Moesin, an ERM protein, negatively regulates RhoA by recruiting Conundrum/Rho GTPase–activating protein 18 to the apical cell cortex, thereby activating its RhoGAP function. Surprisingly, the data also show that activated Conundrum promotes cell proliferation, apparently through its ability to activate Rac1.
Abstract
RhoA, a small GTPase, regulates epithelial integrity and morphogenesis by controlling filamentous actin assembly and actomyosin contractility. Another important cytoskeletal regulator, Moesin (Moe), an ezrin, radixin, and moesin (ERM) protein, has the ability to bind to and organize cortical F-actin, as well as the ability to regulate RhoA activity. ERM proteins have previously been shown to interact with both RhoGEF (guanine nucleotide exchange factors) and RhoGAP (GTPase activating proteins), proteins that control the activation state of RhoA, but the functions of these interactions remain unclear. We demonstrate that Moe interacts with an unusual RhoGAP, Conundrum (Conu), and recruits it to the cell cortex to negatively regulate RhoA activity. In addition, we show that cortically localized Conu can promote cell proliferation and that this function requires RhoGAP activity. Surprisingly, Conu's ability to promote growth also appears dependent on increased Rac activity. Our results reveal a molecular mechanism by which ERM proteins control RhoA activity and suggest a novel linkage between the small GTPases RhoA and Rac in growth control.
INTRODUCTION
One hallmark of polarized epithelial cells is a dense band of actin filaments localized to the apical cortex that is tightly associated with the apical membrane and adherens junctions. The integrity of this apical cytoskeleton is required for junctional organization, apical/basal polarity and epithelial integrity. Ezrin, radixin, and moesin, collectively called the ERM proteins, are among the most extensively studied of the many proteins that regulate interactions between the apical membrane and the cortical cytoskeleton. ERM proteins, through their N-terminal FERM (Four-point-one, Ezrin, Radixin, Moesin) domain, have the ability to associate with the cytoplasmic face of the plasma membrane via interactions with phospholipids and the cytoplasmic tails of transmembrane proteins. ERMs also interact with the cytoskeleton through a C-terminal F-actin binding domain. Together these interactions are thought to organize cortical actin filaments and physically link them to the overlying plasma membrane.
In addition to their role in mediating membrane–cytoskeletal interactions, there is an increasing body of evidence that ERM proteins regulate the cell cortex through effects on activity of the Rho family GTPases, in particular RhoA. In their GTP-bound or active state, Rho family GTPases can activate downstream effectors that in turn regulate the actin cytoskeleton in order to carry out diverse cellular functions (reviewed in Johndrow et al., 2004; Jaffe and Hall, 2005). In Drosophila imaginal epithelia, phenotypes caused by loss of the sole ERM gene, Moesin (Moe), are strongly suppressed by reduction in the dosage of the Rho1 gene (homologous to RhoA), suggesting that Moe negatively regulates Rho1 activity (Speck et al., 2003). Studies in mammalian cells have revealed interactions between ERM proteins and RhoA regulatory proteins, including both positive (RhoGEFs [Rho guanine nucleotide exchange factors]) and negative (RhoGAPs [Rho GTPase activating proteins]) regulators of GTPase activity (Takahashi et al., 1998; Hatzoglou et al., 2007). The functional significance of these interactions has not always been clear, and ERMs have been proposed to both positively and negatively regulate RhoA activity through these interactions (Bretscher et al., 2002; Fehon et al., 2010). In addition, recently there has been evidence for ERM regulation of Rac1 activity, which has been suggested to lead to increased metastatic activity in tumor cells (Valderrama et al., 2012). Although the exact mechanisms by which ERM proteins control the activity of Rho family GTPases remain unclear, there is compelling evidence that the ERM function in organizing the cortical cytoskeleton is mediated both through its ability to bind actin and its ability to regulated Rho family GTPases.
To better understand the role of Moesin in regulating Rho1 activity in Drosophila, we have sought to identify Moe-binding proteins that might be involved in this process. In this study, we present evidence that Moe interacts with and promotes the activity of a previously uncharacterized RhoGAP that we call Conundrum (Conu, CG17082) due to its unexpected phenotypes. We show that Conu has GAP activity toward Rho1, Moe recruits Conu to the cell cortex, and cortical localization activates Conu's function in negatively regulating Rho1. Surprisingly, we also have found that expression of Conu at the cell cortex leads to overproliferation of the epithelium, and that this phenotype is dependent on its GAP activity. In addition, we have found that cortical Conu increases Rac1 activity independent of its GAP activity. These findings suggest that through their ability to regulate Rho1 and Rac1 activity Moe and Conu function together to control proliferation in developing epithelia.
RESULTS
Conu interacts with and is regulated by Moe
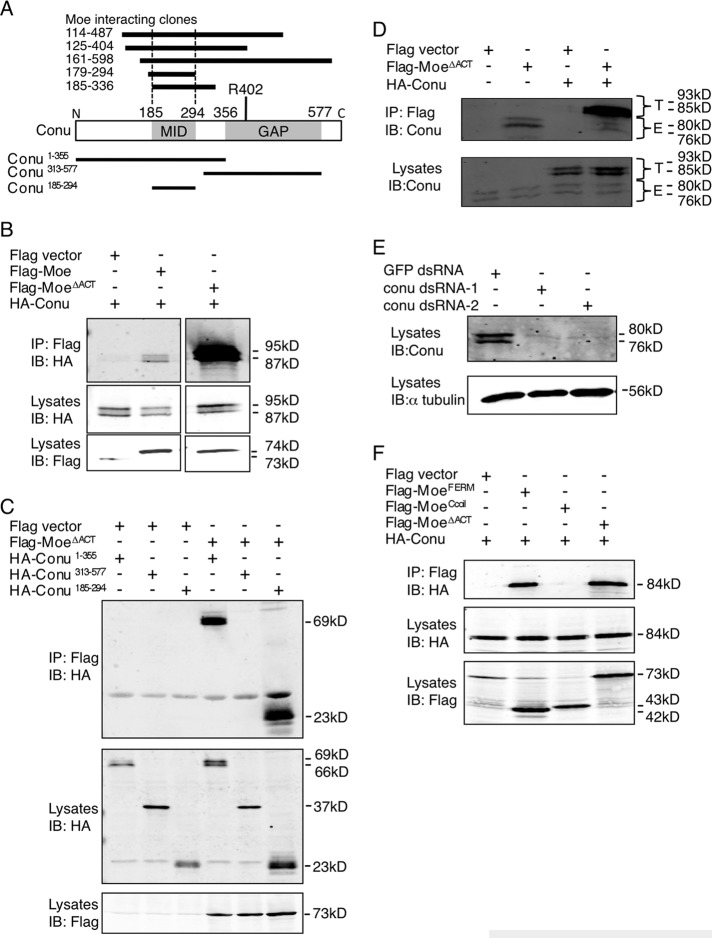
The RhoGAP domain–containing protein, Conu, was identified as a Moe-interacting protein in a yeast two-hybrid screen (Formstecher et al., 2005). A form of Moe that lacks the C-terminal actin-binding domain (Moe∆ACT) interacted with five unique cloned fragments of Conu (Figure 1A). Based on the smallest region of overlap between the clones, the minimal interaction domain of Conu sufficient to bind to Moe maps to residues 185–294, a region with no known domain structure (Figure 1A). A BLAST search identified Rho GTPase–activating protein 18, ARHGAP18, also known as MacGAP (Li et al., 2008; Maeda et al., 2011), to be the closest human homologue to Conu. A pairwise alignment of the two proteins showed that Conu shares 26% identity and 40% similarity to ARHGAP18 (unpublished data).
FIGURE 1:
Conu forms a complex with Moe. (A) Schematic diagram of Conu's structure with amino acid coordinates indicated above the diagram. The Moe interaction domain (MID) and the GAP domain with a predicted arginine finger required for GAP activity at amino acid 402, are indicated. The MID is defined by the overlap of five unique Conu clones that interacted with Moe in the yeast two-hybrid screen, as indicated above the Conu diagram. Conu fragments used for co-IP experiments with Moe are indicated below the schematic. (B–F) S2 cells were cotransfected with expression constructs for the indicated forms of Moe and Conu. (B) Conu coimmunoprecipitates with wild-type Moe, and more strongly with a form of Moe lacking the F-actin binding domain (ΔACT). (C) Moe∆ACT can coimmunoprecipitate N-terminal fragments of Conu (Conu 1–355 and Conu 185–294) that contain the MID, but not the C-terminal GAP domain of Conu (aa 313–577). (D) Endogenous Conu protein (E, endogenous) in S2 cells can be coimmunoprecipitated with Moe∆ACT. Epitope-tagged Conu was used as a positive control (T, tagged). (E) Specificity of the Conu antibody is demonstrated by dsRNA-mediated knockdown against two nonoverlapping regions of Conu. GFP dsRNA serves as a negative control. (F) The Moe FERM domain, but not the coiled-coil region, is sufficient to coimmunoprecipitate Conu.
To determine whether Conu and Moe proteins interact in vivo, we performed coimmunoprecipitation (co-IP) experiments in cultured Drosophila S2 cells. We found that epitope-tagged Conu coimmunoprecipitates robustly with Moe∆ACT and less strongly with the wild-type Moe (Figure 1B). In addition, an N-terminal construct of Conu lacking the GAP domain (residues 1–355), and the minimal interaction domain of Conu (residues 185–294) strongly coimmunoprecipitated with Moe∆ACT when expressed in S2 cells. However, a C-terminal construct comprising the GAP domain (residues 313–577) did not, confirming that the minimal interaction domain in the N-terminus of Conu mediates interactions with Moe (Figure 1C). Additionally, we made a Conu-specific antibody and used it to show that endogenous Conu from cultured cells coimmunoprecipitates with Moe∆ACT (Figure 1D). The specificity of the antibody was validated by double-stranded RNA (dsRNA) knockdown of Conu (Figure 1E).
Moe∆ACT is composed of the N-terminal FERM domain and two predicted coiled-coil domains (Li et al., 2007). Because coiled coils are structural motifs that mediate protein–protein interactions, and because the Moe FERM domain alone did not identify any Conu clones in the two-hybrid screen (Formstecher et al., 2005), we asked whether the coiled-coil domain of Moe is sufficient to mediate interactions with Conu in co-IP experiments. We found that Conu coimmunoprecipitated with the FERM domain of Moe but not with the coiled-coil domain (Figure 1F), suggesting the FERM domain mediates interactions with Conu.
Conu localizes to the cell cortex in a Moe-dependent manner
We next asked whether Conu colocalizes with Moe. Moe is enriched apically at the cell cortex, particularly in the apical junctional region of imaginal disk cells (Speck et al., 2003; Hughes and Fehon, 2006). To investigate the subcellular localization of Conu, we generated a 3× Flag N-terminally tagged conu transgene. Protein expressed by this transgene under the apGal4 driver was found throughout the cytoplasm and was junctionally enhanced in the apical domain (only apical sections shown, Figure 2A). The N-terminal tag did not appear to affect the localization of the protein, as untagged Conu expressed transgenically had a similar localization when stained with a Conu-specific antibody (unpublished data).
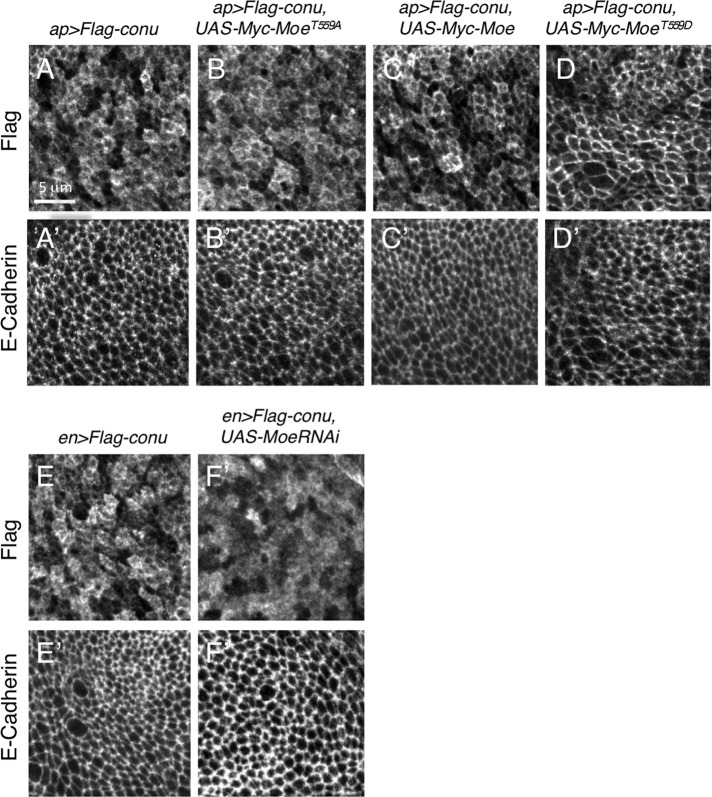
FIGURE 2:
Moe recruits Conu to the cortex, and cortical localization of Conu is dependent on Moe. (A) An epitope-tagged version of Conu has a primarily cortical localization in the wing imaginal epithelium when expressed under the apGal4 driver. (B) An unphosphorylatable form (T559A) of Moe coexpressed with Conu has no obvious effect on its localization. (C) Coexpression of wild-type Moe resulted in more cortically localized Conu. (D) Coexpression of a phosphomimetic form (T559D) of Moe resulted in a strikingly stronger cortical localization of Conu. (E) Under the enGal4 driver, epitope-tagged Conu has a primarily cortical localization, while depletion of Moe by RNAi disrupted this localization (F). E-cadherin, an adherens junction protein, is shown for all images to show that the plane of section cuts through the junctional region (A′ –F′). Scale bar in (A) represents 5 μm in all panels.
Conu localization was examined when Conu was coexpressed with previously characterized Moe mutations that affect its activation state (Speck et al., 2003). All of these transgenes expressed at similar levels (unpublished data). Conu's localization was unchanged when coexpressed with the nonphosphorylatable, inactive MoeT559A mutant, but appeared more junctional when expressed with a wild-type Moe transgene (Figure 2, B and C). Consistent with Conu interacting with active Moe, we found that when coexpressed with phosphomimetic Moe (MoeT559D), Conu displayed more defined junctional staining in the apical cell cortex (Figure 2D). Furthermore, Conu lost its junctional localization when Moe was strongly depleted using a previously published RNA interference (RNAi) transgene (Figure 2, E and F, and Supplemental Figure S1; Karagiosis and Ready, 2004; Hughes and Fehon, 2006). Taken together, these results suggest Moe can recruit Conu to the cell cortex.
Using the Conu antibody, we next examined the localization of the endogenous Conu protein. To confirm antibody specificity in tissues, we expressed a conu RNAi transgene in the wing disk under the enGal4 driver. This allowed us to visualize the subcellular localization of Conu in wild-type cells compared with Conu-depleted cells. Antibody staining in conu RNAi-expressing cells was markedly diminished, confirming specificity of the antibody for endogenous Conu in tissues (Figure 3, A and B). Conu appeared to be expressed uniformly in all tissues examined, including the imaginal epithelia and the follicular epithelium (unpublished data). Cross-sectional views of the epithelium indicated that endogenous Conu is preferentially localized at the apical cortex (Figure 3A). Tangential views through the apical surface of imaginal epithelium showed that, while Moe is preferentially junctionally localized, Conu staining is more broadly spread throughout the apical cortex, though some junctional association was observed (Figure 3, C and C′′). Expression of MoeT559D caused mildly increased Conu staining and enhanced junctional association (Figure 3, D and D′′), while expression of Moe∆ACT induced a dramatic relocalization of endogenous Conu to the apical junctional region (Figure 3, E and E′′), consistent with the strong co-IP we observed between these proteins. Expression of Moe∆ACT also resulted in a marked increase in Conu staining (Figure 3, F and G), suggesting that interaction with Moe stabilizes Conu in imaginal epithelia. We did not observe an obvious effect of Moe depletion by RNAi on Conu localization (unpublished data), suggesting that Moe is not the only means by which Conu can associate with the apical cell cortex.
FIGURE 3:
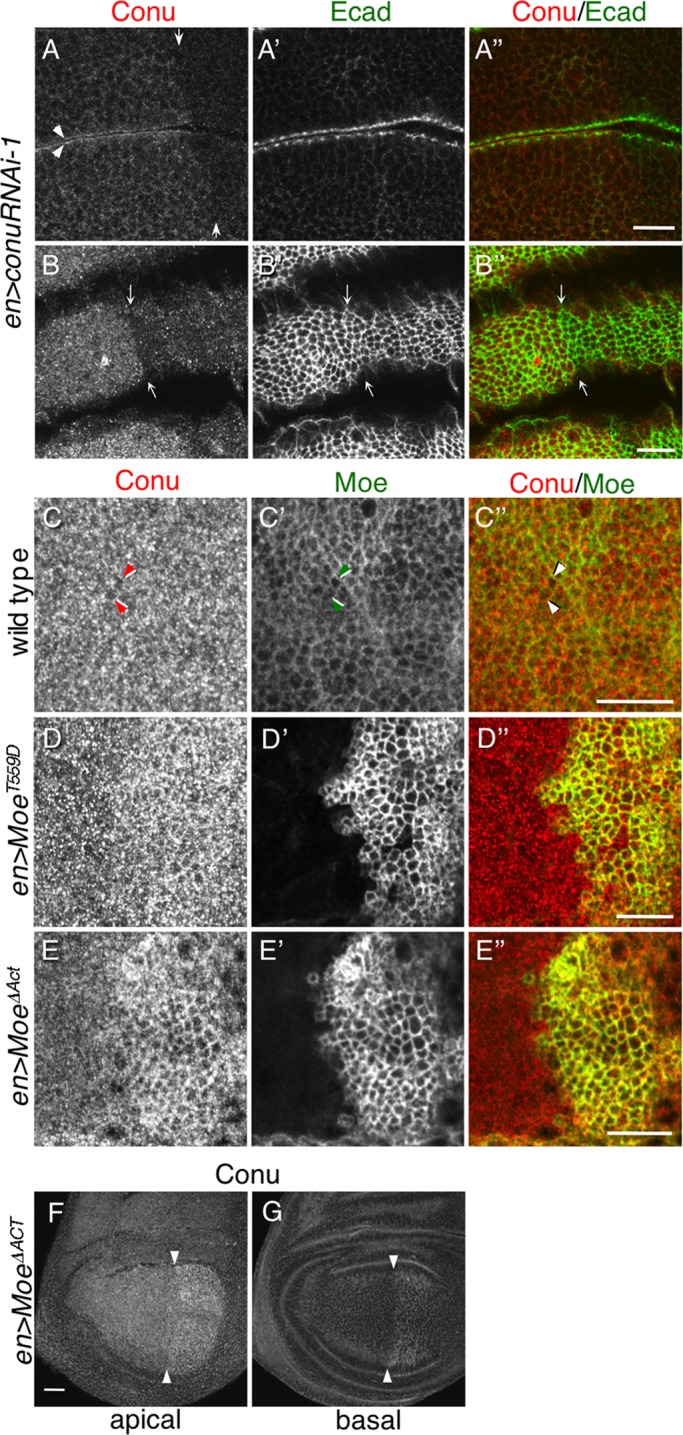
Moe stabilizes Conu at the cell cortex. (A–B′′). Depletion of Conu by expression of conuRNAi under the enGal4 driver (small arrows indicate the boundary of enGal4 expression), shows that the Conu antibody is specific and the conuRNAi transgene effectively knocks down Conu expression. Arrowheads in (A) indicate increased Conu staining at two opposed apical surfaces in an epithelial fold. A slight constriction in the epithelium is also apparent at the boundary of Conu knockdown (B′ and B′′). (C–E′′) Tangential sections through the apical domain of wing disks showing the relationship between Conu and Moe. In wild-type cells (C–C′′) endogenous Conu displays a punctate appearance that is slightly enriched at the apical junctions (arrowheads), while endogenous Moe is primarily localized junctionally (C′). Expression of activated Moe (myc MoeT559D) results in increased endogenous Conu staining and a more obvious colocalization with Moe in the junctional region (D and D′). Expression of myc Moe∆ACT (E–E′′) results in even more obvious colocalization in the apical junctional region. Conu staining is also increased throughout en>Moe Moe∆ACT cells (F and G: expression boundary is indicated by arrowheads). Scale bars: (A–E) 10 μm; (F–G): 20 μm. Transgenically expressed Moe is detected using anti-Myc in (D and E).
Moe activates Conu by recruiting it to the plasma membrane
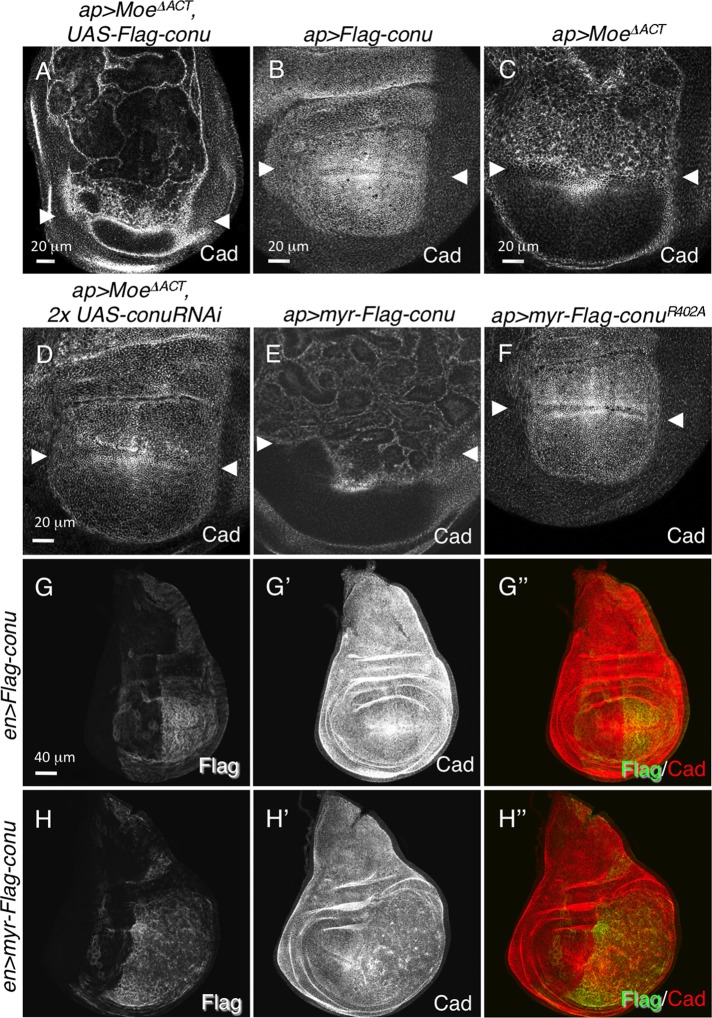
To examine the functional consequences of Moe's ability to recruit Conu to the apical cortex, we examined the effects of expression or depletion of Conu in the presence of Moe∆ACT. Because Conu is a putative RhoGAP, we expected coexpression of Moe∆ACT and Conu to result in phenotypes consistent with a decrease in Rho1 activity. Surprisingly, we found that their coexpression led to overgrowth and convoluted folding of the epithelium (Figure 4A), phenotypes not observed for reduced Rho1 levels or expression of a different RhoGAP, Cv-c, tethered to the membrane (Figure S2, A and B). In contrast, Conu expression on its own has no effect on the epithelium (Figure 4, B and G), while Moe∆ACT expression results in ectopic folding of the epithelium that is similar to, but not as severe as, that seen when Moe∆ACT and Conu are coexpressed (Figure 4C). To test whether the Moe∆ACT phenotype is dependent on endogenously expressed Conu, we coexpressed Moe∆ACT and conu RNAi transgenes. We found that reducing Conu expression suppressed the Moe∆ACT phenotype (Figure 4D), again suggesting that Moe recruits Conu to the cell cortex.
FIGURE 4:
Overgrowth and disruption of the epithelium is a consequence of increased Conu at the cell cortex. (A) Coexpression of Moe∆ACT and epitope-tagged conu under the apGal4 driver in the dorsal portion of the disk (region above arrowheads) results in overproliferation and misfolding of the epithelium. (B) Expression of conu alone under apGal4 has no effect on the epithelium, and expression of Moe∆ACT alone under apGal4 results in a slight disruption of the epithelium (C). (D) Depletion of Conu protein by RNAi in cells expressing Moe∆ACT under apGal4 suppresses overproliferation of the epithelium, suggesting the overproliferation is due to stabilized Conu protein at the cell cortex. (E) Tethering Conu to the membrane using a myr sequence results in overgrowth and overproliferation similar to that seen for coexpression of Moe∆ACT and conu. (F) The overproliferation caused by membrane-tethered Conu is dependent on its GAP activity, because expression of myr-conuR402A, which carries a mutation in the GAP domain, has no effect on the epithelium. (G–G′′) Expression of a Flag-tagged wild-type version of conu under the enGal4 driver in the posterior half of the disk has no effect on the epithelium, while expression of Flag-tagged myr-conu results in overgrowth (H–H′′). Scale bar in (D) corresponds to (E) and (F); scale bar in (G) corresponds to (H).
Taken together, these results suggest that stably maintaining high levels of Conu at the plasma membrane leads to overgrowth. To test this hypothesis, we made a membrane-tethered version of Conu using the myristoylation (myr) sequence tag from Src. Expression of myr-Conu alone caused overgrowth and folding of the epithelium (Figure 4, E and H) similar to that seen when Moe∆ACT and untethered Conu were coexpressed. These phenotypes required the GAP activity of the protein, because expression of a myr-Conu protein that carries a mutation in the arginine finger of the GAP domain (R402A, the predicted catalytic residue) had no phenotype (Figure 4F). Interestingly, expression of the GAP domain alone (residues 313–577) tethered to the membrane was not sufficient to induce overproliferation of the epithelium, but did result in a furrow at the expression boundary and increased apical cell size, both phenotypes that could be associated with reduced Rho1 activity (Figure S2, C and D). Together these results indicate that Conu GAP activity at the cell cortex is necessary but not sufficient for overgrowth.
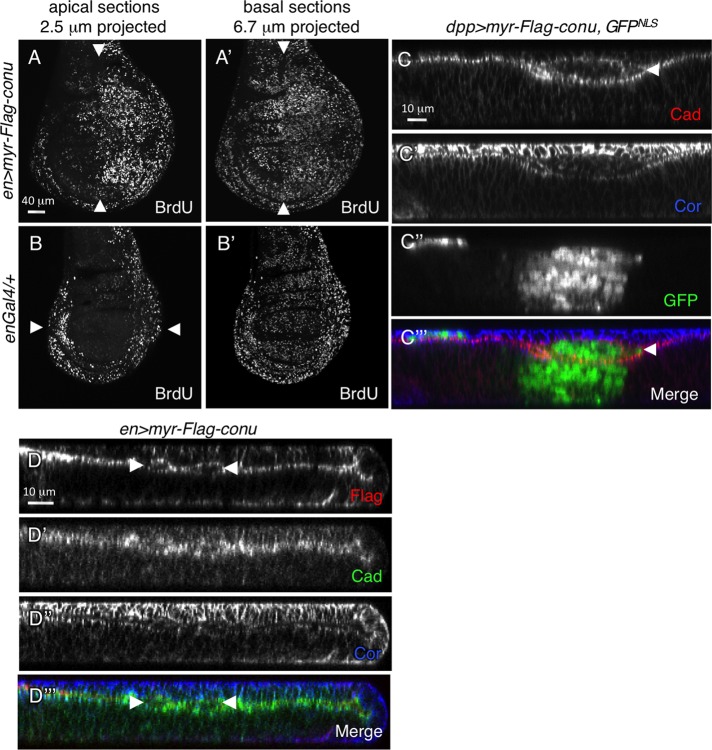
To verify that cells expressing myr-Conu protein were indeed overproliferating, we examined bromodeoxyuridine (BrdU) incorporation in posterior compartment cells expressing myr-conu under the enGal4 driver. In enGal4; myr-conu disks we observed increased numbers of BrdU-positive cells in the posterior compartment compared with the anterior compartment, as well as compared with enGal4/+ control disks (Figure 5, A and A′′ compared with B and B′). Careful examination of these tissues revealed additionally that cells on the peripodial side of the epithelium were abnormal in shape. In control disks, the proximal cells at the edge of the wing disk, called the peripodial and disk margin cells, are cuboidal in shape (McClure and Schubiger, 2005), while peripodial cells over the wing blade are more squamous. Expression of UAS-myr-conu down the middle of the wing epithelium under the dppblnkGal4 driver resulted in a shortening of the disk epithelium and apical extrusion of some cells that became trapped between the disk proper and peripodial layers (arrowhead in Figure 5, C′ and C′′′). When the enGal4 driver was used, both the disk proper and peripodial cells were more cuboidal in shape, and a similar but weaker extrusion phenotype was observed (arrowheads in Figure 5, D and D′′′).
FIGURE 5:
Tethering Conu to the membrane results in increased proliferation and influences cell shape. (A and A′) BrdU incorporation in disks expressing myr-conu under the enGal4 driver in the posterior half of the disk (right of the arrowhead) is increased in the expressing cells compared with wild-type anterior cells. (B–B′′) enGal4/+ was used as a control. Proximal wing cells (indicated by arrowheads in B) normally have high BrdU incorporation, while peripodial cells that overlay the wing blade do not (B), but in disks expressing myr-conu in the posterior compartment (to the right of the arrowheads in A and A′), there is increased BrdU incorporation in both layers (A). (C) Wing imaginal disk cells expressing myr-conu under the dppGal4 driver shorten in height and are extruded apically (arrowhead in C′ and C′′′). Coexpression of GFPNLS was used to mark the expressing cells. (D) Peripodial and disk-proper cells expressing myr-conu under enGal4 are more cuboidal in shape, and some cells are extruded apically from the epithelium and found between the two cell layers (indicated by arrowheads in D and D′′′).
conu is not essential for viability but does affect epithelial morphology
The conundrum gene is located on chromosome 2R at cytological position 41C1. No mutant alleles of conu have been previously identified, probably due to its close proximity to the centromere. A transposable Minos element, conuMB06749, is inserted intronically between the third and fourth coding exons (Figure 6A), but this line is homozygous viable (unpublished data), and immunoblots showed similar protein levels in conuMB06749 and wild-type tissue lysates (Figure 6B), suggesting that the insertion does not affect Conu function. To generate conu mutations, we performed a Minos element imprecise excision screen. Two molecularly defined deficiencies of the region, M(2)41A2 and Nipped-D, that completely uncover conu (Myster et al., 2004) and are adult-lethal were used for complementation tests over the conu excision lines. We tested 405 individual excision lines, but found none to be lethal over one of these deficiencies, suggesting that conu is not essential for viability. However, one semiviable but female sterile excision line, Df(2R)conu6, was found to contain an approximately 200-kbp deletion that removes the 3′ end of conu, leaving the N-terminal portion, amino acids 1–118, intact. This excision also deletes three other genes, CG12547, CG17528, CG14464, as well as the first exon containing the start codon of the gene Ionotropic receptor 41a (Ir41a) (Figure 6A). No Conu protein was detectable in Df(2R)conu6 female ovaries (Figure 6B) or in wing disks from transheterozygous Df(2R)conu6/Df(2R)Nipped-D animals (Figure 6C).
FIGURE 6:
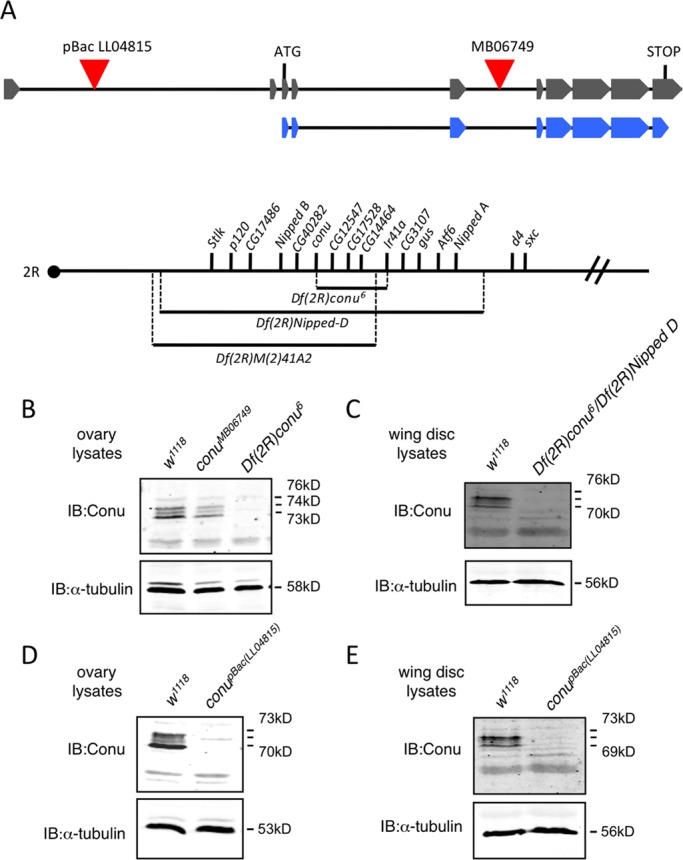
Conu is not required for viability. (A) A schematic of the predicted conu gene transcript RA is shown in gray, with boxes representing exons and lines representing introns. The schematic is not to scale, but relative distances are shown. The coding region is shown underneath in blue. A piggyBac element (LL04815) is inserted in an intron 5′ to the start of translation, while a Minos element (MB06749) is inserted in an intron between coding exons 3 and 4, as indicated by the red triangles. The two deficiencies used to screen the Minos element excision lines (Df(2R)Nipped-D and Df(2R)M(2)41A2) are indicated, along with the small deletion (Df(2R)conu6) produced by excision of the Minos element, with the genes uncovered by each indicated above as reported by Myster et al. (2004) and this study. The dot represents the centromere, and genes are shown in order on the chromosome, but distances between genes are not drawn to scale. (B) Ovary lysates from w1118, conuMB06749, and Df(2R)conu6 homozygous adult females showed that while Conu is expressed at similar levels in conuMB06749 and w1118 ovaries, little or no protein is present in Df(2R)conu6 ovary lysates. (C) Wing disk protein lysates from w1118 and transheterozygous Df(2R)conu6/Df(2R)Nipped-D animals revealed little or no Conu present in transheterozygous animals. (D and E) Little or no Conu is present in ovary or wing disk lysates from animals with a piggyBac insertion (LL04815), inserted 5′ to the conu translation start site. α-tubulin was used as a loading control for all samples analyzed.
A second insertion allele of conundrum (conuLL04815) was previously created using a modified piggyBac element designed to disrupt splicing and therefore to be highly mutagenic when inserted intronically (Schuldiner et al., 2008). This conu insertion is located in an intron 5′ to the start of translation (Figure 6A). This line is homozygous viable and fertile with no obvious defects, suggesting that the fertility defect in the Df(2R)conu6 line is due to one of the other genes deleted. Ovary and imaginal disk lysates from homozygous conuLL04815 females showed significant reduction of Conu protein (Figure 6, D and E). Likewise, two independent RNAi lines that strongly deplete Conu protein in the wing disk did not cause loss of epithelial integrity or apoptosis (unpublished data), phenotypes characteristic of Moe mutations (Speck et al., 2003; Molnar and de Celis, 2006; Neisch et al., 2010). Together these results suggest that Conu is not the sole protein involved in Moe's regulation of Rho1 activity.
Although we did not detect phenotypes when all cells lacked conu function, we did detect a subtle epithelial phenotype in mosaic animals. When the conu RNAi transgene was expressed in a portion of the imaginal epithelium, we commonly observed a subtle constriction of the epithelium in the mutant tissue adjacent to wild-type cells (Figure 3B). We also observed that the apical ends of mutant cells, as indicated by E-cadherin staining, were reduced in size relative to adjacent wild-type cells (Figure 2B′). This phenotype would be expected if loss of Conu resulted in increased apical Rho1 activity and concomitant increased myosin-based contractility in the apical domain. We were not able to confirm this phenotype by somatic mosaic analysis using our mutant alleles, because conu is located proximal to the 42D FRT available for mitotic recombination analysis on chromosome 2R.
Conu functions as a GAP for Rho1
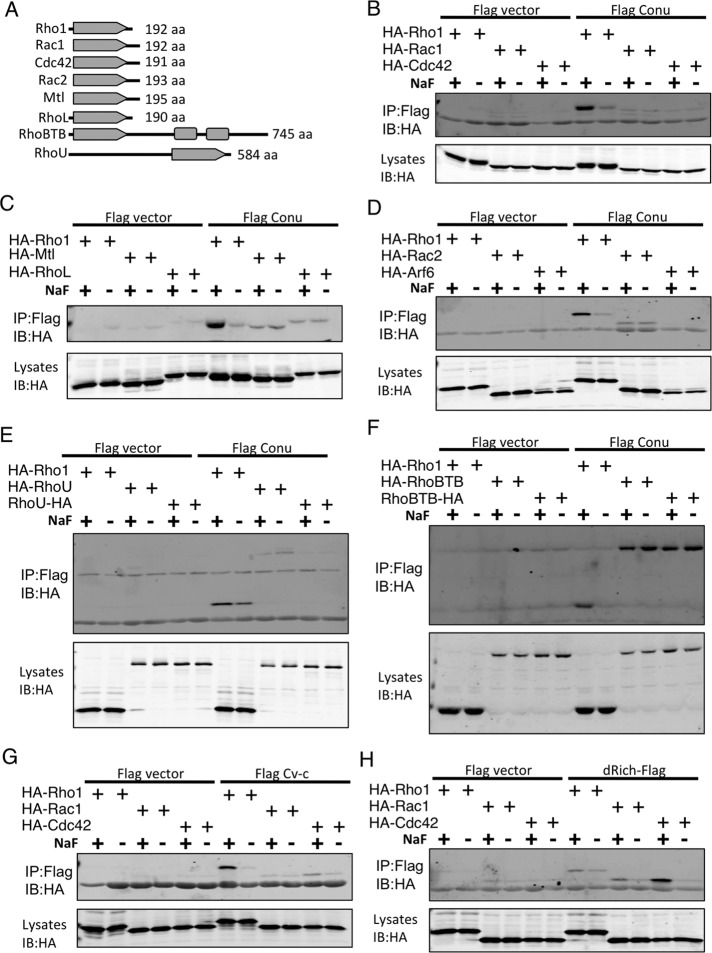
The Drosophila genome encodes 8 Rho family GTPases (Figure 7A). Of these, Rho1, Cdc42, and Rac1, as well as two additional Rac proteins, Rac2 and Mtl, have been well characterized. In addition, there are two GTPases, RhoL and RhoBTB, that are more divergent and have approximately equal similarity to Rho, Rac, and Cdc42 (Johndrow et al., 2004). There is also an uncharacterized GTPase, CG34104, which is most similar to human RhoU/Wrch1 (19% identity, 23% similarity by pairwise alignment, unpublished data). CG34104 will hereafter be referred to as RhoU.
FIGURE 7:
Conu has GAP activity for Rho1. (A) Schematic diagrams of the eight Rho family GTPases in the Drosophila genome. Elongated arrowheads represent the GTPase domains, while boxes represent the BTB (Bric-a-Brac, Tramtrack, Broad-Complex) domains found in RhoBTB. (B–H) S2 cells were transfected with the indicated epitope-tagged DNA constructs, lysed in the presence or absence of NaF, and immunoprecipitated with anti-Flag. (B–F) Conu has GAP activity for Rho1, but not Rac1, Cdc42, Mtl, RhoL, Rac2, RhoU or Arf6, another GTPase that does not belong to the Rho family. In each panel, Rho1 shows greater binding to Conu in the presence of NaF, as expected for the target of its GAP activity. RhoBTB coimmunoprecipitates with Conu (F), but this interaction is not NaF-dependent, indicating that it is not related to Conu's GAP activity. (G and H) Cv-c, a RhoGAP that has specificity for Rho1 and to a much lesser extent Cdc42, and dRich, a RhoGAP that has specificity for Cdc42, were used as positive controls for the specificity of the NaF trapping assay. Each showed the expected GTPase specificity. Protein molecular weights are as follows: HA-Rho1 (35 kDa), HA-Rac1 (32 kDa), HA-Cdc42 (32 kDa), HA-Mtl (34.5 kDa), HA-RhoL (38 kDa), HA-Rac2 (33 kDa), HA-Arf6 (33 kDa), HA-RhoU (81 kDa), RhoU-HA (81 kDa), HA-RhoBTB (97 kDa), RhoBTB-HA (97 kDa).
To investigate whether Conu can function as a Rho family GAP and to determine its GAP specificity, we used a biochemical trapping assay that relies on the ability of a GAP to bind to its cognate GTPase due to the ability of fluoride and magnesium ions to stabilize the GTPase transition state (Vincent et al., 1998). This technique has been used to show that RhoA forms a high-affinity complex with p190 GAP and that Ras interacts with its GAP, NF1 (Vincent et al., 1998; Graham et al., 1999). We developed a variant of this assay in which full-length Conu was expressed together with epitope-tagged constructs of each of the eight GTPases in Drosophila S2 cells; this was followed by co-IP assays in either the presence or absence of NaF. We found that Conu coimmunoprecipitated with Rho1 specifically in the presence of NaF, but not in its absence (Figure 7, B–F). In contrast, Conu did not coimmunoprecipitate any of the seven other GTPases in a fluoride-specific manner, suggesting that, of the Rho family GTPases, Conu has GAP activity specifically for Rho1 (Figure 7, B–F). We found that Conu also coimmunoprecipitated RhoBTB; however, this was not specific to the presence of fluoride (Figure 7F), and we do not know its biological significance. Two additional RhoGAPs with known GTPase specificity, Cv-c and dRich, were used as positive controls to verify the specificity of the NaF assay. In GAP activity assays, Cv-c has been shown to have activity for Rho1 and, to a much lesser extent, Cdc42 (Sato et al., 2010). In the NaF assay, we observed similar results; Cv-c strongly coimmunoprecipitated Rho1 and, to a much lesser extent, Cdc42 in the presence of fluoride but not in its absence (Figure 7G). In contrast, dRich, which has GTPase specificity for Cdc42 (Nahm et al., 2010), strongly coimmunoprecipitated Cdc42 and weakly interacted with Rho1 and Rac1 in a NaF-dependent manner (Figure 7H).
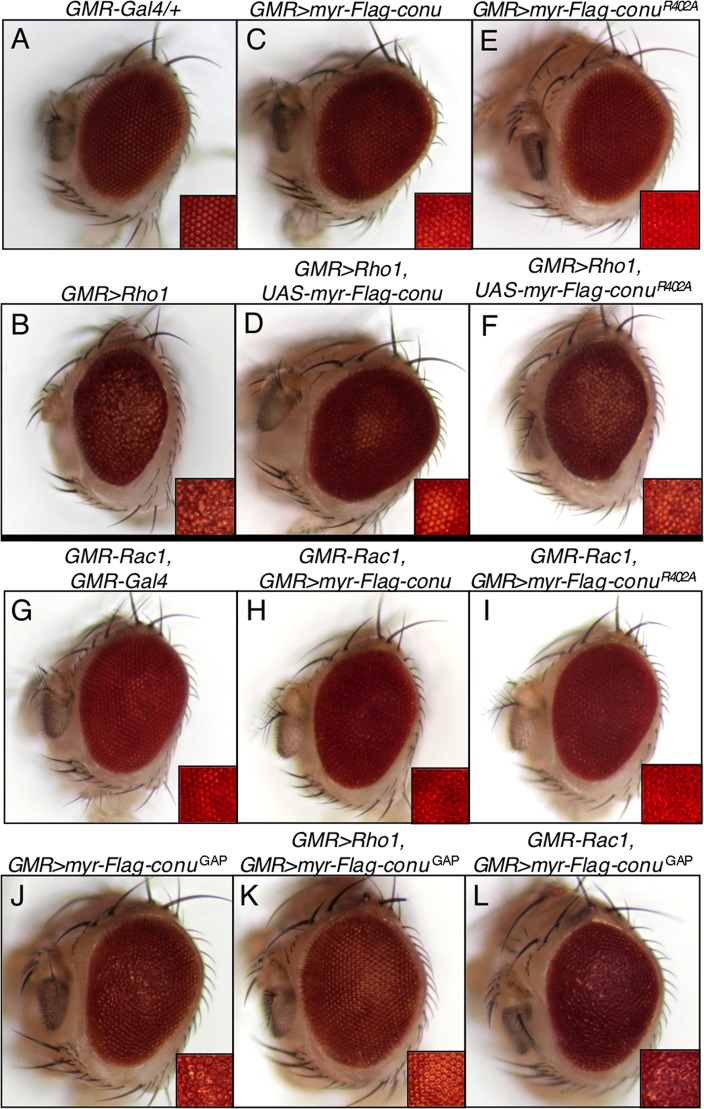
To determine whether Conu functions as a RhoGAP in vivo, we tested its ability to suppress Rho1 overexpression phenotypes in the Drosophila eye. Expression of Rho1 under the GMR promoter results in a rough-eye phenotype in adults due to defects in the ommatidial architecture (Hariharan et al., 1995; Figure 7, A and B). This phenotype has been shown to be suppressible by coexpression of the exotoxin ExoS GAP domain (Avet-Rochex et al., 2005), suggesting that ectopic RhoGAP activity can suppress this phenotype. Expression of a wild-type conu transgene under GMRGal4, which had no phenotype on its own (Figure S3A), caused little if any suppression of the GMR>Rho1 rough-eye phenotype (Figure S3, B and C). In contrast, coexpression of a membrane-tethered version of Conu (myr-conu), which displayed a slight rough-eye phenotype on its own (Figure 8C), strongly suppressed GMR>Rho1 (Figure 8D). Inactivation of the GAP domain (conuR402A) prevented this suppression, as expected if Conu negatively regulates Rho1 activity (Figure 8F). These results suggest that membrane association activates Conu GAP activity and that Conu functions antagonistically to Rho1 in this system.
FIGURE 8:
Conu negatively regulates Rho1 and positively regulates Rac1. (A) In adult eyes, expression of GMRGal4 has no visible ommatidial phenotype, while expression of Rho1 under the GMRGal4 driver results in a rough eye (B). (C) Expression of membrane-tethered Conu, myr-conu, under the GMRGal4 driver results in a slightly rough-eye phenotype, but strongly suppresses the rough-eye phenotype of GMR>Rho1 (D), suggesting that activated Conu functions antagonistically to Rho1. (E and F) This function requires GAP activity, because inactivation of the Conu GAP domain (ConuR402A) strongly inhibits the ability of Conu to suppress Rho1. (G) GMR-Rac1 expression together with GMRGal4 results in a subtle rough-eye phenotype that is enhanced by coexpression of myr-conu (H), suggesting that Conu increases Rac1 activity. (I) A similar enhancement of Rac1 is seen with ConuR402A, indicating that the enhancement is not dependent on GAP activity. (J) Expression of the GAP domain of Conu alone results in a glossy, rough-eye phenotype and fused ommatidia. (K) Coexpression of the GAP domain together with Rho1 under the GMRGal4 driver results in a nearly normal eye, as expected if Conu is a GAP for Rho1. (L) GMR-Rac1 expression together with GMR>myr-conuGAP results in a glossy eye with fused ommatidia.
We also tested genetic interactions between myr-conu and Rac1 or Cdc42 when coexpressed in the eye under the GMR promoter to determine whether Conu GAP activity in this assay was specific to Rho1. Surprisingly, we found that myr-conu did not suppress, but rather mildly enhanced, the GMR-Rac1 eye phenotype (Figure 8, G and H). In contrast, the combined GMR-Cdc42; myr-conu eye phenotype (Figure S3, D and E) appeared similar to GMR>myr-conu alone (Figure 8C), suggesting that Conu does not functionally interact with Cdc42.
It is possible that the observed interactions between Conu and Rho1 and Rac1 reflect Rho/Rac1 cross-talk. To address this possibility, we first asked whether the GAP-deficient myr-conuR402A transgene could enhance the GMR-Rac1 phenotype. Expression of GMR>myr-conuR402A alone led to a slight rough-eye phenotype similar to GMR>myr-conu, in which bristles were misorganized (Figure 8E). Coexpression of GMR>myr-conuR402A with GMR-Rac1 enhanced the rough-eye phenotype (Figure 8I) but had no obvious effect on the phenotype of GMR-Cdc42 (Figure S3F).
We next examined the functions of the isolated Conu GAP domain. Expression of the membrane-tethered GAP domain alone (GMR>myr-conuGAP) results in ommatidial fusions (Figure 8J). Consistent with the idea that this transgene has RhoGAP activity, it strongly suppressed the GMR>Rho1 eye phenotype when coexpressed under the GMRGal4 driver (Figure 8K). We also observed decreased Rho1 staining in cells expressing this construct (unpublished data). In contrast, coexpression of GMR>myr-conuGAP with GMR-Rac1 resulted in a phenotype similar, but not quite identical, to GMR>myr-conuGAP alone (Figure 8L). We do not currently understand the origin of this phenotypic interaction, but think it could result from neomorphic activity of the isolated GAP domain. Taken together, these results suggest that Conu independently negatively regulates Rho1 activity and positively regulates Rac1 activity.
Conu and Arf6 act synergistically in growth control
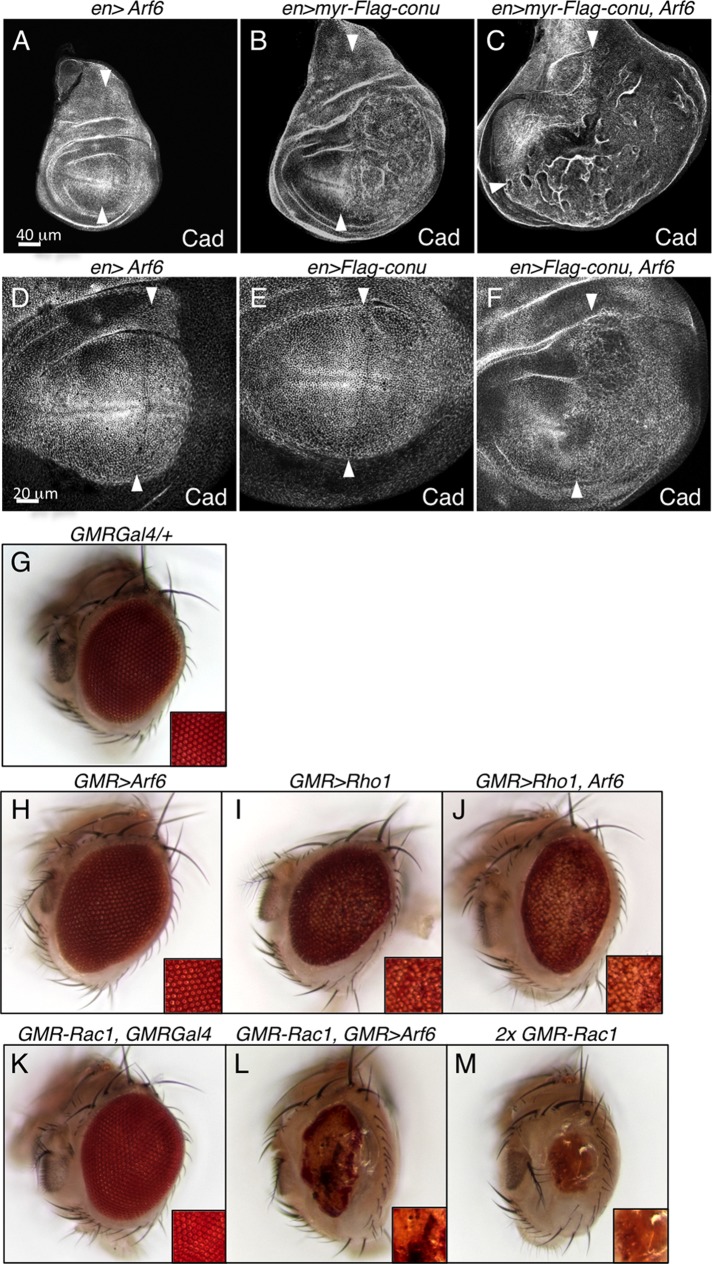
To better determine the functional specificity of Conu, we examined genetic interactions between conu and small GTPases other than Rho1. Specifically, we asked whether coexpression of myr-conu with other Drosophila small GTPases results in enhancement or suppression of the myr-Conu overgrowth phenotype. We started with Rac1, due to the enhancement we observed in the eye, but unfortunately expression of Rac1 under wing-specific drivers resulted in lethality. Parallel experiments gave negative results with Cdc42 and Rac2 (unpublished data), but we observed a strong enhancement with Arf6. Arf6 is an Arf family GTPase that functions to regulate Rac1 activity, phosphatidylinositol 4,5-bisphosphate (PIP2) levels at the plasma membrane, and endocytic membrane trafficking (Chen et al., 2003; Donaldson, 2005; D'Souza-Schorey and Chavrier, 2006; Koo et al., 2007; Bach et al., 2010). While expression of Arf6 alone had very little effect on the epithelium besides slightly altering cell shape (Figures 9, A and D, and S4A), it strongly enhanced the myr-Conu overproliferation phenotype (Figure 9, B and C). However, expression of a dominant-negative Arf6 transgene had no effect on the en>myr-conu phenotype (unpublished data), suggesting myr-Conu does not promote growth via effects on Arf6. Consistent with this idea, we found that Arf6 did not bind to Conu in the NaF GAP assay (Figure 7D). To confirm that this synergy was not unique to the membrane-tethered Conu, we examined interactions between untethered Conu and Arf6, neither of which alone had an overgrowth phenotype (Figure 9, D and E). Coexpression of these proteins resulted, however, in overproliferation and epithelial folding that was similar to myr-Conu, albeit weaker (Figure 9F), suggesting that Arf6 and Conu can act synergistically to promote proliferation. To rule out effects of Arf6 on either Moe or Conu, we examined phospho-Moe and Conu staining in cells expressing Arf6 but did not observe differences (Figure S4, B and C).
FIGURE 9:
Arf6 functions synergistically with Conu and positively regulates Rac1. (A) Expression of Arf6 in the posterior half of the wing disks under enGal4 (to the right of the arrowheads) has no obvious effect on the epithelium, while expression of epitope-tagged myr-conu in the posterior half results in overgrowth of the epithelium (B). (C) Coexpression of Arf6 with myr-conu results in increased overgrowth, (D and E) Expression of either Arf6 or epitope-tagged wild-type conu alone has no effect on the epithelium, but coexpression of the two results in overgrowth of the epithelium (F). (G and H) In adult eyes, GMRGal4/+ has no apparent phenotype, while GMR>Arf6 produces a mildly rough-eye phenotype. (I and J) Coexpression of Arf6 does not appear to enhance the GMR>Rho1 eye phenotype. (K) GMR-Rac1 together with GMRGal4 produces a mildly rough-eye phenotype that is strongly enhanced by coexpression with GMR>Arf6 (L), resulting in an eye that looks similar to two copies of GMR-Rac1 (M) and suggesting that Arf6 increases Rac1 activity. Scale bar in (A) corresponds to (B) and (C); scale bar in (D) corresponds to (E) and (F).
Previous work has shown that Arf6 affects the function of Rho family small GTPases. For example, Arf6 activity can reduce the level of active RhoA in mammalian cells and in an in vitro assay (Boshans et al., 2000). However, Arf6 coexpression did not suppress the GMR>Rho1 adult rough-eye phenotype (Figure 9, G–J), suggesting that it does not regulate Rho1 in Drosophila. Arf6 also has been shown to promote Rac1 activity (D'Souza-Schorey and Chavrier, 2006; Koo et al., 2007; Bach et al., 2010; Ding et al., 2010). Consistent with this idea, we found that Arf6 expression strongly enhanced GMR-Rac1, resulting in an eye phenotype that was similar to expression of two copies of GMR-Rac1 (Figure 9, K–M). In contrast, the GMR-Cdc42 phenotype was unaffected by coexpression with Arf6 (Figure S4, D and E). These results suggest that the ability of Arf6 to promote Rac1 activity is responsible for the synergism between Conu and Arf6, consistent with our observation that Rac1 also enhances the effect of myr-Conu in the eye (Figure 8, E and F).
DISCUSSION
Although previous studies have implicated ERM proteins in the regulation of RhoA activity (Takahashi et al., 1997, 1998; Speck et al., 2003; Hatzoglou et al., 2007; Carreno et al., 2008; Neisch et al., 2010), the molecular mechanisms underlying this function have remained unclear. In this work, we have shown that a previously uncharacterized RhoGAP, Conu, physically and functionally interacts with Moe. Consistent with its sequence similarity to other RhoGAP proteins, we found that Conu has GAP activity for Rho1 in vitro and negatively regulates Rho1 in vivo. Our data further suggest that Moe promotes Conu's RhoGAP activity, and therefore negatively regulates Rho1 by recruiting Conu to the plasma membrane.
Surprisingly, our data suggest that Conu also functions to positively regulate Rac1 activity. Although Conu's ability to promote proliferation is dependent on its RhoGAP activity, this alone is not sufficient to cause overproliferation. Two lines of evidence indicate that this additional function involves positively regulating Rac1. First, coexpression of Conu enhances a rough-eye phenotype associated with Rac1 expression in the eye. This effect is not dependent on Conu GAP activity, indicating that it is not the result of cross-talk between different Rho family small GTPases. Second, Conu acts synergistically with the small GTPase Arf6 in causing overproliferation. Previous studies have shown that Arf6 promotes activation of Rac1 at the plasma membrane (Chen et al., 2003; D'Souza-Schorey and Chavrier, 2006; Koo et al., 2007; Bach et al., 2010). Consistent with the idea that Arf6 promotes Rac1 activity, coexpression of Arf6 strongly enhances the Rac1 eye phenotype. These results suggest that Conu functions to negatively regulate Rho1 activity and positively regulate Rac1 activity, and that the combination of these two effects promotes overproliferation when Conu is activated.
Conu's closest orthologue in the mammalian genome is ARHGAP18, also known as MacGAP, with which it shares 40% sequence similarity. ARHGAP18 has been shown to have GAP activity for the Rho1 homologue RhoA (Maeda et al., 2011). ARHGAP18 was also recently found to promote tumor formation and cell proliferation when ectopically expressed in mammary epithelia (Kim et al., 2011), consistent with our observation that Conu promotes proliferation in Drosophila epithelial tissues. Little is currently known about the details of ARHGAP18 function in mammalian cells, but it is interesting to note that ARHGAP18 is also required for cell spreading (Maeda et al., 2011), a function that is associated with Rac1 activation in many cells. It is also notable that Conu and ARHGAP18 share a region of homology near the N-terminus (aa 45–90) that is separate from the GAP domain and has similarity to the SAM (sterile alpha motif) kazrin repeat 2 domain (unpublished data). SAM domains serve as oligomerization or protein–protein interaction motifs, so it is possible that this domain mediates interactions with one or more Rac1 regulatory proteins. However, our preliminary results indicate that this domain alone does not enhance Rac1 in the eye. It will be interesting to determine whether ARHGAP18 also regulates Rac1 activity, and whether both functions are also involved in growth control in mammals.
Despite the overproliferation phenotypes we have observed from activated Conu, loss-of-function conu mutations are viable and lack obvious imaginal disk phenotypes. Functional redundancy with another RhoGAP seems a likely explanation for this result, because the Drosophila genome encodes 21 RhoGAP proteins in addition to Conu (Greenberg and Hatini, 2011). Thus the loss of a single RhoGAP, such as Conu, may have only a very minor effect on overall Rho1 activity in the imaginal epithelium. In addition, the phenotypes exhibited by Moe mutants, which are much more severe, are probably due to Moe's ability to negatively regulate Rho1 activity via Conu, together with its ability to stabilize the apical cell cortex through interactions with F-actin (Carreno et al., 2008; Kunda et al., 2008). Indeed, we speculate that the severe epithelial defects observed in Moe mutants are the combined result of increased apical actomyosin contractility caused by increased RhoA activity and decreased cortical stability caused by the loss of Moe's membrane–cytoskeletal cross-linking function. In this model, loss of either function alone would have relatively minor effects, but the combined defect would result in the severe cortical disorganization that has been described for Moe mutants. A critical future line of investigation is to identify other Rho1 regulatory proteins that function with Moe to regulate the activity of Rho1 in the apical domain.
A remaining question is how does Conu contribute to growth control in the imaginal disks? The answer to this important question is unclear, but our data suggest that decreased Rho1 activity functions synergistically with Rac1 in this process. Recent studies in Drosophila suggest that Jun N-terminal kinase (JNK) activation downstream of Rac1 activity can promote increased proliferation and metastatic activity in the presence of activated Ras (Brumby et al., 2011). Although attempts to suppress Conu-mediated overgrowth by inhibiting the JNK pathway gave ambiguous results (unpublished data), we observed increased expression of the puc-lacZ reporter for JNK activity in cells expressing activated Conu, consistent with a role of JNK pathway activation in Conu-mediated proliferation. In contrast, other signaling pathways involved in growth control (Notch, TGFβ, Wnt, Hippo, and Hedgehog) were unaffected (Figure S5). Ras activation is thought to protect cells from the proapoptotic effects of JNK pathway activation (Igaki et al., 2006; Wu et al., 2009), and it is possible that decreased Rho1 activity caused by Conu activation has similar effects, especially given that we have previously shown that Rho1 is proapoptotic in these tissues (Neisch et al., 2010). While further studies will be required to more firmly establish the mechanistic basis of Conu function in tissue growth, our findings of a role for Moe and Conu in this process highlight the importance of precise regulation of the apical cell cortex and Rho family small GTPases in growth control.
MATERIALS AND METHODS
Drosophila stocks and crosses
All crosses were carried out at 25ºC. The following lines were obtained from the Bloomington stock center: P{en2.4-Gal4}e16E, P{Gal4}apmd544/Cyo, P{Gal4-ninaE.GMR}12, w; nocSco/SM6a, P{hsILMiT.w+}, Df(2R)M(2)41A2/SM5, Df(2R)Nipped-D/CyO, P{GAL4-Kr.C}, P{UAS-GFP. S65T}, Mi{ET1}CG17082MB06749 (Minos insertion in conu). An enGal4, UAS-MoeRNAi recombinant line was used to deplete Moe levels. The MoeRNAi transgene was described previously (Karagiosis and Ready, 2004). All experiments using the dppGal4 driver were done using a dppblnkGal4, UAS-GFPNLS/TM6B recombinant line. The Moe transgenic lines used were as follows: UAS-Myc-Moe+, UAS-Myc-MoeT559A, UAS-Myc-MoeT559D, which were previously described (Speck et al., 2003); UAS-Myc-Moe∆ACT removes the last 34 amino acids from the C-terminus of Moe (Speck, 2005). Other lines used include UAS-Rho1+ 2.1A (M. Mlodzik, Mount Sinai School of Medicine), UAS-Arf6 and UAS-Arf6DN (E. Chen, Johns Hopkins University), GMR-Rac1+/CyO and GMR-Cdc42+ (J. Settleman, Massachusetts General Hospital), UAS-Rho1RNAi (G. Longmore, Washington University), and E(Spl)Mb-LacZ (D. Bilder, University of California–Berkley). The PiggyBac line conupBAC (dsRed+) LL04815cn bw/CyO cn bw was obtained from the National Institute of Genetics (NIG), Japan.
Yeast two-hybrid methods
The coding sequence for amino acids 1–544 of Moesin (GenBank gi: 386764061) was PCR-amplified, cloned into pB27 as a C-terminal fusion to LexA, and checked by sequencing the entire insert. The library was screened as previously described (Formstecher et al., 2005), except as noted. Sixty-five million clones (sixfold the complexity of the library) were screened, and 227 His+ colonies were selected on a medium lacking tryptophan, leucine, and histidine, supplemented with 5 mM 3-aminotriazole. Sequences of positive clones were used to identify the corresponding interacting proteins in the GenBank database (National Center for Biotechnology Information) using a fully automated procedure.
conu RNAi constructs
Two UAS-conu RNAi transgenes were generated against separate regions of the conu gene. The transgene UAS-conu RNAi-1 was generated by PCR-amplifying the reverse complement of nucleotides 50–800 with primers incorporating a 5′ BamHI and 3′ EcoRV site. The transgene UAS-conu RNAi-2 was generated by PCR-amplifying the reverse complement of nucleotides 1300–1890 with primers to incorporate a 5′ BamHI and 3′ EcoRV. These fragments were then cloned into the pENTR3C vector (Invitrogen, Carlsbad, CA). The subsequent clones were verified by sequencing and then transferred into the pRISE destination vector using a Gateway LR clonase reaction (Invitrogen, Grand Island, NY). The resulting clones were then checked to verify proper recombination. P-element transformation was used to generate transgenic lines (Duke University Model Systems Genomics, Durham, NC). Multiple lines were tested and found to knock down Conu protein levels.
Expression constructs
conu was PCR-amplified from the cDNA clone LD04957 (Drosophila Genomics Resource Center [DGRC], Indiana University, Bloomington, IN) with primers incorporating a 5′ BamHI site but lacking the start codon, and a 3′ XhoI site lacking the stop codon. This PCR fragment was then subcloned into the gateway entry vector pENTR3C (Invitrogen) and verified by sequencing. Site-directed mutagenesis was performed on this clone to make the arginine finger mutant, conuR402A. Wild-type and mutant forms of conu were then transferred to epitope-tagged destination vectors for expression as described below. For making an untagged version of the Conu protein, conu was PCR-amplified from the cDNA clone LD04957 using primers to incorporate a Kozak sequence and a BamHI restriction site at the 5′ end and a stop codon and a XhoI site at the 3′ end. This PCR product was then subcloned into the pENTR3C vector and verified by sequence analysis.
The minimal portion of Conu that interacted with Moe in the yeast two-hybrid screen (nucleotides 553–882), an N-terminal portion of Conu before the GAP domain (nucleotides 1–1065), and a C-terminal portion encompassing the GAP domain (nucleotides 939–1731) were PCR-amplified with primers to incorporate a 5′ BamHI restriction site and a stop codon and a XhoI restriction site at the 3′ end. These PCR fragments were then subcloned into the pENTR3C and verified by sequence analysis.
LR clonase reactions were performed to transfer the conu constructs into the following expression vectors (all obtained from the DGRC): pAFW (actin promoter, 3× N-terminal Flag tag), pAHW (actin promoter, 3× N-terminal hemagglutinin [HA] tag), pTFW (derived from pUAST, 3× N-terminal Flag tag), and pTW (derived from pUAST, untagged).
Moe, MoeT559D, and MoeT559A constructs were made by transferring the coding sequence from previously described constructs (Speck et al., 2003) into pENTR3C, using BamHI and EcoRI sites. Moe∆ACT (aa 1–544), Moe FERM (aa 1–311), and Moe Coiled-coil (aa 299–544) fragments were PCR-amplified with primers incorporating a 5′ BamHI site, a 3′ EcoRI site, and a stop codon. These clones were verified by sequencing, and LR clonase reactions were performed to put all of the Moe constructs into pAFW.
The Rho1, Rac1, and Cdc42 constructs used were described previously (Neisch et al., 2010). Mtl and RhoL were PCR-amplified from a phage cDNA library using gene-specific primers to incorporate 5′ BamHI and 3′ EcoRI sites. RhoU was PCR-amplified from a phage cDNA library with primers to incorporate 5′ XbaI and 3′ EcoRI sites. Rac2 was PCR-amplified from isolated fly DNA of UAS-Rac2.AR (Bloomington Drosophila Stock Center, Indiana University, Bloomington, IN) using pUAST-specific primers followed by amplification with Rac2-specific primers to incorporate 5′ BamHI and 3′ EcoRI sites. RhoBTB was PCR-amplified from the cDNA clone LD24835 (obtained from DGRC) using primers to incorporate 5′ BamHI and 3′ EcoRI sites. Arf6 was PCR-amplified from isolated fly DNA of UAS-Arf6 (E. Chen) using pUAST-specific primers followed by amplification with Arf6-specific primers to incorporate 5′ BamHI and 3′ XhoI sites. The resulting PCR products were then digested and cloned into the pENTR3c vector cut with the same enzymes, and clones were verified by sequencing. LR clonase reactions were performed to transfer the GTPase constructs into the pAHW or pAWH.
The N-terminal fragment of dRich was subcloned from a pAc-dRich–green fluorescent protein (GFP) construct (provided by S. Lee, Nahm et al., 2010) by an EcoRI/SalI restriction digest. The C-terminal portion (∼200 base pairs) was amplified with primers to incorporate 5′ SalI and 3′ EcoRV restriction sites. The N- and C-terminal fragments were then subcloned into the pENTR3c vector digested with EcoRI and EcoRV. The C-terminal portion of the resulting clone was verified by sequencing, and an LR clonase recombination reaction was performed to put dRich into the pAWF vector. Cv-c was PCR-amplified with primers to incorporate 5′ SalI and 3′ EcoRI restriction sites from the cDNA clone RE02250 (obtained from the DGRC). The PCR product was subcloned into the pENTR3c vector cut with the same restriction enzymes. A quick-change reaction was performed on the subsequent clone to change a mutation at base pair 2909 of the coding sequence that resulted in an amino acid substitution and was contained in the original clone. The corrected clone was then used for an LR clonase recombination reaction to put Cv-c into the pAFW and pTFW-myr vectors.
P-element transformation was used to generate transgenic lines (Duke University Model Systems Genomics). Multiple lines of each were tested for expression.
pTFW myr vector
The pTFW myr-tagged vector was generated by cloning the EcoRI fragment of the pTFW vector, which contains the start codon and 3× Flag tag, into pENTR3c. Site-directed mutagenesis was then used to insert the myr sequence directly after the start codon and in front of the 3× Flag tag. The resulting clone was then sequence-verified, and the EcoRI fragment was cloned back into the pTFW vector. Verification of the final pTFW-myr clone was made by digestion.
Immunoprecipitation
S2 cell transfection and Flag immunoprecipitation experiments were carried out as described previously (Neisch et al., 2010). For immunoblotting, 7.5 and 12% SDS–PAGE gels were used and transferred onto nitrocellulose. Antibodies were used at the following concentrations: mouse anti-Flag M2 at 1:20,000 (Sigma-Aldrich, St. Louis, MO), mouse anti–α-tubulin at 1:1000 (Sigma-Aldrich), rabbit anti-HA at 1:5000 (Rockland, Gilbertsville, PA), guinea pig anti-Conu 2151 at 1:5000. Fluorescently labeled IRdye800 (Rockland) and Alexa Fluor 680 (Molecular Probes, Eugene, OR) secondary antibodies were used at 1:5000. Images of the blots were obtained using LI-COR Odyssey with version 2.1 software (Lincoln, NE).
GTPase trapping assays
S2 cells (8.0 × 106) were transfected with the indicated constructs using dimethyldioctadecyl-ammonium bromide (DDAB) at 250 μg/ml (Sigma-Aldrich). Immunoprecipitations were done 2 d posttransfection. Cells were harvested and split into two samples; one sample was lysed in buffer containing 50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 mM EGTA, 10% glycerol, 0.5 mM dithiothreitol, 1% Triton-X 100, and EDTA-free Complete protease inhibitor cocktail (Roche, Indianapolis, IN), and the other was lysed in the same buffer containing 25 mM NaF. Flag IPs were done using anti-Flag M2 agarose beads (Sigma-Aldrich). IP reactions were carried out at 4°C for 1 h. For immunoblotting, 4–20% SDS–PAGE gels were used and transferred onto nitrocellulose. Antibodies were used at the following concentrations: rabbit anti-HA 1:5000 (Rockland) and mouse anti-Flag M2 1:20,000 (Sigma-Aldrich).
Conu antibody production
Polyclonal antibodies recognizing Conu were raised in guinea pigs (Pocono Rabbit Farm and Laboratory, Canadensis, PA) against a glutathione S-transferase (GST) full-length fusion protein. Full-length conu was PCR-amplified with a 5′ BamHI site (lacking the start codon) and a 3′ XhoI site that included the stop codon and was cloned into the pGEX-KText vector. The final clone was confirmed by sequencing. The GST fusion protein was then expressed and purified from BL21 cells.
Tissue lysates
Ovary pairs from eight w1118, w; Mi{ET1}conuMB06749, w; conu6, and w; conuLL04815 females were homogenized in 50 μl of buffer containing 1 mM NaVO4, 50 mM NaF, 180 mM KCl, 10 mM Tris-HCl (pH 6.8), 10 mM glycerol-2-phosphate, 1% Triton-100, 0.1% Tween-20, and Complete protease inhibitor cocktail (Roche). Approximately two ovary equivalents were loaded on a 7.5% low-Bis PAGE gel. Wing disks (35–40) from w1118, w; conu6/Df(2R)Nipped-D, and w; conuLL04815 were homogenized in 30 μl of the buffer used for ovaries. Approximately 25 wing disk equivalents were loaded on a 7.5% low-Bis PAGE gel.
S2 cell lysates
S2 cells (2.7 × 106) in 2 ml of Schneider's insect medium were transfected with the indicated constructs using DDAB at 250 μg/ml (Sigma-Aldrich; Han, 1996). At 2 d posttransfection, 0.5 ml of cells was harvested and boiled in 100 μl of 2X SDS sample buffer. Five microliters of the boiled sample was then loaded on a 7.5% low-Bis PAGE gel.
Immunostaining
Wandering third instar wing disks were dissected in Schneider's medium containing serum and fixed in 2% paraformaldehyde solution for 20 min. BrdU labeling was performed as previously described (Asano et al., 1996). Antibodies were used at the following concentrations: preabsorbed guinea pig anti-Conu 2152 at 1:2000, mouse anti-Flag at 1:20,000 (Sigma-Aldrich), 9B11 mouse anti-Myc at 1:10,000 (Cell Signaling Technology, Danvers, MA), rat anti-cadherin (DCAD2) at 1:250 (Developmental Studies Hybridoma Bank), rabbit anti-cleaved-caspase-3 (Cell Signaling Technology) at 1:1000, guinea pig anti-Coracle at 1:10,000 (Fehon et al., 1994), mouse anti-BrdU at 1:1000 (Invitrogen), rabbit anti-PKCζ 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti–β-galactosidase at 1:1000 (Promega, Madison, WI), guinea pig anti-pMad at 1:500 (E. Laufer, Columbia University), guinea pig anti-Sens at 1:1000 (H. Bellen, Baylor College of Medicine), rat anti-Ci at 1:2 (R. Holmgren, Northwestern University), mouse anti-Dll at 1:500 (D. Duncan, Washington University), and guinea pig anti-Ex at 1:5000 (Maitra et al., 2006). Fluorescent secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were used at 1:1000. Tissues were mounted in ProLong Antifade (Molecular Probes). Confocal images were taken on a Zeiss LSM510 laser-scanning confocal microscope using the LSM acquisition software (Carl Zeiss MicroImaging, Jena, Germany) and either a 40× EC Plan-NeoFluar (numerical aperture [NA] 1.3) objective or a 20× Plan-Apochromat (NA 0.8) objective. Images were then compiled in Adobe Photoshop 7.0 (San Jose, CA).
conuMB06749 Minos element excision screen
The w; Mi{ET1}conuMB06749 line was isogenized before excision screening occurred. conuMB06749 males were crossed to w; nocsco/SM6a, P{hsILMiT.w+} females expressing transposase. Two-day-long collections were heat-shocked every day until pupariation, using the following regimen: 1 h at 37ºC, 1 h at 25ºC, 1 h at 37ºC. Single w; Mi{ET1}conuMB06749/SM6a, P{hsILMiT.w+} adult males were crossed to w; Sco/CyO females, and excision events were scored from this cross by the lack of GFP in the adult eye of non-w+ flies (the Minos element expresses GFP in the adult eye). Four hundred and five such individuals were recovered. For tests for lethality, these individuals were crossed to one of the deficiencies of the conu region, Df(2R)M(2)41A2/SM5 or Df(2R)Nipped-D/CyO, P{GAL4-Kr.C}, P{UAS-GFP.S65T}.
Single-fly genomic preps and PCR analysis
Single homozygous males were collected from w1118, w; Mi{ET1}conuMB06749, and conu6 lines. Flies were ground in 25 μl of buffer containing 10 mM Tris-HCl (pH 8.2), 1 mM EDTA (pH 8.0), 25 mM NaCl, and 200 μg/ml proteinase K and then incubated at 65ºC for 20 min; this was followed by a 2-min incubation at 95ºC to inactivate the proteinase K. Long-range PCRs (to amplify >2 kilobases) were performed using Platinum PCR SuperMix High Fidelity (Invitrogen), according to the manufacturer's instructions.
Inverse PCR analysis of conu6
DNA was extracted from homozygous conu6 flies and inverse PCR was performed (http://flypush.imgen.bcm.tmc.edu/pscreen). Sequencing of products was done using the MI.seq primer. Only a 5′ PCR product was recovered from the conu6 line, and sequencing revealed that this end remained intact, while the 3′ end of the element was deleted.
Conu dsRNA
dsRNA against nucleotides 50–800 (conu dsRNA-1) or nucleotides 1300–1890 (conu dsRNA-2), using PCR products as a template, was prepared using the MEGASCRIPT T7 transcription kit (Ambion, Austin, TX). Primers used were
dsRNA-1 For: 5′-TAATACGACTCACTATAGGGAGAAATTTCTCAATGAGTATTATC-3′
dsRNA-1 Rev: 5′-TAATACGACTCACTATAGGGAGATCATTCTCTGAGGCTACAGC-3′
dsRNA-2 For: 5′-TAATACGACTCACTATAGGGAGAACAGCTACTGTTCATGAACTT-3′
dsRNA-2 Rev: 5′-TAATACGACTCACTATAGGGAGATTAAATAACAAATGGATGTAA-3′
Adult eye images
Eyes from adult males (females were utilized for animals expressing myr-conuGAP) were imaged on a Leica MZFIII dissecting microscope with a Canon Rebel T2i camera. A through-focus series of images was captured of each eye and compiled into an extended focus image using iSolution Lite (iMTechnology, Vancouver, BC, Canada). Images were then processed using Adobe Photoshop 7.0.
Supplementary Material
Acknowledgments
We thank C. Berry for assistance with Minos element excision screening, L. Zeng for technical assistance, and P. Vanderzalm for imaging assistance. We thank M. Nahm and S. Lee for the pAc-dRich-GFP construct and E. Chen, M. Mlodzik, J. Settleman, G. Longmore, D. Bilder, the Bloomington Drosophila Stock Center, and the NIG (Japan) for providing fly stocks, and the DSHB, E. Laufer, H. Bellen, R. Holmgren, and D. Duncan for providing antibodies. Thanks to M. Glotzer, I. Rebay, and G. Marques for comments on the manuscript and the Fehon and Rebay labs for helpful suggestions on this work. This work was funded by grant GM087588 from the National Institutes of Health to R. F. and by a GenHomme Network Grant (02490-6088) to Hybrigenics.
Abbreviations used:
- ARHGAP18
Rho GTPase–activating protein 18
- BrdU
bromodeoxyuridine
- co-IP
coimmunoprecipitation
- Conu
Conundrum
- DDAB
dimethyldioctadecyl-ammonium bromide
- dsRNA
double-stranded RNA
- ERM
ezrin, radixin, and moesin
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HA
hemagglutinin
- JNK
Jun N-terminal kinase
- MID
Moe interaction domain
- Moe
Moesin
- myr
myristoylation
- NA
numerical aperture
- NIG
National Institute of Genetics, Japan
- RNAi
RNA interference
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0800) on March 6, 2013.
*Present address: Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis, MN 55455.
REFERENCES
- Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- Avet-Rochex A, Bergeret E, Attree I, Meister M, Fauvarque MO. Suppression of Drosophila cellular immunity by directed expression of the ExoS toxin GAP domain of Pseudomonas aeruginosa. Cell Microbiol. 2005;7:799–810. doi: 10.1111/j.1462-5822.2005.00512.x. [DOI] [PubMed] [Google Scholar]
- Bach AS, Enjalbert S, Comunale F, Bodin S, Vitale N, Charrasse S, Gauthier-Rouviere C. ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways. Mol Biol Cell. 2010;21:2412–2424. doi: 10.1091/mbc.E09-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshans RL, Szanto S, van Aelst L, D'Souza-Schorey C. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol Cell Biol. 2000;20:3685–3694. doi: 10.1128/mcb.20.10.3685-3694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Brumby AM, Goulding KR, Schlosser T, Loi S, Galea R, Khoo P, Bolden JE, Aigaki T, Humbert PO, Richardson HE. Identification of novel Ras-cooperating oncogenes in Drosophila melanogaster: a RhoGEF/Rho-family/JNK pathway is a central driver of tumorigenesis. Genetics. 2011;188:105–125. doi: 10.1534/genetics.111.127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J Cell Biol. 2008;180:739–746. doi: 10.1083/jcb.200709161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Ding X, et al. Phospho-regulated ACAP4-ezrin interaction is essential for histamine-stimulated parietal cell secretion. J Biol Chem. 2010;285:18769–18780. doi: 10.1074/jbc.M110.129007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Arfs, phosphoinositides and membrane traffic. Biochem Soc Trans. 2005;33:1276–1278. doi: 10.1042/BST0331276. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Dawson IA, Artavanis-Tsakonas S. A Drosophila homologue of membrane-skeleton protein 4.1 is associated with septate junctions and is encoded by the coracle gene. Development. 1994;120:545–557. doi: 10.1242/dev.120.3.545. [DOI] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DL, Eccleston JF, Chung CW, Lowe PN. Magnesium fluoride-dependent binding of small G proteins to their GTPase-activating proteins. Biochemistry. 1999;38:14981–14987. doi: 10.1021/bi991358e. [DOI] [PubMed] [Google Scholar]
- Greenberg L, Hatini V. Systematic expression and loss-of-function analysis defines spatially restricted requirements for Drosophila RhoGEFs and RhoGAPs in leg morphogenesis. Mech Dev. 2011;128:5–17. doi: 10.1016/j.mod.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res. 1996;24:4362–4363. doi: 10.1093/nar/24.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan IK, Hu KQ, Asha H, Quintanilla A, Ezzell RM, Settleman J. Characterization of rho GTPase family homologues in Drosophila melanogaster: overexpressing Rho1 in retinal cells causes a late developmental defect. EMBO J. 1995;14:292–302. doi: 10.1002/j.1460-2075.1995.tb07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzoglou A, Ader I, Splingard A, Flanders J, Saade E, Leroy I, Traver S, Aresta S, de Gunzburg J. Gem associates with Ezrin and acts via the Rho-GAP protein Gmip to down-regulate the Rho pathway. Mol Biol Cell. 2007;18:1242–1252. doi: 10.1091/mbc.E06-06-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SC, Fehon RG. Phosphorylation and activity of the tumor suppressor Merlin and the ERM protein Moesin are coordinately regulated by the Slik kinase. J Cell Biol. 2006;175:305–313. doi: 10.1083/jcb.200608009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006;16:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Johndrow JE, Magie CR, Parkhurst SM. Rho GTPase function in flies: insights from a developmental and organismal perspective. Biochem Cell Biol. 2004;82:643–657. doi: 10.1139/o04-118. [DOI] [PubMed] [Google Scholar]
- Karagiosis SA, Ready DF. Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development. 2004;131:725–732. doi: 10.1242/dev.00976. [DOI] [PubMed] [Google Scholar]
- Kim HH, van den Heuvel AP, Schmidt JW, Ross SR. Novel common integration sites targeted by mouse mammary tumor virus insertion in mammary tumors have oncogenic activity. PLoS One. 2011;6:e27425. doi: 10.1371/journal.pone.0027425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Pelling AE, Liu T, Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18:91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- Li Q, Nance MR, Kulikauskas R, Nyberg K, Fehon R, Karplus PA, Bretscher A, Tesmer JJ. Self-masking in an intact ERM-merlin protein: an active role for the central α-helical domain. J Mol Biol. 2007;365:1446–1459. doi: 10.1016/j.jmb.2006.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu Q, Liu S, Zhang J, Zhang Y. New member of the guanosine triphosphatase activating protein family in the human epididymis. Acta Biochim Biophys Sin (Shanghai) 2008;40:855–863. [PubMed] [Google Scholar]
- Maeda M, et al. ARHGAP18, a GTPase-activating protein for RhoA, controls cell shape, spreading, and motility. Mol Biol Cell. 2011;22:3840–3852. doi: 10.1091/mbc.E11-04-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- McClure KD, Schubiger G. Developmental analysis and squamous morphogenesis of the peripodial epithelium in Drosophila imaginal discs. Development. 2005;132:5033–5042. doi: 10.1242/dev.02092. [DOI] [PubMed] [Google Scholar]
- Molnar C, de Celis JF. Independent roles of Drosophila Moesin in imaginal disc morphogenesis and hedgehog signalling. Mech Dev. 2006;123:337–351. doi: 10.1016/j.mod.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Myster SH, Wang F, Cavallo R, Christian W, Bhotika S, Anderson CT, Peifer M. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics. 2004;166:807–822. doi: 10.1534/genetics.166.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm M, Long AA, Paik SK, Kim S, Bae YC, Broadie K, Lee S. The Cdc42-selective GAP rich regulates postsynaptic development and retrograde BMP transsynaptic signaling. J Cell Biol. 2010;191:661–675. doi: 10.1083/jcb.201007086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch AL, Speck O, Stronach B, Fehon RG. Rho1 regulates apoptosis via activation of the JNK signaling pathway at the plasma membrane. J Cell Biol. 2010;189:311–323. doi: 10.1083/jcb.200912010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D, Sugimura K, Satoh D, Uemura T. Crossveinless-c, the Drosophila homolog of tumor suppressor DLC1, regulates directional elongation of dendritic branches via down-regulating Rho1 activity. Genes Cells. 2010;15:485–500. doi: 10.1111/j.1365-2443.2010.01399.x. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck O. Drosophila ERM Protein Moesin Functions in Epithelia Integrity and Cell Survival. Durham, NC: Duke University Press; 2005. [Google Scholar]
- Speck O, Hughes SC, Noren NK, Kulikauskas RM, Fehon RG. Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature. 2003;421:83–87. doi: 10.1038/nature01295. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Sasaki T, Mammoto A, Hotta I, Takaishi K, Imamura H, Nakano K, Kodama A, Takai Y. Interaction of radixin with Rho small G protein GDP/GTP exchange protein Dbl. Oncogene. 1998;16:3279–3284. doi: 10.1038/sj.onc.1201874. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Sasaki T, Mammoto A, Takaishi K, Kameyama T, Tsukita S, Takai Y. Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J Biol Chem. 1997;272:23371–23375. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- Valderrama F, Thevapala S, Ridley AJ. Radixin regulates cell migration and cell-cell adhesion through Rac1. J Cell Sci. 2012;125:3310–3319. doi: 10.1242/jcs.094383. [DOI] [PubMed] [Google Scholar]
- Vincent S, Brouns M, Hart MJ, Settleman J. Evidence for distinct mechanisms of transition state stabilization of GTPases by fluoride. Proc Natl Acad Sci USA. 1998;95:2210–2215. doi: 10.1073/pnas.95.5.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhuang Y, Han M, Xu T, Deng K. Ras promotes cell survival by antagonizing both JNK and Hid signals in the Drosophila eye. BMC Dev Biol. 2009;9:53. doi: 10.1186/1471-213X-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.