The endoplasmic reticulum (ER) membrane-bound transcription factors ATF6α and ATF6β mediate adjustment of chaperone levels to increased demands in the ER, which is essential for development of the notochord; the latter synthesizes and secretes large amounts of extracellular matrix proteins to serve as the body axis before formation of the vertebra.
Abstract
ATF6α and ATF6β are membrane-bound transcription factors activated by regulated intramembrane proteolysis in response to endoplasmic reticulum (ER) stress to induce various ER quality control proteins. ATF6α- and ATF6β single-knockout mice develop normally, but ATF6α/β double knockout causes embryonic lethality, the reason for which is unknown. Here we show in medaka fish that ATF6α is primarily responsible for transcriptional induction of the major ER chaperone BiP and that ATF6α/β double knockout, but not ATF6α- or ATF6β single knockout, causes embryonic lethality, as in mice. Analyses of ER stress reporters reveal that ER stress occurs physiologically during medaka early embryonic development, particularly in the brain, otic vesicle, and notochord, resulting in ATF6α- and ATF6β-mediated induction of BiP, and that knockdown of the α1 chain of type VIII collagen reduces such ER stress. The absence of transcriptional induction of several ER chaperones in ATF6α/β double knockout causes more profound ER stress and impaired notochord development, which is partially rescued by overexpression of BiP. Thus ATF6α/β-mediated adjustment of chaperone levels to increased demands in the ER is essential for development of the notochord, which synthesizes and secretes large amounts of extracellular matrix proteins to serve as the body axis before formation of the vertebra.
INTRODUCTION
Proteins must gain correct tertiary and quaternary structures to fulfill their functions as assigned by the genetic code. Folding and assembly of newly synthesized secretory and transmembrane proteins occur in the endoplasmic reticulum (ER), the first organelle they encounter after synthesis on the membrane-bound ribosomes, and are assisted or promoted by a number of molecular chaperones and folding enzymes (collectively termed ER chaperones hereafter) constitutively expressed quite abundantly (Bukau et al., 2006). This process of productive folding in the ER is indispensable to life of all eukaryotes, as evidenced by the fact that the major ER chaperone BiP is essential not only to the budding yeast Saccharomyces cerevisiae (Normington et al., 1989; Rose et al., 1989), but also in early mouse embryonic development (Luo et al., 2006). Of importance, proteins still unfolded or misfolded even after assistance from ER chaperones are targeted to the cytosol across the membrane for ubiquitin-dependent degradation by the proteasome, a series of processes collectively termed ER-associated degradation (ERAD; Smith et al., 2011). Thus these two mechanisms—productive folding and ERAD—ensure the quality of proteins that pass through the ER.
Furthermore, all eukaryotic cells have developed a way to adjust the expression levels of ER chaperones and ERAD components according to demands in the ER. Thus, when unfolded proteins accumulate in the ER, this ER stress signal is sensed by a transmembrane protein(s) in the ER and transmitted to the nucleus to induce the transcription of genes coding for ER chaperones and ERAD components, leading to maintenance of the homeostasis of the ER (Mori, 2000; Walter and Ron, 2011). This unfolded protein response (UPR) consists only of transcriptional control in yeast but of both transcriptional and translational controls in metazoans, as the number of ER stress sensors/transducers increased with evolution, from one (IRE1) in S. cerevisiae, to three (IRE1, PERK, and ATF6) in Caenorhabditis elegans and Drosophila melanogaster, to five (IRE1α, IRE1β, PERK, ATF6α, and ATF6β) in mammals (Mori, 2009). Yeast ER expresses IRE1, a type I transmembrane protein, for transcriptional control only (Cox et al., 1993; Mori et al., 1993), whereas PERK, a type I transmembrane protein that emerged in metazoan ER, is able to attenuate translation generally in response to ER stress to decrease the burden on the ER (Harding et al., 1999). Paradoxically, translation of the transcription factor ATF4 is induced under ER stress, resulting in transcriptional induction of its target genes, which are involved in resistance to oxidative stress; amino acid metabolism, including asparagine synthetase; and the proapoptotic transcription factor CHOP (Harding et al., 2000).
ER stress–induced activation of IRE1 results in unconventional splicing of mRNA encoding its downstream transcription factor, namely HAC1 mRNA in yeast and XBP1 mRNA in metazoans, leading to production of active transcription factors pHac1(S) and pXBP1(S), respectively. Transcriptional induction of ER chaperones and ERAD components in response to ER stress is mediated by IRE1 in yeast, worm, and fly cells (Mori, 2009). Of interest and importance, however, transcriptional induction of ER chaperones in response to ER stress is mediated by ATF6α but not by pXBP1(S) in mice (Wu et al., 2007; Yamamoto et al., 2007). ATF6α and ATF6β are constitutively synthesized as type II transmembrane proteins in the ER (Haze et al., 1999, 2001). On ER stress ATF6α and ATF6β are translocated to the Golgi apparatus to be cleaved sequentially by Site-1 protease and Site-2 protease, resulting in liberation of their cytosolic region, designated pATF6α(N) and pATF6β(N), respectively, from the membrane (Ye et al., 2000; Okada et al., 2003; Nadanaka et al., 2004). Because pATF6α(N) and pATF6β(N) contain all domains necessary for an active transcription factor, they enter the nucleus and activate transcription of a limited number of their target genes (Yoshida et al., 2000, 2001b). Because the transcriptional activator activity of pATF6α(N) is much higher than that of pATF6β(N), pATF6α(N) alone is necessary and sufficient for transcriptional induction of ER chaperones in response to ER stress, whereas pATF6α(N) heterodimerizes with pXBP1(S) to up-regulate genes coding for most ERAD components (Okada et al., 2002; Yamamoto et al., 2007; Adachi et al., 2008).
ATF6α and ATF6β single-knockout mice are viable and fertile and show no obvious phenotype unless ATF6α single-knockout mice are challenged with ER stress pharmacologically (Wu et al., 2007; Yamamoto et al., 2010; Egawa et al., 2011) or by other means (Wu et al., 2011; Gade et al., 2012; Usui et al., 2012). However, ATF6α and ATF6β double knockout causes embryonic lethality (Yamamoto et al., 2007). Nonetheless, we have not been able to figure out why ATF6α/β double-knockout mice die before birth, because we are unable to obtain any double-knockout embryos even at embryonic day 8.5. This inconvenience in the mouse system prompted us to take advantage of a fish system in which all developmental stages can be observed directly under the microscope. We chose medaka fish (Oryzias latipes) rather than zebrafish (Danio rerio) as a vertebrate model organism for UPR research because the genome project for medaka fish has been completed (Kasahara et al., 2007) and a reverse genetic approach has been established (Taniguchi et al., 2006), allowing us to identify knockout medaka deficient in various UPR mediators, and because versatile techniques such as morpholino-mediated gene knockdown and mRNA microinjection-mediated overexpression have been established. We previously showed, using a medaka embryonic cell line, that the medaka genome encodes five ER stress sensors/transducers (IRE1α, IRE1β, PERK, ATF6α, and ATF6β), as do mammals, and that three UPR signaling pathways are very well conserved between medaka and mammals (Ishikawa et al., 2011). Namely, XBP1 mRNA is spliced in response to ER stress, resulting in production of pXBP1(S); translation is generally attenuated in response to ER stress, resulting in induction of ATF4 and CHOP; and ATF6α and ATF6β are constitutively synthesized as transmembrane proteins and activated by ER stress–induced proteolysis.
In this article, we identify and characterize ATF6α- and ATF6β single-knockout medaka and then construct and characterize ATF6α and ATF6β double-knockout medaka to find out why ATF6α/β double knockout causes embryonic lethality in medaka.
RESULTS
Point mutation in BiP, as well as ATF6α and ATF6β double knockout, causes embryonic lethality in medaka
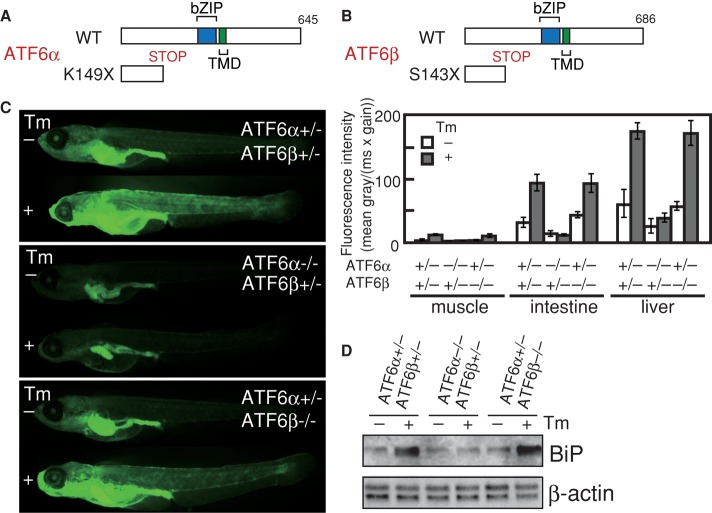
We used the targeting-induced local lesions in genomes (TILLING) method to identify ATF6α- and ATF6β-knockout medaka (Supplemental Figure S1). The N-terminally truncated fragment of ATF6α (K149X) or ATF6β (S143X) produced from the mutated allele must have lost its functionality, as its DNA-binding and transmembrane domains are excluded (Figure 1, A and B). Because both ATF6α and ATF6β single knockouts were born and developed normally, we obtained ATF6α/β double-hetero, ATF6α single-knockout, and ATF6β single-knockout medaka, each carrying the enhanced green fluorescent protein (EGFP) gene under the control of BiP promoter (the PBiP-EGFP reporter gene; see legend to Supplemental Figure S1). In ATF6α/β double hetero, EGFP was ubiquitously expressed, with particularly high expression in the liver and gut, and fluorescence intensity was significantly enhanced by treatment with tunicamycin, which evokes ER stress by inhibiting protein N-glycosylation (Figure 1C), as we previously reported for wild-type (ATF6α+/+ ATF6β+/+) fish (Ishikawa et al., 2011). ATF6β single knockout showed the same phenotype as ATF6α/β double hetero. In marked contrast, constitutive EGFP expression was diminished and tunicamycin treatment did not enhance EGFP expression in ATF6α single knockout. Northern blot analysis of fishes at 1 d posthatching (dph) also showed defective induction of BiP mRNA by tunicamycin treatment in ATF6α single knockout but not in ATF6β single knockout (Figure 1D). These results demonstrated that ATF6α plays a major role in the transcriptional induction of BiP in response to ER stress in medaka, as in mice (Wu et al., 2007; Yamamoto et al., 2007).
FIGURE 1:
Effect of deleting ATF6α or ATF6β on the induction of BiP in response to ER stress. (A) Schematic representation of wild-type and K149X-mutant ATF6α proteins. (B) Schematic representation of wild-type and S143X-mutant ATF6β proteins. (C) ATF6α/β double hetero (ATF6α+/−, ATF6β+/−), ATF6α single knockout (ATF6α−/−, ATF6β+/−), and ATF6β single knockout (ATF6α+/−, ATF6β−/−), each carrying the PBiP-EGFP reporter gene, were grown immediately after hatching in the presence (+) or absence (−) of 2 μg/ml tunicamycin (Tm) for 24 h and then analyzed for EGFP expression by fluorescence microscopy. Fluorescence intensity in muscle, intestine, and liver was determined (n = 3), and means with SDs (error bars) are shown on the right. (D) ATF6α/β double hetero, ATF6α single knockout, and ATF6β single knockout at 1 dph were untreated (–) or treated (+) with 2 μg/ml tunicamycin (Tm) for 8 h. Total RNA was prepared and analyzed by Northern blot hybridization using a digoxigenin-labeled probe specific to BiP or β-actin. The β-actin band becomes doublet from 5 dpf.
We intended to determine the importance of BiP's chaperone function, as well as its expression level for medaka development. Using the TILLING method, we obtained an interesting missense (S38P) mutation in the BiP coding region (see legend to Supplemental Figure S2). Because S38 is present in a region highly conserved among various species and close to the nucleotide-binding site, the S38P mutation was expected to inactivate BiP by inhibiting its nucleotide-binding activity or ATPase activity (Figure 2A). Genotyping revealed that no BiP(S38P) homozygotes were present among 32 fishes hatched upon in-crossing BiP(S38P) heterozygotes (Figure 2B). Thus the chaperone function of BiP is required for early embryonic development in medaka, as in mice (Luo et al., 2006). Similarly, when ATF6α single knockout was crossed with ATF6β single knockout, a total of 44 fishes were hatched, none of which possessed the genotype of ATF6α/β double knockout (Figure 2C). Thus complete loss of ATF6 function causes embryonic lethality in medaka, as in mice (Yamamoto et al., 2007).
FIGURE 2:
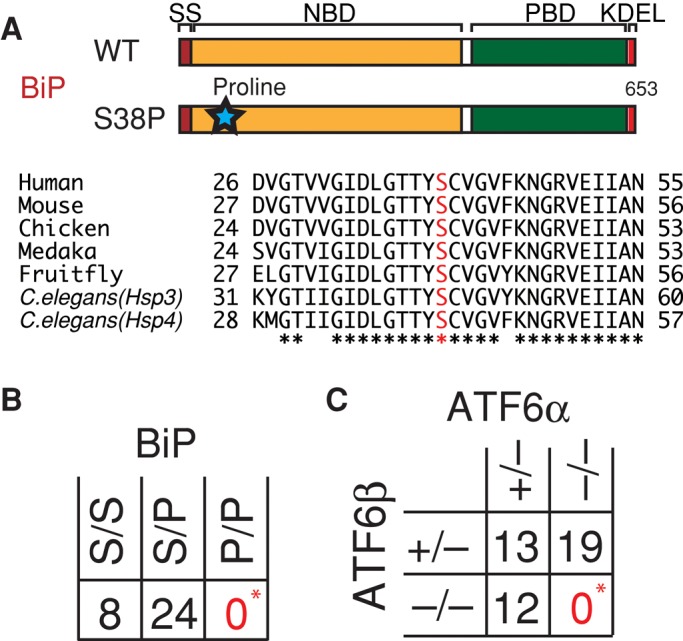
Effect of BiP mutation or ATF6α/β double knockout on medaka embryonic development. (A) Schematic representation of wild-type and S38P-mutant BiP proteins. The regions surrounding S38 in BiP (shown in red) of various species are aligned. Identical amino acids are indicated by asterisks. (B) BiP(S38P) heterozygotes were in-crossed, and the resulting 32 fishes hatched were genotyped. *p < 0.001 (C) ATF6α single knockout was crossed with ATF6β single knockout, and the resulting 44 fishes hatched were genotyped. *p < 0.001.
ER stress occurs physiologically during medaka embryonic development
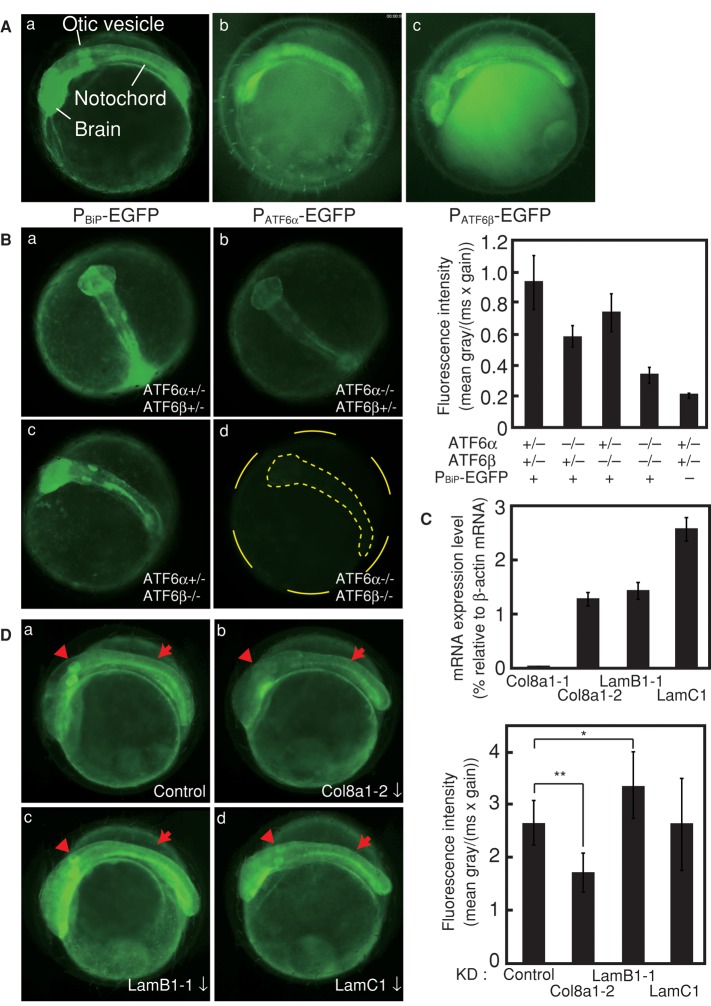
To determine the cause of the embryonic lethality, we examined when and where ER stress occurs physiologically during early embryonic development (1–2 d postfertilization [dpf]; note that medaka hatches at 7 dpf), using ATF6α/β double hetero carrying the PBiP-EGFP reporter gene (Supplemental Figure S3). EGFP expression started to be observed from mid to late gastrula stages 15 and 16 and became restricted to the embryonic body, which began to be clearly visible from stage 17. Uniform expression of EGFP observed in the embryonic body until stage 20 became differentiated from stage 21. Three regions became particularly brightened, namely the brain, otic vesicle, and notochord (Figure 3A, a), and their fluorescence intensity kept increasing until stage 24 (Supplemental Figure S3). This enhancement of EGFP expression in particular regions was specific to the BiP promoter, as EGFP was uniformly expressed in the embryonic body when its expression was driven by the ATF6α or ATF6β promoter (Figure 3A, b and c), which is insensitive to ER stress (Ishikawa et al., 2011). Of importance, EGFP expression level observed at stage 23 was decreased ∼30 and ∼50% by deletion of ATF6β and ATF6α, respectively, and became <20% by deletion of both ATF6β and ATF6α (Figure 3B). Thus physiological ER stress occurred particularly in the brain, otic vesicle, and notochord, which activated ATF6α/β to induce BiP. It is noteworthy that the contribution of ATF6β to BiP induction in response to physiological ER stress (Figure 3B) is more evident than that to pharmacological ER stress (Figure 1C), which can explain no obvious phenotype of ATF6α single knockout under normal growing conditions; ER chaperones must be induced sufficiently by ATF6β in ATF6α single knockout.
FIGURE 3:
Analysis of physiological ER stress occurring during early embryonic development. (A) Embryos at 2 dpf of ATF6α/β double hetero carrying the PBiP-EGFP reporter gene (a), wild-type fish carrying the PATF6α-EGFP gene (b), and wild-type fish carrying the PATF6β-EGFP gene (c) were analyzed by fluorescence microscopy. The positions of the brain, otic vesicle, and notochord are indicated. (B) Embryos at 2 dpf of ATF6α/β double hetero (a), ATF6α single knockout (b), ATF6β single knockout (c), and ATF6α/β double knockout (d), each carrying the PBiP-EGFP reporter gene, or double hetero embryo not carrying the reporter gene (–), were analyzed by fluorescence microscopy. Total fluorescence intensity was determined (n = 8 for PBiP-EGFP+ and n = 5 for PBiP-EGFP−), and means with SDs (error bars) are shown on the right. (C) Total RNA prepared from wild-type embryo at 2 dpf was subjected to quantitative RT-PCR to determine the level of Col8a1-1, Col8a1-2, LamB1-1, and LamC1 mRNAs relative to β-actin mRNA. (D) Embryos of wild-type fish at 2 dpf carrying the PBiP-EGFP reporter gene, into which control morpholino (a) or morpholino targeting Col8a1-2 (b), LamB1-1 (c), or LamC1 (d) had been microinjected at the one-cell stage, were analyzed by fluorescence microscopy. The red arrowheads and arrows indicate the otic vesicle and notochord, respectively. Total fluorescence intensity was determined (at least four embryos were analyzed), and means with SDs (error bars) are shown on the right. *p < 0.05; **p < 0.001.
Given that ER stress occurred in the notochord but not in the somite or neural tube (Figure 3A), we hypothesized that extracellular matrix proteins synthesized and secreted in large amounts by the notochord might be the cause of physiological ER stress. Genetic analysis in zebrafish identified the α1 chain of type VIII collagen (Col8a1; Gansner and Gitlin, 2008), α1 chain of type XV collagen (Col15a1; Pagnon-Minot et al., 2008), laminin β1 and laminin γ1 (Parsons et al., 2002), and fibrillin-2 (Gansner et al., 2008), which are essential for notochord formation. We therefore searched for their counterparts in medaka and found two homologues of Col8a1 (Col8a1-1 and Col8a1-2), two homologues of Col15a1 (Col15a1-1 and Col15a1-2), two homologues of laminin β1 (LamB1-1 and LamB1-2), a single homologue of laminin γ1 (LamC1), and no homologue of fibrillin-2. Among these, information of translational start sites for Col8a1-2, LamB1-1, and LamC1 was sufficient to allow the design of translation-blocking morpholinos. The expression levels of Col8a1-1, Col8a1-2, LamB1-1, and LamC1 mRNAs were determined in comparison with that of β-actin mRNA in wild-type embryos by conducting quantitative reverse transcription (RT)-PCR using a determined number of plasmid molecules carrying the respective gene as calibration standards during amplification. As shown Figure 3C, Col8a1-2, LamB1-1, and LamC1 mRNAs were expressed at the level of 1–3% of β-actin mRNA, but expression of Col8a1-1 mRNA was extremely low in wild-type embryos.
Specific morpholino targeting Col8a1-2, LamB1-1, or LamC1 mRNA was microinjected into one-cell-stage embryos of wild-type fish carrying the PBiP-EGFP reporter gene, and EGFP fluorescence was observed in embryos at 2 dpf. As shown in Figure 3D, the tail became shortened when Col8a1-2 or LamC1 was knocked down, consistent with the phenotype of zebrafish mutants. Because laminin is a heterotrimer of A, B, and C subunits, the lack of effect of LamB1-1 knockdown suggested inefficiency or off-target effect of the designed morpholino, which was implied by our observation that the level of LamB1-1 mRNA was increased by an unknown mechanism in embryos into which LamB1-1–targeting morpholino had been microinjected (Supplemental Figure S4); each designed morpholino was expected to block translation but should not affect expression level of its target mRNA. Of importance, however, EGFP fluorescence in the otic vesicle was greatly mitigated and that in the notochord and brain was significantly (but not completely) reduced when Col8a1-2 was knocked down (Figure 3D, b). Thus synthesis of type VIII collagen causes ER stress during early embryonic development. It is known that productive folding of collagen requires various ER chaperones (Chessler and Byers, 1993; Ferreira et al., 1994; Wilson et al., 1998; Lamande and Bateman, 1999; Nagata, 2003).
BiP's chaperone function, as well as ATF6α/β-mediated induction of ER chaperones, is essential for notochord development
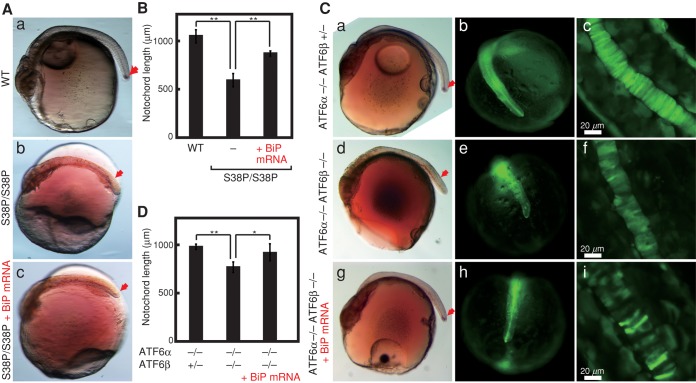
In relation to the foregoing observation, we noticed by optical microscopic analysis that the notochord was severely degenerated in ATF6α/β double knockout as compared with ATF6α single knockout (Figure 4, a and e). This notion was firmly supported by whole-mount in situ hybridization directed to Brachyury, a marker of the notochord (Wilkinson et al., 1990; Schulte-Merker et al., 1992). The notochord extended to the tip of the tail at stage 24 in ATF6α single knockout (Figure 4b), whereas its extension was stopped in the middle, and it did not reach the tip of the tail at the same stage in ATF6α/β double knockout (Figure 4f). Confocal microscopic analysis of EGFP driven by the BiP promoter revealed that disk-like cell structures were smoothly aligned in ATF6α single knockout (Figure 4, c and d), whereas such alignment was heavily disordered in ATF6α/β double knockout (Figure 4, g and h). It should be noted that 20-fold-longer exposure was used for analysis of EGFP expression in ATF6α/β double knockout than ATF6α single knockout, as deletion of both ATF6α and ATF6β greatly decreased EGFP expression level (see Figure 3B). Thus the notochord development was impaired in ATF6α/β double knockout.
FIGURE 4:
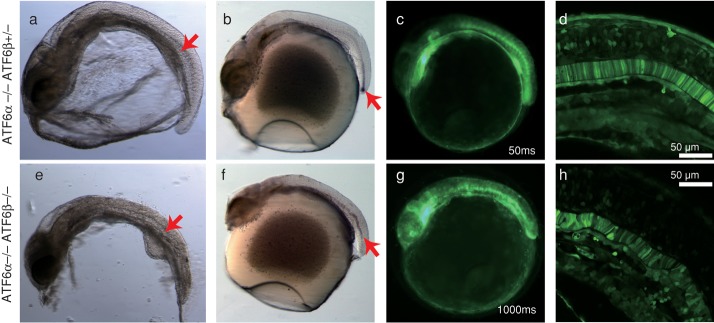
Effect of ATF6α/β double knockout on notochord development. Embryos at 2 dpf of ATF6α single knockout and ATF6α/β double knockout carrying the PBiP-EGFP reporter gene were analyzed by stereomicroscope (a, e), whole-mount in situ hybridization to detect Brachyury mRNA (b, f), fluorescence stereomicroscopy to visualize EGFP expression (c, g), and confocal microscopy to visualize EGFP expression (d, h). The embryo of ATF6α/β double knockout was analyzed for 1000 ms, 20-fold longer than that of ATF6α single knockout (50 ms) for detection of fluorescence. The red arrows indicate the notochord.
We thus monitored the level of ER stress by determining splicing status of XBP1 mRNA, as it is an event downstream of IRE1 activation upon ER stress (Yoshida et al., 2001a). Reverse transcription (RT)-PCR analysis of whole embryos revealed that XBP1 mRNA was spliced constitutively in ATF6α/β double hetero, albeit slightly, consistent with the results shown in Supplemental Figure S3, and that the extent of splicing was markedly enhanced in ATF6α/β double knockout (Figure 5A), indicating that these ATF6α/β double-knockout embryos experienced more profound ER stress than ATF6α/β double-hetero embryos. We considered that this was because the BiP mRNA level was significantly lower in ATF6α/β double knockout than ATF6α/β double hetero (Figure 5B). To confirm this notion further, we conducted microarray analysis using whole embryos at stage 24 (National Center for Biotechnology Information Gene Expression Omnibus accession number GSE37174). We noticed that mRNA levels of canonical target genes of the PERK pathway, namely ATF4, ATF5, CHOP, and asparagine synthetase (Okada et al., 2002; Zhou et al., 2008), as well as mRNA levels of various ERAD-related genes, typical targets of the IRE1 pathway (Yoshida et al., 2003), were higher in ATF6α/β double knockout than ATF6α/β-double hetero (Figures 5, C and D), indicating that the PERK and IRE1 pathways were activated more extensively in the absence of ATF6α/β. In marked contrast, many genes coding for ER chaperones (BiP, GRP94, three calreticulins, and protein disulfide isomerase) were down-regulated in ATF6α/β double knockout (Figure 5E). Note that expression levels of various medaka homologues of genes known to be involved in notochord formation in zebrafish or mice did not differ very significantly between ATF6α/β double hetero and ATF6α/β double knockout (Supplemental Figure S5A). Thus homeostasis of the ER could not be maintained without ATF6α/β-mediated induction of ER chaperones, causing more profound ER stress in ATF6α/β double-knockout embryos. Consistent with this notion, we found that notochord development was markedly sensitive to tunicamycin treatment; notochord development was inhibited by treatment for 24 h with 2 μg/ml tunicamycin, a level that induced BiP mRNA in embryos at 2 dpf, and the brachyury signal was barely detected in embryos treated with 3 or 4 μg/ml tunicamycin (Supplemental Figures S5B and S5C). Although we previously showed that both 2 μg/ml tunicamycin and 300 nM thapsigargin induced splicing of XBP1 mRNA to a similar extent in the medaka cell line OLCAB-e3 (Ishikawa et al., 2011), treatment of embryos with 300 nM thapsigargin for 24 h exhibited no obvious phenotype (our unpublished observations). In contrast, essentially all embryos were killed within 24 h after the addition of 2 mM dithiothreitol (our unpublished observations), which potently evoked ER stress in OLCAB-e3 (Ishikawa et al., 2011). Thus thapsigargin and dithiothreitol are not suitable as ER stress inducers in medaka fish.
FIGURE 5:
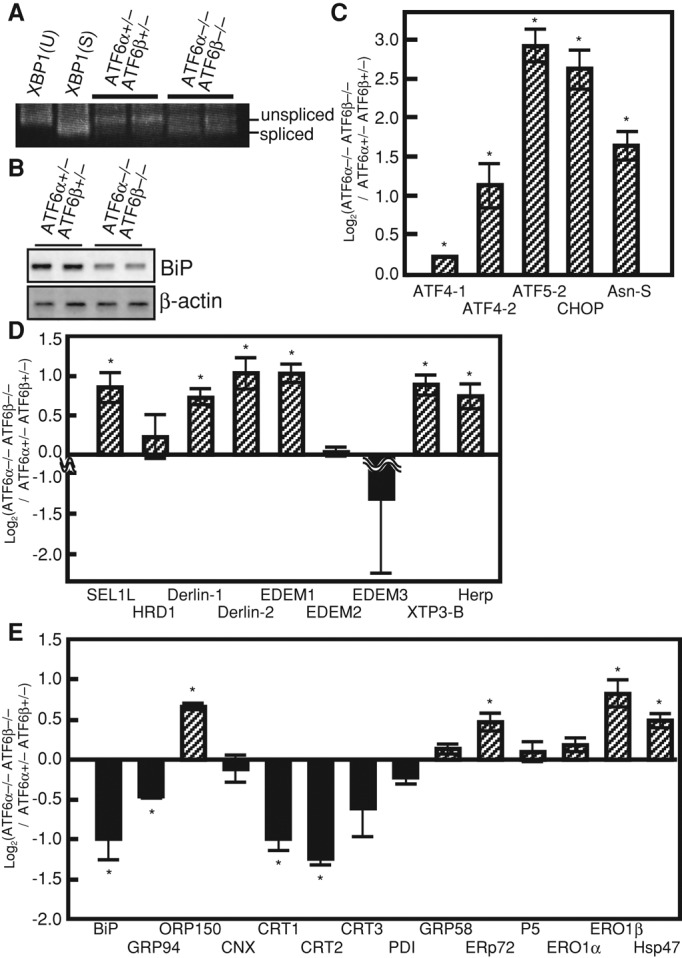
Effect of ATF6α/β double knockout on splicing of XBP1 mRNA and expression levels of various genes. (A) Total RNA extracted from embryos at 2 dpf of ATF6α/β double hetero and double knockout was subjected to RT-PCR to detect splicing of XBP1 mRNA. pCMV-myc-medaka pXBP1(S) and pCMV-myc-medaka pXBP1(U) (Ishikawa et al., 2011) were used for control. Positions of the RT-PCR products corresponding to unspliced and spliced XBP1 mRNA are indicated. (B) Total RNA extracted as in A was analyzed by Northern blot hybridization as in Figure 1D. (C) Expression levels of canonical target genes of the PERK pathway in ATF6α/β double knockout determined by microarray analysis are shown in comparison with those in ATF6α/β-double hetero (n = 2). *p < 0.05. (D) Expression levels of various ERAD components are shown as in C. *p < 0.05. (E) Expression levels of various ER chaperones are shown as in C. *p < 0.05.
We next examined whether BiP(S38P) mutation also affects notochord development and found that this was indeed the case. In situ hybridization analysis of Brachyury revealed that the notochord did not reach the tip of the tail in BiP(S38P) homozygotes (Figure 6A, b). Of importance, when BiP mRNA synthesized in vitro was microinjected into one-cell-stage embryos, notochord development progressed so that it was extended to the tip of the tail as expected (Figure 6A, c), although the length of the tail was still slightly shorter than in wild-type fish (Figure 6A, a; see Figure 6B for quantified data).
FIGURE 6:
Effect of overexpression of BiP mRNA on the defect in notochord development observed in BiP(S38P) homozygote or ATF6α/β double knockout. (A) Embryos at 2 dpf of wild-type fish (a) and BiP(S38P)-homozygote (b), as well as an embryo of BiP(S38P) homozygote into which in vitro–synthesized BiP mRNA had been microinjected at the one-cell stage (c), were analyzed by whole-mount in situ hybridization to detect Brachyury mRNA. The red arrow indicates the notochord. (B) Lengths of the notochord obtained in A were measured (n = 3), and means with SDs (error bars) are shown. **p < 0.01. (C) Embryos at 2 dpf of ATF6α single knockout (a–c) and ATF6α/β double knockout (d–f), as well as an embryo of ATF6α/β double knockout into which in vitro–synthesized BiP mRNA had been microinjected at the one-cell stage (g–i), were analyzed by whole-mount in situ hybridization to detect Brachyury mRNA (a, d, g), fluorescence microscopy to visualize EGFP expression (b, e, h), and confocal microscopy to visualize EGFP expression (c, f, i). Length of fluorescence detection was similar to that in Figure 4. The red arrow indicates the notochord. (D) Lengths of the notochord obtained in C were measured (n = 3), and means with SDs (error bars) are shown. **p < 0.01, *p < 0.05.
Because a limited number of ER chaperones were identified to be ATF6α/β targets by microarray analysis (Figure 5E), we further examined whether microinjection-mediated overexpression of BiP mRNA could rescue the defect in notochord development observed in ATF6α/β double knockout. We found that the notochord was able to reach the tip of the tail in ATF6α/β double knockout after microinjection (Figure 6C, g and h), although the length of the tail was still slightly shorter than in the wild-type fish (Figure 6C, a and b; see Figure 6D for quantified data). The regularity of alignment of disk-like cell structures was also restored, albeit partially (Figure 6Ci). We thus concluded that both the chaperone function of BiP and ATF6α/β-mediated adjustment of expression levels of ER chaperones are essential to notochord development in medaka because the notochord synthesizes and secretes large amounts of extracellular matrix proteins to serve as the body axis.
DISCUSSION
Our present results unravel highly conserved features in the molecular mechanism and physiological importance of the UPR between medaka and mice, particularly with the ATF6 pathway, as well as in the essentiality of not only the function, but also the expression level of the major ER chaperone BiP. First, ATF6α is primarily responsible for transcriptional induction of BiP in response to ER stress in medaka, as in mice, which is in marked contrast to the case in nonvertebrates, in which the IRE1 pathway is essential to the induction of BiP. Second, the chaperone function of BiP is essential to medaka early embryonic development, as in mice. Third, ATF6α and ATF6β single knockout develop normally, but ATF6α/β double knockout causes embryonic lethality in medaka, as in mice. Medaka is hardy and prolific, and rearing costs are reasonable, allowing us to construct and characterize various multiple knockouts with relative ease, as we did here for ATF6α and ATF6β double knockout. Thus medaka is identified as a highly useful vertebrate model for comprehensive analysis of the biology and physiology of the UPR.
We showed here that ER stress occurs physiologically during medaka early embryonic development, which activates ATF6α/β to induce transcription of BiP, and which activates IRE1α/β to induce splicing of XBP1 mRNA constitutively. Double deletion of ATF6α and ATF6β significantly increased the extent of splicing of XBP1 mRNA. Microarray analysis revealed that more profound ER stress experienced in the embryos of ATF6α/β double knockout indeed activated the PERK pathway and the IRE1 pathway more extensively. The same microarray analysis revealed that GRP94, calreticulins, and protein disulfide isomerase are also targets of the ATF6 pathway in addition to BiP, as their expression levels were down-regulated in the embryos of ATF6α/β double knockout. We noticed that target genes of medaka ATF6α/β do not completely overlap with those of mouse ATF6α. The ER chaperones ORP150, ERp72, and ERO1β are certainly targets of ATF6α in mouse embryonic fibroblasts (Adachi et al., 2008) but not in medaka embryos. It remains to be determined whether this difference can be ascribed to the experimental conditions used, namely pharmacological ER stress (tunicamycin treatment) in mice and physiological ER stress in medaka. In addition, although various ERAD components were upregulated by the heterodimer of pXBP1(S) and pATF6α(N) in mice (Yamamoto et al., 2007; Adachi et al., 2008), they seem to be controlled solely by the IRE1-XBP1 pathway in medaka, because their expression levels rather increased in the embryos of ATF6α/β double knockout. We can thus envision a scenario in which the major regulator of BiP and some other but not all ER chaperones has been switched from the IRE1 pathway to the ATF6 pathway during evolution, probably with development of the vertebra but for yet undetermined reasons, and that the ATF6 pathway became responsible for the induction of more ER chaperones and most ERAD components during evolution from teleosts to mammals. Dual regulation of ERAD components by the IRE1 and ATF6 pathways probably gives rise to safer methods of destroying proteins that have been synthesized by consuming numerous amounts of ATP molecules.
Physiological ER stress occurs particularly in the brain, otic vesicles, and notochord, and ATF6α/β double knockout showed a severe defect in notochord development. We reasoned that the notochord synthesizes and secretes large amounts of extracellular matrix proteins to serve as the body axis before formation of the vertebra. Indeed, various extracellular matrix proteins, such as laminins (Parsons et al., 2002), collagens (Gansner and Gitlin, 2008; Pagnon-Minot et al., 2008), and fibrillins (Gansner et al., 2008), are known to be essential for the notochord formation in zebrafish. Of importance, morpholino-mediated knockdown of Col8a1-2 significantly but not completely mitigated ATF6α/β-mediated activation of BiP promoter in the notochord. Consistent with this observation, it was previously shown that Col8a1 is robustly expressed in the notochord of zebrafish (Gansner and Gitlin, 2008). Because Col8a1 was also shown to be strongly expressed in the jaw cartilages of zebrafish (Gansner and Gitlin, 2008), strong EGFP expression observed in the brain region of medaka fish most likely represents ATF6α/β-mediated activation of BiP promoter in the cartilages, and thereby morpholino-mediated knockdown of Col8a1-2 significantly mitigated EGFP expression in the brain region.
Thus productive folding of these extracellular matrix proteins requires not only correct functioning of the ER chaperone BiP but also ATF6α/β-mediated transcriptional induction of ER chaperones, and the notochord cannot develop normally in the absence of such adjustment to the increased demands in the ER. This is reminiscent of the requirement of the IRE1-XBP1 pathway for the development of the liver (Reimold et al., 2000), the terminal differentiation of B cells into plasma cells (Reimold et al., 2001), and the development of exocrine glands (Lee et al., 2005), as well as that of the PERK pathway for maintenance of pancreatic β cells (Harding et al., 2001); all of these tissues synthesize and secrete large amounts of proteins. Nonetheless, for the first time to our knowledge we were able to track down a particular protein, Col8a1-2, as one of the contributing factors to ER stress that occurs physiologically and specifically activates the ATF6 pathway. An interesting question is what causes such differential tissue-dependent requirements for a particular pathway of the UPR, despite the fact that the three pathways function ubiquitously. It also remains to be determined in the near future whether the lack of transcriptional induction of ER chaperones in ATF6α/β double knockout affects the function of the brain or otic vesicle.
In summary, we discovered the reason for the embryonic lethality caused by ATF6α/β double knockout by maximally using the advantages of the medaka fish system. Synthesis of large amounts of extracellular matrix proteins gives rise to physiological ER stress in the developing notochord, which cannot extend to the tip without ATF6α/β-mediated adjustment of ER chaperone levels and thus without maintenance of the homeostasis of the ER.
MATERIALS AND METHODS
Fish
Medaka southern strain cab was used as wild-type fish. Fishes were maintained in a recirculating system with a 14:10–h light:dark cycle at 27.5°C. All experiments were performed in accordance with the guidelines and regulations established by the Animal Research Committee of Kyoto University. EGFP imaging was performed under a fluorescence stereomicroscope (M205FA; Leica, Wetzlar, Germany) using a GFP3 filter (470/40–nm excitation filter, 525/50–nm barrier filter) with a camera (Leica DFX310FX) and acquisition software (Leica las AF) or under a confocal microscopic system (Leica TCS SP2 system) using a 63×/numerical aperture 1.40 objective lens at room temperature.
TILLING method
Previously ∼100 male fishes were randomly mutagenized and then crossed with wild-type female fishes, giving rise to a library of 5771 mutated male fishes of N1, whose sperms and genomic DNA were cryopreserved (Taniguchi et al., 2006). PCR fragments amplified from this library were directly sequenced or subjected to high-resolution melting curve analysis (Ishikawa et al., 2010) to identify desired mutations. ATF6α and ATF6β single knockouts were obtained as described in the legend to Supplemental Figure S1. All ATF6-knockout experiments were conducted by in-crossing ATF6α (N10) and ATF6β (N3) double heterozygotes carrying the PBiP-EGFP reporter gene. However, the phenotypes of ATF6α (N10) and ATF6β (N3) double homozygotes were not changed when ATF6α heterozygotes (N10) and ATF6β heterozygotes (N10) were used for crossing. BiP(S38P) mutant was obtained as described in the legend to Supplemental Figure S2. All BiP experiments were conducted by in-crossing BiP(S38P) heterozygotes (N10).
Genotyping
Embryos or hatched fishes were suspended in 50 μl of lysis buffer (400 mM Tris/HCl, pH 8.0, containing 150 mM NaCl, 5 mM EDTA, 0.1% Tween 20, and 1 mg/ml Proteinase K), incubated at 55°C for 30 min, and then boiled for 10 min to inactivate Proteinase K. The DNA fragment containing the mutation site of ATF6α(K149), ATF6β(S143), or BiP(S38) was amplified by PCR directly from lysates using the primers 5′-AGTTTTCCCGTCTGCTCATC-3′ and 5′-TAACTGCAGTGCGTGCCTAT-3′ for ATF6α, 5′-TACATGTACGGAGACGTGCTG-3′ and 5′-GTCTGTGTCTGAATGCTGCTGAT-3′ for ATF6β, and 5′-TGAGCATCGAGAATAGAAGTAGTCC-3′ and 5′-GTATGCAGGGACAGTGACGA-3′ for BiP, and amplified fragments were directly sequenced.
Northern blot hybridization, RT-PCR, quantitative RT-PCR, and microarray analysis
Total RNA was extracted from embryos at 2 dpf or fishes at 1 dph by the acid guanidinium/phenol/chloroform method using Isogen (Nippon Gene, Tokyo, Japan). Northern blot hybridization was carried out as described previously (Ishikawa et al., 2011). For quantitative RT-PCR analysis, total RNA was purified by RNeasy MinElute (Qiagen, Valencia, CA), reverse transcribed using oligo dT primer, and then subjected to the ABI StepOne Real-Time PCR System using the primers 5′-CGGTATCCATGAGACCACCT-3′ and 5′-AGCACAGTGTTGGCGTACAG-3′ for β-actin, 5′-CTGCTTCATCTCCTGACCTTG-3′ and 5′-TGCCACTTTTGGAGGGTTT-3′ for Col8a1-1, 5′-CGCTTTCACAGCCATAGTGA-3′ and 5′-GCGGCCATTGTACAAGAGTT-3′ for Col8a1-2, 5′-TAATGAGGCCTGGACCAATC-3′ and 5′-CAGGGAGGTCATCTGGTTGT-3′ for LamB1-1, and 5′-CGACATGAGAGACGACCTGA-3′ and 5′-TGCAGTCTGCTGCTCACTCT-3′ for LamC1. For microarray analysis, total RNA from four embryos each of ATF6α/β double hetero and double knockout was mixed, purified through RNeasy micro kit (Qiagen), and checked for quality with an RNA 6000 Nano Assay using an Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA). We subjected 100 and 500 ng of total RNA to RT-PCR to detect ER stress–induced splicing of XBP1 mRNA and Northern blot hybridization, respectively, as described previously (Ishikawa et al., 2011). We converted 150 ng of total RNA to cDNA, which we then transcribed with cyanine 3-CTP using a Low Input Quick Amp labeling kit, One-Color (Agilent Technologies). After purification through an RNeasy mini kit (Qiagen), 1.65 μg samples of the labeled cRNA probes (two each from ATF6α/β double hetero and double knockout) were hybridized separately with a 4×44K Agilent oligo microarray designed by Minoru Tanaka and his colleagues, on which 44,000 medaka genes were spotted. Cyanine 3 fluorescence intensity of each spot was determined after subtraction of the respective background intensity using an Agilent G2565BA Microarray Scanner. The raw data were normalized using GeneSpringGX (Agilent Technologies, version 11.5.1).
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed according to the standard procedure (Kinoshita et al., 2009) using a digitonin-labeled RNA probe. Total RNA prepared from embryos at 6 dpf was subjected to RT-PCR to amplify a Brachyury cDNA fragment using the primers 5′-AAGTACGTGAACGGGGAGTG-3′ and 5′-taatacgactcactatagggAGTTGGGTGTGGAGTTGGAG-3′ (lowercase letters denote the sequence of the T7 promoter), which was then used as template to synthesize the RNA probe with a DIG RNA labeling kit (Roche, Indianapolis, IN).
Microinjection of mRNA synthesized in vitro, as well as morpholino, into embryo
The 5′-capped BiP mRNA was transcribed in vitro from BiP cDNA obtained previously (Ishikawa et al., 2011) with a mMESSAGE mMACHINE kit (Ambion, Austin, TX) and then microinjected into one-cell-stage embryos at the concentration of 1 μg/μl. Standard control morpholino or specific morpholino targeting the translational start site in Col8a1-2, LamB1-1, or LamC1 was microinjected into one-cell-stage embryos at the concentration of 1 mM. mRNA or morpholino dissolved in 0.5× Yamamoto's buffer, 0.5× I-SceI buffer, and 0.05% phenol red was microinjected using a FemtoJet (Eppendorf, Hauppauge, NY). Morpholino oligonucleotide sequences were 5′-GGGAAGACATGGCCAAAGCCTTTTC-3′ for Col8a1-2, 5′-ACCAAGACCAGAGCAAGCATGGAAC-3′ for LamB1-1, and 5′-ACAGTCCGCACAAAACTCTGCATCC-3′ for LamC1. All morpholinos were purchased from Gene Tools (Philomath, OR).
Supplementary Material
Acknowledgments
We thank Kaoru Miyagawa and Yayoi Yamamoto for technical and secretarial assistance. We are grateful to Kiyoshi Naruse at the National Institute for Basic Biology (Okazaki, Japan) for providing materials of NBRP medaka. This work was financially supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (19058009 to K.M.) and in part by a grant from the Mitsubishi Foundation. T.I. was a recipient of a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists.
Abbreviations used:
- dpf
day postfertilization
- dph
day posthatching
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- RT
reverse transcription
- UPR
unfolded protein response
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0830) on February 27, 2013.
Present addresses: *Comparative Genomics Laboratory, National Institute of Genetics, Mishima 411-8540, Japan
†Toyohashi University of Technology, Toyohashi 441-8580, Japan
‡Department of Preventive Medicine and Public Health, School of Medicine, Keio University, Tokyo 160-8582, Japan.
REFERENCES
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chessler SD, Byers PH. BiP binds type I procollagen pro alpha chains with mutations in the carboxyl-terminal propeptide synthesized by cells from patients with osteogenesis imperfecta. J Biol Chem. 1993;268:18226–18233. [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Egawa N, Yamamoto K, Inoue H, Hikawa R, Nishi K, Mori K, Takahashi R. The endoplasmic reticulum stress sensor, ATF6α, protects against neurotoxin-induced dopaminergic neuronal death. J Biol Chem. 2011;286:7947–7957. doi: 10.1074/jbc.M110.156430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LR, Norris K, Smith T, Hebert C, Sauk JJ. Association of Hsp47, Grp78, and Grp94 with procollagen supports the successive or coupled action of molecular chaperones. J Cell Biochem. 1994;56:518–526. doi: 10.1002/jcb.240560412. [DOI] [PubMed] [Google Scholar]
- Gade P, Ramachandran G, Maachani UB, Rizzo MA, Okada T, Prywes R, Cross AS, Mori K, Kalvakolanu DV. An IFN-gamma-stimulated ATF6-C/EBP-beta-signaling pathway critical for the expression of death associated protein kinase 1 and induction of autophagy. Proc Natl Acad Sci USA. 2012;109:10316–10321. doi: 10.1073/pnas.1119273109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansner JM, Gitlin JD. Essential role for the alpha 1 chain of type VIII collagen in zebrafish notochord formation. Dev Dyn. 2008;237:3715–3726. doi: 10.1002/dvdy.21779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansner JM, Madsen EC, Mecham RP, Gitlin JD. Essential role for fibrillin-2 in zebrafish notochord and vascular morphogenesis. Dev Dyn. 2008;237:2844–2861. doi: 10.1002/dvdy.21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa II, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in Perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, et al. High-resolution melting curve analysis for rapid detection of mutations in a medaka TILLING library. BMC Mol Biol. 2010;11:70. doi: 10.1186/1471-2199-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Taniguchi Y, Okada T, Takeda S, Mori K. Vertebrate unfolded protein response: mammalian signaling pathways are conserved in medaka fish. Cell Struct Funct. 2011;36:247–259. doi: 10.1247/csf.11036. [DOI] [PubMed] [Google Scholar]
- Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Murata K, Naruse K, Tanaka M. Medaka: Biology, Management, and Experimental Protocols. Ames, IA: Wiley-Blackwell; 2009. [Google Scholar]
- Lamande SR, Bateman JF. Procollagen folding and assembly: the role of endoplasmic reticulum enzymes and molecular chaperones. Semin Cell Dev Biol. 1999;10:455–464. doi: 10.1006/scdb.1999.0317. [DOI] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Yoshida H, Kano F, Murata M, Mori K. Activation of mammalian unfolded protein response is compatible with the quality control system operating in the endoplasmic reticulum. Mol Biol Cell. 2004;15:2537–2548. doi: 10.1091/mbc.E03-09-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K. HSP47 as a collagen-specific molecular chaperone: function and expression in normal mouse development. Semin Cell Dev Biol. 2003;14:275–282. doi: 10.1016/j.semcdb.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Okada T, Haze K, Nadanaka S, Yoshida H, Seidah NG, Hirano Y, Sato R, Negishi M, Mori K. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J Biol Chem. 2003;278:31024–31032. doi: 10.1074/jbc.M300923200. [DOI] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnon-Minot A, et al. Collagen XV, a novel factor in zebrafish notochord differentiation and muscle development. Dev Biol. 2008;316:21–35. doi: 10.1016/j.ydbio.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Pollard SM, Saude L, Feldman B, Coutinho P, Hirst EM, Stemple DL. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129:3137–3146. doi: 10.1242/dev.129.13.3137. [DOI] [PubMed] [Google Scholar]
- Reimold AM, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Ho RK, Herrmann BG, Nusslein-Volhard C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development. 1992;116:1021–1032. doi: 10.1242/dev.116.4.1021. [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, et al. Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol. 2006;7:R116. doi: 10.1186/gb-2006-7-12-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui M, Yamaguchi S, Tanji Y, Tominaga R, Ishigaki Y, Fukumoto M, Katagiri H, Mori K, Oka Y, Ishihara H. Atf6alpha-null mice are glucose intolerant due to pancreatic beta-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metabolism. 2012;61:1118–1128. doi: 10.1016/j.metabol.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Wilson R, Lees JF, Bulleid NJ. Protein disulfide isomerase acts as a molecular chaperone during the assembly of procollagen. J Biol Chem. 1998;273:9637–9643. doi: 10.1074/jbc.273.16.9637. [DOI] [PubMed] [Google Scholar]
- Wu J, et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13:160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6α-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001a;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Mori K. ATF6 activated by proteolysis directly binds in the presence of NF-Y (CBF) to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol Cell Biol. 2001b;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.