Abstract
Dielectrophoresis was used to stretch and suspend actin filaments across a trench etched between two electrodes patterned on a glass slide. Optical tweezers were used to bring a motor protein-coated bead into close proximity to a pre-selected, suspended actin filament, facilitating the attachment of the myosin-coated bead to the filament. The clearance beneath the filament allowed the bead to move freely along and around its filamentous track, unhindered by solid surfaces. Using defocused images, the three-dimensional position of the bead was tracked as a function of time to obtain its trajectory. Experiments were carried out with myosin V and myosin X. Both motor proteins followed left-handed helical paths with the myosin X motor exhibiting a shorter pitch than the myosin V. The combined use of electrostatic and optical tweezers facilitates the preparation of motility assays with suspended tracks. Variants of this technique will enable higher complexity experiments in vitro to better understand the behavior of motors in cells.
1. Introduction
Molecular motors are cellular energy transducers involved in determining cell shape and motions. They power muscle contraction, transport cargo along tracks of cytoskeletal actin or microtubule filaments, and are implicated in many disease processes.1–4
For the most part, mechanical studies of myosin and other molecular motors have utilized surface-immobilized motors5–9 or filaments,10–16 which impact on the range of motion of a motor. For example, myosin V has been shown to have an average step size of 36 nm, coinciding with the half-pitch of the actin helix.6,14 However, in these studies, the actin track was attached to a glass microscope slide that restricts motions to the demi-cylindrical domain on one side of the filamentous track, possibly constraining the motor to particular binding sites. The classic three-bead optical trap assay—an actin “dumbbell” lowered onto a myosin-coated polymeric or ceramic bead—has yielded valuable information on the force production, step size, and kinetics of molecular motors.5–9 In this case, immobilization of the motor onto the bead may again adversely impact on the range of motion of the motor. To enable greater freedom of motion, it is desirable to develop motility assays that enable the transport activity to take place away from any surfaces.
One way of avoiding surface immobilization of the filament and motor is to suspend filaments from fixed supports, giving the motor or the motor-coated bead unimpeded freedom of motion about its filamentous track. To this end, Ali et al. suspended actin filaments (randomly) between two immobilized 4.5 µm-diameter beads and, using optical tweezers, brought 1 µm-diameter, myosin-coated beads into close proximity to the filament.17,18 They were able to detect the path of the myosin-coated bead along and around actin for two myosin isoforms in the absence of constraints imposed by surface attachment. This technique is laborious since just a small fraction of the filaments falls in or next to a horizontal plane and few are sufficiently stretched between the two supporting beads to facilitate accurate tracking of the attached bead.
Here, we describe an attractive alternative that facilitates the control of placement and tightness of actin filaments or microtubules. The method consistently yields tightly suspended filaments positioned in a single horizontal plane, allowing us to keep the entire length of a filament in focus during motility experiments. Dielectrophoresis, which enables positioning the filaments at predetermined locations and controlling their tautness,19 is combined with an optical trap (laser tweezer), which is used to bring a motor protein-coated bead near the filament. Once the bead is positioned next to the filament and a motor protein binds to the filament, the optical trap is turned off, and the bead is carried along the filament by the molecular motors. The coordinates of the center of the bead, x̄, ȳ, and z̄, are then tracked by processing images obtained with an optical microscope. Although the use of electric fields to trap and position actin filaments and microtubules is not new,19,20 the combined use of dielectrophoresis and optical traps, to the best of our knowledge, has not been previously reported.
2. Experimental procedure
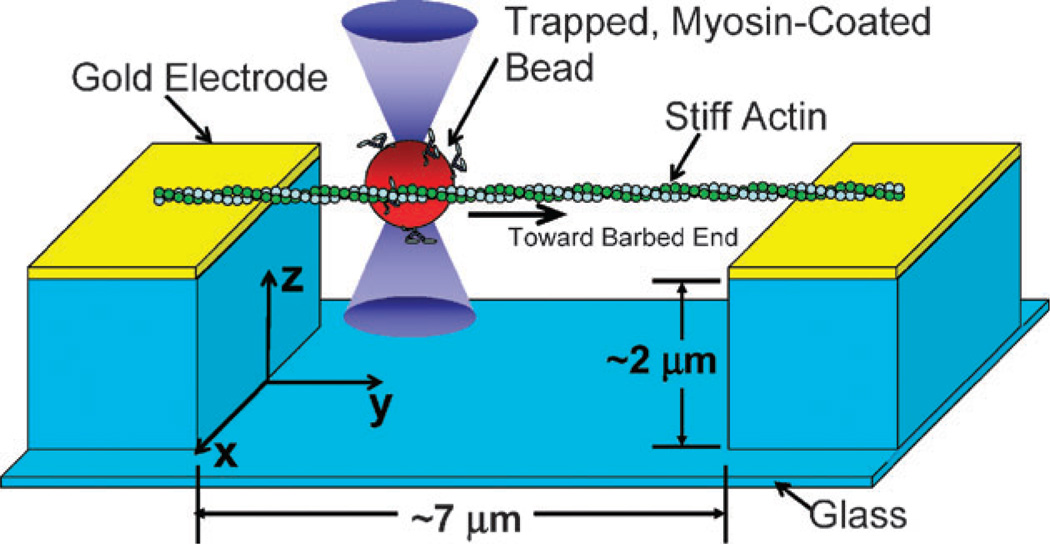
The electrode arrangement, a filament, and a bead are depicted schematically in Fig. 1. Electrodes with a 7 µm gap and a 2 µm-deep trench were patterned on a glass coverslip using standard photolithography techniques. The coverslip containing the electrodes was assembled into a flow cell using a second coverslip and double-sided adhesive tape. See Experimental Methods for additional information on the patterning of the electrodes and the other materials.
Fig. 1.
A schematic depiction of a dielectrophoretically positioned and tightened actin filament suspended across a trench between two gold electrodes. A myosin-coated bead is being positioned near the filament with optical tweezers.
A solution of actin filaments was infused into the flow cell. An AC potential was applied across the electrodes. The electric field polarized the suspended filaments and their adjacent electric double layer and induced AC electro-osmosis in the cell, which was observed by seeding the solution with 900 nm diameter tracer beads. It consisted of two counter-rotating vortices with the flow at the electrode surfaces directed away from the center of the gap and the flow velocity next to the edges of the electrode of the order of µm s−1. Thus, the electric field served two purposes. It facilitated the positioning of the filaments across the gap between the electrodes, and it controlled the tension of the filaments.19,21 Once the filaments settled across the gap, their ends adhered non-specifically to the gold surfaces of the electrode. When several filaments were suspended tightly across the gap (<~200 nm rms lateral displacement at the center of the gap), the potential was removed, and the free actin was washed out of the flow cell.
The myosin-coated beads were infused into the flow cell, and a 1064 nm optical trap9 was used to position an individual bead next to the center of a pre-selected actin filament. Single filaments were identified by their relative intensity. Once a motor-coated bead attached to the actin, the bead was released from the trap. The motors traveled along, and rotated around, the actin filament [ESI†: myosin V (Movie S1) and myosin X (Movie S2)]. The bead motion was recorded using bright-field illumination in an inverted microscope and a 60×, 1.2 NA water immersion objective (Olympus plan apo). Fluorescent images of the filament were periodically taken to ensure that, throughout the experiment, the filament remained securely attached to the gold electrodes and remained approximately in focus. The three-dimensional coordinates of the position of the bead were determined using the defocused images and a custom-written algorithm in MatLabt™.
3. Results and discussion
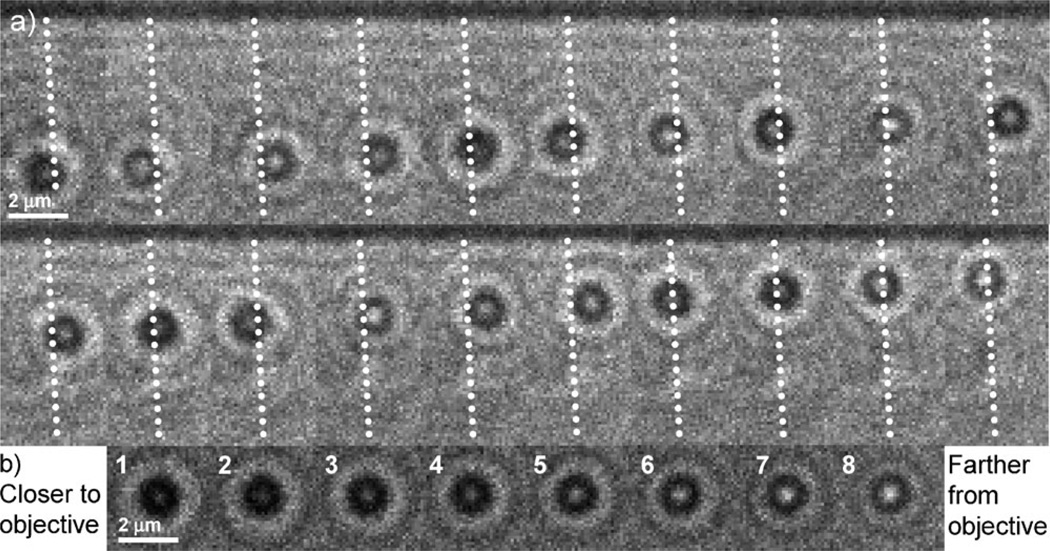
Fig. 2a shows a series of images of a myosin-V-coated bead traveling along a tightly suspended actin filament. The white, dotted lines identify the location of the filament. Bright and dark images of the center of the bead indicate, respectively, that the bead is above and below the focal plane. In other words, the motor followed a helical path around the actin filament. In Fig. 2, the path is left-handed as explained below.
Fig. 2.
(a) A series of bright-field optical micrographs of a myosin V-coated bead traveling along a tightly suspended actin filament. The illuminating condenser is stopped down from its maximum to increase contrast. The viewing objective is located below the sample. The time interval between frames is 850 ms and the exposure time is 80 ms. For clarity, dotted lines are overlaid on the actin filament. (b) Images of a stationary bead on the microscope slide surface moved in 150 nm increments along the optical axis, z. Images 1 and 8 are, respectively, closer to and farther from the imaging objective.
When a bead is displaced out of the focal plane of the imaging optics, spherical aberrations and diffraction form ring patterns centered on the bead.22,23 The unique topologies of these images can be used to estimate the out-of-plane position of the bead. A bead fixed to each slide was used to calibrate the z-position data. These calibration images were taken as the slide was translated at 50 nm increments along the optical axis and were fit to the triple Gaussian function (one Gaussian peak and two Gaussian rings)
| (1) |
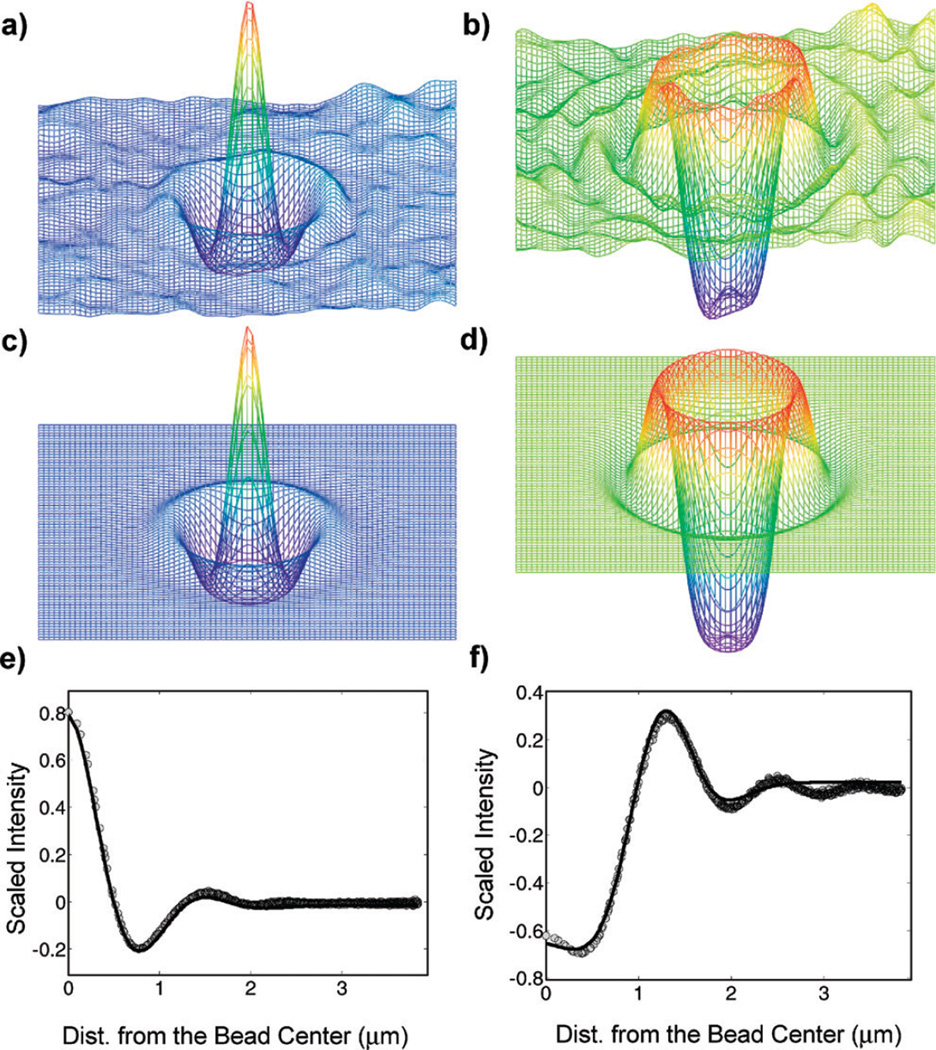
similar to Gosse and Croquette22 In the above equation, AB is the background intensity, (x̄, ȳ) is the lateral position of the center of the bead, σi (i = 0, 1, 2) is a width factor, and Ri (i = 1,2) is the radius of the Gaussian ring. Fig. 2b shows the defocused images of a stationary, calibration bead at 150 nm increments along the optical axis, where image 1 is closest to the microscope objective lens, located below the focal plane. z-movement of the stage is equivalent to z-motion of a bead in solution, because the objective lens is a water immersion type. Fig. 3 depicts the intensity distributions of the images for a bead located farther from the objective lens (a, above the focal plane) and closer to the objective lens (b, below the focal plane). The images had their background intensities subtracted using a two-dimensional moving average, taken over a square region with sides approximately equal to the diameter of the outer-most Gaussian ring, and were smoothed using a Gaussian convolution kernel of half width = 1 pixel.23 Eqn (1) was fitted to the smoothed images by minimizing the square of the difference between the fitted function (Fig. 3c and d) and the smoothed image. Fig. 3e and f compare A(x̄, ȳ) as a function of x (solid line) and the experimental data (symbols) of Fig. 3a and b.
Fig. 3.
(a) and (b) Smoothed surface plots of images similar to those in Fig. 2. (c) and (d) Fits of eqn (1) to images (a) and (b), respectively. (e) and (f) Average radial line profiles from the center of the images shown in (a) and (b), respectively (symbols), and the corresponding radial slices of the axisymmetric fits shown in (c) and (d), respectively (solid lines).
The fits of the experimental data were normalized with (max[A(x,y)] − min[A(x,y)]). To determine the z-position of a bead, we compared the normalized fits Anorm (eqn (1)) of the experimental data with the k normalized fits of the calibration data Acal,norm(k) (Fig. 2b). In other words, the z-position of a bead was determined by minimizing
| (2) |
over k, where k = 1, 2, 3… correspond to the fitted surfaces of the calibration bead intensities that were located at known z-positions and i and j are integers designating individual pixels.
Additionally, as checks of this procedure, estimates of the z-positions were obtained using the fitted radius of the outer-most Gaussian ring and using the zeroth and second moments of the light intensity.23 All these techniques yielded similar estimates for z (Fig. S5, ESI†).
After beads were brought into proximity with the suspended actin filament and released from the optical trap, many of the beads remained in contact with and moved along the actin filament with helical paths. The diameter of the helix was approximately the diameter of the bead, 1 µm, as expected, making the helical path readily observable. The trajectory, velocity, pitch, and handedness of the path of each bead were determined from its three-dimensional position as a function of time. Twenty-one myosin-V-coated beads were bound to single actin filaments, and all were observed to travel along their filaments. The average pitch was calculated using only beads that rotated for more than half a turn (Table 1). Rotations of less than half a turn were a result of short runs. Only one bead was observed to travel an appreciable distance without rotating. The average velocities in the direction of the filament axis at 500 nM, 2 µM, and 5 µM MgATP were very similar to those reported previously.13
Table 1.
Summary of myosin-V-coated bead experiments (variabilities are presented as ±1 standard deviation)
| [MgATP] | 500 nM | 2 µM | 5µM |
| Myosin–bead ratio | 104 | ||
| No. of trials | 2 | 11 | 8 |
| No. of beads that rotated >0.5 turns | 1 | 5 | 5 |
| Linear velocity/nm s−1 | 21 ± 3 | 49 ± 11 | 116 ± 46 |
| Pitch/µm | 2 | 1.9 ± 0.9 | 1.7 ± 0.4 |
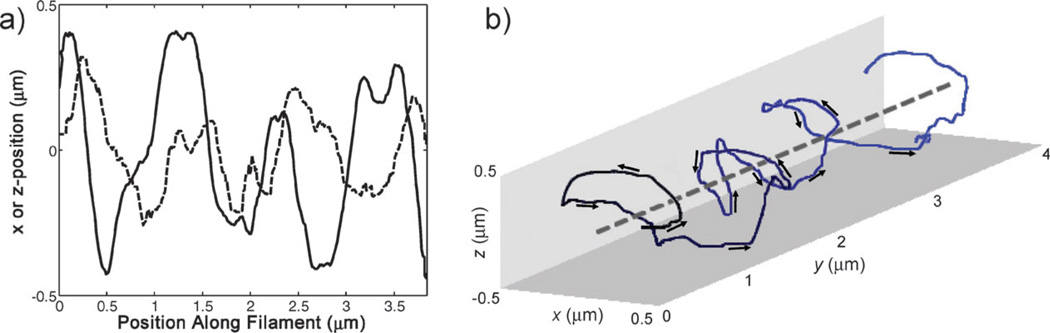
Displacements in the transverse direction (x) and along the optical axis (z), depicted as functions of the position along the filament (y) (Fig. 4a), showed that the x-position (solid line) is roughly 90° out-of-phase with the z-position (dashed line), as expected for a helical path. The bead moves in the positive y-direction. Since, in the first quadrant (x > 0, z > 0), , the path is left-handed (Fig. 4b).
Fig. 4.
(a) The displacement in the transverse direction, x (solid line) and the displacement in the optical axis, z (dashed line) of a myosin-V-coated bead, vs. the position along the filament, y. The z-position lags the x-position by roughly 90°, as expected for a left-handed helical path. (b) The left-handed, helical motion of the same myosin-V-coated bead as it traveled along an actin filament.
All helical paths taken by myosin-V-coated beads were left-handed with an average pitch of 1.8 µm without significant dependence on [MgATP] (Table 1). The handedness and pitch agree with a previous report that found the average pitch of myosin-V-coated beads on suspended actin filaments to be 2.2 µm.17
Similar experiments with myosin X at 50 µM MgATP yielded left-handed helical motion with a pitch of 1.5 µm (calculated from 4 beads – all the beads that were tested rotated). These observations are new. The path, handedness, and pitch of myosin X have not been previously reported. Our results are compatible with a recent report indicating torque generation by myosin X in gliding filament assays.24 In the latter study, the pitch and the handedness of the motion resulting from this torque were not determined.
With processive myosins, a helical path along actin can be explained by the motor preferentially binding to actin monomers that do not coincide with the pitch of the actin helix. The helical geometry of the actin filament can be defined in several ways. The tightest helical disposition, termed the ‘genetic’ helix, is a left-handed 5.5 nm pitch helix, with an axial spacing of 2.75 nm per monomer and left-handed rotation angle around the filament axis of 166° per monomer. Because this rotation per subunit along the genetic helix is close to 180°, the structure is often described as two 72–74 nm pitch, right-handed helices that coil around each other.
Actin filaments are often approximated as a 13/6 helix when interpreting the path of myosins on actin.17 The 13/6 nomenclature defines an actin filament with monomers located along the left-handed short-pitch (5.5 nm) genetic helix rotating 6 full turns in 13 monomers. Taking the origin as the zeroth actin monomer, the 13th monomer along this short-pitch helix is oriented at the same azimuth around the axis of the filament as the zeroth monomer. A motor binding to successive sites 13 monomers apart would walk straight. Based on the left-handed helical motions of the beads along the suspended actin filaments, it was surmised that myosin V occasionally binds the 11th monomer, resulting in a gradual, left-handed rotation about the actin filament that was insensitive to the number of myosin motors bound to the bead.17 On the 13/6 actin helix, myosin V binding to the 11th monomer more often than the 15th would produce left-handed twirling. Since the filament is flexible, other known actin helical indices would also be consistent with left-handed rotation.25 For instance, on a 28/13 helix (generated by a slight untwist of the 13/6 helix), monomer 13 is not disposed on a straight path, but azimuthally 13° to the left of monomer zero. Monomer 15 is disposed 13° to the right. Thus myosin binding to monomer 13 more often than monomer 15 would still result in a left-handed path. We have also found that beads coated densely with myosin X track a left-handed helix with a shorter pitch than myosin V. In contrast, myosin VI has been shown to twirl with a right-handed pitch according to a different assay (fluorescent labeled actin monomers in a gliding assay).16
Whenever a comparison is possible between data on single molecule trajectories and unconstrained paths of cargos transported by several motors, the paths have been similar in handedness and pitch,16–18,26 indicating that the helical paths are generated by intrinsic properties of the motor mechanism. For processive motors, the main determinants of their paths seem to be the step size distribution and the actin monomer indices selected by the stepping molecule. Helical paths, possibly resulting from off-axis forces or attachment biased towards sites before or after the azimuthally optimal binding site, have also been observed with myosin II, a non-processive motor.26,27 The techniques presented here may help to better our understanding of the various factors determining the functional mechanisms of molecular motions.
4. Experimental methods
Electrodes were fabricated on sulfuric–peroxide (H2SO4–H2O2, 3 : 1)-cleaned, 150 µm-thick glass coverslips. 10 nm of NiCr was evaporated to act as an adhesion layer, upon which 100 nm of gold was evaporated. The electrode pattern was defined in positive photoresist (Shipley 1813), and the gold and NiCr were wet-etched using a potassium-iodide-based etchant and nichrome etchant, respectively. Hydrofluoric acid (HF) was used to etch a ~2 µm-deep trench in the glass between the electrodes, using the patterned electrodes as a mask. Electric fields (20 kHz, 2 Vrms AC) were applied across the electrodes, and the signals (and currents) were monitored using an oscilloscope (Tektronix 3036-B).
G-actin was obtained from rabbit skeletal muscle and purified.28 F-actin was prepared from G-actin, at 1 µM total actin monomer concentration, and stabilized with 1.1 µM rhodamine-phalloidin (Molecular Probes, Carlsbad, CA). F-actin was suspended in 37 mM KCl, 2 mM MgCl2, 1 mM K-EGTA, 20 mM Hepes, and 1 mM DTT. Polystyrene beads (1 µm diameter, Polysciences, Inc.) were incubated in 30 mM KCl, 2 mM MgCl2, 1 mM K-EGTA, 20 mM Hepes, 10 mM DTT, 6 µM CaM (expressed in bacteria29), 100 ng mL−1 tetramethyl rhodamine (TMR) BSA, and then with myosin V or X at a molar ratio of ~104 myosin molecules per bead. The TMR-coated beads were easily observable under low-light conditions before attachment to actin. Chick brain myosin V was purified from tissue.30 Myosin X was a kind gift of Dr Mitsuo Ikebe and Dr Osamu Sato, University of Massachusetts. The motility buffer (pH = 7.4) also contained 0.2 mg mL−1 casein, 5 mM phosphocreatine (Sigma P7936), 0.1 mg mL−1 creatine phosphokinase (Sigma C3755), 0.5 mg mL−1 unlabeled BSA, 7.2 mg mL−1 glucose, 9 units per mL catalase, 4 mg mL−1 glucose oxidase, and various concentrations of MgATP.
5. Conclusions
In summary, we have developed an improved method for studying the path and azimuthal rotation of molecular motors in vitro that uses two non-contact techniques: dielectrophoresis and optical trapping. Although both methods have been previously used independently, their combined application is new. The ability to place tracks for molecular motors at predetermined locations and to control their tension with electrostatic tweezers overcomes a significant technical hurdle – searching for tracks that are fortuitously suspended between fixed beads with a sufficient degree of tension.
We describe an assay that facilitates the study of the motility of motors unhindered by solid surfaces, enabling the motors to rotate around their tracks as they may be able to do in vivo. The study indicates that when they are afforded the opportunity, myosin V and myosin X motors, indeed, rotate.
The left-handedness of myosin V reported earlier was reproduced, and we found an average pitch of 1.8 µm without significant dependence on [MgATP]. Additionally, we found that myosin X also follows a left-handed helical path, with an average pitch of 1.5 µm, as it proceeds along actin.
Our method makes progress toward the goal of generating in vitro assays that reproduce some of the complexity present in cells. Using other electrode and flow arrangements will allow more complex filamentous networks to be generated for molecular motor studies.
Acknowledgements
We thank Dr Mitsuo Ikebe and Dr Osamu Sato for the gift of myosin X and Dr John C. Crocker for helpful discussions pertaining to the image analysis. We acknowledge support from the National Science Foundation through the Nano/Bio Interface Center (NSF NSEC DMR-0425780) and NIH grant AR26846.
Footnotes
Electronic supplementary information (ESI) available: Bead twirling movies and image processing details. See DOI: 10.1039/b821861g
References
- 1.Schliwa M. Molecular Motors. Weinheim: Wiley-VCH; 2003. [Google Scholar]
- 2.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Hasson T, Mooseker MS. The growing family of myosin motors and their role in neurons and sensory cells. Curr. Opin. Neurobiol. 1997;7:615–623. doi: 10.1016/s0959-4388(97)80080-3. [DOI] [PubMed] [Google Scholar]
- 4.Hirokawa N, Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr. Opin. Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics—piconewton forces and nanometer steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 6.Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 7.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 9.Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys. J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block SM, Goldstein LSB, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 11.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 12.Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forkey JN, Quinlan ME, Shaw MA, Corrie JET, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 14.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 15.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur ELF. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Schroeder HW, Beausang JF, Homma K, Ikebe M, Goldman YE. Myosin VI walks “Wiggly” on actin with large and variable tilting. Mol. Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali MY, Uemura S, Adachi K, Itoh H, Kinosita K, Jr, Ishiwata S. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 2002;9:464–467. doi: 10.1038/nsb803. [DOI] [PubMed] [Google Scholar]
- 18.Ali MY, Homma K, Iwane AH, Adachi K, Itoh H, Kinosita K, Jr, Yanagida T, Ikebe M. Unconstrained Steps of Myosin VI Appear Longest among Known Molecular Motors. Biophys. J. 2004;86:3804–3810. doi: 10.1529/biophysj.103.037416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arsenault ME, Zhao H, Purohit PK, Goldman YE, Bau HH. Confinement and Manipulation of Actin Filaments by Electric Fields. Biophys. J. 2007;93:L42–L44. doi: 10.1529/biophysj.107.114538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asokan SB, Jawerth L, Carroll RL, Cheney RE, Washburn S, Superfine R. Two-Dimensional Manipulation and Orientation of Actin-Myosin Systems with Dielectrophoresis. Nano Lett. 2003;3:431–437. [Google Scholar]
- 21.Uppalapati M, Huang YM, Jackson TN, Hancock WO. Microtubule Alignment and Manipulation Using AC Electrokinetics. Small. 2008;4:1371–1381. doi: 10.1002/smll.200701088. [DOI] [PubMed] [Google Scholar]
- 22.Gosse C, Croquette V. Magnetic Tweezers: Micromanipulation and Force Measurement at the Molecular Level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crocker JC, Grier DG. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 24.Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilfan A. Influence of fluctuations in actin structure on myosin V step size. J. Chem. Inf. Model. 2005;45:1672–1675. doi: 10.1021/ci050182m. [DOI] [PubMed] [Google Scholar]
- 26.Beausang JF, Schroeder HW, Nelson PC, Goldman YE. Twirling of Actin by Myosins II and V Observed via Polarized TIRF in a Modified Gliding Assay. Biophys. J. 2008;95:5820–5831. doi: 10.1529/biophysj.108.140319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishizaka T, Yagi T, Tanaka Y, Ishiwata S. Right-handed rotation of an actin filament in an in vitro motile system. Nature. 1993;361:269–271. doi: 10.1038/361269a0. [DOI] [PubMed] [Google Scholar]
- 28.Pardee JD, Spudich JA. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 29.Putkey JA, Slaughter GR, Means AR. Bacterial expression and characterization of proteins derived from the chicken calmodulin cDNA and a calmodulin processed gene. J. Biol. Chem. 1985;260:4704–4712. [PubMed] [Google Scholar]
- 30.Cheney RE. Purification and assay of myosin V. Methods Enzymol. 1998;298:3–18. doi: 10.1016/s0076-6879(98)98003-x. [DOI] [PubMed] [Google Scholar]