Sp100 isoforms differentially regulate a CMV-promoter–regulated transcription site, which can be visualized in single cells. Sp100A promotes decondensation and increases lysine acetylation. However, it cannot overcome Daxx- and ATRX-mediated repression, indicating that PML-NB/ND10 factors function within a regulatory hierarchy.
Abstract
Promyelocytic leukemia nuclear bodies (PML-NBs)/nuclear domain 10s (ND10s) are nuclear structures that contain many transcriptional and chromatin regulatory factors. One of these, Sp100, is expressed from a single-copy gene and spliced into four isoforms (A, B, C, and HMG), which differentially regulate transcription. Here we evaluate Sp100 function in single cells using an inducible cytomegalovirus-promoter–regulated transgene, visualized as a chromatinized transcription site. Sp100A is the isoform most strongly recruited to the transgene array, and it significantly increases chromatin decondensation. However, Sp100A cannot overcome Daxx- and α-thalassemia mental retardation, X-linked (ATRX)–mediated transcriptional repression, which indicates that PML-NB/ND10 factors function within a regulatory hierarchy. Sp100A increases and Sp100B, which contains a SAND domain, decreases acetyl-lysine regulatory factor levels at activated sites, suggesting that Sp100 isoforms differentially regulate transcription by modulating lysine acetylation. In contrast to Daxx, ATRX, and PML, Sp100 is recruited to activated arrays in cells expressing the herpes simplex virus type 1 E3 ubiquitin ligase, ICP0, which degrades all Sp100 isoforms except unsumoylated Sp100A. The recruitment Sp100A(K297R), which cannot be sumoylated, further suggests that sumoylation plays an important role in regulating Sp100 isoform levels at transcription sites. This study provides insight into the ways in which viruses may modulate Sp100 to promote their replication cycles.
INTRODUCTION
Promyelocytic leukemia nuclear bodies (PML-NBs)/nuclear domain 10s (ND10s) are nuclear structures implicated in a range of cellular processes, including intrinsic immunity, senescence, apoptosis, oncogenesis, and tumor suppression, yet their molecular functions are poorly understood (Lallemand-Breitenbach and de The, 2010; Geoffroy and Chelbi-Alix, 2011). Their importance for proper cellular function, however, is highlighted by the fact that their dissociation is linked to oncogenesis and viral infection. For example, the PML–retinoic acid receptor α (PML-RARA) fusion protein, which drives acute promyelocytic leukemia, disperses PML-NBs/ND10s (Lallemand-Breitenbach and de The, 2010). In addition, ICP0, the herpes simplex virus type 1 (HSV-1) E3 ubiquitin ligase required for efficient entry into the lytic cycle and latent genome reactivation (Everett, 2000; Hagglund and Roizman, 2004), disperses PML-NBs/ND10s by degrading PML protein, which is required for their assembly (Ishov et al., 1999; Zhong et al., 2000). However, the exact molecular events disrupted by the dissolution of PML-NBs/ND10s are unknown.
A large number of transcription and chromatin regulatory factors localize to PML-NBs/ND10s, suggesting that these processes occur within them. Consistent with this hypothesis, the PML-NB/ND10 proteins, Daxx and α-thalassemia mental retardation, X-linked (ATRX), were recently reported to regulate replication-independent histone H3.3 chromatin assembly at heterochromatic sites including, embryonic stem cell (ESC) telomeres, pericentromeres, and a cytomegalovirus (CMV)-promoter–regulated transgene array (Szenker et al., 2011; Newhart et al., 2012). Daxx is a histone H3.3–specific chaperone, and ATRX is an Snf2-type chromatin-remodeling factor, mutated in the intellectual disability syndrome α-thalassemia mental retardation, X-linked (Picketts et al., 1996). Previously, both were known to be transcriptional repressors and inhibitors of HSV-1, CMV, and Epstein–Barr virus replication (Lukashchuk and Everett, 2010; Tsai et al., 2011; Tomtishen, 2012). Knocking down ATRX in ESCs induces telomere dysfunction (Wong et al., 2010). Loss-of-function Daxx and ATRX mutations have also been identified in brain and pancreatic neuroendocrine tumors (PanNETs; Heaphy et al., 2011; Jiao et al., 2011; Molenaar et al., 2012; Schwartzentruber et al., 2012). The finding that all PanNETs with ATRX or DAXX mutations are positive for the alternative lengthening of telomeres (ALT) pathway (Heaphy et al., 2011) and PML protein is enriched at ALT telomeres (Henson et al., 2002) suggests that PML-NBs/ND10s play important roles in heterochromatin regulation.
Sp100 is another core PML-NB/ND10 protein linked to viral gene regulation (Negorev et al., 2006, 2009). It is a single-copy gene that gives rise to four alternatively spliced isoforms (Weichenhan et al., 1998; Seeler et al., 2001), three of which, Sp100B, C, and HMG (see discussion of Figure 3A later in the paper), contain the SAND domain (named after Sp100, AIRE-1, NucP41/45, and DEAF-1), which preferentially binds DNA composed of unmethylated CpGs (Bottomley et al., 2001; Isaac et al., 2006). These isoforms repress transcription from viral promoters (Negorev et al., 2006, 2009). In addition to the SAND domain, Sp100C contains a bromodomain, which binds acetylated lysines (Filippakopoulos and Knapp, 2012), and a plant homeodomain, which binds methylated lysines in histones (Baker et al., 2008). Sp100 high-mobility group (Sp100HMG) contains a domain homologous to the HMG-1/2 family of nonhistone chromosomal DNA-binding proteins (Seeler et al., 1998). The presence of these domains in Sp100B, C, and HMG suggests that these isoforms interact with DNA and chromatin.
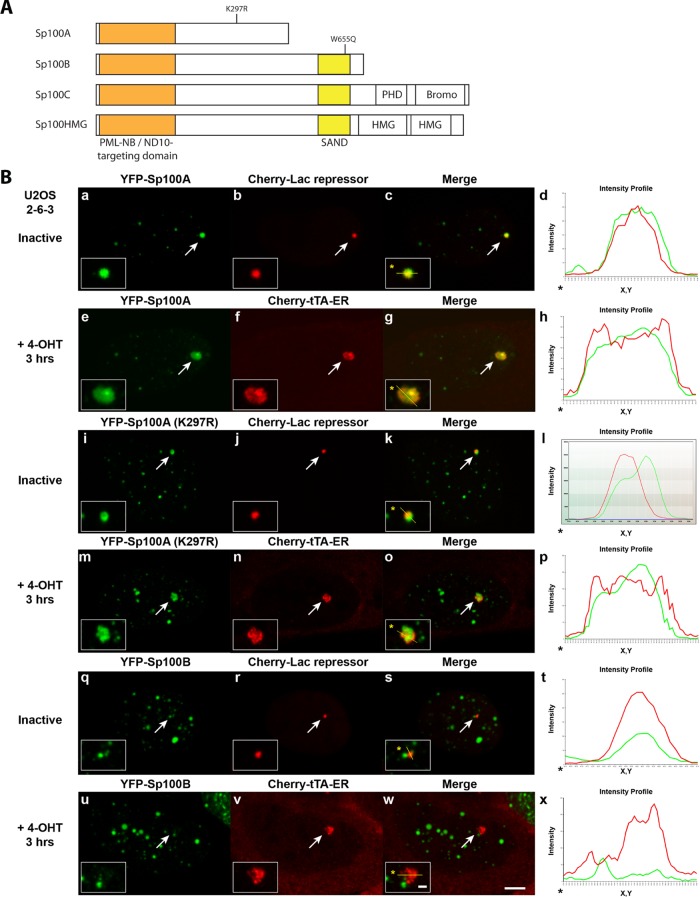
FIGURE 3:
Analysis of Sp100 isoform recruitment to inactive and activated transgene arrays in U2OS cells. (A) Diagram of the domain organization of the Sp100 isoforms and the locations of the K297R sumoylation mutation in Sp100A and the W655Q mutation in the SAND domains of Sp100B, C, and HMG. (B) Localization of (a–h) YFP-Sp100A, (i–p) YFP-Sp100A(K297R), and (q–x) YFP-Sp100B at inactive arrays, marked by Cherry-lac repressor, and activated arrays, marked by Cherry-tTA-ER, in U2OS (2-6-3) cells. Arrows indicate the transgene array locations. Yellow lines in enlarged merge insets show the path through which the red and green intensities were measured in the intensity profiles (d, h, l, p, t, and x). Asterisks mark the start of the measured lines. Scale bar, 5 μm. Scale bars in enlarged inset, 1 μm.
In contrast to the repressive effects of the SAND-domain isoforms, Sp100A, which shares 477 of its 480 amino acids with Sp100B, C, and HMG, promotes transcription (Wasylyk et al., 2002; Negorev et al., 2006, 2009). Of interest, Sp100B, with a point mutation in the SAND domain (W655Q) predicted to disrupt its interaction with DNA (Bottomley et al., 2001), has also been reported to promote transcription (Negorev et al., 2006, 2009). This suggests that the SAND domain masks a transcriptional-promoting activity in the Sp100 N-terminus. It also highlights the importance of alternative splicing in regulating the differential functions of Sp100 in transcription.
Much of what we know about the way specific factors, including Sp100 (Negorev et al., 2006, 2009), regulate transcription was learned through the use of transiently expressed reporter constructs. However, these assays typically only measure the total output of mRNA and protein in a cell population and therefore cannot provide insight into the mechanisms through which factors interact and regulate chromatin in single cells. In addition, transcription requires the concerted actions of multiple regulatory factors, whose coordinated interactions need to be determined. The development of single-cell imaging systems, which allow the visualization of regulatory factor recruitment to molecularly defined transcription sites, now makes it possible to attempt investigations into this coordination (Rafalska-Metcalf and Janicki, 2007).
Studies focused on identifying cellular factors that interact with viruses during infection revealed many of the PML-NB/ND10 factors degraded and disorganized by viruses in their efforts to evade intrinsic immunity (Geoffroy and Chelbi-Alix, 2011; Van Damme and Van Ostade, 2012). However, because PML-NB/ND10 function is incompletely understood, it has been difficult to fully discern how virally induced changes in the levels and organization of these factors affect gene expression. Here we report experiments examining the dynamic behavior of PML-NB/ND10 factors at a chromatinized transcriptional reporter construct in single cells. We recently showed that PML, Daxx, and ATRX are enriched at the CMV promoter of this transgene and that the Daxx and ATRX pathway is required for its transcriptional repression (Newhart et al., 2012). The presence of PML protein technically classifies this transgene array site as a PML-NB/ND10 and suggests that we can use this single-cell gene-imaging system to study how PML-NB/ND10 proteins regulate transcription. Here we use it to investigate the function of Sp100 and define its place in the regulatory hierarchy of PML-NB/ND10 factors. Our results advance the understanding of PML-NB/ND10 function and also provide insight into viral latency.
RESULTS
Fluorescent fusion proteins allow the visualization of specific gene elements at a transcription site
To study the temporal and spatial organization of gene expression, we use a CMV-promoter–regulated inducible transgene that can be visualized in single cells (Figure 1A; Janicki et al., 2004). The transgene is constructed from sequence elements that allow the spatial dissection of DNA and RNA in living cells (Shav-Tal et al., 2004; Rafalska-Metcalf and Janicki, 2007; Rafalska-Metcalf et al., 2010). The transgene-binding and gene-regulatory factors being studied can be fused to cyan, yellow, and cherry fluorescent proteins (CFP, YFP, and Cherry, respectively) and coexpressed in different combinations to investigate the way in which transcription and chromatin organization are coordinately regulated at a transcription site (Shaner et al., 2005). Figure 1A shows all of the studied factors labeled with YFP, but to dissect functions, factors are labeled with the various fluorescent tags as described in the figures and legends.
FIGURE 1:
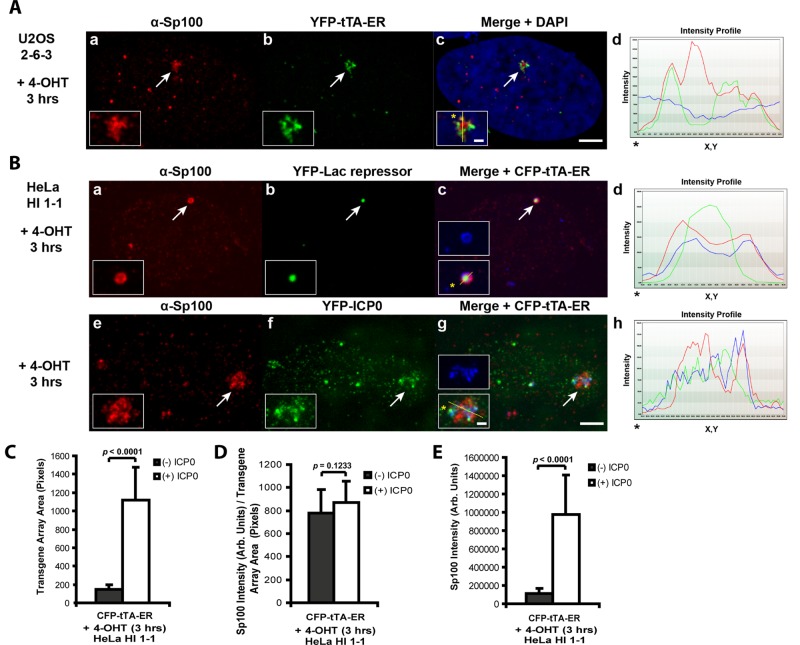
Sp100 is recruited to the CMV promoter of the inducible transgene in the U2OS and HeLa cell lines. (A) Diagram of the inducible transgene drawn to scale. Expression of YFP-lac repressor allows the transgene to be visualized in both the inactive and the active state. Transcription is induced from the minimal CMV promoter by the activators, YFP-tTA-ER or ER-tTA, in the presence of 4-OHT and rtTA in the presence of Dox. The transcribed RNA encodes CFP fused to a peroxisomal targeting signal (SKL). The RNA is visualized by YFP-MS2, which binds to the stem loops in the transcript. The 3′ end of the transcription unit is composed of the intron 2 splicing unit of the rabbit β-globin gene. YFP-tagged regulatory factors can be monitored for recruitment to the array by coexpression with the fluorescently tagged lac repressor and/or activator proteins. All of the factors are shown in YFP-tagged form. However, Cherry- and CFP-tagged versions are also used in different combinations in the assays as described in figures and legends. (B) Immunofluorescence localization of Sp100 at the inactive array, marked by YFP-lac repressor, in (a–d) U2OS (2-6-3) and (e–h) HeLa (HI 1-1) cells. Arrows indicate the location of the transgene array. Yellow lines in enlarged merge insets show the path through which the red, green, and blue intensities were measured in the intensity profiles (d, h). Asterisks mark the start of the measured lines. Scale bar, 5 μm. Scale bars in enlarged inset, 1 μm. (C) Diagram of the transgene showing the location of the primer pairs used for real-time PCR in the Sp100 ChIP assays with chromatin lysates prepared from inactive U2OS (2-6-3) and HeLa (HI 1-1) cell lines. Results are the average of at least three independent experiments. SDs are given in the form of error bars, and p values are calculated using unpaired t test.
This transgene integrates into cellular genomes as a multicopy array, which can be visualized when the lac repressor protein is expressed (Figure 1, A and B, b and f). We fused the tetracycline transcriptional activator (tTA) to both an autofluorescent protein and the estrogen receptor (ER) hormone–binding domain (Figure 1A; Rafalska-Metcalf et al., 2010). In the absence of tetracycline, tTA binds constitutively to the tetracycline response elements (TREs) and induces transcription. However, the protein product of this activator construct, YFP-tTA-ER, remains in the cytoplasm until 4-hydroxytamoxifen (4-OHT) induces it to enter the nucleus, where it initiates transcription and permits the visualization of the transcription site. The reverse tet transcriptional activator (rtTA), which activates transcription in the presence of the tetracycline analogue doxycycline (Dox), can also be used.
The transcription unit in the transgene is composed of CFP fused to a peroxisomal targeting signal (SKL), 24 repeats of the MS2 translational operator, and the intron 2 splicing module from rabbit β-globin (Figure 1A). When the RNA is expressed, the MS2 protein binds to the stem loop repeats in the transcript and allows the RNA at the transcription site to be visualized. When the CFP-SKL protein is translated, it accumulates in the peroxisomes in the cytoplasm and confirms that the mRNA has been properly processed and exported. The fusion of CFP to the peroxisomal targeting signal (SKL) prevents it from interfering with the visualization of the CFP-tagged factors used to monitor events at the transcription site in the nucleus.
The CMV-promoter–regulated transgene (Figure 1A) has been stably integrated into U2OS (cell line, 2-6-3) and HeLa (cell line, HI 1-1) cells (Janicki et al., 2004; Newhart et al., 2012). U2OS cells are a human osteosarcoma cell line, which is ALT positive, does not express ATRX, and can be infected with ICP0-null HSV-1 (Yao and Schaffer, 1995; Lukashchuk and Everett, 2010; Heaphy et al., 2011). HeLa cells, derived from cervical carcinoma, express both Daxx and ATRX. In the HeLa cell line, the transgene is transcriptionally repressed; however, ICP0 permits activation. Specifically, ICP0 accumulates at the array and removes Daxx, ATRX, and PML from the site (Newhart et al., 2012). Both the U2OS and HeLa cell lines are used in this study to analyze the function(s) of Sp100 in transcription.
Sp100 is enriched at the CMV promoter
We found that several constituents of PML-NBs/ND10s (Daxx, ATRX, and PML) are recruited to the transgene described in Figure 1A and that Sp100 isoforms differentially regulate viral promoters (Negorev et al., 2006, 2009; Newhart et al., 2012). To determine whether we could use this single-cell imaging system to investigate the function of Sp100 in transcription, we used an antibody that recognizes all four isoforms to evaluate its recruitment to the array. Given that Sp100 colocalizes with YFP-lac repressor, which marks the inactive site, in both the U2OS and HeLa cell lines, this indicates that Sp100, like Daxx, ATRX, and PML, is also recruited to the inactive transcription site (Figure 1B). The intensity profiles (Figure 1B, d and h) graph the pixel intensity measurements along the line drawn through the array (note the yellow line in enlarged insets in the merged images, Figure 1B, c and g). 4′,6-Diamidino-2-phenylindole (DAPI) staining of the DNA is represented by the blue line in the graph, which is uniform across the region. The significant overlap between the red (Sp100) and green (YFP-lac repressor) signals indicates the significant enrichment of Sp100 at the site in relation to YFP-lac repressor, which binds to the 10 kb of lac operator repeats upstream of the transcription unit (Figure 1A). The lack of complete overlap between the red and green signals suggests that substructural features of the chromatinized array can be detected.
Chromatin immunoprecipitation (ChIP) analyses of the inactive transgene from both the U2OS and HeLa cell lines indicate that Sp100, like Daxx, ATRX, and PML, is also enriched at the CMV promoter (Figure 1C). Taken together, these results suggest that sequences in the CMV promoter seed the accumulation of PML-NB/ND10 proteins, including Sp100, at the array and that this system can be used to study how they regulate transcription.
Sp100 is enriched at the activated transgene array
To determine whether Sp100 is also enriched at the transcriptionally activated arrays, we first evaluated it in the ATRX-null U2OS cell line expressing the 4-OHT–inducible-activator YFP-tTA-ER, which both marks and activates the transcription site (Figure 1A). The enrichment of endogenous Sp100 at the activated array in U2OS cells (Figure 2A) indicates that it is not displaced by transcriptional events. The enrichment of Sp100 at both the inactive (Figure 1B, a–d) and activated (Figure 2A) arrays in U2OS cells also indicates that ATRX is not required for its recruitment.
FIGURE 2:
Sp100 is enriched at the activated transgene arrays in U2OS and HeLa cells. (A) Immunofluorescence staining of Sp100 at the YFP-tTA-ER–activated transgene array in (a–d) U2OS (2-6-3) cells. (B) Immunofluorescence staining of Sp100 at the CFP-tTA-ER–activated transgene array in (a–d) control and (e–h) YFP-ICP0–expressing HeLa (HI 1-1) cells. YFP-lac repressor (b) is expressed in control cells to identify the array. Arrows indicate the location of the transgene array. Yellow lines in enlarged merged insets show the path through which the red, green, and blue intensities were measured in the intensity profiles (d, h). Asterisks mark the start of the measured lines. Scale bar, 5 μm. Scale bars in enlarged inset, 1 μm. (C) Average pixel areas of the activated arrays in HeLa (HI 1-1) cells (+/−) ICP0 as demarcated by the array-binding proteins. (D) Average intensities (total intensity/transgene array area) of Sp100 at the activated arrays in HeLa (HI 1-1) cells (+/−) ICP0. (E) Total intensity levels of Sp100 at the activated arrays in HeLa (HI 1-1) cells (+/−) ICP0. SDs are given in the form of error bars, and p values are calculated using the unpaired t test; (−) ICP0 (n = 20); (+) ICP0 (n = 28).
In HeLa cells, which express both Daxx and ATRX, the transgene array is refractory to activation; however, ICP0 permits transcription (Newhart et al., 2012). To evaluate ICP0’s effects on Sp100, we immunostained HeLa cells activated with CFP-tTA-ER (Figure 2B). In the absence of ICP0, Sp100 is enriched at the array, which remains condensed (Figure 2B, a–d). The state of chromatin decondensation is evaluated by measuring the pixel area of the region occupied by the transgene-binding proteins (Figure 2C; note the smaller average array area in (–) ICP0 cells). In fact, to identify the site in the absence of ICP0, it is necessary to coexpress YFP-lac repressor because the activator, CFP-tTA-ER, forms only a faint ring around the array, suggesting that it is unable to access its binding sites (Figure 2B, c, enlarged inset of blue signal).
Both the average (Figure 2D; total intensity/array area) and total (Figure 2E) intensities of Sp100 at the array in the presence and absence of ICP0 are presented. Although the average intensity is the same (+/–) ICP0, Sp100’s total intensity is higher in the presence of ICP0. The increase in total intensity is likely due to an increase in the number of Sp100-binding sites from array decondensation in the ICP0-expressing cells (Figure 2, B, e–h, and C). ICP0 degrades Sp100B, C, and HMG and sumoylated Sp100A (Everett et al., 2009). This suggests that it is the unsumoylated form of Sp100A that accumulates at the array in ICP0-expressing cells.
Sp100A is the isoform most strongly recruited to the transgene array
Our previous reports on the effects of the Sp100 isoforms on transcription used transiently expressed reporter constructs driven by viral promoters (Negorev et al., 2006, 2009). To evaluate the effects of the Sp100 isoforms on the chromatinized CMV-promoter–regulated reporter, we first examined the recruitment of YFP-tagged constructs (Figure 3A) to arrays in the U2OS cell line (Figure 3B and Supplemental Figure S1). Sp100A was the most strongly recruited isoform at both the inactive site, marked by Cherry-lac repressor (Figure 3B, a–d), and the activated site, marked by Cherry-tTA-ER (Figure 3B, e–h). To determine whether sumoylation is required for Sp100A recruitment, we examined the localization of Sp100A(K297R), in which the sumoylated lysine is converted to an arginine (Figure 3B, i–p; Sternsdorf et al., 1999). The enrichment of Sp100A(K297R) at both inactive and activated arrays indicates that sumoylation is not required for Sp100A recruitment. This is consistent with a report showing that sumoylation is not required for Sp100A targeting to PML-NBs/ND10s (Sternsdorf et al., 1999). It also validates the hypothesis that the Sp100 isoform enriched at the ICP0-activated array in HeLa cells is unsumoylated Sp100A (Figure 2B, e–h).
In contrast to Sp100A, only minimal accumulation of Sp100B (Figure 3B, q–x), Sp100C and Sp100HMG (Supplemental Figure S1, i–x) was detected at both inactive and activated arrays in the U2OS cell line (compare the green signals in the intensity profiles). Previously, we reported that Sp100B, with a mutation in the SAND domain (W655Q; Figure 3A), predicted to disrupt its interaction with DNA (Bottomley et al., 2001), promotes transcription at viral promoters in a manner similar to Sp100A (Negorev et al., 2006, 2009). To evaluate the localization pattern of Sp100B(W655Q) and determine whether it is different from wild-type Sp100B, we examined its recruitment to the transgene array in U2OS cells. Indeed, Sp100B(W655Q) accumulated strongly in a manner more similar to Sp100A than to wild-type Sp100B (Supplemental Figure S1, a–h). This suggests that there may be a correlation between the localization patterns of the Sp100 isoforms and their effects on transcription.
Sp100A shares 477 of its 480 amino acids with Sp100B, C, and HMG, suggesting that the signal, which mediates strong recruitment, is present in all four isoforms; the increased accumulation of Sp100B(W655Q) compared with Sp100B suggests that the SAND domain inhibits it. To confirm this hypothesis, we introduced the W655Q mutation into Sp100C and HMG. Indeed, the increased accumulation of Sp100C(W655Q) and Sp100HMG(W655Q) at activated arrays in U2OS cells compared with wild-type Sp100C and HMG (Supplemental Figure S2) supports the conclusion that the SAND domain inhibits a signal in the first 477 amino acids of the Sp100 isoforms, which mediates strong accumulation.
Sp100A promotes decondensation of the transgene array chromatin
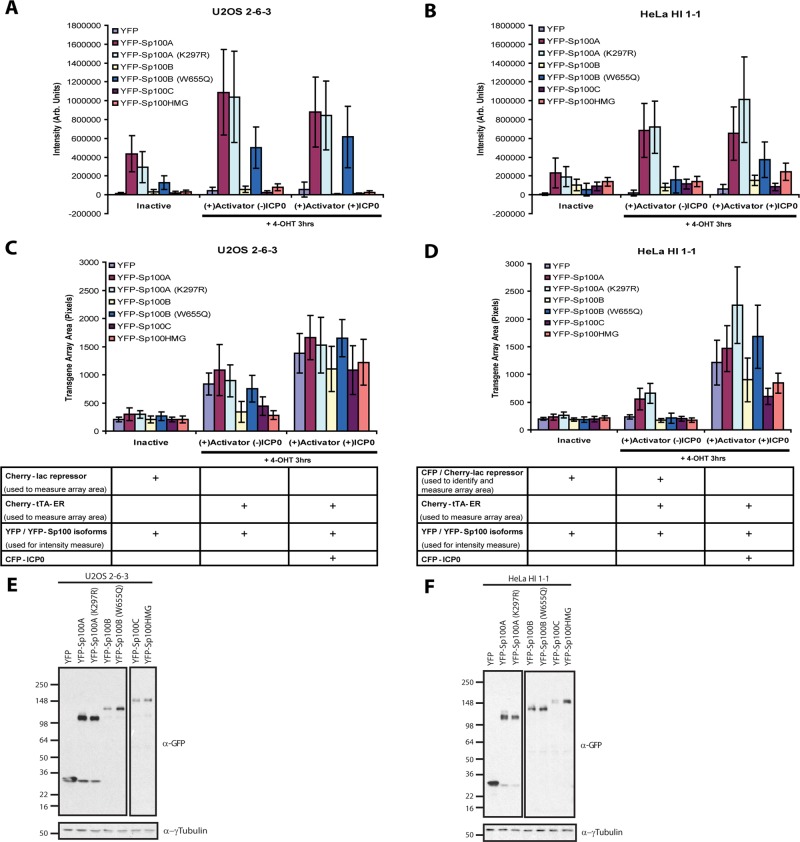
Measurements of the total intensity levels of each isoform at transgene arrays in U2OS and HeLa cells, in the inactive and activated states (+/− ICP0 expression), confirmed that Sp100A, including the K297R sumoylation mutant, is the most strongly recruited isoform (Figure 4, A and B, and Table 1). Furthermore, the strong accumulation of Sp100A at activated arrays in both U2OS and HeLa cells (Figure 4, A and B, middle and right data sets; E and F) correlates with increased chromatin decondensation (Figure 3, C and D, middle and right data sets). This suggests that Sp100A promotes chromatin decondensation during transcriptional activation. Of interest, in HeLa cells, the levels of Sp100A(K297R) were significantly higher than for Sp100A at ICP0-activated arrays (Figure 4B, data set on the right, and Table 1), a difference that also correlates with increased chromatin decondensation (Figure 4D, data set on the right, and Table 1). It is possible that because ICP0 cannot degrade Sp100A(K297R), it is able to promote decondensation for a longer period of time. This idea is further supported by the dispersed accumulation of Sp100A at the activated array compared with Sp100A(K297R), which also colocalizes in a concentrated accumulation with CFP-ICP0 (Supplemental Figure S3).
FIGURE 4:
Measurements of Sp100 isoform recruitment and effects on transgene array decondensation in U2OS and HeLa Cells. YFP and YFP-tagged Sp100 isoform intensity levels at transgene arrays in (A) U2OS (2-6-3) and HeLa (HI 1-1) (B) cells. Levels were measured at inactive transgene arrays and transgene arrays activated in the presence and absence of ICP0. Measurements of transgene array pixel area in inactive and transcriptionally activated transgene arrays (+/−) ICP0 in (C) U2OS (2-6-3) and (D) HeLa (HI 1-1) cells expressing YFP and YFP-tagged Sp100 isoforms. SDs are given in the form of error bars, and p values are calculated using unpaired t test and listed in Table 1. Western blots of transiently expressed, YFP-tagged Sp100 isoforms in (E) U2OS (2-6-3) and (F) HeLa (HI 1-1) cells detected using α-GFP antibody. γ-Tubulin is used as a loading control. A higher exposure of YFP-Sp100C and HMG in U2OS cells and YFP-Sp100B, B(W655Q), C, and HMG in HeLa cells was needed to detect these isoforms. The different exposures are outlined separately.
TABLE 1:
Statistical analysis of Sp100 isoform recruitment to transgene arrays in U2OS and HeLa cells using unpaired t test.
| U2OS 2-6-3 intensity p values | ||||||
|---|---|---|---|---|---|---|
| Inactive | (+) Activator (−) ICP0 | (+) Activator (+) ICP0 | ||||
| n | YFP-Sp100A (n = 26) vs. | n | YFP-Sp100A (n = 39) vs. | n | YFP-Sp100A (n = 23) vs. | |
| YFP | 17 | <0.0001 | 17 | <0.0001 | 17 | <0.0001 |
| YFP-Sp100A (K297R) | 26 | 0.0058 | 24 | 0.6833 | 45 | 0.6871 |
| YFP-Sp100B | 24 | <0.0001 | 21 | <0.0001 | 20 | <0.0001 |
| YFP-Sp100B (W655Q) | 25 | <0.0001 | 34 | <0.0001 | 25 | 0.0115 |
| YFP-Sp100C | 25 | <0.0001 | 25 | <0.0001 | 25 | <0.0001 |
| YFP-Sp100HMG | 23 | <0.0001 | 21 | <0.0001 | 20 | <0.0001 |
| YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | ||||
| <0.0001 | <0.0001 | <0.0001 | ||||
| HeLa HI 1-1 intensity p values | ||||||
| Inactive | (+) Activator (−) ICP0 | (+) Activator (+) ICP0 | ||||
| n | YFP-Sp100A (n = 20) vs. | n | YFP-Sp100A (n = 41) vs. | n | YFP-Sp100A (n = 21) vs. | |
| YFP | 17 | <0.0001 | 28 | <0.0001 | 20 | <0.0001 |
| YFP-Sp100A (K297R) | 24 | 0.3102 | 29 | 0.6136 | 29 | 0.0024 |
| YFP-Sp100B | 20 | 0.0017 | 38 | <0.0001 | 22 | <0.0001 |
| YFP-Sp100B (W655Q) | 33 | <0.0001 | 31 | <0.0001 | 24 | 0.0003 |
| YFP-Sp100C | 23 | 0.0002 | 35 | <0.0001 | 22 | <0.0001 |
| YFP-Sp100HMG | 38 | 0.0011 | 36 | <0.0001 | 22 | <0.0001 |
| YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | ||||
| 0.0063 | 0.0019 | <0.0001 | ||||
| U2OS 2-6-3 area p values | ||||||
| Inactive | (+) Activator (−) ICP0 | (+) Activator (+) ICP0 | ||||
| n | YFP-Sp100A (n = 25) vs. | n | YFP-Sp100A (n = 37) vs. | n | YFP-Sp100A (n = 23) vs. | |
| YFP | 26 | 0.0003 | 20 | 0.0214 | 25 | 0.0161 |
| YFP-SP100A (K297R) | 26 | 0.8967 | 20 | 0.1000 | 45 | 0.2631 |
| YFP-Sp100B | 26 | 0.0004 | 24 | <0.0001 | 23 | <0.0001 |
| YFP-Sp100B (W655Q) | 24 | 0.2171 | 33 | 0.0002 | 26 | 0.9570 |
| YFP-Sp100C | 26 | 0.0004 | 29 | <0.0001 | 31 | <0.0001 |
| YFP-Sp100HMG | 26 | 0.0008 | 26 | <0.0001 | 25 | 0.0005 |
| YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | ||||
| 0.0030 | <0.0001 | <0.0001 | ||||
| HeLa HI 1-1 area p values | ||||||
| Inactive | (+) Activator (−) ICP0 | (+) Activator (+) ICP0 | ||||
| n | YFP-Sp100A(n = 20) vs. | n | YFP-Sp100A (n = 42) vs. | n | YFP-Sp100A (n = 30) vs. | |
| YFP | 24 | 0.0225 | 28 | <0.0001 | 27 | 0.0231 |
| YFP-Sp100A (K297R) | 24 | 0.0375 | 29 | 0.0271 | 29 | <0.0001 |
| YFP-Sp100B | 20 | 0.0020 | 38 | <0.0001 | 32 | <0.0001 |
| YFP-Sp100B (W655Q) | 37 | 0.0026 | 34 | <0.0001 | 26 | 0.1123 |
| YFP-Sp100C | 22 | 0.0394 | 35 | <0.0001 | 33 | <0.0001 |
| YFP-Sp100HMG | 38 | 0.1422 | 36 | <0.0001 | 31 | <0.0001 |
| YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | YFP-Sp100B vs. YFP-Sp100B(W655Q) | ||||
| 0.7675 | 0.0263 | <0.0001 | ||||
This analysis indicates that Sp100A is the only isoform that promotes chromatin decondensation. In fact, Sp100B, C, and HMG inhibit array decondensation in activated U2OS and HeLa cells (+/−) ICP0 (Figure 4, C and D, middle and right data sets), which is consistent with our previous reports that the SAND domain–containing isoforms repress immediate-early viral promoters (Negorev et al., 2006, 2009). To determine whether the SAND domain is the element required to promote chromatin condensation, we measured array areas in Sp100B(W655Q)-expressing cells (Figure 4, C and D, and Table 1). In activated U2OS cells, Sp100B(W655Q) failed to inhibit decondensation in a manner similar to Sp100B (Figure 4C, middle data set, and Table 1), supporting the hypothesis that a wild-type SAND domain is required for the promotion of chromatin condensation. However, the W655Q SAND-domain mutation is not sufficient to convert Sp100B into Sp100A because the mutant does not promote decondensation in activated HeLa cells (Figure 4D, middle data set).
Sp100A cannot overcome Daxx- and ATRX-mediated transcriptional repression
Our results indicate that Sp100A promotes chromatin decondensation at the activated CMV-promoter–regulated transgene array. However, from this analysis, it is unclear whether Sp100A does so by increasing transcription or boosting a mechanism that unwinds activated chromatin. Although Sp100A promotes chromatin decondensation in activated U2OS cells (Figure 4C, middle and right data sets), this cell line cannot be used to answer this question because the activator, on its own, is able to induce transcription due to its ATRX-null status. In contrast, the CMV-promoter–regulated array in the HeLa cell line is refractory to activation and therefore can be used to address this question (Newhart et al., 2012). In the presence of a functional Daxx and ATRX pathway, the activator is unable, on its own, to induce the changes in chromatin organization needed to expose its binding sites (Figure 2B, a–d).
To determine whether the Sp100A-induced increase in chromatin decondensation at the activated array in HeLa cells is accompanied by transcription, we used the Cherry-MS2–binding protein to measure RNA accumulation at the transcription site (Figure 5, A and B). In control cells coexpressing YFP-tTA-ER (the activator) and CFP-lac repressor (to identify the array), Cherry-MS2 does not significantly accumulate (Figure 5, A, a–d, and B). Of interest, Cherry-MS2 also does not accumulate at activated arrays in YFP-Sp100A–expressing cells (Figure 5A, e–h), which indicates that despite its ability to promote chromatin decondensation (Figure 5A; note the accumulation of CFP-tTA-ER at the activated array; g, enlarged inset), Sp100A cannot overcome Daxx- and ATRX-mediated transcriptional repression (Figure 5B). Consistent with our previous report that ICP0 permits transcriptional activation in HeLa cells, Cherry-MS2 accumulates strongly at the activated site in YFP-ICP0–expressing cells (Figure 5, A, i–l, and B).
FIGURE 5:
Sp100A cannot overcome Daxx- and ATRX-mediated transcriptional repression. Images of Cherry-MS2 accumulation at activated transgene arrays in (a–h) control, (e–h) YFP-Sp100A–, and (i–l) YFP-ICP0–expressing HeLa (HI 1-1) cells. Arrows indicate the locations of the transgene arrays. Yellow lines in enlarged merge insets show the path through which the red, green, and blue intensities were measured in the intensity profiles (d, h, l). Asterisks mark the start of the measured lines. Scale bar, 5 μm. Scale bars in enlarged inset, 1 μm. (B) Single-cell analysis of Cherry-MS2 intensity levels at inactive and activated transgene arrays in HeLa (HI 1-1) cells. Factors were transiently expressed from plasmids. Constructs expressed and N values are shown in the chart below the graph. (C) qRT-PCR analysis of mRNA levels in activated HeLa (HI 1-1) cells after lentiviral transduction of Sp100A alone or in combination with ICP0 or ICP0-FxE, deleted of the RING finger zinc-binding motif, which is required for activity. Constructs expressed are shown in the chart below the graph. Results are the average of at least three independent experiments. SDs are given in the form of error bars, and p values are calculated using the unpaired t test.
To measure the effects of Sp100A and ICP0 on the levels of total mRNA expressed in HeLa cells, we used quantitative real-time PCR (qRT-PCR; Figure 5C). Because this analysis was done on RNA isolated from cell populations, we expressed these factors and the activator using lentiviruses in order to achieve high expression efficiency (>90% of cells). Consistent with the single-cell analysis (Figure 5, A and B), the increase in transcription induced by Sp100A was small compared with ICP0 (Figure 5C). Of interest, coexpression of Sp100A and ICP0 did not significantly increase transcription above the level induced by ICP0 alone, which indicates that, in this assay, their effects are not additive (Figure 5C). It is possible that transcription plateaus in ICP0-expressing cells because RNA pol II saturates the gene template or another limiting factor is exhausted. Taken together, these results indicated that Sp100A does not promote transcription in HeLa cells, which have a functional Daxx and ATRX pathway.
Sp100A promotes lysine acetylation at the activated transgene array
The increase in chromatin decondensation induced by Sp100A in HeLa cells is not accompanied by increased transcription (Figure 5), which indicates that they are separable events. This led us to speculate that Sp100A may promote decondensation by recruiting proteins that unwind activated chromatin, such as acetyl-lysine regulatory factors. To test this hypothesis, we measured YFP-tagged histone acetyltransferases (GCN5, PCAF, and p300) and acetyl-lysine–binding proteins (Brd2 and Brd4) at activated arrays in U2OS and HeLa cells coexpressing Cherry, Cherry-Sp100A, or Cherry-Sp100B (Figure 6, A and B, and Table 2). All of the factors were enriched at activated arrays in U2OS cells (Figure 6A), consistent with our previous report that they accumulate during activation (Rafalska-Metcalf et al., 2010). Of interest, Sp100B did not prevent their accumulation at activated arrays in U2OS cells (Figure 6A) despite its inhibitory effects on chromatin decondensation (Supplemental Figure S4A, and see discussion of Table 4 later in the paper). In fact, the concentration of Brd2 and Brd4 was the highest in Sp100B-expressing U2OS cells (Figure 6A) due to the compaction of the arrays (Supplemental Figure S4A and see later discussion of Table 4).
TABLE 2:
Statistical analysis of YFP-tagged acetyl-lysine regulatory factor recruitment to transgene arrays in U2OS and HeLa cells using unpaired t test.
| U2OS 2-6-3 intensity/area p values | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ch-Sp100A vs. Cherry | Ch-Sp100A vs. Ch-Sp100B | Ch-Sp100A vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100B | Ch-Sp100B vs. Ch-Sp100A (inactive) | Cherry, n | Ch-Sp100A, n | Ch-Sp100B, n | Ch-Sp100A (inactive), n | |
| YFP-GCN5 | 0.1162 | 0.8076 | <0.0001 | <0.0001 | 0.0397 | <0.0001 | 18 | 17 | 15 | 23 |
| YFP-PCAF | 0.3716 | 0.0823 | <0.0001 | <0.0001 | 0.0026 | <0.0001 | 16 | 18 | 16 | 29 |
| YFP-p300 | 0.2614 | 0.0062 | <0.0001 | <0.0001 | 0.1929 | <0.0001 | 15 | 16 | 17 | 28 |
| YFP-BRD2 | 0.9121 | < 0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 19 | 17 | 19 | 24 |
| YFP-BRD4 | 0.7914 | 0.0231 | <0.0001 | <0.0001 | 0.0039 | <0.0001 | 18 | 18 | 14 | 25 |
| HeLa HI 1-1 intensity/area p values | ||||||||||
| Ch-Sp100A vs. Cherry | Ch-Sp100A vs. Ch-Sp100B | Ch-Sp100A vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100B | Ch-Sp100B vs. Ch-Sp100A (inactive) | Cherry, n | Ch-Sp100A, n | Ch-Sp100B, n | Ch-Sp100A (inactive), n | |
| YFP-GCN5 | <0.0001 | <0.0001 | <0.0001 | 0.0719 | 0.0097 | 0.5296 | 14 | 24 | 23 | 19 |
| YFP-PCAF | <0.0001 | <0.0001 | <0.0001 | 0.2427 | 0.1661 | 0.0181 | 18 | 22 | 23 | 19 |
| YFP-p300 | <0.0001 | <0.0001 | <0.0001 | 0.1874 | 0.0184 | 0.1970 | 15 | 25 | 22 | 19 |
| YFP-BRD2 | 0.0005 | <0.0001 | <0.0001 | <0.0001 | 0.0013 | 0.0233 | 16 | 26 | 21 | 20 |
| YFP-BRD4 | <0.0001 | <0.0001 | <0.0001 | 0.0005 | 0.0031 | 0.1205 | 14 | 26 | 22 | 20 |
FIGURE 6:
Sp100A promotes lysine acetylation at the activated CMV-promoter–regulated transgene array. Average intensity levels of YFP-tagged acetyl-lysine regulatory factors at transgene arrays in (A) U2OS (2-6-3) and (B) HeLa (HI 1-1) cells. Factors were expressed by transient transfection. For activated U2OS (2-6-3) cells, the activator (CFP-tTA-ER) was coexpressed with Cherry, Cherry- Sp100A, or Cherry-Sp100B and a YFP-tagged acetyl-lysine regulatory factor. For activated HeLa (HI 1-1) cells, the activator (tTA-ER) was coexpressed with Cherry, Cherry- Sp100A, or Cherry-Sp100B, a YFP-tagged acetyl lysine regulatory factor, and CFP-lac repressor (to identify the array). For analysis of the Sp100A's effects on acetyl-lysine regulatory factor recruitment to the inactive array, YFP-tagged factors were coexpressed with Cherry-Sp100A and CFP-lac repressor (to identify the array). SDs are given in the form of error bars, and p values are calculated using the unpaired t test and listed in Table 2. (C, D) Average intensity levels of endogenous Brd4 and histone H4 lysine 5 acetylation (H4AcK5) levels, detected by immunofluorescence staining, at CFP-tTA-ER activated arrays in (C) U2OS (2-6-3) and (D) HeLa (HI 1-1) cells coexpressing YFP, YFP-Sp100A, or YFP-Sp100B. SDs are given in the form of error bars, and p values are calculated using the unpaired t test and listed in Table 3. (E) Immunofluorescence localization of Brd4 at the activated array in (a–d) control and (e–h) YFP-Sp100A– and (i–l) YFP-Sp100B–expressing HeLa (HI 1-1) cells. YFP could not be imaged in control samples because it was extracted by the immunofluorescence staining protocol. Arrows indicate the locations of the transgene arrays. Yellow lines in enlarged merge insets show the path through which the red, green, and blue intensities were measured in the intensity profiles (d, h, l). Asterisks mark the start of the measured lines. Scale bar, 5 μm. Scale bars in enlarged inset, 1 μm.
TABLE 4:
Statistical analysis of YFP-tagged and endogenous acetyl-lysine regulatory factor recruitment to transgene arrays in U2OS and HeLa cells using unpaired t test.
| U2OS 2-6-3 area p values | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ch-Sp100A vs. Cherry | Ch-Sp100A vs. Ch-Sp100B | Ch-Sp100A vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100B | Ch-Sp100B vs. Ch-Sp100A (inactive) | Cherry, n | Ch-Sp100A, n | Ch-Sp100B, n | Ch-Sp100A (inactive), n | |
| YFP-GCN5 | <0.0001 | <0.0001 | <0.0001 | 0.0150 | 0.6624 | 0.1459 | 18 | 17 | 15 | 23 |
| YFP-PCAF | 0.0004 | <0.0001 | <0.0001 | 0.2320 | 0.2044 | 0.7672 | 16 | 18 | 16 | 30 |
| YFP-p300 | 0.0036 | <0.0001 | <0.0001 | 0.0021 | <0.0001 | 0.3817 | 15 | 16 | 17 | 28 |
| YFP-BRD2 | 0.0677 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.9247 | 19 | 17 | 19 | 24 |
| YFP-BRD4 | 0.0184 | <0.0001 | <0.0001 | <0.0001 | 0.0003 | 0.8920 | 18 | 18 | 14 | 25 |
| HeLa HI 1-1 area p values | ||||||||||
| Ch-Sp100A vs. Cherry | Ch-Sp100A vs. Ch-Sp100B | Ch-Sp100A vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100A (inactive) | Cherry vs. Ch-Sp100B | Ch-Sp100B vs. Ch-Sp100A (inactive) | Cherry, n | Ch-Sp100A, n | Ch-Sp100B, n | Ch-Sp100A (inactive), n | |
| YFP-GCN5 | <0.0001 | <0.0001 | <0.0001 | 0.1525 | 0.0102 | <0.0001 | 14 | 24 | 23 | 19 |
| YFP-PCAF | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.3986 | <0.0001 | 18 | 22 | 23 | 19 |
| YFP-p300 | <0.0001 | <0.0001 | 0.0002 | 0.0006 | 0.0103 | <0.0001 | 15 | 25 | 22 | 19 |
| YFP-BRD2 | <0.0001 | <0.0001 | <0.0001 | 0.0514 | 0.0159 | <0.0001 | 16 | 26 | 21 | 20 |
| YFP-BRD4 | <0.0001 | <0.0001 | <0.0001 | 0.0291 | 0.0003 | <0.0001 | 14 | 26 | 22 | 20 |
| U2OS 2-6-3 area p values | ||||||||||
| YFP-Sp100A vs. YFP | YFP-Sp100A vs. YFP-Sp100B | YFP vs. YFP-Sp100B | YFP, n | YFP-Sp100A, n | YFP-Sp100B, n | |||||
| Anti-BRD4 | 0.0693 | <0.0001 | <0.0001 | 22 | 24 | 22 | ||||
| Anti-H4AcK5 | 0.2342 | <0.0001 | <0.0001 | 27 | 27 | 25 | ||||
| HeLa HI 1-1 area p values | ||||||||||
| YFP-Sp100A vs. YFP | YFP-Sp100A vs. YFP-Sp100B | YFP vs. YFP-Sp100B | YFP, n | YFP-Sp100A, n | YFP-Sp100B, n | |||||
| Anti-BRD4 | <0.0001 | <0.0001 | 0.0084 | 20 | 24 | 24 | ||||
| Anti-H4AcK5 | <0.0001 | <0.0001 | 0.0004 | 21 | 24 | 24 | ||||
In HeLa cells, in contrast, the acetyl-lysine regulatory factors were only highly enriched at activated arrays in Sp100A-expressing cells (Figure 6B and Table 2). This result indicates that, in the presence of a functional Daxx and ATRX pathway, only Sp100A is able to promote acetyl-lysine regulatory factor recruitment and chromatin decondensation during activation. Of interest, Sp100B prevented their accumulation in HeLa cells (Figure 6B and Table 2). The higher and more variable increases in acetyl-lysine regulatory factor levels at arrays in HeLa control cells suggest that the SAND domain–containing isoforms may function to stabilize transcriptionally silent expression states by preventing lysine acetylation. The fact that Sp100B fails to inhibit the accumulation of these factors at activated arrays in U2OS cells (Figure 6A and Table 2) suggests that it requires the Daxx and ATRX pathway to do so.
Taken together, these results suggest that Sp100A promotes chromatin decondensation at transcription sites by increasing lysine acetylation. To determine whether increasing the concentration of Sp100A at the site is sufficient for this effect, we measured acetyl-lysine regulatory factor levels at inactive arrays (marked by CFP-lac repressor) in Cherry-Sp100A–expressing cells. The low accumulation of these factors at inactive arrays in both Sp100A-expressing U2OS and HeLa cells indicates that Sp100A only promotes their recruitment in conjunction with the activator (Figure 6, A and B, and Table 2). This suggests that Sp100A functions to accelerate transcription by promoting lysine acetylation and chromatin decondensation during activation.
Using antibodies against endogenous Brd4 and histone H4 lysine 5 acetylation, we show that their levels also increase at activated arrays in Sp100A-expressing cells (Figure 6, C–E, Tables 3 and 4, and Supplemental Figure S4, C and D), further supporting the conclusion that Sp100A promotes chromatin decondensation by increasing lysine acetylation at activated transcription sites. This result also indicates that YFP-tagged acetyl-lysine regulatory factor accumulation at the activated arrays (Figure 6, A and B) is not due to overexpression. Figure 7 summarizes the functions of PML-NB/ND10 and transcription factors in the regulation of CMV promoter activity.
TABLE 3:
Statistical analysis of endogenous BRD4 and H4AcK5 accumulation at activated transgene arrays in U2OS and HeLa cells using unpaired t test.
| U2OS 2-6-3 intensity/area p values | ||||||
|---|---|---|---|---|---|---|
| YFP-Sp100A vs. YFP | YFP-Sp100A vs. YFP-Sp100B | YFP vs. YFP-Sp100B | YFP, n | YFP-Sp100A, n | YFP-Sp100B, n | |
| Anti-BRD4 | 0.0710 | 0.0003 | <0.0001 | 22 | 24 | 22 |
| Anti-H4AcK5 | 0.9210 | 0.0103 | 0.0302 | 27 | 27 | 25 |
| HeLa HI 1-1 intensity/area p values | ||||||
| YFP-Sp100A vs. YFP | YFP-Sp100A vs. YFP-Sp100B | YFP vs. YFP-Sp100B | YFP, n | YFP-Sp100A, n | YFP-Sp100B, n | |
| Anti-BRD4 | <0.0001 | <0.0001 | <0.0001 | 19 | 25 | 24 |
| Anti-H4AcK5 | <0.0001 | <0.0001 | <0.0001 | 21 | 24 | 24 |
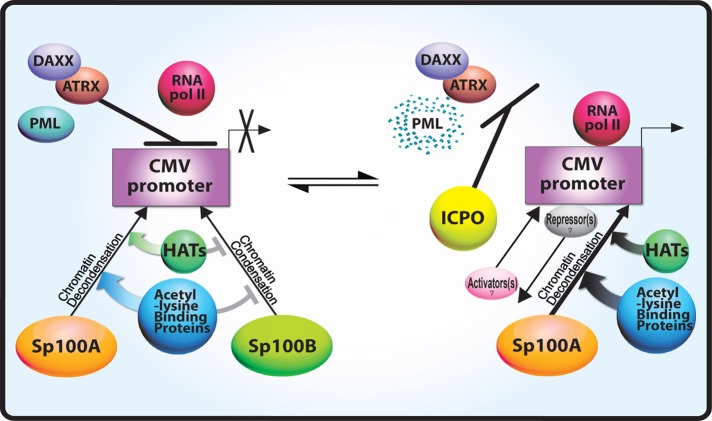
FIGURE 7:
Model of PML-NB/ND10-factor–mediated transcription regulation of the CMV promoter. Daxx and ATRX repress transcription from the CMV promoter and prevent RNA pol II recruitment. In the presence of Daxx and ATRX, Sp100A promotes chromatin decondensation and acetyl-lysine regulatory factor recruitment, and Sp100B prevents these events. ICP0 degrades PML and causes Daxx and ATRX to be displaced from the CMV-promoter–regulated transcription site. Under these conditions, RNA pol II is able to transcribe from the CMV promoter. ICP0 likely also displaces and/or recruits additional, as-yet-unidentified repressors and activators to the site.
DISCUSSION
Many studies examined the effects of the Sp100 isoforms on viral and cellular gene expression and found both activation and repression (Zong et al., 2000; Wasylyk et al., 2002; Yordy et al., 2004, 2005; Ling et al., 2005; Wilcox et al., 2005; Isaac et al., 2006; Negorev et al., 2006, 2009). Although many of these reports indicate that Sp100A potentiates transcription and that the SAND domain–containing isoforms repress it, interpretations are complicated by differences in the experimental systems used. An incomplete understanding of the relationship between PML-NBs/ND10s and transcription also makes it unclear how PML-NB/ND10 factors regulate different viruses, promoters, regulatory factors, and cell types. Although it was proposed that PML-NBs/ND10s are transcription sites (Xie et al., 1993), there is still a great deal of debate about whether transcription occurs within or apart from them.
In this study, we examined the effects of the Sp100 isoforms on transcription and chromatin organization using a single-cell imaging system that allows a stably integrated array of a CMV-promoter–regulated transcription unit to be directly visualized. Recently we reported that Daxx, ATRX, and PML are enriched at this site, specifically at the CMV promoter (Newhart et al., 2012). Here we show that Sp100 is also enriched at this transgene array and, like these other PML-NB/ND10 factors, is also associated with the CMV promoter. The accumulation of PML protein at these arrays technically classifies them as PML-NBs/ND10s (Ishov et al., 1999; Zhong et al., 2000) and suggests that at least some PML-NBs/ND10s are transcription sites. The specific enrichment of Sp100, PML, Daxx, and ATRX at the CMV promoter also suggests that sequence elements within promoters can seed the assembly of PML-NBs/ND10s.
A major finding of this study is that Sp100A, in conjunction with the transcriptional activator, increases chromatin decondensation at the transcriptionally repressed array in HeLa cells by promoting the recruitment of acetyl-lysine regulatory factors. However, despite the ability of Sp100A to increase chromatin decondensation, it is unable to overcome Daxx- and ATRX-mediated repression. This indicates that lysine acetylation is not sufficient to promote transcription. This is consistent with the finding that trichostatin A, an inhibitor of histone deacetylation, had no effect on the replication efficiency of ICP0-null mutant HSV-1 in either fibroblasts or HepaRG cells (Everett et al., 2008). It is not known how Sp100A promotes acetyl-lysine regulatory factor recruitment and chromatin decondensation. However, Sp100A only increases the concentration of these factors at the site in the presence of the activator, indicating that it is not sufficient for their recruitment. Perhaps Sp100A functions to promote gene activation by accelerating the conversion of chromatin to a transcriptionally permissive state through the acetylation of lysine residues in histones and transcriptional regulators.
Although Sp100B, C, and HMG are not strongly recruited to the transgene array, they significantly repress decondensation. However, because they include 477 of Sp100A's 480 amino acids, this suggests that the SAND-domain isoforms also contain the strong recruitment and chromatin-decondensing properties of Sp100A. The finding that Sp100B, C, and HMG containing the W655Q SAND-domain mutation accumulate strongly in a manner similar to Sp100A indicates that the SAND domain is the dominant recruitment signal and that it blocks strong accumulation. Part of the mechanism through which the SAND domain isoforms may promote array condensation is through the prevention of acetyl-lysine regulatory factor recruitment. However, this requires a functional Daxx and ATRX pathway because Sp100B fails to prevent their accumulation at activated arrays in ATRX-null U2OS cells. This suggests that the function of the SAND-domain isoforms may be to prevent lysine acetylation and stabilize transcriptionally repressed chromatin. Because transcription is highly stochastic (Kaern et al., 2005), even a small increase in lysine acetylation could significantly increase a gene's chances of becoming activated. The differential effects of the Sp100 isoforms on chromatin organization also suggest that Sp100 alternative splicing could have a significant effect on development and viral latency.
A surprising finding of this study is that Sp100 is enriched at the activated array in ICP0-expressing cells. Given the many reports detailing the disaggregating effects of ICP0 on PML-NB/ND10 organization (Chelbi-Alix and de The, 1999; Lopez et al., 2002; Smith et al., 2011), we expected that ICP0 would also displace Sp100 from the array in a manner similar to Daxx, ATRX, and PML (Newhart et al., 2012). Of interest, the accumulation of Sp100 at the ICP0-activated array indicates that PML, Daxx, and ATRX are not required for its recruitment. The finding that Sp100A(K297R), which cannot be sumoylated, is recruited to both inactive and activated arrays in U2OS and HeLa cells further supports the conclusion that it is the unsumoylated form of Sp100A that accumulates in the presence of ICP0. This is also consistent with reports that ICP0 degrades Sp100B, C, and HMG and sumoylated Sp100A (Everett et al., 2009) and that sumoylation is not required for Sp100A to be targeted to PML-NBs/ND10s (Sternsdorf et al., 1999). It is possible that HSV-1 evolved a strategy to maintain unsumoylated Sp100A at its transcription sites because its ability to promote lysine acetylation is beneficial to the HSV-1 life cycle.
Viral replication requires the precise expression of viral genes. Therefore virally induced changes in cellular regulators of transcription, such as Sp100, are likely to be critical to viral life cycles (Adler et al., 2011; Kim et al., 2011; Tavalai et al., 2011; Full et al., 2012). It is difficult to decipher mechanisms of Sp100-mediated transcriptional regulation using viruses because of the complicating effects of infection and host response. However, the single-cell imaging method used in this article makes it is possible to directly evaluate the effects of each Sp100 isoform on transcription and chromatin organization. Taken together, the results presented here suggest that a complex and highly dynamic set of regulatory interactions occurs between PML-NB/ND10 factors at transcription sites. In addition, PML-NB/ND10 factors function within a regulatory hierarchy (Figure 7). Daxx and ATRX elicit a dominant effect on transcriptional repression, whereas modulation of lysine acetylation by the Sp100 isoforms has important implications for the timing of transcriptional events. This study therefore provides important new insight into the mechanisms by which viruses may selectively regulate PML-NB/ND10 factors to promote their replication cycles.
MATERIALS AND METHODS
Cell culture
The U2OS cell line, 2-6-3, and the HeLa cell line, HI 1-1, and their growth conditions were previously described (Janicki et al., 2004; Rafalska-Metcalf et al., 2010; Newhart et al., 2012).
Plasmids
SV2-CFP/Cherry/YFP-lac repressor, Cherry-MS2, Cherry/YFP/CFP-tTA-ER, pLU-YFP/Cherry-tTA-ER, YFP/CFP-ICP0, pLU-YFP-ICP0, pLU-YFP-ICP0-FxE; YFP-GCN5, YFP-p300, YFP-PCAF; YFP-BRD4, and YFP-BRD2 were previously described (Janicki et al., 2004; Rafalska-Metcalf et al., 2010; Newhart et al., 2012). YFP-Sp100A was constructed by introducing the PCRed cDNA (BglII/NheI) into YFP-C1 (BamHI/XbaI). Sp100B, Sp100C, Sp100HMG, and Sp100B(W655Q) were made by introducing the PCRed cDNAs (HindIII/KpnI) into YFP-C3. pLU-Cherry was made by cloning Cherry (NheI/BamHI) into the pLU vector (XbaI/BamHI). pLU-Cherry-Sp100A was made by cloning the Cherry-tagged Sp100A-NLS construct (NheI/BclI; Negorev et al., 2006) into pLU (XbaI/BamHI). YFP-Sp100A(K297R), YFP-Sp100C(W655Q), and YFP-Sp100HMG(W655Q) were made by introducing point mutations into the wild-type constructs using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA).
Protein expression
For the single-cell imaging experiments in the U2OS and HeLa cell lines, transient transfections were performed using FuGENE6 transfection reagent (Promega, Madison, WI). These experiments include those in which we analyzed YFP-tagged Sp100 isoform recruitment, the effects of Sp100A and ICP0 on Cherry-MS2 accumulation, YFP-tagged acetyl-lysine regulatory factor recruitment, and endogenous Brd4 and H4AcK5 accumulation. Cells plated at ∼70% confluency on coverslips in six-well dishes were transfected the next day with combinations of CFP-, YFP-, and Cherry-tagged factors using ∼0.6 μg of each plasmid not exceeding 2 μg of total DNA in a 100-μl reaction volume (3 μl of FuGENE + 97 μl of serum-free media). After overnight expression, transcription was induced for 3 h using 4-OHT (1 μm) as previously described (Rafalska-Metcalf et al., 2010). Cells were then fixed and mounted on slides as described in the section describing microscope image acquisition.
For qRT-PCR analysis of mRNA expression, (8–8.5) × 105 HI 1-1 HeLa cells were plated in 10-cm dishes and infected with lentiviruses (1:4 ratio of virus for each construct expressed) using polybrene (8 μg/ml) to ensure expression in >90% of cells. After 24 h of expression, activation was induced by 4-OHT (1 μm) for 3 h, and RNA was isolated using TRIzol (Life Technologies, Grand Island, NY).
RT-PCR and quantitative PCR analysis
HeLa HI 1-1 cells were infected with factors expressed from the pLU lentiviral vectors, described in the Plasmids section, for ∼24 h before transcription was induced. RT-PCR and quantitative PCR (qPCR) analysis were done as previously described (Newhart et al., 2012). Briefly, 1 μg TRIzol (Life Technologies)-isolated RNA was treated with DNase I (Promega), heated to 65°C (15 min), cooled on ice (1 h), heated again to 65°C (15 min), cooled on ice (5 min), and purified with the RNeasy Kit (Qiagen, Valencia, CA). Transgene-specific RT reaction was done using OmniScript (Qiagen), followed by qPCR with SYBR Green (Sigma-Aldrich, St. Louis, MO) using the 7500 Fast Real-Time PCR system (Applied Biosystems, Life Technologies, Carlsbad, CA). qPCR data were analyzed using the ddCt method according to Applied Biosystems guidelines, using the tetracycline activator primers for normalization. The following primer pairs were used: 5′-GGTGCAGGCTGCCTATCAG-3′ and 5′-TTTGTGAGCCAGGGCATTG-3′ (rabbit β-globin exon 3) and 5′-ACAGCGCATTAGAGCTGCTTAAT-3′ and 5′-GGCGAGTTTACGGGTTGTTAAA-3′ (tetR of tTA activator).
Cross-linked chromatin immunoprecipitation
Cross-linked ChIP was done as previously described (Zeng et al., 2006; Rafalska-Metcalf et al., 2010; Newhart et al., 2012). Briefly, cells grown to ∼80% confluency were trypsinized, resuspended in 1× phosphate-buffered saline (PBS), and incubated with freshly prepared 1.5 mM ethylene glycolbis[succinimidyl succinate] (Pierce Biotechnology, Rockford, IL) dissolved in dimethyl sulfoxide (Sigma-Aldrich) for 25 min at room temperature with gentle shaking. Cells were cross-linked by incubating in 1% formaldehyde (Sigma-Aldrich) for 10 min, followed by quenching with glycine-PBS (50 mM) for 10 min. Cells were centrifuged (1500 rpm, 5 min, 4°C), washed twice with 1× PBS, resuspended in 1 ml SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.0, freshly added protease inhibitors), and incubated on ice for 20 min and sonicated until DNA was uniformly sheared to ∼500 base pairs in length. Chromatin samples were incubated overnight with 2 μg of rabbit α-Sp100 (ab43151; Abcam, Cambridge, MA) and 2 μg of rabbit immunoglobulin G (Abcam).
Primer pairs for ChIP analysis of CMV-promoter–regulated transgene are as follows:
5′-CCACCTGACGTCTAAGAAACCAT-3′ (For)5′-GATCCCTCGAGGACGAAAGG-3′ (Rev)
5′-GTTATCCGCTCACAATTCCACA-3′ (For)5′-TTCTCTATCACTGATAGGGAGTGG-3′ (Rev)
5′-TGTACGGTGGGAGGCCTATATAA-3′ (For)5′-GCGTCTCCAGGCGATCTG-3′ (Rev)
5′-GGAAGATGTCCCTTGTATCACCAT-3′ (For)5′-TGGTTGTCAACAGAGTAGAAAGTGAA-3′ (Rev)
5′-GGCATTTCAGTCAGTTGCTCAA-3′ (For)5′-TTGGCCGATTCATTAATGCA-3′ (Rev)
Microscope image acquisition
For image analysis, cells were plated overnight on No. 1.5 coverslips, transfected using FuGENE6 transfection reagent (Promega), and transcriptionally activated as indicated ∼24 h later. Cells were then fixed for 15 min in 3% paraformaldehyde in 1× PBS, and for cells expressing autofluorescent proteins, coverslips were immediately mounted in antifade fluorescence mounting medium (1 mg/ml p-phenylenediamine, 90% glycerol in PBS, pH 8.0–9.0 adjusted with sodium carbonate/bicarbonate buffer, pH 9.2; Janicki et al., 2004). Images were acquired at room temperature using a Leica DMI 6000 B inverted automated microscope (Leica, Wetzlar, Germany) with an HCX PL APO 100×/1.40–0.70 oil objective lens using a 514-nm/50-mW solid-state laser for YFP, a 561-nm/50-mW solid-state laser for mCherry, and a 442-nm diode laser for CFP imaging (Melles Griot, Albuquerque, NM) and a Yokogawa CSU-10 real-time spinning disk confocal attachment (Yokogawa, Tokyo, Japan) with Nipkow and microlens disks. Single images or image stacks (0.4-μm increments) were acquired using a Hamamatsu ORCA-AG camera (1 × 1 binning; 1344 × 1024 pixels) and COMPIX SimplePCI software (Hamamatsu, Sewickley, PA).
Image analysis
Images of autofluorescently tagged Sp100 isoforms were taken by selecting cells in which the factors localized in the expected patterns for PML-NB/ND10 proteins. Cells in which the proteins were highly overexpressed and formed aggregates were excluded from the analysis. For image stacks, maximum projections were compiled, and intensity profiles for merged pictures were generated using COMPIX SimplePCI software. Image contrast adjustments were performed using COMPIX SimplePCI and Photoshop (Adobe, San Jose, CA) software. Image analysis was done using ImageJ software (National Institutes of Health, Bethesda, MD) on images taken with exposure and gain settings such that images had maximum intensity values between 2000 and 3000 on a scale of 0 to 4095. Intensity measurements were normalized by subtracting the background value as determined by averaging three independent regions in the cell nucleus measured in regions distinct from the transgene array site. Images, which included DAPI staining, were acquired at room temperature using a Leica TCS SP5 II confocal microscope with an HCX PL APO 63×/1.40–0.60 oil CS lens (Leica Microsystems, Wetzlar, Germany).
Immunofluorescence and Western blotting
For immunofluorescence, antibody staining was done as previously described (Rafalska-Metcalf et al., 2010). The following antibodies were used on cells that were preextracted: Sp100 (1:500; ab43151; Abcam) and histone H4 acetyl K5 (1:2000; 07-327; Millipore, Temecula, CA). The following antibody was used without preextraction: Brd4 (1:40,000; Wang et al., 2012; gift of Jianxin You (University of Pennsylvania). Western blotting was done with the antibodies GFP (1:1000; Roche, Indianapolis, IN) and γ-tubulin (1:1000; Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank Jianxin You for the Brd4 antibody; Michael Showe, Louise Showe, Dario Altieri, and Aswathy Rai for their critical comments on the manuscript; and Sylvie Shaffer for her work on the model. This work was funded by start-up funding from the Wistar Institute, funding from the Pennsylvania Department of Health (CURE), a Beckman Young Investigator Award, the Mallinckrodt Foundation, the Emerald Foundation, a March of Dimes Basil O'Connor Award, National Institutes of Health Grant R01 GM 093000-02, and the Imaging and Genomics/Sequencing and Research Supply Facilities in Wistar Cancer Center Core (Grant P30 CA10815).
Abbreviations used:
- 4-OHT
4-hydroxytamoxifen
- ATRX
α-thalassemia mental retardation, X-linked
- CMV
cytomegalovirus
- HSV-1
herpes simplex virus type 1
- ND10
nuclear domain 10
- PanNET
pancreatic neuroendocrine tumor
- PML-NB
promyelocytic leukemia-nuclear body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-09-0669) on March 13, 2013.
REFERENCES
- Adler M, Tavalai N, Muller R, Stamminger T. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J Gen Virol. 2011;92:1532–1538. doi: 10.1099/vir.0.030981-0. [DOI] [PubMed] [Google Scholar]
- Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol. 2001;8:626–633. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- Chelbi-Alix MK, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22:761–770. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Everett RD, Parada C, Gripon P, Sirma H, Orr A. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol. 2008;82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Parsy ML, Orr A. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol. 2009;83:4963–4977. doi: 10.1128/JVI.02593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- Full F, Reuter N, Zielke K, Stamminger T, Ensser A. Herpesvirus saimiri antagonizes nuclear domain 10-instituted intrinsic immunity via an ORF3-mediated selective degradation of cellular protein Sp100. J Virol. 2012;86:3541–3553. doi: 10.1128/JVI.06992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res. 2011;31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- Hagglund R, Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol. 2004;78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Isaac A, Wilcox KW, Taylor JL. SP100B, a repressor of gene expression preferentially binds to DNA with unmethylated CpGs. J Cell Biochem. 2006;98:1106–1122. doi: 10.1002/jcb.20841. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kim YE, Lee JH, Kim ET, Shin HJ, Gu SY, Seol HS, Ling PD, Lee CH, Ahn JH. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J Virol. 2011;85:11928–11937. doi: 10.1128/JVI.00758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling PD, et al. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 2005;24:3565–3575. doi: 10.1038/sj.emboj.7600820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P, Jacob RJ, Roizman B. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J Virol. 2002;76:9355–9367. doi: 10.1128/JVI.76.18.9355-9367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol. 2010;84:4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Negorev DG, Vladimirova OV, Ivanov A, Rauscher F 3rd, Maul GG. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J Virol. 2006;80:8019–8029. doi: 10.1128/JVI.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negorev DG, Vladimirova OV, Maul GG. Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J Virol. 2009;83:5168–5180. doi: 10.1128/JVI.02083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A, Rafalska-Metcalf IU, Yang T, Negorev DG, Janicki SM. Single cell analysis of Daxx and ATRX-dependent transcriptional repression. J Cell Sci. 2012;125:5489–5501. doi: 10.1242/jcs.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf IU, Janicki SM. Show and tell: visualizing gene expression in living cells. J Cell Sci. 2007;120:2301–2307. doi: 10.1242/jcs.008664. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM. Single cell analysis of transcriptional activation dynamics. PLoS One. 2010;5:e10272. doi: 10.1371/journal.pone.0010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Losson R, Desterro JM, Hay RT, Chambon P, Dejean A. Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol Cell Biol. 2001;21:3314–3324. doi: 10.1128/MCB.21.10.3314-3324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Boutell C, Davido DJ. HSV-1 ICP0: paving the way for viral replication. Future Virol. 2011;6:421–429. doi: 10.2217/fvl.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T, Jensen K, Reich B, Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, modification by small ubiquitin-like modifiers. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T. Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J Virol. 2011;85:9447–9458. doi: 10.1128/JVI.00870-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomtishen JP 3rd. Human cytomegalovirus tegument proteins (pp65, pp71, pp150, pp28) Virol J. 2012;9:22. doi: 10.1186/1743-422X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Thikmyanova N, Wojcechowskyj JA, Delecluse HJ, Lieberman PM. EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 2011;7:e1002376. doi: 10.1371/journal.ppat.1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E, Van Ostade X. Crosstalk between viruses and PML nuclear bodies: a network-based approach. Front Biosci. 2012;17:2910–2920. doi: 10.2741/3889. [DOI] [PubMed] [Google Scholar]
- Wang R, Li Q, Helfer CM, Jiao J, You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem. 2012;287:10738–10752. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Schlumberger SE, Criqui-Filipe P, Wasylyk B. Sp100 interacts with ETS-1 and stimulates its transcriptional activity. Mol Cell Biol. 2002;22:2687–2702. doi: 10.1128/MCB.22.8.2687-2702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenhan D, Kunze B, Traut W, Winking H. Evolution by fusion and amplification: the murine Sp100-rs gene cluster. Cytogenet Cell Genet. 1998;80:226–231. doi: 10.1159/000014985. [DOI] [PubMed] [Google Scholar]
- Wilcox KW, Sheriff S, Isaac A, Taylor JL. SP100B is a repressor of gene expression. J Cell Biochem. 2005;95:352–365. doi: 10.1002/jcb.20434. [DOI] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Lambie EJ, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Schaffer PA. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy JS, Li R, Sementchenko VI, Pei H, Muise-Helmericks RC, Watson DK. SP100 expression modulates ETS1 transcriptional activity and inhibits cell invasion. Oncogene. 2004;23:6654–6665. doi: 10.1038/sj.onc.1207891. [DOI] [PubMed] [Google Scholar]
- Yordy JS, Moussa O, Pei H, Chaussabel D, Li R, Watson DK. SP100 inhibits ETS1 activity in primary endothelial cells. Oncogene. 2005;24:916–931. doi: 10.1038/sj.onc.1208245. [DOI] [PubMed] [Google Scholar]
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41:694, 696, 698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- Zong RT, Das C, Tucker PW. Regulation of matrix attachment region-dependent, lymphocyte-restricted transcription through differential localization within promyelocytic leukemia nuclear bodies. EMBO J. 2000;19:4123–4133. doi: 10.1093/emboj/19.15.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.