Abstract
In their seminal publication describing the structure of the DNA double helix1, Watson and Crick wrote what may be one of the greatest understatements in the scientific literature, namely that “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.” Half a century later, we more fully appreciate what a huge challenge it is to replicate six billion nucleotides with the accuracy needed to stably maintain the human genome over many generations. This challenge is perhaps greater than was realized 50 years ago, because subsequent studies have revealed that the genome can be destabilized not only by environmental stresses that generate a large number and variety of potentially cytotoxic and mutagenic lesions in DNA, but also by various sequence motifs of normal DNA that present challenges to replication. Towards a better understanding of the many determinants of genome stability, this chapter reviews the fidelity with which undamaged and damaged DNA is copied, with a focus on the eukaryotic B and Y family DNA polymerases, and considers how this fidelity is achieved.
Introduction
In order to pass genetic information from one generation to the next, all organisms must accurately replicate their genomes during each cell division. This includes the nuclear genome and mitochondrial and chloroplast genomes. These are normally replicated with high fidelity that is achieved through the combined action of accurate DNA polymerases and DNA mismatch repair. The major replicative DNA polymerases have evolved mechanisms to strongly favor correct over incorrect dNTP incorporation. In addition, several DNA polymerases contain an associated 3′→5′ exonuclease activity that can excise incorrect bases from the growing DNA chain, allowing another attempt at correct synthesis. In the event that the polymerase makes an error that escapes this proofreading activity, post-replication DNA mismatch repair monitors the DNA for errors, excises the error in the newly synthesized strand and then re-synthesizes DNA. In total, these three discrimination steps result in an in vivo mutation rate estimated to be lower than 1 ×10−9, i.e., less than one error for every billion (or more) bases pairs copied (Figure 1A). Moreover, at each step of the process, there are competing forces (Figure 1B) that can affect the fidelity with which DNA is replicated. In this review, we focus on the contributions and mechanisms of DNA polymerase selectivity and proofreading. Readers interested in DNA mismatch repair (MMR)a can consult recent comprehensive reviews of that subject2, 3. The focus of this chapter will be on eukaryotic DNA polymerases in the B and Y families, with discussion of links to human disease where possible.
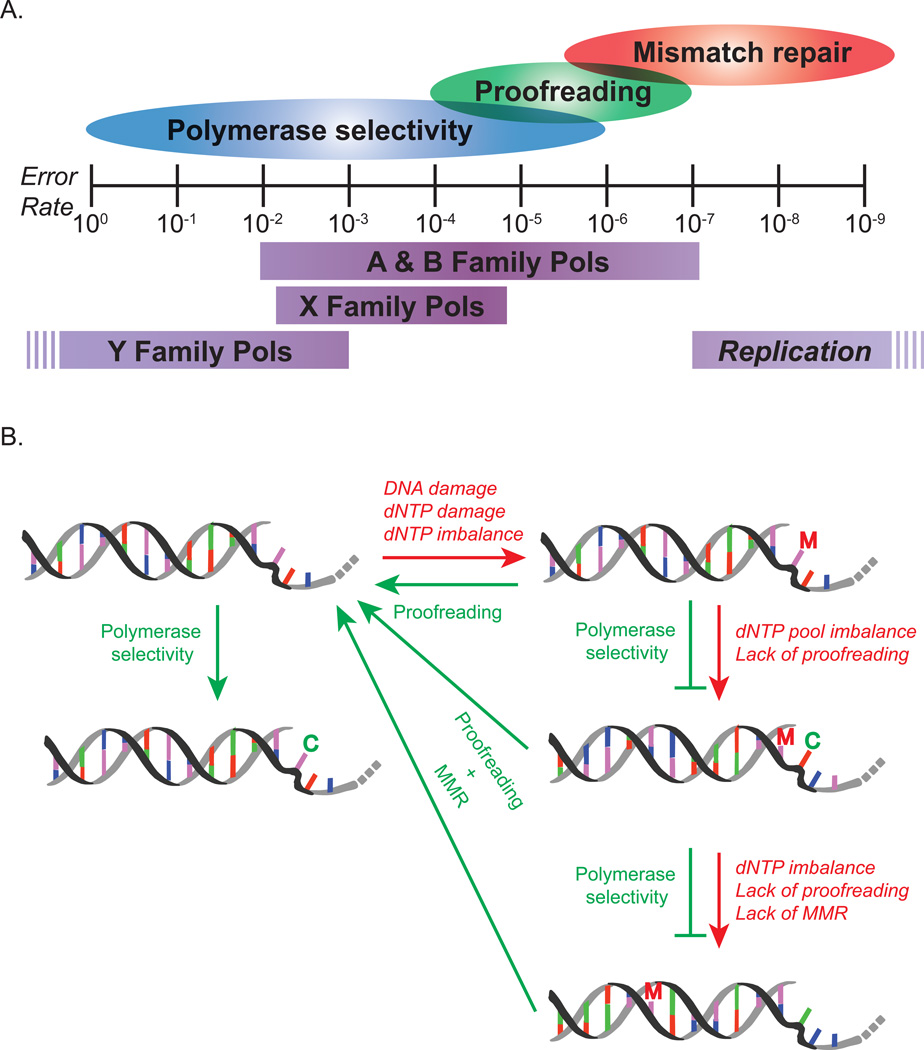
Figure 1. Determinants of replication fidelity.
A. The relative contribution levels of the three main components of replication fidelity are shown above the scale, estimated from the mutation rates of systems defective in one or more of the components. The overlapping ovals represent the fact that there is a range of possible increases in the level of fidelity that each mechanism provides dependent on many factors. The range of fidelity that a given mechanism is capable of providing is the critical factor (i.e. MMR can still provide up to 4 orders of magnitude increase in fidelity for polymerase errors that occur at a frequency of 10−2). The horizontal bars below the graph show the ranges of in vitro determined error rates for the different families of polymerases and the estimated mutation rate range of the in vivo complete replication complex. Within each family, the error rates can differ widely between polymerases and type of error. The broken bars at the left and right ends indicate that the rates could be even higher and lower than indicated. B. Graphic depicting the various means by which DNA replication can be modulated. DNA is shown as a stylized double helix (backbone is black and gray), with purine-pyrimidine base pairs indicated as red-green and blue-purple bars. The single strand region is meant to depict the unwound DNA at a replication fork, with the kink in the DNA representing the bend in the template strand identified by crystallography119. Red arrows and text indicate conditions that lead to lower fidelity. Green arrows and text indicate conditions that promote higher fidelity, green bars indicate conditions that block mutations. M = Mutation; C = Correct.
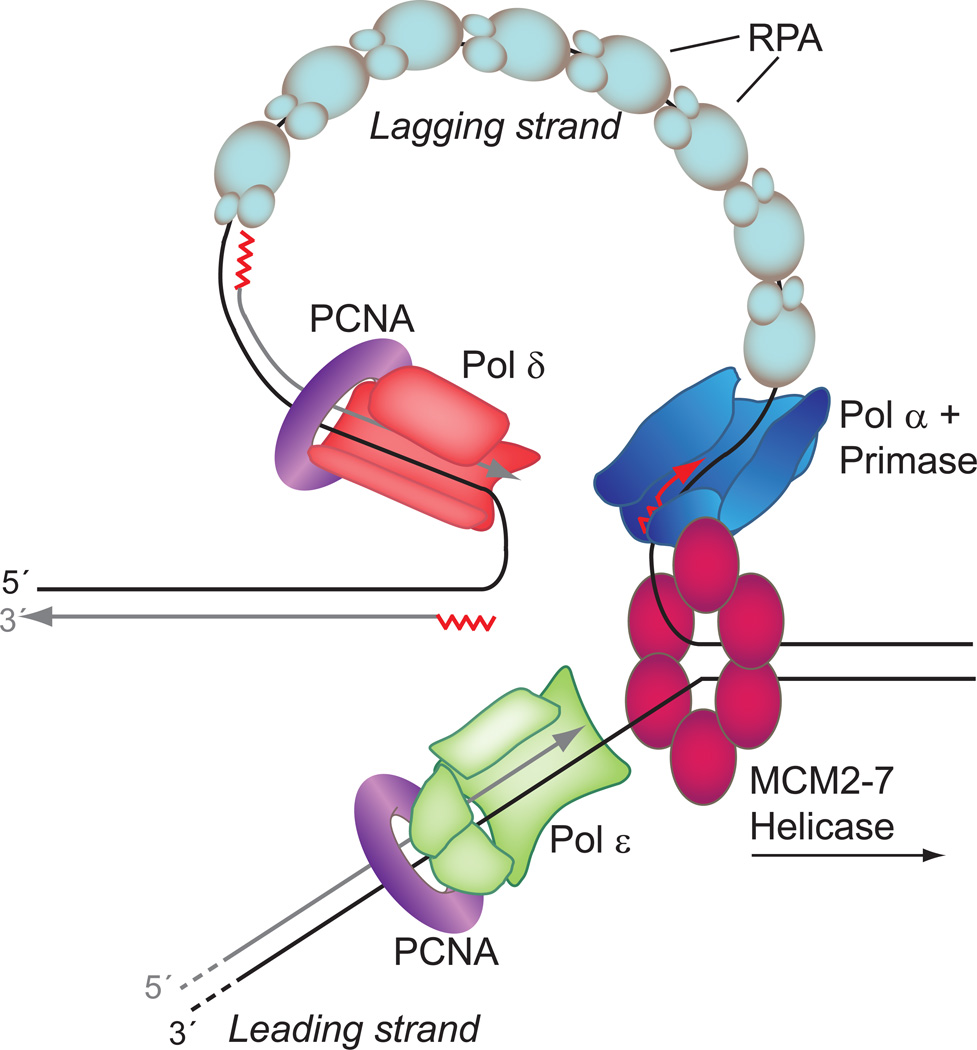
DNA replication requires the combined activity of dozens of proteins4, a subset of which are shown in Figure 2. Three members of the B-family5 of polymerases are involved in the bulk of DNA replication, pols α, δ, and ε. After the DNA duplex is unwound, likely by the MCM2-7 helicase complex6, synthesis is initiated on both the leading and lagging strand by the four subunit pol α-primase complex that synthesizes a short RNA-DNA hybrid primer. For leading strand synthesis, a polymerase then binds and extends the primer in a continuous fashion for as long as the polymerase is able to stay bound. For replication of the lagging strand, a discontinuous mode of synthesis occurs in patches of ~250 base pairs called Okazaki fragments, each of which must be initiated by pol α-primase activity4. The complexity of the system is illustrated by the fact that five decades after the discovery of the structure of DNA, uncertainty still remains as to the identity of major leading and lagging strand DNA polymerase(s)4, 7. In mitochondria, pol γ is responsible for all DNA synthesis activities8, 9. The importance of accurate replication of mitochondrial DNA will be discussed below.
Figure 2. Simplified model of replication fork.
A cartoon model of eukaryotic replication fork. Protein depictions are based on currently accepted sub-unit composition of S. cerevisiae proteins but are not meant to be accurate structure-based models. The assignment of pol ε to the leading strand is based on a recent report120, but has not been definitively established for all replication. Pol δ is consequently assigned to the lagging strand, consistent with earlier reports121–123. Helicase hexamer (magenta); Replication protein A (RPA; light blue ovals); Proliferating Cell Nuclear Antigen (PCNA; purple torus); pol α-primase complex (blue); RNA-DNA hybrid primer (red zig-zag and arrow); pol δ (red); pol ε (green); Template strand DNA (black lines); Newly synthesized DNA (gray lines). Figure inspired by and adapted from Figure 1 in reference7 and Figure 7 reference4.
In addition to the major replicative polymerases, there are a number of other DNA polymerases that have specialized roles in replicating the nuclear genome. Several of these, including the B-family member pol ζ, and multiple members of the Y-family (η,κ, ι, Rev1) are involved in bypassing DNA lesions that otherwise impede the major replicative polymerases. At least one of these, pol η, not only has the remarkable ability to copy damaged DNA more efficiently than the equivalent undamaged sequence, it can also ‘sense’ that the lesion has been bypassed, triggering a lesion dependent dissociation from the DNA10. This results in the simple model for translesion synthesis (TLS) shown in Figure 3A. Other results using different combinations of polymerases and lesions give rise to the “multiple-polymerase” model shown in Figure 3B11, wherein one polymerase may insert opposite a lesion and another extends from the resulting primer terminus. Given the range of possible lesions and number of TLS polymerases, it is possible that both models occur, depending on both the lesion and polymerase involved. Adding another layer of complexity, it may be that TLS can in some instances occur at the replication fork, whereas for other lesions, bypass may occur during gap-filling after the fork has moved on (Figures 3C and D)12. There is currently no data indicating that either the 1 or 2 polymerase model would be specific to a certain timing of TLS. In addition to their roles in TLS, many of the specialized polymerases have also been implicated in other DNA transactions, including somatic hypermutation (SHM; pols ζ, η, ι, Rev1)13, 14, homologous recombination (pol η)15, nucleotide excision repair (pol κ)16, and base excision repair (pol ι)17. The Y-family member Rev1 is G-template specific deoxycytidyl transferase and it has a non-catalytic role in TLS as well18, 19. Rev1p interacts with multiple polymerases20 and is required for in vivo mutagenesis, although the transferase activity of the enzyme is dispensable21. In addition, although not covered in detail here (except for family A pol γ), there exist several mammalian family A and X polymerases with widely varying fidelities and whose in vivo functions are the subject of intense investigation5.
Figure 3. Models of translesion synthesis.
A. The 1-polymerase model of TLS, shown here for a thymine-thymine dimer, states that a single polymerase is responsible for the complete bypass of a lesion, including insertion opposite all lesion bases and extension from the primer termini opposite a damaged template base. B. The 2-polymerase model of TLS, shown here for a thymine-thymine 6-4 photoproduct, states that different polymerases are responsible for the insertion steps at the various lesion positions. In the example given, note that while pol ζ is responsible for extension from the template-3´T primer terminus, it is also an insertion at the 5´ T position of the lesion. For a single base lesion, the insertion step would be opposite undamaged DNA. A more comprehensive listing of 2-polymerase/lesion combinations is given elsewhere11. Note that for both examples given the actual TLS reaction is flanked relatively closely both upstream (1–2 bases) and downstream (1–5 bases) of the lesion by replicative polymerase synthesis. C. Model for TLS that occurs at a replication fork during the process of ongoing synthesis. D. Model for TLS that takes place as a ‘gap-filling’ reaction, away from the main replication machinery. Note that both of these models are consistent with either the 1- or 2-polymerase model of TLS given in Panels A and B. In both cases, post-translational modification of PCNA and possible other proteins is critical for the polymerase switch. Note that Panels A and B are models of the actual TLS process while Panels C and D depict models for the timing of TLS. As such (and as noted in the text), there is overlap between the panels.
With this brief background, we first consider DNA replication fidelity and how defects in the steps required for high fidelity lead to genome instability (Figure 1B). After describing the fidelity of the major A and B-family replicative polymerases, we then discuss how nucleotide selectivity and proofreading help maintain genome stability during DNA synthesis. We then consider the role that replication accessory proteins have in modulating DNA synthesis fidelity, followed by a discussion of the fidelity of the translesion synthesis polymerases and the lesion bypass process. We conclude with a discussion of how defects in several of these pathways are associated with human disease.
High fidelity of eukaryotic replicative DNA polymerases when copying undamaged DNA
The bulk of DNA synthesis in a eukaryotic cell occurs during replication of undamaged DNA templates. This synthesis is catalyzed by the B-family polymerases α, δ, and ε for nuclear DNA and by the A-family polymerase γ for mitochondrial DNA. These four DNA polymerases are highly accurate, generating on average less than one single base substitution or single base insertion/deletion (indel) for every 10,000 correct incorporation events (Table 1). Such low error rates are consistent with other reports using homologous polymerases and different methods of analysis22–25. The rates listed in Table 1 are averages for all 12 possible single base-base mismatches and for a variety of single base indels in different sequence contexts. Rates for individual base substitutions and indels can vary by more than 100-fold, depending on the composition of the mismatch and the DNA sequence flanking the mismatch (error rate ranges are shown as purple boxes in Fig. 1A). Each of the different polymerases listed in Table 1 also differs to some degree from the others regarding error specificity (see references listed in Table 1 for examples).
Table 1.
Error Rates125 of Selected A, B and Y Family DNA Polymerases
| Error Rate (× 10−5) | |||||
|---|---|---|---|---|---|
| Origina | Polymeraseb | Family | Single base substitution | Single base indel | Reference |
| Replicative polymerases | |||||
| Exonuclease Deficient | |||||
| Hs | γ | A | 4.5 | 1.7 | 126 |
| Sc | α | B | 9.6 | 3.1 | 127, 128c |
| Sc | δ | B | 1.3 | 5.7 | 129 |
| Sc | ε | B | 24 | 5.6 | 128 |
| Exonuclease Proficient | |||||
| Hs | γ | A | 1.0 | ≤ 0.12 | 126 |
| Sc | δ | B | ≤ 1.3 | 1.3 | 129 |
| Sc | ε | B | ≤ 0.2 | ≤ 0.05 | 128 |
| Translesion synthesis polymerases (Exonuclease Deficient) | |||||
| Sc | ζ | B | 110 | 10 | 76 |
| Sc | η | Y | 950 | 93 | 92 |
| Hs | η | Y | 3500 | 240 | 130 |
| Hs | κ | Y | 580 | 180 | 131 |
| Hs | ιd | Y | 72,000 (T·dGTP) | - | 69 |
Hs = Homo sapiens; Sc = Saccharomyces cerevisiae.
Values are average error rates obtained using M13mp2 gap-filing DNA synthesis assays.
Pol γ, heterodimer of 140 and 55 kDa subunits
Pol α, heterotetramer of 180, 72, 48, and 48 kDa subunits
Pol δ, heterotrimer of 125, 55, and 40 kDa subunits
Pol ε, heterotetramer of 256, 80, 34, and 29 kDa subunits
Pol ζ, heterodimer of 173 and 29 kDa subunits.
Determined using a 5-base gap filling assay, since pol ι does not fill long gaps.
High nucleotide selectivity
Consistent with the need to accurately replicate billions of base pairs during every cell division cycle, the major replicative polymerases almost always initially insert the correct dNTP onto properly aligned primer-templates. Among the three major steps, high selectivity against misincorporation provides the single greatest contribution to replication fidelity. This is illustrated by the low base substitution and indel error rates of pol α, which naturally lacks proofreading activity, and by the low error rates of derivatives of pols δ, ε and γ that have been engineered to inactivate their intrinsic proofreading activities (Table 1, top). The fidelity of all four enzymes is much higher than that of polymerases involved in translesion DNA synthesis, which are also naturally exonuclease-deficient (Table 1, bottom). The importance of high nucleotide selectivity to genome stability, and its relationship to disease outcomes, is illustrated by recent studies of the effects of conservative amino acid substitutions in the active site of replicative DNA polymerases. When a highly conserved residue in the active site of S. cerevisiae B-family polymerases that is predicted to interact with the incoming dNTP is replaced with a different amino acid, the mutant enzymes have decreased DNA synthesis fidelity in vitro and they generate mutator phenotypes in vivo26–30. The amino acid changes introduced (L868F in pol α, L612M in pol δ and M644F in pol ε) do not greatly affect the overall activity of the polymerases, and error rates are elevated for only certain types errors that differ among the polymerases. Mice with replacements for the homologous residue (L604G or L604K) in murine pol δ display homozygous lethality, and heterozygotes have a decreased lifespan, increased genomic instability and accelerated tumorigenesis31. A second example of the importance of polymerase selectivity is the Y955C substitution in human pol γ. This change reduces pol γ fidelity in vitro when copying undamaged DNA and when bypassing template 8-oxo-dG32, 33, and the Y955C substitution is associated with autosomal dominant progressive external ophthalmoplegia (PEO)8.
How is high nucleotide selectivity achieved? Hydrogen bonding between template bases and incoming dNTPs is clearly important for replication fidelity34. However, this alone is unlikely to explain high selectivity, since the free energy difference between correct and incorrect base pairs in solution35–37 accounts for error rates of ~1:100, while the selectivity of the exonuclease-deficient major replicative polymerases is much greater than this (Table 1). Several ideas have been put forth to account for high selectivity. One is enthalpy-entropy compensation36, 38. In order for the incoming dNTP to hydrogen bond to a template base, water molecules that are hydrogen bonded to the base of the incoming dNTP must be stripped away, thereby decreasing the entropy of the system. This magnifies the contribution of enthalpy to the free energy difference (ΔΔG° = ΔΔH°−TΔΔS°), thereby increasing nucleotide selectivity. Consistent with this idea, the error rates of Y-family enzymes (Table 1, bottom), which have solvent accessible active sites (see below), are in the range predicted if enthalpy-entropy compensation was not contributing to selectivity. Kinetic analysis of insertion of non-polar base analogs by yeast pol η further support that they do not use enthalpy-entropy compensation to increase selectivity39.
A wealth of evidence supports the idea that the high nucleotide selectivity of accurate DNA polymerases results partly from the exquisite shape complementarity of their nascent base pair binding pockets34, 36, 37, 40. The four canonical Watson-Crick base pairs are nearly identical in size and shape, and numerous structural studies reveal that these correct base pairs fit snugly with the binding pocket, without steric clashes34, 41. Among many beautiful and informative structures now available (alas, none yet with pols α, δ, ε or γ), Figure 4 (panels B and D) shows the structure of T7 DNA polymerase, a highly accurate family A homolog of Pol γ, with a correct base pair bound in its nascent base pair binding pocket. While the correct pair fits snugly in the active site, the presence of mismatches, which have different and variable geometries40, 42, are predicted to create steric clashes that would (1) reduce incorrect dNTP binding affinity, (2) affect subsequent conformational changes needed to set up the proper geometry for catalysis, and/or (3) reduce the rate of phosphodiester bond formation, i.e., chemistry. The relative contribution of these three parameters to the fidelity of DNA replication has and continues to be the subject of many structural and kinetic investigations that elegantly employ mutant and wild type DNA polymerases and modified dNTPs. Of particular recent interest in the field is the extent to which incorrect incorporation is limited by chemistry or by a dNTP-induced conformational change that has been inferred from kinetic studies. The latter possibility also raises the important issue of the nature of the relevant conformational change among many that occur to assemble the active site, and whether the rate limiting conformational change differs for different replication errors. These issues are not considered in further detail here because they have been discussed at length in recent articles that interested readers can consult43–45.
Figure 4. Open and closed active sites of low and high fidelity polymerases.
A and B. Shown are the molecular surfaces of Dpo4 from S. solfataricus and T7 DNA polymerase with blue representing positively charged regions and red representing negatively charged regions. Note the tighter fit of DNA (ball and stick model) to the higher fidelity T7 DNA polymerase, evidenced by closer contact with polymerase regions. In Dpo4, the DNA is located at further distance from the polymerase, and a ‘hole’ in the polymerases structure is visible, indicating a much looser association of the DNA with the polymerase. C and D. Closeup of the active site showing the templating base and nucleotide. Again, note the relatively open and solvent accessible region in Dpo4 compared to the snug fit in T7 DNA polymerase. Also note the increased amount of neutral region of protein in T7 DNA polymerase, indicating that it is the geometry of the replicative polymerase active site that plays a role in fidelity, as noted in the text. This figure appears originally as Figure 7 in124 and has been reproduced with the permission of the authors.
Polymerization errors due to substrate misalignments
In addition to direct misincorporation of an incorrect dNTP resulting in a base substitution error, all four major eukaryotic replicative polymerases (and TLS polymerases) also insert and delete one or more nucleotides during DNA synthesis. These errors result from strand misalignments that generate one or more unpaired bases, either in the primer strand, leading to additions, or in the template strand, leading to deletions. Several ideas have been proposed to account for how these misalignments initiate and are stabilized for continued synthesis to generate a mutation, including DNA strand slippage46, misinsertion followed by primer relocation47, melting-misalignment48, and misalignment of a nucleotide at the active site49, also referred to as dNTP stabilized misalignment50. Extensive biochemical and initial structural support for several of these three models has recently been comprehensively reviewed51.
The exonuclease-deficient major eukaryotic replicative DNA polymerases generate single base deletions at rates that are similar to those for single base substitutions (Table 1, top). Single base deletion error rates are typically substantially (10-fold or more) higher than for single base insertions, with possible explanations as discussed in51. It is also important to note that the “average” error rates in Table 1 are perhaps somewhat misleading, because the indel error rates of the replicative polymerases are highly dependent on sequence context, being higher in repetitive rather than in non-repetitive sequences and increasing as the length of a repetitive sequence increases51, 52, just as predicted by Streisinger over 40 years ago46. For this reason (and see proofreading section below), long repetitive sequences in eukaryotic genomes are “hot spots” for replication errors (reviewed in51).
Contribution of proofreading by intrinsic 3´ exonuclease activity to genome stability
Once an incorrect dNTP is incorporated into DNA, the mismatched primer terminus is more difficult to extend than is a correctly paired and properly aligned primer terminus. Extension of such aberrant termini occurs with lower efficiency than does extension of a matched terminus. For mismatch extension, the geometric and kinetic considerations mentioned above are important. The delay in extension caused by a mismatch allows the primer terminus to fray and move to the 3′ exonuclease active site (if present) for excision of the incorrect nucleotide37, 38. Interestingly, among many mammalian DNA polymerases, only those responsible for the bulk of chain elongation during replication (pols δ, ε, and γ) contain intrinsic 3´ exonucleolytic proofreading activity. The contribution of proofreading to base substitution fidelity is illustrated by the lower error rates of the exonuclease-proficient pols δ, ε, and γ as compared to their exonuclease-deficient derivatives (Table 1; see also25 and references therein that demonstrate similar high fidelity for pol δ and pol ε isolated from mammalian sources). Although not obvious from the “less than or equal to” error rates of the exonuclease-proficient wild type polymerases in Table 1, a variety of in vitro studies indicate that proofreading improves replication fidelity by factors ranging from a few-fold to more than 100-fold, depending on the mismatch, the sequence context and the polymerase. The critical role of proofreading in maintaining eukaryotic genome stability is illustrated by genetic studies of yeast strains harboring genes for exonuclease deficient pol δ, ε and γ, all of which have a mutator phenotype53–57. The importance of proofreading to suppressing tumorigenesis is suggested by seminal studies showing that mice harboring exonuclease-deficient pol δ have a shortened life span and increased susceptibility to several types of cancer58, 59. Also in mice, inactivating the 3´ exonuclease of pol γ elevates levels of mitochondrial DNA mutations and leads to loss of mitochondria and premature ageing60, 61.
Proofreading also corrects misaligned intermediates containing extra bases in one strand or the other near the primer terminus, as illustrated by the higher indel error rates of exonuclease-deficient pols δ, ε and γ (Table 1, top) when compared to their proofreading proficient counterparts (Table 1, middle). However, the efficiency of proofreading of indels decreases as the length of a repetitive sequence increases, both in vitro62 and in vivo63. This is because the extra base in the misalignment substrate is protected from excision, since it can be located far upstream of the polymerase active site. Such diminished proofreading further contributes to the observation that long repetitive sequences are at risk for a high rate of replication errors, as evidenced by the now well known “microsatellite instability” phenotype of eukaryotic cells defective in DNA mismatch, especially including tumors from humans and mice with mutations that inactivate mismatch repair.
“Extrinsic proofreading” may also contribute to genome stability
Assuming that pol α, which lacks its own proofreading activity, synthesizes 10 nucleotides of each ~250-nucleotide Okazaki fragment on the lagging strand, it synthesizes about 2% of the human genome. Given its base substitution error rate of ~10−4 (Table 1, top), this amount of replication would generate 12,000 mismatches during each replication cycle. This leads to the issue of whether such a potentially heavy load of replication errors might be offset by proofreading of pol α errors by a separate exonuclease, a process referred to as extrinsic proofreading. This possibility was recently examined in a genetic study of yeast pol α with a Leu868Met substitution at the polymerase active site64. L868M pol α copies DNA in vitro with normal activity and processivity but with reduced fidelity. In vivo, the pol1-L868M allele confers a mutator phenotype. This mutator phenotype is strongly increased upon inactivation of the 3´ exonuclease of pol δ but not that of pol ε. Among several non-exclusive explanations that were considered, the results support the hypothesis that the 3´ exonuclease of pol δ proofreads errors generated by pol α during initiation of Okazaki fragments. Given that eukaryotes encode many other specialized, naturally proofreading-deficient DNA polymerases with even lower fidelity than pol α5, extrinsic proofreading could be relevant to several other DNA transactions that control genome stability65, such as base excision repair and possibly translesion synthesis by pol η (see below).
Modest contribution of accessory proteins to eukaryotic replication fidelity
In addition to polymerases, DNA replication requires the coordinated action of a large number of other proteins4 (Figure 2). Thus, it is reasonable to ask whether these proteins influence replication fidelity. Although relatively few investigations have been performed to investigate this subject with eukaryotic replication accessory proteins, experiments to date (see66 and references therein) suggest that replication accessory proteins like polymerase clamps and single stranded DNA binding proteins generally have relatively small effects on fidelity (i.e., typically a few-fold) in comparison to the large contribution of the polymerases themselves. In maintaining the focus of this review on the eukaryotic replication, one notable exception is the ability of RPA and PCNA to strongly suppress formation of large deletion errors occurring between direct repeat sequences during synthesis in vitro by pol δ67. PCNA and RPA may suppress large deletions by preventing the primer terminus from fraying and/or by preventing primer relocation and annealing to the downstream repeat.
Fidelity of TLS polymerases when copying undamaged DNA
Complete replication of the nuclear genome occasionally requires translesion synthesis by specialized polymerases, including family B pol ξ and family Y pols η, κ, and ι. These polymerases all lack proofreading activity and they also have lower nucleotide selectivity than the major replicative polymerases, as indicated by their higher error rates for base substitutions and indels (Table 1, bottom). The extreme case is for pol ι, which although reasonably accurate in preventing some mismatches (e.g., error rate of ~10−4 for A•dCTP), preferentially inserts dGTP more often than correct dATP opposite template T68, 69. Structural studies suggest that the low fidelity of family Y enzymes is partly due to relaxed geometric selectivity in the nascent base pair binding pocket, which is more open and solvent accessible (e.g., see Sso Dpo4, Fig. 4A and C) than those of more accurate DNA polymerases. This fact is likely relevant to the ability of family Y polymerases to bypass lesions that distort helix geometry more efficiently than can the major replicative polymerases (see below). Pols η and κ are not only error-prone for base substitution, but also for indels (Table 1, bottom). Indeed, over-expression of pol κ in cultured cells increases indel mutations70. While not the focus of this review, much of the work on family Y polymerases has been performed using bacterial enzymes, whose functions and properties are reviewed elsewhere71, 72.
Another TLS polymerase is the B-family member pol ζ. When copying undamaged DNA, pol ξ has somewhat higher fidelity than the Y-family polymerases, but lower fidelity than the other B-family members (Table 1, bottom). The ability of pol ξ to generate both base substitutions and indels at relatively high rates is consistent with its known role in generating a large majority of spontaneous mutations, as well as mutations induced by a variety of DNA damaging agents21. The high base substitution error rate of pol ζ clearly demonstrates that it has low nucleotide selectivity, consistent with a possible direct role in mutagenic misinsertion of dNTPs in vivo. Also relevant are kinetic studies demonstrating that pol ξ efficiently extends terminal mismatches21, 73, 74. This is true with undamaged DNA as well as for extending primer termini opposite damaged template bases, the latter being consistent with a role for pol ξ in the extension step of TLS in a 2-polymerase model (Figure 3B). A similar role has also been proposed for pol κ, which like pol ξ, is promiscuous for mismatch extension75. During in vitro DNA synthesis, pol ξ also generates “complex” mutations that contain multiple substitutions and indels within a short tract of DNA76. Consistent with this property, pol ξ also generates complex errors in vivo77, 78. Most recently, two other eukaryotic DNA polymerases have been shown to have low fidelity and are implicated in TLS, pol ν79, 80 and pol θ81, 82. Interestingly, both are members of the family A group of polymerases, other members of which are high fidelity enzymes (e.g., T7 DNA polymerase, pol γ).
The fidelity of translesion synthesis
There are numerous reports describing the TLS ability and fidelity of various Y-family (and other) polymerases when encountering a wide range of structurally diverse lesions5, 83–85. These studies show that for a given lesion, the bypass efficiency and fidelity is polymerase specific. Many of these reports focus on the specificity and kinetics with which polymerases insert individual correct or incorrect dNTPs opposite lesions, and/or their ability to extend matched and mismatched termini opposite damaged bases. These studies have been extremely valuable towards understanding two of the multiple steps needed to completely bypass lesions. Rather than exhaustively review the details of these studies, here we focus on what is known about error rates for complete bypass reactions that require insertion and multiple extensions and are performed in the presence of all four dNTPs. Since such studies are fewer in number, we discuss three common and biologically important lesions with different coding abilities: an abasic site (non-coding), a cis-syn cyclobutane thymine-thymine (TTD, which retains base coding potential), and 8-oxo-dG (highly ambiguous coding potential).
Since an abasic site contains no base coding information for a polymerase to use to direct dNTP insertion, it is not surprising that the “fidelity”, or more appropriately the “specificity”, of abasic site bypass by multiple polymerases is low. Like a number of other polymerases86, the archeabacterial DinB homolog Dpo4 of Sulfolobus solfataricus preferentially inserts dAMP opposite an abasic site, and like many other DinB homologs, also generates a significant level of −1 deletions87. Structurally, this has been suggested to occur because the polymerase ‘loops out’ the abasic site and instead copies the next available base88. Human pol η displays very similar abasic site bypass characteristics87. By comparison, a crystal structure of the B-family polymerase from bacteriophage RB69 (family B) with a template abasic site in the active site shows that while dAMP is inserted opposite the abasic site, the enzyme was unable to extend the resulting primer-terminus, with no translocation of the DNA occurring and no ‘closing’ of the polymerase89. This helps explain why the efficiency of bypass of abasic sites by replicative polymerases is low.
One of the most well studied lesions to date has been the TTD, one of the major DNA lesions resulting from exposure to UV radiation. TTD’s contain two covalently linked thymines, both of which retain base coding potential. The early characterization of TTD bypass by pol η was termed “error-free” because dAMP was inserted more frequently than other nucleotides90. Subsequent studies have determined that the error rate for TTD bypass by yeast and human pol η is actually quite low, e.g., one dGMP incorporated opposite the 3´-T of the dimer for ~30 dAMP incorporations. This high rate is similar to that observed when pol η copies the corresponding undamaged thymine91, 92. The error rate of homologous Sso Dpo4 is even lower (approaching 1 error in every 10 bypass events), and in this case is far higher than for copying the equivalent undamaged base91. Interestingly, both enzymes are more accurate when copying the 5′-T of the TTD. A possible explanation for higher fidelity at the 5´-T is provided by crystal structures of Dpo4 bound to TTD-containing templates93. Incoming ddATP is paired with the 3′-T of the dimer in a normal Watson-Crick pair, while ddATP orients in a syn configuration to accommodate the template distortion resulting from covalent linkage to the 3´-T, such that the ddATP is paired with the 5′-T of the dimer in a Hoogsteen pair. It may be that Hoogsteen bonded mispairs are less stable, thereby disfavoring errors at this position.
Another well studied lesion is 7,8-dihydro-8-oxo-guanine (8-oxo-dG), a common lesion generated by oxidative stress. 8-oxo-dG in an anti configuration can pair correctly with dCMP, while in a syn configuration it pairs incorrectly with dAMP94, 95. Kinetic studies of mammalian pol δ have shown that although dCMP is inserted more efficiently opposite the lesion, it is the 8-oxo-dG:dAMP pair that is preferentially extended96. Moreover, the 8-oxo-dG:dAMP pair interacts with bacteriophage T7 DNA polymerase in a manner that allows this mispair to escape proofreading97, thereby further increasing the mutagenic potential of this lesion. Readers interested in further information of mutagenesis resulting from 8-oxo-dG can consult excellent reviews on this subject98, 99.
The biological relevance of TLS is best illustrated by the observation that loss of pol η function in humans and in mice results in sensitivity to sunlight and predisposition to skin cancer100, 101. Knocking out mouse pol ι in a pol η deficient background further increased susceptibility to UV light-induced skin cancer, suggesting that pol ι also has a role in bypassing UV photoproducts102. Studies in cultured cells also implicate pol ι in UV-induced mutagenesis either when pol η is absent or present103, 104. Pol ι has also been linked to susceptibility of urethane induced lung cancers105. The importance of 8-oxo-dG bypass fidelity is illustrated by the pol γ mutant described above (Y955C), which has a higher propensity for inserting 8-oxo-dGTP and for bypassing template 8-oxo-dG in a mutagenic manner33. Patients with this mutation have an autosomal dominant form of PEO that manifests with a large number of clinical phenotypes. Finally, while the loss of pol ξ results in embryonic lethality in mice106–108, cells lacking pol ξ display severe chromosomal instability109. These examples clearly demonstrate the importance of the TLS polymerases in maintaining genomic stability. While these examples predict an anti-mutagenic effect of the polymerases, the process of somatic hypermutation during immunoglobulin diversity requires the mutagenic properties of these polymerases for normal functioning. In fact, a recent model of SHM invokes both pols η and ξ, possibly in the bypass of abasic sites13, 14. Clearly, the mechanisms that control where, when and which TLS polymerases have access to primer-templates are critical to genomic stability.
Controlling TLS
The low fidelity of family Y polymerases when copying undamaged DNA implies the need to strictly limit their activities to specific circumstances, or too much error-prone synthesis could lead to error catastrophe and cell death. One means of control is through the polymerase clamp PCNA. Studies have shown that all three eukaryotic Y-family polymerases interact with PCNA110–112, and it is known that upon replication fork stalling induced by DNA damage, PCNA becomes mono-ubiquitylated113. It is currently thought that this post-translational modification is important for delivering TLS polymerases to sites of damage. However, no studies to date have indicated that PCNA, whether unmodified or mono-ubiquitylated, affects the fidelity of TLS polymerases. The eukaryotic TLS polymerases also interact with Rev120, which as mentioned above plays a critical role in mutagenesis independent on its DNA synthetic capacity. It is possible that Rev1, which binds to single-stranded DNA and primer termini114, could help deliver the TLS polymerases to gaps or sites of stalled replication forks, in effect becoming a polymerase accessory protein. Again however, there is currently no evidence that Rev1/polymerase interactions alters the fidelity of TLS. Thus the few studies that have been done so far come to the same conclusions as the results with the major replicative polymerases, namely that the polymerases themselves are the prime determinants of the fidelity of TLS92, 111.
Concluding remarks
It has been hypothesized that multistage carcinogenesis requires a mutator phenotype115. This idea is now supported by examples wherein defects in several pathways that determine DNA replication fidelity result in decreased genome stability and increased susceptibility to cancer. These defects include reduced dNTP selectivity31, loss of the 3´ exonuclease activity of a replicative polymerase58, 59, defects in other nucleases involved in DNA replication116, defects in MMR2, 3, and defects in translesion synthesis101, 102. While cancer is the most cited example, a number of other diseases have some connection to defects in replication fidelity and genomic stability. These include (but are not limited to) Alzheimers, Cockayne’s Syndrome, Parkinson’s disease, Friedreich’s ataxia, Huntington’s disease, tricothiodystrophy, a number of progressive neuropathies, and multiple mitochondrial wasting diseases117, 118. Clearly, much work remains to be done in this area, with the goal of not only better understanding, but ultimately of preventing and/or treating these diseases.
Acknowledgements
We thank Drs. Stephanie Nick-McElhinny and Zachary Pursell for constructive comments on the manuscript. This work was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (TAK) and by the College of Agriculture and Life Sciences at North Carolina State University (SDM).
Footnotes
Abbreviations used: MMR, mismatch repair; pol, polymerase; TLS, translesion synthesis; SHM, somatic hypermutation; indel, insertion/deletion; 8-oxo-dG, 7,8-dihydro-8-oxo-guanine; PEO, progressive external ophthalmoplegia; Dpo4, DNA polymerase 4 from Sulfolobus solfataricus; TTD, cis-syn thymine-thymine cyclobutane pyrimidine dimer.
References
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 4.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 5.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 6.Forsburg SL. Eukaryotic MCM Proteins: Beyond Replication Initiation. Microbiol Mol Biol Rev. 2004;68(1):109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40(2):115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 8.Graziewicz MA, Longley MJ, Copeland WC. DNA Polymerase γ in Mitochondrial DNA Replication and Repair. Chem Rev. 2006;106(2):383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 9.Falkenberg M, Larsson N-G, Gustafsson CM. DNA Replication and Transcription in Mammalian Mitochondria. Annual Review of Biochemistry. 2007;76(1):679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 10.McCulloch SD, Kokoska RJ, Kunkel TA. Efficiency, fidelity and enzymatic switching during translesion DNA synthesis. Cell Cycle. 2004;3(5):580–583. [PubMed] [Google Scholar]
- 11.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16(15):1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5(12):1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26(4):215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends in Immunology. 2006;27(7):313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamoto T, Araki K, Sonoda E, et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20(5):793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8(6):640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 17.Prasad R, Bebenek K, Hou E, et al. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase ι by controlled proteolysis. J Biol Chem. 2003;278(32):29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 18.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277(18):15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 19.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382(6593):729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 20.Guo C, Fischhaber PL, Luk-Paszyc MJ, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22(24):6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence CW, Gibbs PE, Murante RS, et al. Roles of DNA polymerase ζ and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb Symp Quant Biol. 2000;65:61–69. doi: 10.1101/sqb.2000.65.61. [DOI] [PubMed] [Google Scholar]
- 22.Einolf HJ, Guengerich FP. Kinetic analysis of nucleotide incorporation by mammalian DNA polymerase delta. J Biol Chem. 2000;275(21):16316–16322. doi: 10.1074/jbc.M001291200. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto K, Shimizu K, Nakashima N, Sugino A. Fidelity of DNA polymerase delta holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42(48):14207–14213. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel TA, Sabatino RD, Bambara RA. Exonucleolytic proofreading by calf thymus DNA polymerase δ. Proc Natl Acad Sci U S A. 1987;84(14):4865–4869. doi: 10.1073/pnas.84.14.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas DC, Roberts JD, Sabatino RD, et al. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30(51):11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 26.McElhinny SAN, Stith CM, Burgers PMJ, Kunkel TA. Inefficient Proofreading and Biased Error Rates during Inaccurate DNA Synthesis by a Mutant Derivative of Saccharomyces cerevisiae DNA Polymerase δ. J Biol Chem. 2007;282(4):2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niimi A, Limsirichaikul S, Yoshida S, et al. Palm Mutants in DNA Polymerases α and η Alter DNA Replication Fidelity and Translesion Activity. Mol Cell Biol. 2004;24(7):2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pursell ZF, Isoz I, Lundstrom E-B, Johansson E, Kunkel TA. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase ε. Nucl Acids Res. 2007;35(9):3076–3086. doi: 10.1093/nar/gkm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator Phenotypes Caused by Substitution at a Conserved Motif A Residue in Eukaryotic DNA Polymerase δ. J Biol Chem. 2006;281(7):4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase delta in Saccharomyces cerevisiae. Genetics. 2005;170(2):569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatesan RN, Treuting PM, Fuller ED, et al. Mutation at the Polymerase Active Site of Mouse Polymerase δ Increases Genomic Instability and Accelerates Tumorigenesis. Mol Cell Biol. 2007;27(21):7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponamarev MV, Longley MJ, Nguyen D, Kunkel TA, Copeland WC. Active Site Mutation in DNA Polymerase gamma Associated with Progressive External Ophthalmoplegia Causes Error-prone DNA Synthesis. J Biol Chem. 2002;277(18):15225–15228. doi: 10.1074/jbc.C200100200. [DOI] [PubMed] [Google Scholar]
- 33.Graziewicz MA, Bienstock RJ, Copeland WC. The DNA polymerase γ Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2'-deoxyguanosine. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 35.Petruska J, Goodman MF, Boosalis MS, et al. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 37.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 38.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28(2):83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 39.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick Hydrogen Bonding for DNA Synthesis by Yeast DNA Polymerase η. Mol Cell Biol. 2003;23(14):5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman MF. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci U S A. 1997;94(20):10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11(5):489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 42.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116(6):803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 43.Rothwell PJ, Waksman G, John MS, David ADP. Advances in Protein Chemistry. Academic Press; 2005. Structure and mechanism of DNA polymerases; pp. 401–440. [DOI] [PubMed] [Google Scholar]
- 44.Tsai YC, Johnson KA. A New Paradigm for DNA Polymerase Specificity. Biochemistry. 2006;45(32):9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothwell PJ, Waksman G. A Pre-equilibrium before Nucleotide Binding Limits Fingers Subdomain Closure by Klentaq1. J Biol Chem. 2007;282(39):28884–28892. doi: 10.1074/jbc.M704824200. [DOI] [PubMed] [Google Scholar]
- 46.Streisinger G, Okada Y, Emrich J, et al. Frameshift mutations and the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263(29):14784–14789. [PubMed] [Google Scholar]
- 48.Fujii S, Akiyama M, Aoki K, et al. DNA Replication Errors Produced by the Replicative Apparatus of Escherichia coli. Journal of Molecular Biology. 1999;289(4):835–850. doi: 10.1006/jmbi.1999.2802. [DOI] [PubMed] [Google Scholar]
- 49.Kunkel TA. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986;261(29):13581–13587. [PubMed] [Google Scholar]
- 50.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase β violates the "A-rule". Novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J Biol Chem. 1997;272(4):2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem Sci. 2006;31(4):206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase λ. Implications for function. J Biol Chem. 2003;278(36):34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 53.Foury F, Vanderstraeten S. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. EMBO J. 1992;11(7):2717–2726. doi: 10.1002/j.1460-2075.1992.tb05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12(4):1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrison A, Sugino A. The 3'-->5' exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242(3):289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 56.Pavlov YI, Maki S, Maki H, Kunkel TA. Evidence for interplay among yeast replicative DNA polymerases α, δ and ε from studies of exonuclease and polymerase active site mutations. BMC Biol. 2004;2(1):11. doi: 10.1186/1741-7007-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanderstraeten S, Van den Brule S, Hu J, Foury F. The role of 3'–5' exonucleolytic proofreading and mismatch repair in yeast mitochondrial DNA error avoidance. J Biol Chem. 1998;273(37):23690–23697. doi: 10.1074/jbc.273.37.23690. [DOI] [PubMed] [Google Scholar]
- 58.Goldsby RE, Lawrence NA, Hays LE, et al. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat Med. 2001;7(6):638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 59.Goldsby RE, Hays LE, Chen X, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc Natl Acad Sci U S A. 2002;99(24):15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D, Mott JL, Chang SW, et al. Construction of transgenic mice with tissue-specific acceleration of mitochondrial DNA mutagenesis. Genomics. 2000;69(2):151–161. doi: 10.1006/geno.2000.6333. [DOI] [PubMed] [Google Scholar]
- 61.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 62.Kroutil LC, Register K, Bebenek K, Kunkel TA. Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry. 1996;35(3):1046–1053. doi: 10.1021/bi952178h. [DOI] [PubMed] [Google Scholar]
- 63.Tran HT, Gordenin DA, Resnick MA. The 3'-->5' Exonucleases of DNA Polymerases delta and varepsilon and the 5'-->3' Exonuclease Exo1 Have Major Roles in Postreplication Mutation Avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(3):2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pavlov YI, Frahm C, Nick McElhinny SA, et al. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16(2):202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 65.McElhinny SAN, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5(9):958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bebenek A, Carver GT, Dressman HK, et al. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics. 2002;162(3):1003–1018. doi: 10.1093/genetics/162.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34(16):4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tissier A, McDonald JP, Frank EG, Woodgate R. pol ι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14(13):1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 69.Bebenek K, Tissier A, Frank EG, et al. 5'-Deoxyribose phosphate lyase activity of human DNA polymerase ι in vitro. Science. 2001;291(5511):2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 70.Ogi T, Kato T, Jr, Kato T, Ohmori H. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells. 1999;4(11):607–618. doi: 10.1046/j.1365-2443.1999.00289.x. [DOI] [PubMed] [Google Scholar]
- 71.Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 72.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15(2):70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase ζ is an efficient extender of primer ends opposite from 7,8-dihydro-8-oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23(4):1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrence CW, Hinkle DC. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- 75.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc Natl Acad Sci U S A. 2002;99(25):16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong X, Garg P, Stith CM, et al. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34(17):4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Minesinger BK, Abdulovic AL, Ou TM, Jinks-Robertson S. The effect of oxidative metabolism on spontaneous Pol zeta-dependent translesion synthesis in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5(2):226–234. doi: 10.1016/j.dnarep.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 78.Harfe BD, Jinks-Robertson S. DNA polymerase γ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6(6):1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 79.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase nu. DNA Repair (Amst) 2007;6(2):213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takata K-i, Shimizu T, Iwai S, Wood RD. Human DNA Polymerase N (POLN) Is a Low Fidelity Enzyme Capable of Error-free Bypass of 5S-Thymine Glycol. J Biol Chem. 2006;281(33):23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 81.Seki M, Marini F, Wood RD. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucl Acids Res. 2003;31(21):6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seki M, Wood RD. DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair. 2007 doi: 10.1016/j.dnarep.2007.08.005. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 84.Shcherbakova PV, Bebenek K, Kunkel TA. Functions of eukaryotic DNA polymerases. Sci Aging Knowledge Environ. 2003;2003(8):RE3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- 85.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 86.Strauss BS. The "A" rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst) 2002;1(2):125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- 87.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J Biol Chem. 2003;278(50):50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 88.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13(5):751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 89.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23(7):1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc Natl Acad Sci U S A. 2001;98(15):8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCulloch SD, Kokoska RJ, Masutani C, et al. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428(6978):97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 92.McCulloch SD, Wood A, Garg P, Burgers PM, Kunkel TA. Effects of accessory proteins on the bypass of a cis-syn thymine-thymine dimer by Saccharomyces cerevisiae DNA polymerase eta. Biochemistry. 2007;46(30):8888–8896. doi: 10.1021/bi700234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424(6952):1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 94.Lipscomb LA, Peek ME, Morningstar ML, et al. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc Natl Acad Sci U S A. 1995;92(3):719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McAuley-Hecht KE, Leonard GA, Gibson NJ, et al. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33(34):10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 96.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J Biol Chem. 2001;276(6):3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 97.Brieba LG, Eichman BF, Kokoska RJ, et al. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23(17):3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends in Genetics. 1993;9(7):246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 99.Boiteux S, Gellon L, Guibourt N. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radical Biology and Medicine. 2002;32(12):1244–1253. doi: 10.1016/s0891-5849(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 100.Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399(6737):700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 101.Lin Q, Clark AB, McCulloch SD, et al. Increased susceptibility to UV-induced skin carcinogenesis in polymerase η-deficient mice. Cancer Res. 2006;66(1):87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- 102.Dumstorf CA, Clark AB, Lin Q, et al. Participation of mouse DNA polymerase {iota} in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proceedings of the National Academy of Sciences. 2006;103(48):18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Feraudy S, Limoli CL, Giedzinski E, et al. Pol [eta] is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene. 2007;26(39):5713–5721. doi: 10.1038/sj.onc.1210385. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Woodgate R, McManus TP, et al. Evidence that in Xeroderma Pigmentosum Variant Cells, which Lack DNA Polymerase {eta}, DNA Polymerase {iota} Causes the Very High Frequency and Unique Spectrum of UV-Induced Mutations. Cancer Res. 2007;67(7):3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 105.Wang M, Devereux TR, Vikis HG, et al. Pol ι is a candidate for the mouse pulmonary adenoma resistance 2 locus, a major modifier of chemically induced lung neoplasia. Cancer Res. 2004;64(6):1924–1931. doi: 10.1158/0008-5472.can-03-3080. [DOI] [PubMed] [Google Scholar]
- 106.Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol. 2000;10(19):1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- 107.Esposito G, Godindagger I, Klein U, et al. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr Biol. 2000;10(19):1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 108.Wittschieben J, Shivji MK, Lalani E, et al. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10(19):1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 109.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA Polymerase ζ Causes Chromosomal Instability in Mammalian Cells. Cancer Res. 2006;66(1):134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 110.Haracska L, Acharya N, Unk I, et al. A single domain in human DNA polymerase ι mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005;25(3):1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haracska L, Johnson RE, Unk I, et al. Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol. 2001;21(21):7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Haracska L, Unk I, Johnson RE, et al. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol Cell Biol. 2002;22(3):784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3(8):1011–1013. [PubMed] [Google Scholar]
- 114.Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J Biol Chem. 2006;281(34):24314–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- 115.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51(12):3075–3079. [PubMed] [Google Scholar]
- 116.Zheng L, Dai H, Zhou M, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13(7):812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 117.Bohr VA, Ottersen OP, Tonjum T. Genome instability and DNA repair in brain, ageing and neurological disease. Neuroscience. 2007;145(4):1183–1186. doi: 10.1016/j.neuroscience.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 118.DePamphilis ML. DNA replication and human disease. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 119.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391(6664):251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 120.Pursell ZF, Isoz I, Lundstrom E-B, Johansson E, Kunkel TA. Yeast DNA Polymerase ε Participates in Leading-Strand DNA Replication. Science. 2007;317(5834):127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18(22):2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jin YH, Obert R, Burgers PM, et al. The 3'-->5' exonuclease of DNA polymerase δ can substitute for the 5' flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci U S A. 2001;98(9):5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PMJ. Okazaki Fragment Maturation in Yeast. II. Cooperation between the polymerase and 3'-->5'-exonuclease activities of pol δ in the creation of a ligatable nick. J Biol Chem. 2003;278(3):1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 124.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 125.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 126.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase γ with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276(42):38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 127.Kunkel TA, Hamatake RK, Motto-Fox J, Fitzgerald MP, Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9(10):4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shcherbakova PV, Pavlov YI, Chilkova O, et al. Unique error signature of the four-subunit yeast DNA polymerase ε. J Biol Chem. 2003;278(44):43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 129.Fortune JM, Pavlov YI, Welch CM, et al. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280(33):29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 130.Matsuda T, Bebenek K, Masutani C, et al. Error rate and specificity of human and murine DNA polymerase η. J Mol Biol. 2001;312(2):335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 131.Ohashi E, Bebenek K, Matsuda T, et al. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J Biol Chem. 2000;275(50):39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]