransfected with plasmids encoding HA-talin and GFP-Vh, GFP-Vh (A50I), GFP-vinculin full length, or GFP-vinculin full length (T12) as indicated. Integrin activation was measured by flow cytometry (mean ± SE; n ≥ 3), and protein expression was verified by immunoblotting as in (a). The differences between Vh and Vh(50I) (p = 0.039) and vinculin and vinculin T12 (p = 0.002) were statistically significant. (c) Vinculin-driven integrin activation is independent of Rap1 activity. CHO-A5 cells were transfected with cDNA encoding hemagglutinin (HA)-talin in combination with GFP-Vh or GFP-RIAM (1–301) in the presence or absence of Rap1GAP as indicated. Integrin activation was measured by flow cytometry (mean ± SE; n ≥ 3), and protein expression was confirmed by immunoblotting as in (a). The difference between RIAM (1–301) and RIAM (1–301) + Rap1GAP was statistically significant (p = 0.036), whereas that between Vh and Vh + Rap1GAP was not (p = 0.44). (d) Vinculin increases the interaction of talin with integrin β3 cytoplasmic tail in vitro. Immobilized integrin cytoplasmic tails were incubated in the presence or absence of recombinant talin in the presence or absence of recombinant Vh. Bound proteins were separated by SDS–PAGE and analyzed by immunoblotting. SDS–PAGE of integrin tails was stained by Coomassie blue to verify equivalent loading. (e) Vinculin increases talin binding to integrin β3 in vivo. CHO-A5 cells were transfected with cDNAs encoding HA-talin alone or in combination with GFP-Vh or GFP-Vh(A50I) and lysed, and β3 integrins were immunoprecipitated with anti-β3 antibody. Coimmunoprecipitated proteins and whole-cell lysate (WCL) were separated by SDS–PAGE and analyzed by immunoblotting for talin (α-HA), Vh (α-GFP), and integrin αIIb.
RIAM-mediated integrin activation is Rap1 GTPase dependent (Lafuente et al., 2004; Han et al., 2006). To test whether vinculin-stimulated integrin activation is controlled by Rap1, we examined the effect of Rap1GAP in cells transfected with cDNA encoding GFP-Vh. Rap1GAP had no effect on Vh-induced Pac1-binding (Figure 6c), indicating that vinculin-mediated integrin activation is independent of Rap1 activity. In contrast, expression of Rap1GAP prevented integrin activation induced by RIAM (Figure 6c). Thus, in contrast to RIAM-driven integrin activation, activation in mature, vinculin-driven adhesions is independent of Rap1 activity.
We next addressed the mechanism of integrin activation by vinculin. Talin binding to integrins is the final essential step in inducing integrin activation (Shattil et al., 2010). Recombinant talin was incubated with immobilized recombinant integrin β3 or β3(L746A), which has reduced affinity for talin (Garcia-Alvarez et al., 2003) cytoplasmic domain, in the absence or presence of purified recombinant Vh. The integrin β3–talin interaction was enhanced in the presence of Vh (Figure 6d). As expected, integrin β3(L746A) exhibited negligible interaction with talin in the presence or absence of Vh (Figure 6d). We examined the capacity of vinculin to induce talin-integrin interaction in cells by using coimmunoprecipitation. We cotransfected αIIbβ3-expressing cells with plasmids encoding HA-talin in combination with GFP-Vh or GFP-Vh(A50I) and immunoprecipitated integrin αIIbβ3 with an anti-β3 antibody. Coexpression with Vh markedly increased the quantity of talin that coimmunoprecipitated with the integrin (Figure 6e). In contrast, Vh(A50I) failed to increase the quantity of coimmunoprecipitated talin. Thus vinculin binding to talin increases the interaction of talin with the integrin β tail, leading to Rap1-independent integrin activation.
DISCUSSION
Assembly of adhesions from integrin activation modules
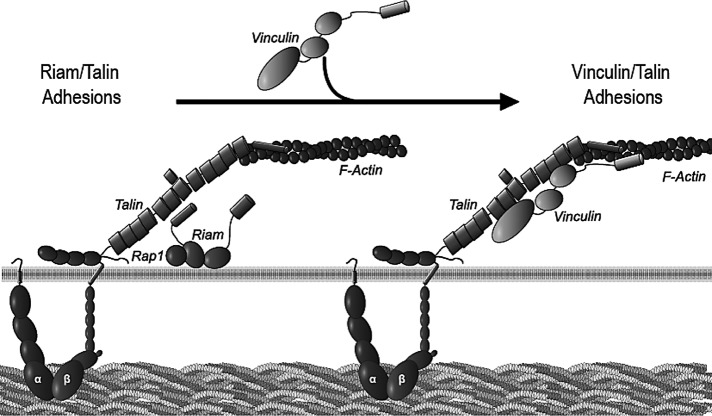
Both adhesion formation and integrin activation require talin–integrin interactions; we now delineate two distinct mechanisms to promote these events. Previous work (Han et al., 2006) showed that RIAM can form a quaternary complex with active Rap1, talin, and the integrin; hence this complex can be viewed as the core of a module in which the activated integrin is primed to enter into adhesions. We now find that vinculin promotes the talin–integrin association and integrin activation; hence the complex of vinculin, talin, and integrin forms the core of another module (Figure 7). Of importance, these modules differ in their abundance in different cellular locales, in regulation of their formation, and in their major functions. Specifically, we show that RIAM is most abundant at the cell edge and lamellipodium, where it supports protrusion. Protrusive activity is likely because of RIAM's capacity to increase actin polymerization, probably via its interactions with ENA/VASP (Lafuente et al., 2004). In contrast, vinculin is enriched in maturing FAs, which begin at the lamellum/lamellipodial border, where vinculin reinforces the ability of the adhesion to transmit and bear force (Ziegler et al., 2006). Furthermore, the biochemical mechanisms that regulate formation of these modules differ markedly: formation of the RIAM module requires GTP-Rap1 and is therefore subject to rapid modulation by regulators of Rap1 guanine nucleotide exchange. In contrast, we show that the vinculin module does not directly require GTP-Rap1 or indeed energy input once the vinculin's autoinhibition is overcome either structurally, as shown here, or potentially by a combination of tension (del Rio et al., 2009), PIP2 and F-actin (Bakolitsa et al., 2004), and paxillin phosphorylation (Pasapera et al., 2010) in cells. Thus, in mature FAs, the vinculin–talin–integrin complex can act as a stable link between the ECM and the actin cytoskeleton, whereas in the lamellipodium the RIAM module turns over rapidly under control of regulators of Rap1 and drives lamellipodial protrusion.
FIGURE 7:
Model comparing RIAM and vinculin-driven adhesion modules. RIAM induces integrin activation for focal complex formation during initial protrusion, a process that is dependent on Rap1 activity and drives membrane protrusion. Vinculin competes with RIAM for binding sites on talin, leading to stabilization of adhesions.
Temporal utilization of modules in adhesion maturation
The capacity of vinculin to displace RIAM from talin is a binary molecular switch that can account for some of the changes in composition of maturing adhesions. In particular, we show that RIAM deposition precedes vinculin in maturing adhesions and that vinculin directly competes with RIAM both in vitro and in vivo. This binary switch–mediated temporal progression in abundance of the two modules accounts for some of the changes in composition and function as adhesions mature into FAs. Of importance, formation of the RIAM module requires Rap activation; this is can be achieved by soluble agonist–stimulated activation of calcium and diacyl-glycerol–regulated guanine nucleotide exchange factors such as CalDAG-GEF1 (Gloerich and Bos, 2011). In matrix-driven mesenchymal cell migration small FAK-containing nascent adhesions (Lawson et al., 2012), signaling via CAS-Crk-C3G, may provide the activated Rap1 (Gloerich and Bos, 2011) at the leading edge that induces assembly of the RIAM modules. Thus adhesions can be viewed as an evolving assembly of modules; each module is composed of integrins in complex with specific elements that imbue each module with distinct assembly mechanisms and functional outputs.
There is great interest in defining the molecular composition, signaling properties, and ultimately the molecular structure of integrin-based cellular adhesions (Kanchanawong et al., 2010; Byron et al., 2011; Kuo et al., 2011). Critical barriers to progress are the insolubility and the heterogeneity of these adhesions. Their insolubility results in extensive contamination of purified adhesions with other membrane structures, ECM components, and other ECM-binding proteins. Heterogeneity is due to the continuum of evolving forms of adhesions, which exhibit marked variability in composition, structure, and properties. The modular nature of adhesions implies that efforts to isolate and determine the protein composition of such modules in soluble form and to visualize the behavior of individual modules in cells may provide valuable insights into the assembly and function of integrin-based adhesions.
MATERIALS AND METHODS
Plasmids and antibodies
cDNAs encoding hexahistidine (His6)-human talin and chicken vinculin were provided by Robert Liddington (Burnham Institute, La Jolla, CA). Plasmids encoding glutathione S-transferase (GST)– and GFP-vinculin and the vinculin D1 domain (amino acid residues 1–258; referred to here as Vh) were generated by PCR-based cloning. For construction of vinculin mutants (A50I and T12), GFP-vinculin wild-type cDNA was mutagenized by QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). The mutations were verified by DNA sequencing. Viral constructs in FG12 encoding RIAM short hairpin RNA (shRNA; 2am and 3am) and control have been previously described (Han et al., 2006). A cDNA encoding shRNA-resistant RIAM was generated by PCR and cloned into pTagBFP-N (Evrogen, Moscow, Russia). Viral constructs encoding RIAM-specific shRNA were cotransfected into 293T cells with gag-pol, VSV-G, and Rev constructs (Dull et al., 1998). Virus-containing supernatant was concentrated by ultrafiltration (EMD Millipore Corporation, Billerica, MA). mCherry-Vh and mCherry-talin were produced by replacing the GFP portion of cDNA encoding GFP-Vh or GFP-talin with mCherry. Monoclonal antibodies, anti-RIAM (Novus Biologicals, Littleton, CO), anti-talin 8d4 ascites (Sigma-Aldrich, St. Louis, MO), and anti-vinculin hVIN-1 ascites (Sigma-Aldrich) were purchased, and anti-RIAM rabbit polyclonal antibody was generously provided by V. Boussiotis (Dana-Farber Cancer Institute, Boston, MA). Fluorescein isothiocyanate, tetramethylrhodamine isothiocyanate (Santa Cruz Biotechnology, Santa Cruz, CA), and Alexa 647 (Invitrogen, Carlsbad, CA) conjugates of goat anti-mouse and anti-rabbit antibodies were purchased.
Cell lines and transfections
Vinculin-deficient and wild-type mouse embryonic fibroblasts (MEFs) were a gift from W. Ziegler (University of Leipzig, Leipzig, Germany). CHO cells stably expressing integrin αIIbβ3 (CHO-A5; Han et al., 2006) or integrin α4β1 (CHO-α4; Liu et al., 1999) and NIH 3T3 fibroblasts were cultured in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum, nonessential amino acids, and glutamine. Transient transfections were conducted using Lipofectamine-Plus Reagent (Invitrogen) or by FuGeneHD Transfection Reagent (Promega, Madison, WI), according to manufacturer's suggested protocols, or by nucleoporation with the Amaxa system (Lonza, Cologne, Germany), according to the manufacturer's recommended conditions.
Microscopy
For fixed sample imaging, cells were adhered to the indicated substrate for 1–2 h at 37°C, rinsed once in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, pH 7.4) and fixed with 3.7% formaldehyde in PBS for 10 min. Cells were permeabilized with 0.1% Triton X-100/PBS for 5 min and stained with primary antibodies in 0.01% TX-100/2% bovine serum albumin (BSA)/2% normal goat serum/PBS and counterstained with secondary antibodies. Coverslips were mounted on glass slides in Prolong Gold antifade reagent (Invitrogen) or Fluosave mounting reagent (Calbiochem, La Jolla, CA) for epifluorescence imaging. Epifluorescence images of cells were acquired with a 60× oil immersion objective on either an Eclipse TE2000U microscope (Nikon, Melville, NY) equipped with the appropriate excitation and emission filter sets (Semrock, Rochester, New York), 14-bit cooled charge-coupled device (CCD) camera (CoolSnap HQ, Photometrics, Tucson, AZ), electronically controlled z-stage (Ludl, Hawthorne, NY) and software controlled by QED InVivo (Media Cybernetics, Bethesda, MD) or an IX81 inverted microscope (Olympus, Tokyo, Japan) equipped with a CSU-X1-A1 spinning-disk head (Yokogawa, Tokyo, Japan), electron-multiplying CCD 14 bit 1K × 1K camera (Hamamatsu, Hamamatsu, Japan), four laser lines (405, 488, 560, 640 nm), and software controlled by Volocity (PerkinElmer, Waltham, MA). Some images as shown are maximal projections of images acquired at 0.1-μm z-section intervals and deconvolved using the 3D Blind Deconvolution algorithm of AutoQuantX (Media Cybernetics).
TIRFM imaging was performed on either an Olympus IX-81 cell-TIRF microscope equipped with a 60×/numerical aperture (NA) 1.49 Apo TIRF objective, electronically controlled shutters and filter wheels (Sutter, Novato, CA), a CoolSnap HQ2 cooled CCD camera (Photometrics) controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA), and solid-state 488-nm (10 mW) and 561-nm (20 mW) lasers (Melles Griot, Albuquerque, NM), or the Applied Precision (Issaquah, WA) OMX Super Resolution System (equipped with a ring TIRF module) with 100× objective (NA 1.40). Samples were typically illuminated at incident laser angles that achieve a calibrated evanescent wave penetration depth of ∼75–85 nm. For live-cell imaging, cells were seeded onto substrate-coated coverglass mounted on a Chamlide chamber (Quorum Technologies, Guelph, Canada) for 1 h before imaging. During imaging, cells were maintained in a Weather Station (Precision Control, Sammamish, WA) environmental control chamber (37°C, 5% CO2 and humidity) fitted onto the cell-TIRF microscope. For TIRFM imaging of fixed samples, cells fixed on coverslips were immunostained as described, then mounted on a Chamlide chamber and imaged while submerged in PBS. Additional postacquisition processing of images was performed using Volocity, ImageJ (National Institutes of Health, Bethesda, MD), and Photoshop (Adobe, San Jose, CA). The Manders coefficient and the Pearson coefficient were calculated by plug-ins for ImageJ from www.uhnresearch.ca/facilities/wcif/imagej/colour_analysis.htm and http://rsbweb.nih.gov/ij/plugins/colocalization-finder.html, respectively.
Cell-edge tracking
Live-cell imaging was performed on a Olympus IX81 inverted microscope with a 63×/NA 1.42 objective fitted with a Weather Station environmental control chamber maintained at 37°C and 5% CO2 and humidity. The instrument control and image acquisition were done by Volocity software. Subsequently, fluorescence images of cells transfected with mCherry Ras-Caax were input into a custom-built software package written in Matlab (MathWorks, Natick, MA; Machacek and Danuser, 2006) to detect cell edges. The overlaid mask for the cell edge at each time point was pseudocolored to indicate the time of acquisition. Similarly, the software calculated the changes in protrusion area with respect to time, which were displayed using Prism 5 (GraphPad Software, La Jolla, CA), and statistical comparisons used Student's t test.
Flow cytometry
As previously described (Han et al., 2006), CHO-A5 cells were transfected with the indicated plasmids and harvested after 24 h. Cells were incubated with PAC1, an integrin αIIbβ3–activation specific antibody. After being washed, the cells were incubated with R-phycoerythrin–conjugated goat anti-mouse immunoglobulin M secondary antibody. PAC1 binding to live and GFP-positive cells was then measured with two-color FACS Calibur (BD Biosciences, San Diego, CA), and the obtained data were analyzed with CellQuest software (BD Biosciences). Activation status of integrin was quantified by calculating the activation index, defined as (F − Fo)/(Fmax − Fo), where F is the median fluorescence intensity (MFI) of PAC1 binding, Fo is the MFI of PAC1 binding in the presence of a competitive inhibitor, integrilin (10 μM), and Fmax is the MFI of PAC1 binding in the presence of the activating antibody anti-LIBS6 (2 μM).
In vitro protein interaction assay and immunoprecipitation
Purification of recombinant His6-talin, His6- and GST-vinculin head (Vh), and GST-RIAM (1–301) for protein interaction assay was performed per manufacturer's protocols (GE Healthcare Biosciences, Piscataway, NJ). For competition experiments, progressively increasing amounts of purified His6-Vh or Vh(A50I) were added to a mixture of matrix-immobilized GST-RIAM and soluble His6-talin in binding buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, protease inhibitor [complete mini; Roche, Indianapolis, IN], E-64 [10 μM; Sigma-Aldrich], and phenylmethylsulfonyl fluoride [1 mM; Sigma-Aldrich] in ethanol with BSA [2 mg/ml]) and incubated at 4°C for 2 h. After washing, bound proteins were fractionated by SDS–PAGE and detected by Coomassie blue staining. RIAM peptides (6-30 WT and 6-30-4E mutant) used for the competition experiment have been described (Lee et al., 2009). RIAM peptide was added to the mixture of soluble His6-talin and immobilized GST-Vh in binding buffer and incubated at 4°C for 2 h. After washing, bound proteins were separated by SDS–PAGE and stained by Coomassie blue. For affinity chromatography with integrin cytoplasmic tails, recombinant biotinylated His6-Avi-tagged integrin tail (Arias-Salgado et al., 2005) was incubated with NeutrAvidin agarose (Thermo Scientific, Waltham, MA) overnight at 4ºC. After washing, integrin tail–coated beads were mixed with His6-talin in the presence or absence of His6-Vh in reaction buffer and incubated at 4°C for 2–4 h. After washing, bound proteins were fractionated by SDS–PAGE and detected by immunoblotting. Loading of affinity matrix with integrin tails was confirmed by Coomassie blue staining of SDS–PAGE–fractionated proteins. For immunoprecipitations, CHO-A5 cells were transfected with the designated plasmids and grown at 37°C. At 24 h after transfection, cell lysates were prepared in a lysis buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% Igepal CA-630), and the lysates were mixed with rabbit polyclonal anti–β3 integrin antibody for 4 h at 4°C. The immune complexes were captured by protein G–Sepharose (Invitrogen) and washed with lysis buffer. Bound proteins were eluted in SDS-sample buffer, fractionated by SDS–PAGE, and analyzed by immunoblotting.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health to M.H.G. P.A. was supported by a grant from a Post Doctoral Research Fellowship (PDF) from the Agency for Science, Technology and Research (A*STAR), Singapore.
Abbreviations used:
- BFP
blue fluorescent protein
- BSA
bovine serum albumin
- CalDAG-GEF
calcium and diacylglycerol binding guanine nucleotide exchange factor
- CAS-Crk-C3G
Crk-associated kinase/Crk/Crk-SH3 domain binding guanine nucleotide releasing factor
- ECM
extracellular matrix
- FA
focal adhesions
- FAK
focal adhesion kinase
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- MFI
median fluorescence intensity
- PIP2
phosphatidylinositol 4,5-bisphosphate
- Rap1
Ras-related protein 1
- Rap1GAP
Rap1 GTPase-activating protein
- Riam
Rap1 Interacting adaptor molecule
- shRNA
short hairpin RNA
- TIRFM
total internal reflection fluorescence microscopy
- VASP
vasodilator-stimulated phosphoprotein
- Vh
vinculin 1–258aa
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-09-0695) on March 6, 2013.
*These authors contributed equally to this work.
H.S.L., C.J.L., P.A., and C.C.L. performed experiments, analyzed data, and wrote the Materials and Methods section. M.H.G. conceived the project, directed experiments, analyzed data, and wrote the remainder of the article.
REFERENCES
- Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- Arias-Salgado EG, Lizano S, Shattil SJ, Ginsberg MH. Specification of the direction of adhesive signaling by the integrin beta cytoplasmic domain. J Biol Chem. 2005;280:29699. doi: 10.1074/jbc.M503508200. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Banno A, Goult BT, Lee H, Bate N, Critchley DR, Ginsberg MH. Subcellular localization of talin is regulated by inter-domain interactions. J Biol Chem. 2012;287:13799–13812. doi: 10.1074/jbc.M112.341214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC, Miller DE, Bertagnolli ME, Locke SJ. Activation-dependent redistribution of the adhesion plaque protein, talin, in intact human platelets. J Cell Biol. 1989;109:3333. doi: 10.1083/jcb.109.6.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Brown NH, Gregory SL, Rickoll WL, Fessler LI, Prout M, White RA, Fristrom JW. Talin is essential for integrin function in Drosophila. Dev Cell. 2002;3:569. doi: 10.1016/s1534-5807(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Byron A, Humphries JD, Bass MD, Knight D, Humphries MJ. Proteomic analysis of integrin adhesion complexes. Sci Signal. 2011;4:pt2. doi: 10.1126/scisignal.2001827. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J Biol Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- Colo GP, Lafuente EM, Teixido J. The MRL proteins: adapting cell adhesion, migration and growth. Eur J Cell Biol. 2012;91:861–868. doi: 10.1016/j.ejcb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez B, de Pereda JM, Calderwood DA, Critchley DR, Ulmer TS, Campbell ID, Ginsberg MH, Liddington RC. Structural determinants of integrin interaction with talin. Mol Cell. 2003;11:49. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, Emsley J, Barsukov IL, Roberts GC, Critchley DR. Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem. 2010;285:29577–29587. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011;21:615–623. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Goksoy E, Ma YQ, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16:1796. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Hanein D, Horwitz AR. The structure of cell-matrix adhesions: the new frontier. Curr Opin Cell Biol. 2012;24:134–140. doi: 10.1016/j.ceb.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MJ, Akiyama SK, Komoriya A, Olden K, Yamada KM. Neurite extension of chicken peripheral nervous system neurons on fibronectin: relative importance of specific adhesion sites in the central cell-binding domain and the alternatively spliced type III connecting segment. J Cell Biol. 1988;106:1289. doi: 10.1083/jcb.106.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7:571. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente EM, et al. RIAM, an Ena/VASP and profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7:585. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Lawson C, Lim ST, Uryu S, Chen XL, Calderwood DA, Schlaepfer DD. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J Cell Biol. 2012;196:223–232. doi: 10.1083/jcb.201108078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to Ras GTPase membrane-targeting sequences. J Biol Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Thomas SM, Woodside DG, Rose DM, Kiosses WB, Pfaff M, Ginsberg MH. Paxillin binding to alpha 4 integrins modifies integrin-dependent biological responses. Nature. 1999;402:676. doi: 10.1038/45264. [DOI] [PubMed] [Google Scholar]
- Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders MM, Verbeek PJ, Aten JA. Measurement of co-localization of objects in dual colour confocal images. J Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patla I, Volberg T, Elad N, Hirschfeld-Warneken V, Grashoff C, Fassler R, Spatz JP, Geiger B, Medalia O. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat Cell Biol. 2010;12:909–915. doi: 10.1038/ncb2095. [DOI] [PubMed] [Google Scholar]
- Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, Boussiotis VA, Ginsberg MH, Shattil SJ. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin aIIbb3. J Cell Biol. 2008;181:1211–1222. doi: 10.1083/jcb.200803094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Calderwood DA, Yaspan B, Ginsberg MH. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J Biol Chem. 2001;276:28164. doi: 10.1074/jbc.M104161200. [DOI] [PubMed] [Google Scholar]
- Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.