Abstract
Current screening practices have been able to identify PMS2 mutations in 78% of cases of colorectal cancer from the Colorectal Cancer Family Registry (Colon CFR) which showed solitary loss of the PMS2 protein. However the detection of large-scale deletions in the 3′ end of the PMS2 gene has not been possible due to technical difficulties associated with pseudogene sequences. Here, we utilised a recently described MLPA/long-range PCR-based approach to screen the remaining 22% (n = 16) of CRC-affected probands for mutations in the 3′ end of the PMS2 gene. No deletions encompassing any or all of exons 12 through 15 were identified; therefore, our results suggest that 3′ deletions in PMS2 are not a frequent occurrence in such families.
Keywords: Lynch syndrome, PMS2, germline testing, large deletions, pseudogenes, colorectal cancer
In a recent publication, Vaughn et al. 1 described a combinatorial approach of long-range PCR and MLPA (multiplex ligation-dependant PCR amplification) to detect deletions specific to PMS2 at the 3′ end of the gene. The accurate screening for mutations in PMS2 has, until fairly recently, been plagued by problems associated with the large number of highly homologous sequences within pseudogenes that flank the functional gene.2,3 Previous studies have led to long-range PCR becoming the method of choice for the detection of point mutations as it circumvents these pseudogene-associated problems.4–6 The detection of large-scale deletions has, however, not been so reliable, particularly at the 3′ end of the gene due to technical issues caused by the high degree of homology between the real PMS2 gene and its pseudogenes. The technique most commonly used for large scale deletion detection is a multiplex ligation-dependant PCR amplification method more commonly referred to as MLPA.7 For PMS2, this technique has taken advantage of small differences between the PMS2 gene and the pseudogenes for the positioning of its analytical probes. This has proved successful for the majority of exons within PMS2 but towards the 3′ end of the gene purported differences between the real gene and the pseudogenes (paralogous sequence variants (PSVs)) have turned out to be shared polymorphism sites (SPSs),8 i.e. they are polymorphic in both sequences, which has resulted in un-interpretable findings at these locations.9
MRC Holland recently addressed this issue by modifying their PMS2 kit to include non-specific probes as well as probes designed to hybridise to both alleles of these SPSs. When this kit is used in conjunction with gene and pseudogene specific sequencing, Vaughn et al. found that it can be used to accurately assess the deletion status at the 3′ end of the gene.1 In their methodological study, sample selection was biased towards cases likely to harbour a 3′ deletion, therefore the true extent of such deletions still remains unclear in a clinical setting.
In this study, we utilise this new method (Supplementary Document 1) to screen for 3′ deletions in a cohort of CRC-affected cases from the Colon Cancer Family Registry10 who were suspected of carrying deleterious mutations in the PMS2 gene, based on the loss of expression of the PMS2 protein by immunohistochemistry (IHC), but for whom no mutation could be identified using the standard long-range PCR approach and the previous MLPA method for large deletion detection of exons 1–11.
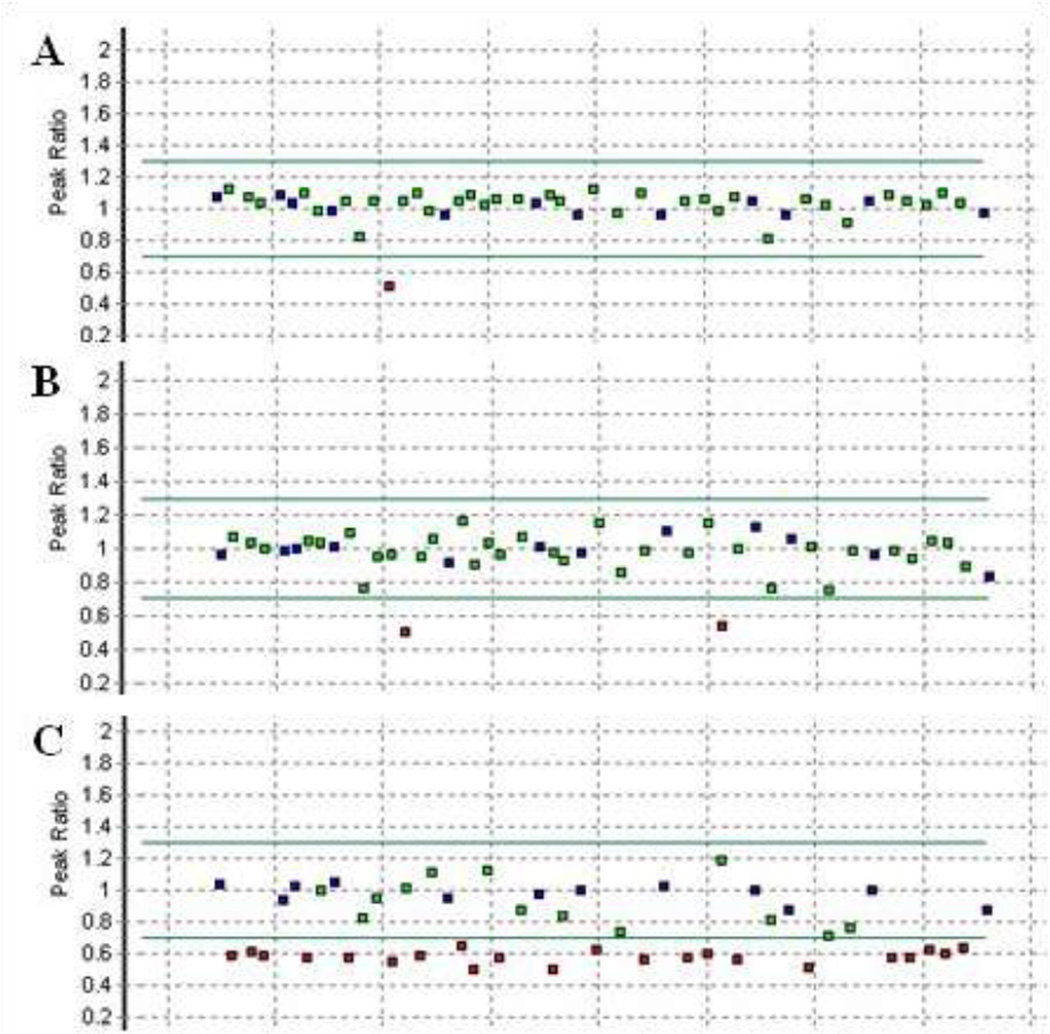
Because of the complexity of the method, we first utilised it to screen a small reference set of cases with colorectal cancer from other sources who were predicted to have a 3′ deletion. As in Vaughn et al, they had been selected because they had a deletion of at least exon 11, or the previous MLPA kit suggested a deletion in conjunction with suggestive sequence data (homozygosity) for any of the terminal 3 exons. Among this reference set, we identified an exon 14 deletion in PMS2, a deletion of exons 14 and 15 from the pseudogene and confirmed the deletion of the complete PMS2 gene in two subjects (Figure 1). Further investigation of the exon 14 deletion enabled us to determine the location of the breakpoint (c.2276-113_c.2245+1596del) and allowed for the design of a breakpoint specific PCR which simplifies mutation detection (Supplementary Document 2). This mutation has previously been identified amongst a Dutch cohort through an RNA based screening approach.9
Figure 1.
MLPA findings for 3 reference DNAs which showed abnormal probe levels. A) Individual with a deletion of PMS2 exon 14. B) Individual with an apparent deletion of exons 14 and 15 from PMS2-CL. C) Confirmation that the entire PMS2 coding region is deleted in an individual previously classified as having an exon 1 – 12 deletion with the older version of the MLPA kit. Amounts of specific probes, relative to a set of control samples which have two copies of each probe, are represented by squares. A red square denotes a probe whose copy number is suggestive of a gain or a deletion. Sequencing across the probe binding sites in both PMS2 and PMS2-CL allows for deletions and duplications to be assigned to specific loci. A detailed description of the analyses can be found in Vaughn et al. (2011).1
The test set consisted of 76 CRC-affected cases that were identified as candidates for a mutation in PMS2 based on the solitary loss of expression of PMS2 following IHC analysis, and microsatellite instability (MSI-H phenotype) as determined by the analysis of a 10 microsatellite marker panel.11 These 76 CRC-affected cases were identified from a total of 4402 CRC-affected cases whose tumours had undergone IHC analysis for all four mismatch repair proteins. Standard mutation screening via long-range PCR and large deletion detection for the 5′ end of the PMS2 gene identified pathogenic mutations in 59 cases (59/76; 78%). Of the remaining 17 cases whose tumours showed solitary loss of PMS2 expression but did not have a pathogenic mutation detected in PMS2, 16 had sufficient DNA available to test for large deletions in the 3′ end of the PMS2 gene using the method described by Vaughn et al.1 No deletions in exons 12–15 were detected from these 16 CRC-affected cases and so the causative mutation still remains to be identified.
In this study, we have confirmed that an integration of MLPA and long-range PCR can be used to detect aberrations in the 3′ end of the gene; however, we did not identify any deletions of exons 12–15 in our test set of 16 CRC-affected cases. Our data therefore suggest that, whilst plausible, large scale deletions at the 3′ end of the PMS2 gene are not a substantive cause of disease in cases for which a mutation cannot be identified by previous methods. In contrast to our findings, Vaughn et al. have recently followed up on their original methods paper with a study similar to the one outlined here, wherein they identified 7 cases with a 3′ deletion.12 In this study, they screened a total of 58 samples with unexplained loss of PMS2 emanating from an initial cohort of 117 samples. When compared to our own, the findings of Vaughn et al. highlight a considerable disparity between the type of mutations as well as the total percentage of mutations identified. Even in the absence of any 3′ deletions, our own study identifies mutations in 78% (59/76) of cases, where as Vaughn et al. find a mutation in 55% (64/117) of cases, with 11% (7/64) of these being due to 3′ terminal deletions. Both of these cohorts have been screened using very similar methods and so we would expect that the ability to detect PMS2 mutations is the same in each study, which leaves a significant proportion (particularly in the Utah cohort) of PMS2 suspected cases which have not had a causative mutation identified in the coding regions of the gene.
With this in mind, it seems likely that mutation-screening strategies will have to be broadened in order to pinpoint the causative mutation in such cases. One possible source for these elusive mutations might be intronic sequences that are not routinely examined; an example of which we have recently identified in the context of the MSH2 gene.13 For many genes the screening of these large genomic regions could eventually become cost-effective with the decreasing cost of next generation sequencing (NGS). It should be noted, however, that the pseudogene sequences which have caused many of the exonic screening problems to date will also impact on the reliability of NGS of the PMS2 loci.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all study participants of the Colon Cancer Family Registry and study co-coordinators and technical staff for their contributions to this project, in particular Judi Maskiell, Belinda Nagler, Sally-Ann Pearson, Rhiannon Walters, David Packenas and Erika Pavluk and participant interviewers for their contributions to this project. We thank individual participants in the study who made this work possible and the contribution of the Jeremy Jass Memorial Pathology Collection.
Funding
This work was supported by the National Cancer Institute, National Institutes of Health under RFA #CA-95-011 and through cooperative agreements with members of the Colon Cancer Family Registry and Principal Investigators. Collaborating centres include Australasian Colorectal Cancer Family Registry (U01 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01 CA074799) [USC], Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), Seattle Colorectal Cancer Family Registry (U01 CA074794), and University of Hawaii Colorectal Cancer Family Registry (U01 CA074806). MAJ is an NHMRC Senior Research Fellow. JLH is an NHMRC Australia Fellow. During this work JY was supported by a Cancer Council Queensland Senior Research Fellowship.
Footnotes
Publisher's Disclaimer: Disclaimer
The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the CFRs, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government or the CFR. Authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
Disclosure
The authors have no conflict of interest to declare with respect to this manuscript.
References
- 1.Vaughn CP, Hart KJ, Samowitz WS, Swensen JJ. Avoidance of pseudogene interference in the detection of 3' deletions in PMS2. Hum Mutat. 2011 doi: 10.1002/humu.21540. [DOI] [PubMed] [Google Scholar]
- 2.De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa H, et al. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–4727. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 4.Senter L, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaughn CP, et al. Clinical analysis of PMS2: mutation detection and avoidance of pseudogenes. Hum Mutat. 2010;31:588–593. doi: 10.1002/humu.21230. [DOI] [PubMed] [Google Scholar]
- 6.Clendenning M, et al. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–495. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- 7.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavlicek AR, et al. Traffic of genetic information between segmental duplications flanking the typical 22q11.2 deletion in velo-cardio-facial syndrome/DiGeorge syndrome. Genome Res. 2005;15:1487–1495. doi: 10.1101/gr.4281205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Klift HM, et al. Quantification of sequence exchange events between PMS2 and PMS2CL provides a basis for improved mutation scanning of Lynch syndrome patients. Hum Mutat. 2010;31:578–587. doi: 10.1002/humu.21229. [DOI] [PubMed] [Google Scholar]
- 10.Newcomb PA, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 11.Lindor NM, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 12.Vaughn CP, et al. The frequency of previously undetectable deletions involving 3' Exons of the PMS2 gene. Genes Chromosomes Cancer. 2012;52:107–112. doi: 10.1002/gcc.22011. [DOI] [PubMed] [Google Scholar]
- 13.Clendenning M, et al. Mutation deep within an intron of MSH2 causes Lynch syndrome. Fam Cancer. 2011;10:297–301. doi: 10.1007/s10689-011-9427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Klift HM, et al. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC) Genes Chromosomes Cancer. 2005;44:123–138. doi: 10.1002/gcc.20219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.