Abstract
Purpose
Recurrent glioblastoma multiforme (GBM) is characterized by resistance to radiotherapy and chemotherapy and a poor clinical prognosis. In this study, we investigated the role of the oncogenic transcription factor FoxM1 in GBM cells’ resistance to TMZ and its potential molecular mechanism.
Experimental Design
FoxM1 expression levels were measured by immunohistochemical analysis in 38 pairs of primary and recurrent GBM tumor samples. Expression levels were also measured in primary recurrent GBM cell lines, and their responses to TMZ were characterized. In a mechanistic study, an siRNA array was used to identify downstream genes, and a chromatin immunoprecipitation assay was used to confirm transcriptional regulation.
Results
Recurrent tumors that were TMZ resistant expressed higher levels of FoxM1 than did primary tumors. Recurrent GBM cell lines expressed higher levels of FoxM1 and the DNA damage repair gene Rad51 and were resistant to TMZ. TMZ treatment led to increased FoxM1 and Rad51 expression. FoxM1 knockdown inhibited Rad51 expression and sensitized recurrent GBM cells to TMZ cytotoxicity. FoxM1 directly regulated Rad51 expression through two FoxM1-specific binding sites in its promoter. Rad51 re-expression partially rescued TMZ resistance in FoxM1-knockdown recurrent GBM cells. A direct correlation between FoxM1 expression and Rad51 expression was evident in recurrent GBM tumor samples.
Conclusion
Targeting the FoxM1-Rad51 axis may be an effective method to reverse TMZ resistance in recurrent GBM.
Keywords: FoxM1, glioblastoma, Rad51, resistance, temozolomide
INTRODUCTION
Glioblastoma multiforme (GBM), the most malignant glioma, is characterized by rapid progression, high resistance to radiotherapy and chemotherapy, and an extremely poor clinical prognosis. Despite radical surgical resection and standard radiation therapy and chemotherapy, the 2-year survival rate of GBM patients remains less than 25%, and few survive beyond 5 years(1).
Clinical trials have shown that patient survival can be extended by adding the alkylator temozolomide (TMZ) to the treatment regimen (1). TMZ methylates the O6, N7 position of guanine and the N3 position of adenine (2, 3). O6-methylguanine lesions account for most TMZ-induced cytotoxicity; this effect can be reversed with O6-methylguanine-DNA methyltransferase (MGMT) (4–7). However, studies have shown that more than 40% of gliomas have MGMT downregulation and are still resistant to TMZ, indicating that a high MGMT level is only one mechanism (11, 12).
Rad51 is one of the central components of the DNA double-strand break repair gene, whose expression can be induced by DNA damage (14, 15). After a DNA double-strand break occurs, Rad51 foci form at the sites of single-stranded DNA in lesions that promote homologous recombination (HR) (16–18). Rad51 overexpression, but not gene amplification or mutation, has been reported in human malignancies, such as pancreatic adenocarcinoma, soft tissue sarcoma, prostate cancer, and ovarian cancer (19–22). High Rad51 levels are associated with increased HR, which accounts for resistance to DNA-damaging reagents such as chemotherapy (21–24). Several studies have shown that Rad51 expression levels are elevated in GBM cell lines and that targeting Rad51 with Rad51 antisense or imatinib can effectively sensitize cancer cells to radiation therapy (25–28). More recently, targeting Rad51 was found to increase GBM cells’ sensitivity to TMZ (29). However, the molecular mechanisms of Rad51 overexpression in GBM cells remain unclear.
The proliferation-specific oncogenic transcription factor FoxM1 is widely overexpressed in human tumors, including mammary, gastrointestinal, lung, and reproductive organ tumors (31–33). In a previous study, we found that FoxM1 is overexpressed in GBM and that its expression level is correlated with pathological grade and patients’ cumulative prognosis (34). The results of other previous studies revealed that FoxM1 mediates the resistance of a diverse spectrum of anti-cancer drugs, such as doxorubicin, lapatinib, gefitinib, imatinib, and cisplatin in breast cancer (40–43). A recent investigation confirmed that FoxM1 stabilization after DNA damage response is critical for stimulating the expression of DNA damage repair genes such as XRCC1 and BRCA2 in resistant breast cancer cells (41). However, whether FoxM1 overexpression is associated with GBM chemotherapy resistance is unknown.
In this study, we found that FoxM1 expression levels were significantly elevated in recurrent GBM and that targeting FoxM1 sensitized resistant tumor cells to TMZ. In addition, FoxM1 transcriptionally regulated Rad51, and re-expressing Rad51 in FoxM1-knockdown cells partially rescued TMZ resistance. FoxM1 expression was highly correlated with Rad51 expression in recurrent GBM tumor samples, suggesting that the FoxM1-Rad51 axis contributes to chemotherapy resistance in GBMs.
MATERIALS AND METHODS
Cell lines and primary cell culture
The human glioma U87 cell line was obtained from the American Type Culture Collection; normal human astrocyte (NHA) cells were purchased from Lonza. The GBM primary culture cell lines primary GBM1 and recurrent GBM1 were established from a paired primary and recurrent GBM surgical specimens, respectively. The GBM primary culture cell line recurrent GBM2 was established from another recurrent GBM surgical specimens, using method described by Peñuelas S et al (44). In brief, fresh human glioma samples were digested with 200 U/ml collagenase I (Sigma) plus 500 U/ml DNase I (Sigma) in PBS for 2 hours at 37°C with constant vigorous agitation. The single-cell suspension was filtered through a 70-μm cell strainer (BD Falcon) and washed with PBS 3 times. Cells were suspended and maintained in neurosphere medium (neurobasal medium [Gibco] supplemented with B27 [Invitrogen], L-glutamine [Gibco], penicillin/streptomycin, and growth factors [20 ng/ml EGF and 20 ng/ml basic fibroblast growth factor, from Invitrogen]). Before each experiment, the GBM cells were cultured in Dulbecco’s modified eagle medium, supplemented with 10% fetal bovine serum, sodium pyruvate, nonessential amino acids, L-glutamine, and a vitamin solution (Invitrogen). Only early passages of GBM cells were used in the experiments (less than 5 passages). Clinical information on the patient and tumor characteristics of the GBM1 and GBM2 tumors is summarized in Supplementary Table 1. The study was approved by the ethics committee of Sun Yat-sen University, and informed consent was obtained from all subjects.
Antibodies and oligonucleotides
Antibodies against FoxM1 (K-19), Chk2 (B-4), Rad51 (3C-10), and MGMT (C-5) were from Santa Cruz Biotechnology; antibodies against BRCA2 (ab90541) were from Abcam. The antibody against actin was from Sigma. Fluorescent antibodies Alexa Fluor 488 and Alexa Fluor 596 were from Invitrogen.
The FoxM1 or Rad 51shRNA lentivirus vectors were generated as previously described (33) The target sequences of FoxM1 shRNA are sh-FoxM1-1, 5′-CTCTTCTCCCTCAGATATA-3′; and sh-FoxM1-2, 5′-GGACCACTTTCCCTACTTT-3′. The target sequence of Rad51 shRNA are sh-Rad51-1, 5′-CTAATCAGGTGGTAGCTCATT-3′; and sh-Rad51-2, 5′-CTTTGGCCCACAACCCAT-3′. A hairpin shRNA with no homology to any human, mouse, and rat mRNA sequence in the NCBI RefSeq database (GenScript) was also cloned into the lentivirus vector and used as a negative control (sh-control). The target sequences of two FoxM1 siRNA are the same as those of the FoxM1 shRNA shown above. A hairpin siRNA with no homology to any known mRNA sequences in the human, mouse, and rat genomes was used as a negative control (control siRNA) (Ambion). Oligonucleotides used for real-time polymerase chain reaction (RT-PCR) and chromatin immunoprecipitation (ChIP) assays are described in Supplementary Table 2. The expression plasmid of Rad51 was from Fulengen, PR China.
Microarray analysis
Gene expression was analyzed using Human Genome U133 Plus 2.0 Array (Bohao, Shanghai, China). In brief, total RNA was extracted using Trizol Reagent (Life technologies) according to the manufacturer’s instructions and purified using RNeasy micro kit (Qiagen). Total RNA was then amplified, labeled and purified using GeneChip 3′IVT Express Kit (Affymetrix) to obtain biotin-labeled cRNA. Arrays were hybridized and washed using GeneChip® Hybridization, Wash and Stain Kit (Affymetrix) following the manufacturer’s instructions. Slides were scanned by GeneChip® Scanner 3000 (Affymetrix) and Command Console Software 3.1 (Affymetrix) with default settings. Raw data were normalized using Gene Spring Software 11.0 (Agilent technologies). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE40051 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40051).
Cell transfection and virus infection
All transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). To establish stable cell lines of different shRNA, shRNA constructs were transduced into glioma cells using lentivirus in the presence of polybrene (6 μg/ml, Sigma). Cells were selected with 500 μg/ml neomycin for 14 days.
Patient tissue specimens
Our study included 38 randomly selected pairs of primary and recurrent GBM tumor samples (each pair was from the same patient) that had been histopathologically diagnosed in our department from 2000 to 2010. All 38 patients had received TMZ plus radiation therapy after surgery. The study was approved by the ethics committee of Sun Yat-sen University, with informed consent obtained from all subjects. Clinical information on the samples is summarized in Supplementary Table 3.
TMZ treatment
TMZ was obtained from Sigma-Aldrich (T2577) and dissolved in dimethyl sulfoxide. Cells were pre-incubated with various concentrations of TMZ (from 20 to 120 μM) for 1 to 5 days before cell lysates were collected or viable cells were counted. Each experiment was performed in triplicate.
Flow cytometry analysis
Cells were harvested and fixed in cold ethanol before being stained with propidium iodide (Sigma, 0.45 mg/mL). Resuspended cells were analyzed for DNA content on a fluorescence-activated cell sorter (FACSVantage), and data were processed using FACS CellQuest software (Becton Dickinson).
Colony formation assays
Neurospheres were disassociated, and single cells in 0.6% top agar-medium were plated in triplicate onto 60-mm dishes (300 cells per dish) that had been pre-coated with 1.2% agar-medium before being treated with different concentrations of TMZ. Cells were incubated at 37°C, and the medium was replaced with fresh medium for every 3–4 days. Survived colonies were stained with crystal violet 14 days later and counted.
Promoter reporters and dual-luciferase assay
The Rad51 promoter and truncated promoters with different lengths were cloned into PGL3-Basic vector (Promega). The Rad51 mutant promoter constructs were generated using the QuikChange site-directed mutagenesis kit (Stratagene). In the dual-luciferase assays, cells were cultured for 36 hours after transfection, and cell lysate was used to measure luciferase reporter gene expression using the dual-luciferase reporter assay system (Promega). Luciferase activities were normalized to the co-transfected pRL-TK plasmid. All experiments were performed at least twice, in triplicate.
ChIP assay
ChIP assays were performed using the ChIP assay kit (Cell Signaling). The resulting precipitated DNA samples were analyzed by semi-quantitative RT-PCR using primers that amplify the four regions of the Rad51 promoter. The PCR products were resolved electrophoretically on a 2% agarose gel and visualized by ethidium bromide staining.
Immunohistochemical analysis
An immunohistochemical analysis of human GBM specimens was performed using anti-FoxM1 or anti-Rad51 antibody. Nonspecific immunoglobulin (IgG) was used as the negative control. We quantitatively scored tissue sections according to the percentage of positive cells and staining intensity, as previously described (34).
Immunofluorescence staining
Cells or frozen glioma sections were fixed with 4% paraformaldehyde, permeabilized with phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (PBS-T), and blocked with 1% bovine serum albumin in PBS-T. Immunostaining was performed using the appropriate primary antibodies and stained with 4′, 6-diamidino-2-phenylindole, anti-rabbit IgG conjugated with Alexa Fluor 488 or anti-mouse IgG conjugated with Alexa Fluor 596. Images were acquired using a scanning confocal microscope (Olympus FluoView FV1000).
Real-time RT-PCR
Total RNA from cultured cells or frozen glioma tissues was extracted using the Trizol reagent (Invitrogen). Real-time RT-PCR assays were performed as described previously (34). β-actin was used as an internal control. All experiments were performed in triplicate.
Western blotting
Cells were harvested in sample buffer (62.5 mmol/L Tris-HCl [pH 6.8], 10% glycerol, and 2% sodium dodecyl sulfate) and boiled for 5 minutes. A Western blot analysis of cell lysates was carried out using the antibodies described earlier. The membranes were stripped and re-probed with an anti-beta-actin monoclonal antibody as a loading control.
Intracranial tumor assay
All mouse experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. GBM cells were injected intracranially into nude mice as described previously (34). Animals were killed when they were moribund; the remaining animals were killed 160 days after GBM-cell injection. Tumor formation was determined by histologic analysis of H&E-stained sections.
Statistical analysis
We used Pearson’s correlation test to identify correlations in the human GBM data. We used Log-rank analysis for the in vivo survival study. In addition, we evaluated significant differences in vitro data using Student’s t-test (two-tailed). A significance level set at p<0.05 was considered significant for all the tests.
RESULTS
FoxM1 expression is upregulated in recurrent GBMs
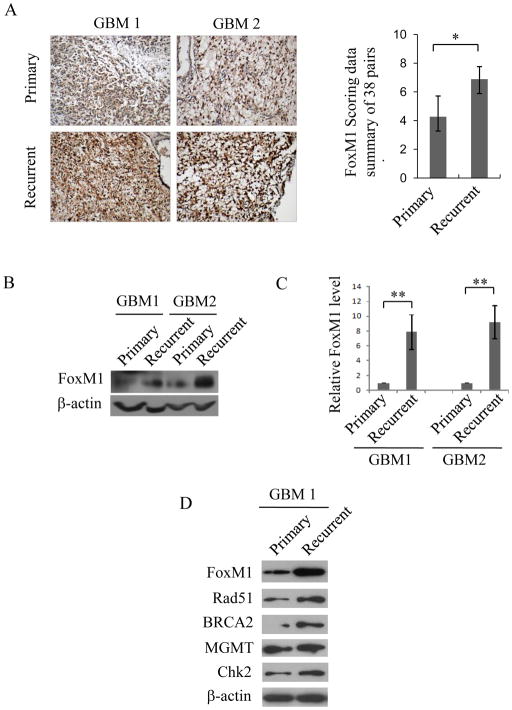
Immunohistochemical staining of FoxM1 protein was performed in 38 randomly selected paraffin-embedded GBM primary and recurrent tumor sample pairs. The patients had been treated with radiation therapy and chemotherapy after surgery. Compared with primary tumors, most recurrent tumors exhibited higher FoxM1 staining intensity and more nuclear distribution (Fig. 1A, left). To quantify FoxM1 expression in the 38 pairs of specimens, we scored FoxM1 expression (Supplementary Table 4), as we described previously (34). The FoxM1 expression level in recurrent samples was markedly increased compared with that in primary samples (Fig. 1A, right panel and Supplementary Fig. 1A). Next, we determined FoxM1 expression levels in two pairs of frozen tumor specimens, including primary tumors and the corresponding recurrent tumors. As shown in Fig. 1B and C, FoxM1 was upregulated in recurrent tumor samples, both at the mRNA and protein levels.
Figure 1. FoxM1 and DNA repair genes are upregulated in recurrent GBMs.
A, Immunohistochemical staining of FoxM1 in 38 pairs of GBM samples. Left panel, representative images of FoxM1 expression in primary and recurrent tumors. Right panel, scoring of FoxM1 expression levels in 38 pairs of primary and recurrent tumors. B and C, Two GBM sample pairs, including primary and recurrent tumors, were subjected to Western blot (B) and real-time RT-PCR analyses (C). The real-time PCR data are from three independent experiments. *p<0.05 and **p<0.001. D, Western blot analysis shows the expression levels of FoxM1, Rad51, BRCA2, Chk2, and MGMT in primary cultured cell lines from the paired primary and recurrent GBM1 samples.
To exclude the possibility that FoxM1 overexpression in recurrent GBMs was due to other types of cells rather than tumor cells, we established two primary cultured cell lines from the paired primary and recurrent GBM1 samples, respectively. As shown in Fig. 1D, compared with the primary GBM cells, the recurrent GBM cells showed significant overexpression of FoxM1. Moreover, DNA damage genes, such as BRCA2, Chk2, and Rad51, were all upregulated in recurrent GBM cells more so than in primary GBM cells (Fig. 1D). However, no obvious difference in MGMT expression was detected, indicating that TMZ resistance in recurrent GBMs is not due to higher MGMT levels. We also detected the expression of FoxM1 and the DNA damage genes in U87, NHA and another primary cultured recurrent GBM cell line (GBM2). Likewise, FoxM1 and the DNA damage genes were highly expressed in primary cultured recurrent GBM cells but lower in NHA and U87 cells (Supplementary Fig. 1B and C). These results suggest that FoxM1, along with DNA damage genes, are upregulated in recurrent GBMs.
FoxM1 and DNA repair gene are upregulated by TMZ in recurrent GBMs and correlated with TMZ resistance
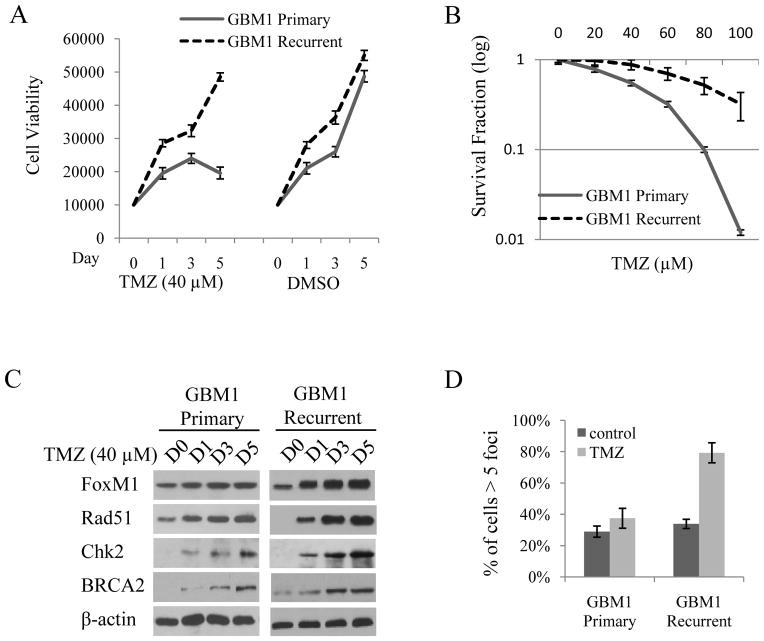
Tumor recurrence in these patients indicates that the recurrent GBM cells may be TMZ resistant. We therefore analyzed the response of the two primary cultured cell lines from the primary and the corresponding recurrent GBM sample to TMZ treatment. TMZ had little effect on the viability of the recurrent GBM cells compared with primary GBM cells after 5 days (Fig. 2A). Furthermore, clonogenic survival of the primary GBM cells was significantly reduced along with a different TMZ concentration gradient, but not in the recurrent GBM cells (Fig. 2B). We also analyzed cell viability and clonogenic survival in NHA, U87, and two recurrent GBM cells after treating them with TMZ. The results revealed that NHA and U87 cells were sensitive but recurrent GBM cells were resistant to TMZ treatment (Supplementary Fig. 2). Moreover, after TMZ treatment, more U87 cells were arrested in G2 stage (Supplementary Fig. 3). Together with the data shown in Supplementary Fig. 1C, these results suggest that the TMZ sensitivities of the above cell lines are inversely correlated with their FoxM1 expression levels.
Figure 2. Recurrent GBM cells are resistant to TMZ treatment by upregulating FoxM1 and Rad51.
A, Cell viability assay shows the cell growth of primary and recurrent GBM1 cells after being treated with 40 μM TMZ for 1 to 5 days. DMSO was used as a treatment control. B, Colony formation assay of the primary and recurrent GBM1 cells after being treated with different concentrations of TMZ for 14 days. C, Western blot analysis shows the expression of FoxM1 and DNA repair genes after being treated with 40 μM TMZ for 1 to 5 days in primary and recurrent GBM1 cells. β-actin was used as a loading control. D, Immunofluorescence staining of Rad51 foci in primary and recurrent GBM1 cells. Percentage of cells with more than five Rad51 foci in 10 random microscopic fields was calculated after TMZ treatment for 5 days.
Next, we determined the expression of FoxM1 and the DNA damage genes in primary and recurrent GBM cells after treating them with TMZ for 1 to 5 days. As shown in Fig. 2C, compared with the primary GBM cells, FoxM1 and Rad51 expression was dramatically increased in recurrent GBM cells. However, no obvious difference in BRCA2 and Chk2 was found between these two cell lines by TMZ treatment. Previous studies indicated that Rad51 forms repair-associated foci after DNA damage at sites of replication fork collapse (14, 15) and that high Rad51 levels are predictive of stronger radiation therapy or chemotherapy resistance in human cancers (25–27). Indeed, after TMZ treatment, the percentage of cells with more than 5 Rad51 foci was higher in recurrent GBM cells than in primary GBM cells (Fig. 2D). These results suggest that high FoxM1 and Rad51 levels contribute to TMZ resistance in recurrent GBM cells.
FoxM1 regulates Rad51 promoter activity in GBM cells
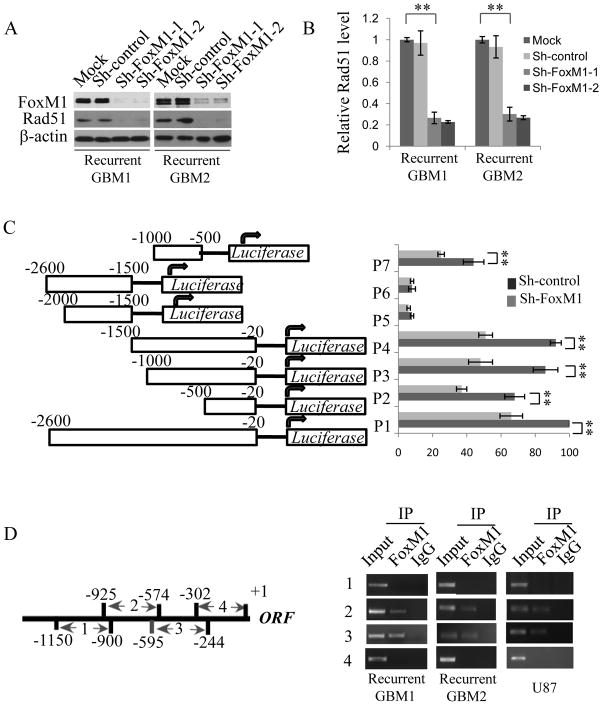
To screen the downstream genes of FoxM1, a microarray analysis was performed after FoxM1 knockdown by siRNA in the U87 glioma cell line. We found that Rad51 is a potential downstream target of FoxM1, as the Rad51 mRNA level was decreased in the microarray analysis (Supplementary Table 7). To further test this result, we generated two stable cell lines of FoxM1 knockdown using recurrent GBM1 and GBM2 cells, referred to as shFoxM1-1 and shFoxM1-2, respectively. As shown in Fig. 3Aand B, Rad51 was decreased in FoxM1-stable knockdown cell lines, both at the protein and mRNA levels. These data suggest that FoxM1 regulates Rad51 expression.
Figure 3. FoxM1 transcriptionally regulates Rad51 expression by directly binding to the promoter region.
A, Western blot analysis reveals Rad51 protein levels in the two recurrent GBM cells stably expressing sh-FoxM1. ShRNA, with a sequence that targets no genes, was used as a negative control. B, Real-time PCR assay shows Rad51 mRNA levels in recurrent GBM1 and GBM2 cells expressing sh-FoxM1. Data were from three independent assays. **P<0.001. C, Dual luciferase assay shows the relative promoter activities of constructs harboring different fragments of the human Rad51 promoter in sh-FoxM1 or control recurrent GBM1 cells. The values were normalized to a Renilla transfection control. Three independent assays were performed. **P<0.001. D, ChIP assay shows the direct binding of FoxM1 to Rad51 promoter. Left panel, schematic illustration of four PCR-amplified fragments of Rad51 promoter; right panel, ChIP assays were performed using FoxM1 antibody in recurrent GBM1 and GBM2, and in U87 cells. IgG was used as a negative control.
To determine whether FoxM1 directly regulates Rad51 expression at the transcriptional level, we amplified and cloned a 2.6-kb Rad51 promoter into the PGL3 reporter plasmid and generated several other plasmids harboring different truncated fragments of the promoter (Fig. 3C, left). By using the 2.6-kb Rad 51 promoter plasmid, we found that silencing FoxM1 in GBM1 and GBM2 cells dramatically reduced Rad51 promoter activity (Fig. 3C). Promoter activities driven by the truncated fragments, including those covering regions −20 to −500 (P2), −20 to −1000 (P3), −20 to −1500 (P4), −1000 to −1500 (P5), −1500 to −2600 (P6), and −500 to −1000 (P7), were also analyzed. The results revealed that promoter activities driven by P2, P3, P4, and P7 fragments were dramatically decreased by FoxM1 ShRNA as the P1 fragment, whereas no obvious response was found in fragments P5 and P6 after FoxM1 knockdown(Fig. 3C). These data demonstrate that FoxM1 is involved in the transactivation of Rad51 through the promoter region from −20 to −1000.
We further determined whether FoxM1 activates Rad51 expression through the direct binding of the promoter region by performing a ChIP assay with four pairs of primers covering four regions of the Rad51 promoter: −900 to −1150, −574 to −925, −244 to −595, and +1 to −302. FoxM1 binds to regions 2 (nucleotides −595 to −500) and 3 (nucleotides −244 to −595) of the Rad51 promoter in GMB1, GBM2, and U87 cells (Fig. 3D). Overall, these results indicate that FoxM1 transactivates Rad51 expression by directly binding to Rad51 promoter in GBM cells.
FoxM1-binding sites are critical for activating the Rad51 promoter in GBM cells
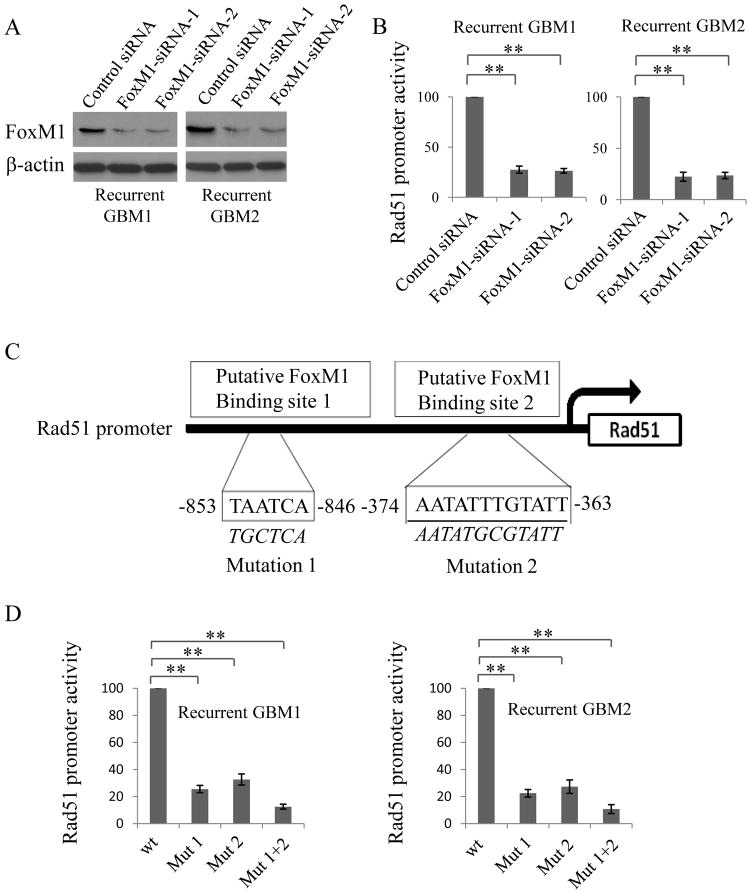
To determine the functional role of FoxM1-binding sites in Rad51 gene regulation, various mutant reporters harboring different predicted mutant binding sites of FoxM1 were generated from the wild-type Rad51 promoter construct, including site 1 (mutation 1) and site 2 (mutation 2) and site 1 plus site 2 (Fig. 4C). Compared with the wild-type Rad51 promoter, disruption of site 1 or site 2 significantly attenuated Rad51 promoter activity (Fig. 4D). Moreover, disruption of both site 1 and site 2 simultaneously further decreased Rad51 promoter activity (Fig. 4D), suggesting that both of the binding sites are critical for Rad51 promoter activation mediated by FoxM1 in GBM cells.
Figure 4. FoxM1 binding sites in the Rad51 promoter are critical for Rad51 transactivation.
A, Western blotting showing the knockdown of FoxM1 using specific siRNAs in recurrent GBM1 and GBM2 cells. SiRNA, with a sequence that does not target genes, was used as a negative control. B, Relative promoter activity of Rad51 after transfecting FoxM1 siRNAs in GBM1 and GBM2 cells. C, Schematic structure of the putative FoxM1 binding sites in Rad51 promoter. FoxM1-binding site sequences are shown in both wild-type (WT) and mutant (Mut) forms. D, Rad51 promoter activities of the promoter, with or without mutations, in the predicted FoxM1 binding sites. All experiments were repeated in triplicate, **P<0.001.
FoxM1 inhibition increased chemosensitivity to TMZ-resistant GBM cells
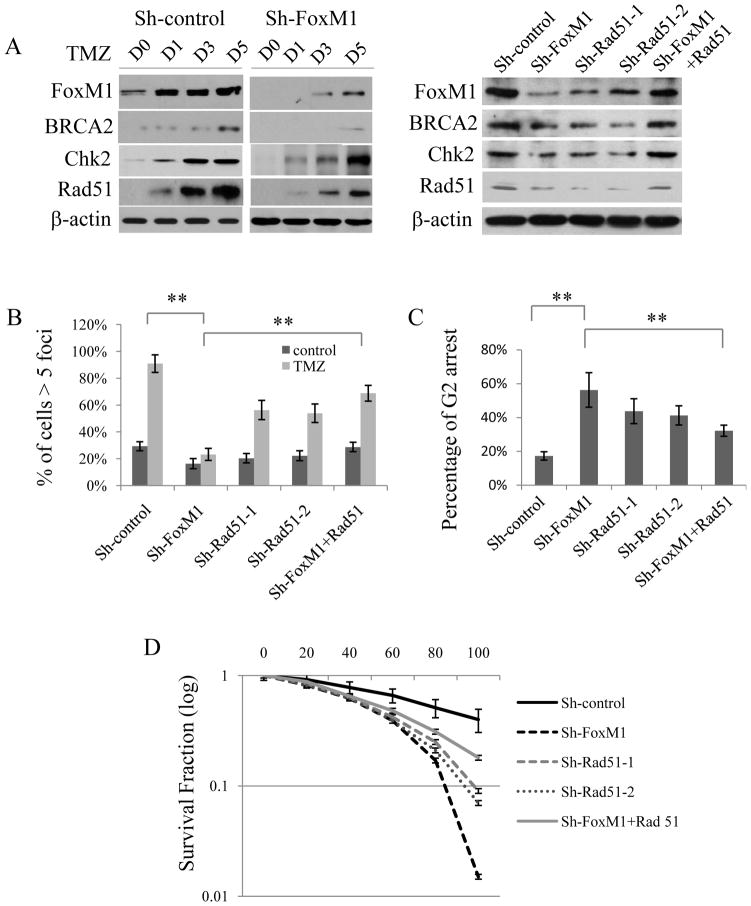
We next determined whether FoxM1 is involved in TMZ resistance in recurrent GBM cells using GBM stable cells overexpressing FoxM1 shRNA. GBM1 and GBM2 stable cells and the corresponding control stable cells were treated with 60 μM of TMZ for 1 to 5 days. FoxM1 induction by TMZ was significantly attenuated in FoxM1 knockdown cells (Fig. 5A). TMZ-induced expression levels of Rad51, BRCA2, and Chk2 genes were also attenuated in FoxM1 stable knockdown cells (Fig. 5A, left panel). Moreover, FoxM1 knockdown significantly decreased the number of Rad51 foci (Fig. 5B), increased TMZ-induced G2 arrest (Fig. 5C), decreased cell viability after 5 days of TMZ treatment (Supplementary Fig. 4), and decreased colony survival after longer TMZ treatment (Fig. 5D). Together, these results indicate that FoxM1 decreases TMZ chemosensitivity in recurrent GBM cells.
Figure 5. Silencing FoxM1 sensitized TMZ-induced cytotoxicity in recurrent GBM cells, and Rad51 overexpression rescued chemoresistance in FoxM1-silenced recurrent GBM cells.
A, Left, Western blot analysis reveals different responses of DNA repair genes to TMZ in sh-FoxM1 and control recurrent GBM1 cells. Right, Western blot analysis reveals the expression of different DNA repair genes in recurrent GBM1 cells expressing sh-FoxM1, sh-Rad51, or sh-FoxM1 plus Rad51. B, Immunofluorescence staining of Rad51 foci in recurrent GBM1 cells expressing sh-FoxM1, sh-Rad51, or sh-FoxM1 plus Rad51. Percentage of cells with more than five Rad51 foci in 10 random microscopic fields was calculated. C, Flow cytometry analysis shows the percentage of G2 arrest in recurrent GBM1 stable cells expressing sh-FoxM1, sh-Rad51, or sh-FoxM1 plus Rad51 after TMZ treatment. D, Colony formation assay shows cell survival in recurrent GBM1 cells expressing sh-FoxM1, sh-Rad51, or sh-FoxM1 plus Rad51 treated by TMZ. All experiments were performed in triplicate, **P<0.001.
Rad51 rescues TMZ resistance in FoxM1-silenced GBM cells
To determine whether TMZ resistance in recurrent GBM tumor cells is dependent on Rad51, we inhibited Rad51 expression by sh-Rad51 in GBM cells (Fig. 5A, left). Rad51 knockdown significantly decreased the number of Rad51 foci induced by TMZ treatment (Fig. 5B), increased TMZ-induced G2 arrest (Fig. 5C), decreased cell viability after 5 days of TMZ treatment (Supplementary Fig. 4), and decreased colony survival after longer TMZ treatment (Fig. 5D). Moreover, re-expression of Rad51 in FoxM1-silenced GBM cells resulted in increased levels of BRCA2 and Chk2 expression (Fig. 5A, right). Interestingly, when Rad51 was overexpressed, FoxM1 expression levels were also slightly increased (Fig. 5A, right). We further analyzed the cell responses to TMZ after Rad51 re-expression in FoxM1-silenced GBM 1 cells. Rad51 re-expression increased the number of Rad51 foci (Fig. 5B). Further, Rad51 overexpression partially rescued cells from G2 arrest induced by TMZ (Fig. 5C) and attenuated TMZ-induced cytotoxicity in FoxM1-knockdown GBM cells (Fig. 5D and supplementary Fig. 4). These data suggest that Rad51 partially rescues TMZ resistance after FoxM1 knockdown.
We further performed an in vivo tumor formation assay. Glioma cells expressing sh-FoxM1, sh-Rad51, or sh-FoxM1 with re-expression of Rad51 were intracranially injected into nude mice. The nude mice were then treated with TMZ or control vehicle. As shown in Supplementary Fig. 5, FoxM1 or Rad51 knockdown led to decreased tumor formation and prolonged mouse survival and Rad51 re-expression partially rescued the processes mediated by knockdown of FoxM1. Moreover, TMZ treatment increased the mouse survival of Rad51 knockdown group as compared with untreated group, whereas TMZ treatment did not shown any effect in FoxM1 knockdown plus Rad51 re-expression group (Supplementary Fig. 5).
FoxM1 and Rad51 expression levels were correlated in recurrent GBM specimens and were independently predictive of poor prognosis
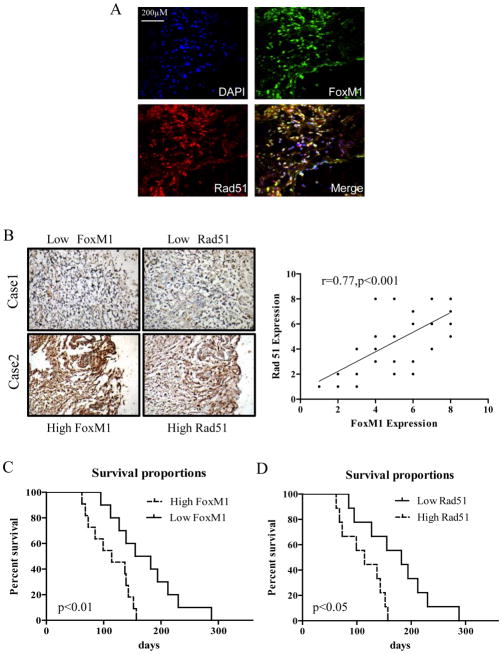
Co-localization of FoxM1 and Rad51 in recurrent GBM samples was analyzed by immunofluorescence staining. FoxM1 and Rad51 showed overlapping expression in recurrent GBM cells, mostly in the nucleus (Fig. 6A). We further performed an immunohistochemical analysis of FoxM1 and Rad51 proteins in 38 recurrent GBM samples. FoxM1 and Rad51 expression levels were positively correlated in recurrent GBM samples, as determined by Pearson’s correlation test (Fig. 6B, r=0.77, p<0.001). Moreover, FoxM1 and Rad51 protein levels were significantly correlated with survival duration in recurrent GBM (P < 0.05) (Fig. 6C, D). However, the FoxM1 protein level was not correlated with MGMT promoter methylation status or IDH1 R132 mutation in patient samples (Supplementary Tables 5 and 6).
Figure 6. FoxM1 and Rad51 expression levels were highly correlated with each other and were predictive of poor prognosis in recurrent GBM.
A, Immunofluorescence assay showing the co-localization of FoxM1 with Rad51 in recurrent glioblastoma specimens. Images shown are representative of 10 frozen glioblastoma specimens. Scale bar, 200 μM. B, Co-expression of FoxM1 and Rad51 in recurrent GBM surgical specimens. Left panel, representative image of immunohistochemical staining of FoxM1 and Rad51 in two recurrent GBM surgical specimens. Right panel, statistical analysis of FoxM1 and Rad51 expression correlation in 38 recurrent GBM specimens (r=0.77, P<0.001). C, Survival curves of recurrent GBM patients with high and low FoxM1 expression levels (P <0.01, log-rank test). D. Survival curves of recurrent GBM patients with high and low Rad51 expression levels (P <0.05, log-rank test).
DISCUSSION
In this study, we found that FoxM1 expression levels were higher in recurrent than in original GBM tumors and that targeting FoxM1 sensitized recurrent GBM cells to TMZ cytotoxicity. Mechanistically, FoxM1 directly regulated the DNA damage repair gene Rad51 at the transcriptional level. Knocking down FoxM1 inhibited Rad51 expression, and re-expression of Rad51 partially rescued FoxM1 knockdown’s inhibitory effect on TMZ resistance.
FoxM1 promotes tumorigenesis by activating a series of cell cycle genes. As FoxM1 may be a therapeutic target for malignant tumors, its role in chemotherapy has become the focus of recent research. Carr et al (41) confirmed that FoxM1 mediates breast cancer cells’ resistance to trastuzumab and paclitaxel by directly regulating the expression of the tubulin-destabilizing protein stathmin. Kwok et al (43) also reported that acquired cisplatin resistance in breast cancer cells occurs through induction of FoxM1 and its proposed downstream targets, BRCA2 and XRCC1. Our immunohistochemical analyses revealed that FoxM1 expression was elevated in recurrent GBMs. In primary culture GBM cell lines, FoxM1 expression was upregulated compared with in U87 cells and NHA cells, both at the mRNA and protein levels. In accordance with clinical data, at least in some aspect, primary GBM cells derived from recurrent GBM also exhibited relatively higher resistance to TMZ. Primary GBM cells were sensitized to TMZ cytotoxicity after FoxM1 knockdown.
Reports have shown that elevated Rad51 expression protects human head and neck tumor cells from apoptosis and enhances chemotherapy resistance by decreasing DNA damage and overcoming G2 arrest (45). The results of a recent study showed that Rad51 protein was elevated in 53% of GBM primary tumor patients but 70% of recurrent GBM patients (29). Rad51 levels were also inversely correlated with sensitivity to TMZ cytotoxicity (30). However, the mechanism of Rad51 upregulation in recurrent GBMs is unknown. We found that FoxM1 crucially regulated Rad51 expression by directly interacting with the promoter through FoxM1 binding sites; mutation of the two sites significantly decreased Rad51 promoter activity, both alone and in combination. Further, when Rad51 was overexpressed in FoxM1-knockdown GBM cells, the cells became resistant to TMZ, indicating that the FoxM1-Rad51 axis plays a critical role in TMZ resistance. However, Rad51 overexpression has only a partial rescue effect in vitro, suggesting that other FoxM1 target genes, such as BRCA2, are involved in TMZ resistance. Furthermore, Rad51 re-expression partially rescued the tumor inhibition mediated by FoxM1 knockdown and Rad51 knockdown increased the TMZ sensitivity of recurrent GBM cells in vivo, although animal experiments with different doses of TMZ are needed in future studies. Finally, we found that FoxM1 expression levels are directly correlated with Rad51 levels in relapsed GBM specimens, and both are independent prognostic markers for survival duration in recurrent GBM patients.
Our findings are consistent with previously published results that Rad51 overexpression is correlated with clinical outcome, such as in lung, head, neck, and breast carcinomas (45–47). Other studies have reported that inhibition of several FoxM1 downstream genes, such as survivin and PLK1, can sensitize breast cancer cells to chemotherapy (48, 49). In our recently published paper, we demonstrated that FoxM1 levels are much higher in glioma-initiated cells, which are more resistant than other cancer cells to current therapies (33). Moreover, FoxM1 inhibition can impair the self-renewal of glioma-initiated cells (33). These results indicate that FoxM1 plays a key role in tumor cell resistance and that targeting it is an effective method of increasing tumor cells’ chemosensitivity. In summary, our study provides clinical and molecular evidence that FoxM1 mediates TMZ resistance in GBMs by directly regulating Rad51 expression and describes a novel potential therapeutic target for recurrent GBMs.
Supplementary Material
Translational Relevance.
Drug resistance remains a major clinical challenge in recurrent GBM treatment. We found that the transcription factor FoxM1 was significantly upregulated in recurrent GBM, and its expression level was inversely correlated with the treatment effects of TMZ. The primary cultured GBM cells derived from recurrent specimens displayed TMZ resistance, and FoxM1 inhibition sensitized them to TMZ. A mechanistic investigation revealed that FoxM1 directly upregulated the expression of the DNA damage repair gene Rad51 through two FoxM1 binding sites in its promoter. Further, Rad51 re-expression in FoxM1-knockdown GBM cells partially rescued their TMZ resistance. Our findings demonstrate that the FoxM1-Rad51 axis plays an important role in GBM chemotherapy resistance and that targeting FoxM1 may be an efficient method to enhance TMZ sensitivity.
Acknowledgments
We thank Ann M. Sutton for editing this article.
Grant Support
This work was supported in part by NCI grants R01 CA157933 and R21 CA152623, the National Natural Science Foundation of China (81001119), Grant Awarded to New Teacher From Chinese Ministry of Education (20110171120112), The Fundamental Research Funds for the Central Universities (11ykzd06), and the Natural Science Foundation of Guangdong Province, China (10451008901004231 and 2009B060700031) Science and Technology Planning Project of Guangdong Province, China (2010B031600057).
Footnotes
Conflicts of interest: There are no conflicts of interest for all authors.
References
- 1.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Tisdale MJ. Antitumor imidazotetrazines–XV. Role of guanine O6 alkylation in the mechanism of cytotoxicity of imidazotetrazinones. Biochem Pharmacol. 1987;36:457–62. doi: 10.1016/0006-2952(87)90351-0. [DOI] [PubMed] [Google Scholar]
- 3.Sobol RW. Temozolomide. In: Schwab M, editor. Encyclopedia of cancer. 2. Berlin: Springer; 2009. [Google Scholar]
- 4.Wang JY, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006;9:417–8. doi: 10.1016/j.ccr.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Friedman HS, Johnson SP, Dong Q, Schold SC, Rasheed BK, Bigner SH, et al. Methylator resistance mediated by mismatch repair deficiency in a glioblastoma multiforme xenograft. Cancer Res. 1997;57:2933–6. [PubMed] [Google Scholar]
- 6.Caporali S, Falcinelli S, Starace G, Russo MT, Bonmassar E, Jiricny J, et al. DNA damage induced by temozolomide signals to both ATM and ATR: role of the mismatch repair system. Mol Pharmacol. 2004;66:478–91. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 7.Quiros S, Roos W, Kaina B. Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell Cycle. 2010;9:168–78. doi: 10.4161/cc.9.1.10363. [DOI] [PubMed] [Google Scholar]
- 8.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–99. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 9.Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–8. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1001. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 11.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–45. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–9. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sathornsumetee S, Rich JN. New treatment strategies for malignant gliomas. Expert Rev Anticancer Ther. 2006;6:1087–104. doi: 10.1586/14737140.6.7.1087. [DOI] [PubMed] [Google Scholar]
- 14.Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, et al. Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Yuan SS, Liu W, Xu Y, Trujillo K, Song B, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto A, Taki T, Yagi H, Habu T, Yoshida K, Yoshimura Y, et al. Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol Gen Genet. 1996;251:1–12. doi: 10.1007/BF02174338. [DOI] [PubMed] [Google Scholar]
- 17.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–70. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 19.Maacke H, Jost K, Opitz S, Miska S, Yuan Y, Hasselbach L, et al. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–5. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 20.Hannay JA, Liu J, Zhu QS, Bolshakov SV, Li L, Pisters PW, et al. Rad51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation. Mol Cancer Ther. 2007:61650–60. doi: 10.1158/1535-7163.MCT-06-0636. [DOI] [PubMed] [Google Scholar]
- 21.Rapakko K, Heikkinen K, Karppinen SM, Winqvist R. Screening for RAD51 and BRCA2 BRC repeat mutations in breast and ovarian cancer families. Cancer Lett. 2006;236:142–7. doi: 10.1016/j.canlet.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 22.Collis SJ, Tighe A, Scott SD, Roberts SA, Hendry JH, Margison GP, et al. Ribozyme mini-gene-mediated RAD51 down-regulation increases radiosensitivity of human prostate cancer cells. Nucleic Acids Res. 2001;29:1534–1538. doi: 10.1093/nar/29.7.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu ZY, Loignon M, Han FY, Panasci L, Aloyz R. Xrcc3 induces cisplatin resistance by stimulation of Rad51-related recombinational repair, S-phase checkpoint activation, and reduced apoptosis. J Pharmacol Exp Ther. 2005;314:495–505. doi: 10.1124/jpet.105.084053. [DOI] [PubMed] [Google Scholar]
- 24.Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–25. [PubMed] [Google Scholar]
- 25.Taki T, Ohnishi T, Yamamoto A, Hiraga S, Arita N, Izumoto S, et al. Antisense inhibition of the RAD51 enhances radiosensitivity. Biochem Biophys Res Commun. 1996;223:434–38. doi: 10.1006/bbrc.1996.0911. [DOI] [PubMed] [Google Scholar]
- 26.Ohnishi T, Taki T, Hiraga S, Arita N, Morita T. In vitro and in vivo potentiation of radiosensitivity of malignant gliomas by antisense inhibition of the RAD51 gene. Biochem Biophys Res Commun. 1998;245:319–24. doi: 10.1006/bbrc.1998.8440. [DOI] [PubMed] [Google Scholar]
- 27.Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, et al. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–83. [PubMed] [Google Scholar]
- 28.Saydam O, Saydam N, Glauser DL, Pruschy M, Dinh-Van V, Hilbe M, et al. HSV-1 amplicon-mediated post-transcriptional inhibition of Rad51 sensitizes human glioma cells to ionizing radiation. Gene Ther. 2007;14:1143–51. doi: 10.1038/sj.gt.3302967. [DOI] [PubMed] [Google Scholar]
- 29.Welsh JW, Ellsworth RK, Kumar R, Fjerstad K, Martinez J, Nagel RB, et al. Rad51 protein expression and survival in patients with glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2009;15:741251–5. doi: 10.1016/j.ijrobp.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Short SC, Giampieri S, Worku M, Alcaide-German M, Sioftanos G, Bourne S, et al. Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro Oncol. 2011;13:487–99. doi: 10.1093/neuonc/nor010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, et al. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385–400. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- 32.Pilarsky C, Wenzig M, Specht T, Detlev H, Grutzmann R. Identification and validation of commonly over-expressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–50. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–42. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 35.Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol. 2008;28:5162–71. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci USA. 2002;99:16881–6. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Fernández M, Halim VA, Krenning L, Aprelia M, Mohammed S, Heck AJ, et al. Recovery from a DNA-damage-induced G2 arrest requires Cdk-dependent activation of FoxM1. EMBO Rep. 2010;11:452–8. doi: 10.1038/embor.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson MS, Brosens JJ, Schwenen HD, Lam EW. FOXO and FOXM1 in cancer: the FOXO-FOXM1 axis shapes the outcome of cancer chemotherapy. Curr Drug Targets. 2011;12:1256–66. doi: 10.2174/138945011796150244. [DOI] [PubMed] [Google Scholar]
- 40.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29:2983–95. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–63. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henning W, Sturzbecher HW. Homologous recombination and cell cycle checkpoints: Rad51 in tumour progression and therapy resistance. Toxicology. 2003;193:91–109. doi: 10.1016/s0300-483x(03)00291-9. [DOI] [PubMed] [Google Scholar]
- 43.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peñuelas S, Anido J, Prieto-Sánchez RM, Folch G, Barba I, Cuartas I, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–27. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 45.Connell PP, Jayathilaka K, Haraf DJ, Weichselbaum RR, Vokes EE, Lingen MW. Pilot study examining tumor expression of RAD51 and clinical outcomes in human head cancers. Int J Oncol. 2006;28:1113–19. [PubMed] [Google Scholar]
- 46.Qiao GB, Wu YL, Yang XN, Zhong WZ, Xie D, Guan XY, et al. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br J Cancer. 2005;93:137–43. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Scodan R, Cizeron-Clairac G, Fourme E, Meseure D, Vacher S, Spyratos F, et al. DNA repair gene expression and risk of locoregional relapse in breast cancer patients. Int J Radiat Oncol Biol Phys. 2010;78:328–36. doi: 10.1016/j.ijrobp.2009.07.1735. [DOI] [PubMed] [Google Scholar]
- 48.Krishnan A, Hariharan R, Nair SA, Pillai MR. Fluoxetine mediates G0/G1 arrest by inducing functional inhibition of cyclin dependent kinase subunit (CKS)1. Biochem Pharmacol. 2008;75:1924–34. doi: 10.1016/j.bcp.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Spänkuch B, Heim S, Kurunci-Csacsko E, Lindenau C, Yuan J, Kaufmann M, et al. Down-regulation of Polo-like kinase 1 elevates drug sensitivity of breast cancer cells in vitro and in vivo. Cancer Res. 2006;66:5836–46. doi: 10.1158/0008-5472.CAN-06-0343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.