The planarian chromatin regulator HP1, at least in part acting through increased expression of Mcm5, inhibits differentiation of adult stem cells and triggers their proliferation in response to injury.
Abstract
Adult stem cells (ASCs) capable of self-renewal and differentiation confer the potential of tissues to regenerate damaged parts. Epigenetic regulation is essential for driving cell fate decisions by rapidly and reversibly modulating gene expression programs. However, it remains unclear how epigenetic factors elicit ASC-driven regeneration. In this paper, we report that an RNA interference screen against 205 chromatin regulators identified 12 proteins essential for ASC function and regeneration in planarians. Surprisingly, the HP1-like protein SMED–HP1-1 (HP1-1) specifically marked self-renewing, pluripotent ASCs, and HP1-1 depletion abrogated self-renewal and promoted differentiation. Upon injury, HP1-1 expression increased and elicited increased ASC expression of Mcm5 through functional association with the FACT (facilitates chromatin transcription) complex, which consequently triggered proliferation of ASCs and initiated blastema formation. Our observations uncover an epigenetic network underlying ASC regulation in planarians and reveal that an HP1 protein is a key chromatin factor controlling stem cell function. These results provide important insights into how epigenetic mechanisms orchestrate stem cell responses during tissue regeneration.
Introduction
Adult stem cells (ASCs) in tissues constitute a long-lived reservoir with the ability for self-renewal and to give rise to multiple cell types during tissue homeostasis and regeneration (Weissman, 2000; Li and Clevers, 2010). Detailed mechanistic understanding of how ASCs are maintained and are regulated in response to injury is likely to have important implications for regenerative medicine. Planarians can regenerate missing body parts, owing to a population of pluripotent ASCs called neoblasts (Newmark and Sánchez Alvarado, 2002; Wagner et al., 2011), representing a powerful system for investigating stem cells and regeneration (Agata, 2003; Reddien and Sánchez Alvarado, 2004; Sánchez Alvarado, 2006). Upon injury, neoblasts undergo extensive cell division to form the regenerating blastema in which they differentiate into the needed cell types (Saló and Baguna, 1984; Newmark and Sánchez Alvarado, 2000; Wenemoser and Reddien, 2010). Expression profiling and lineage tracing experiments have defined genes specifically expressed in either neoblasts or their descendants (Eisenhoffer et al., 2008), providing an entry point to study the cellular basis of regeneration processes. Gene perturbation by RNAi (Newmark et al., 2003) facilitates the identification of genes controlling stem cell function and/or regeneration (Reddien et al., 2005a; Guo et al., 2006; Rouhana et al., 2010; Wagner et al., 2012). However, the molecular cascade that triggers regenerative proliferation is currently unclear.
Typically, the process of regeneration requires the potential of stem cells to coordinate proliferation and differentiation programs to form the new tissue (Barrero and Izpisua Belmonte, 2011). Chromatin regulation has emerged as a key epigenetic mechanism to modulate stem cell behaviors by contributing to activation or silencing subsets of genes in a rapid and reversible manner and by maintaining their expression status during subsequent cell divisions (Orkin and Hochedlinger, 2011). Increasing evidence from higher animal species has suggested that, similar to embryonic stem (ES) cells (Azuara et al., 2006; Bernstein et al., 2006), ASCs also maintain bivalent chromatin domains, which consist of overlapping repressive and active histone modifications, to keep silenced genes poised for activation (Mikkelsen et al., 2007; Cui et al., 2009). Thus, it is plausible that tissues might use such an epigenetic plasticity to maintain stem cell states and enable coordinate and rapid induction of gene expression under injury stress. Chromatin factors contribute to neoblast function and planarian regeneration (Reddien et al., 2005a; Bonuccelli et al., 2010; Scimone et al., 2010; Wagner et al., 2012). However, we still lack a complete picture of chromatin regulation in neoblasts. A global survey of chromatin genes essential for neoblast function would not only advance our understanding of how chromatin factors modulate neoblast properties but should also help to discover novel epigenetic mechanisms controlling stem cell biology.
Here, using an RNAi screen against chromatin factors, we identified 12 genes essential for stem cell functions and regeneration, including components of six chromatin complexes (nucleosome remodeling and deacetylase [NuRD], CAF-1, BRG1/Brm-associated factor [BAF], facilitates chromatin transcription [FACT], Cdk-activating kinase, and minichromosome maintenance [MCM] complex). Interestingly, an HP1 family protein, HP1-1, is expressed exclusively in ASCs, controls stem cell self-renewal during homeostatic maintenance, and contributes to the trigger for regenerative proliferation upon injury. Moreover, in contrast to the commonly appreciated role of HP1 homologues in gene silencing, HP1-1–mediated stem cell mobilization requires interaction with SSRP1 and active RNA polymerase II to induce expression of the proliferation gene Mcm5. These data expand the repertoire of chromatin genes controlling ASC activity, reveal an unexpected role for an HP1 protein in stem cell regulation and tissue regeneration, and present a framework to analyze in vivo chromatin regulation in stem cells.
Results
An RNAi screen unveils the chromatin signature of ASCs in planarians
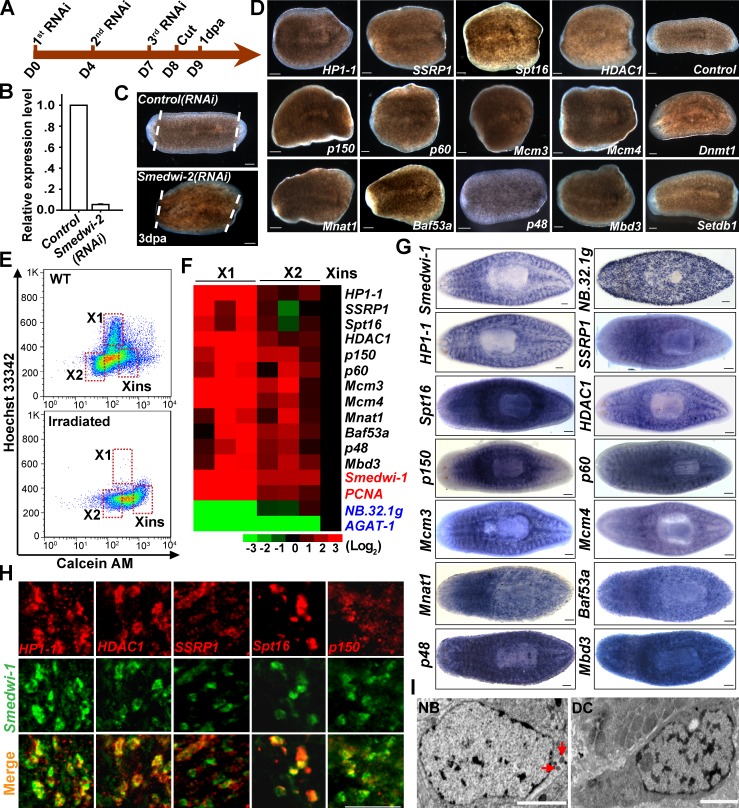
To identify candidate chromatin regulators of planarian neoblasts, we conducted an RNAi screen for genes essential for neoblast-driven regeneration. Feeding RNAi reduced smedwi-2 mRNA levels by 95% (Fig. 1, A and B) and abolished regenerative capacity (Fig. 1 C). These results are consistent with a previous study (Reddien et al., 2005b) demonstrating the effectiveness of RNAi. We then searched for genes potentially encoding proteins with motifs common to chromatin regulators in the planarian draft genome (Zayas et al., 2005; Robb et al., 2008) and obtained ∼210 chromatin gene candidates. Among them, 205 genes were successfully cloned for RNAi assays, and 12 genes exhibited various degrees of regeneration defects upon depletion (Fig. S1 A). The 12 candidates were further retested with a different double-stranded RNA (dsRNA) sequence, and all of them reproduced the phenotypes observed in the initial RNAi screen (Fig. 1 D). Furthermore, knockdown of individual components from the same protein complex, such as CAF-1 and NuRD, displayed a similar phenotype. These data suggest that few false positives were identified.
Figure 1.
Identification of chromatin regulators for neoblasts. (A) Flowchart of dsRNA feeding and amputation schedules. D, days after first dsRNA feeding. dpa, day postamputation. (B) qRT-PCR to measure mRNA levels of smedwi-2. Error bars show SDs, n = 3. (C) Phenotypic analysis of regenerating worms (n = 10). White dotted lines indicate the amputation site. (D) Representative regeneration phenotypes after knockdown of the indicated genes (n ≥ 12 for each condition) at 6 dpa. (E) Flow cytometry results of wild-type (WT) and irradiated animals. Shown are representative results from three independent experiments. (F) Heat map illustrating comparisons of relative mRNA levels in FACS-purified X1, X2, and Xins cells. Expression levels in Xins cells were set as 1. n = 3. (G) WISH showing the expression pattern of 12 identified genes. Smedwi-1, neoblast marker; NB.32.1g, early progeny marker. n = 6 for each gene. (H) Expression of five genes (top row) was analyzed by double fluorescent WISH (FISH) with smedwi-1 (middle row). Shown are representative dorsal views (n = 4 for each gene). (I) Transmission electron microscopy analysis showing nuclear structure of a neoblast (NB) and differentiated cell (DC). Neoblasts were identified by the chromatoid bodies (red arrows). Bars: (C, D, and G) 0.1 mm; (H) 50 µm; (I) 2 µm.
We next sought to test whether these identified genes are enriched in neoblasts by comparing their expression levels in neoblasts and postmitotic progeny isolated by FACS (Fig. 1 E). All 12 genes were highly enriched in the neoblast population (Fig. 1 F, greater than threefold on average in X1 cells compared with irradiation-insensitive [Xins] cells). Whole-mount in situ hybridization (WISH) further validated that expression of nearly all genes displayed a neoblast-like pattern (Fig. 1 G), although three genes (Baf53a, p48, and Mbd3) also extended to the region anterior to the photoreceptors (Fig. S1 B). Of the five genes detected by double FISH, all showed overlapping expression with neoblast marker smedwi-1 (Fig. 1 H). These data suggest that the 12 genes are expressed preferentially in neoblasts, and the regeneration defects we observed may largely result from dysfunction of neoblasts. In accord with this speculation, individual knockdown of all genes severely decreased expression of neoblast markers smedwi-1 (Fig. S1, C and D) or smedwi-2 (Fig. S1 E). Thus, the in vivo RNAi screen identified 12 chromatin factors important for ASC function in planarian.
Several genes identified have been well documented in murine ES cells. For instance, p150 knockdown rapidly depleted neoblasts and eventually their progeny (Fig. S1, E and F), suggesting that p150 is required for neoblast maintenance, which is consistent with the requirement of p150 in sustaining ES cell viability (Houlard et al., 2006). Furthermore, NuRD components HDAC1 and Mbd3 primarily maintain the expression of the early progeny marker (Fig. S1, C–F), which is in agreement with their role in pluripotency (Kaji et al., 2006; Dovey et al., 2010). Moreover, neoblasts, like mammalian ES cells (Gaspar-Maia et al., 2011), have an open chromatin structure largely devoid of heterochromatin (Fig. 1 I). These data suggest that neoblasts unexpectedly share common features in chromatin regulation with murine ES cells. Thus, our results unveil a conserved chromatin network that is potentially involved in controlling planarian ASCs (Fig. S1 G).
HP1-1 is required for blastema development during regeneration
Several genes identified are not known to be involved in regulating stem cells, demonstrating the utility of the screen. Among them, we focused on an HP1 family gene because of its strong effect on neoblast maintenance (Fig. S1 E) and because little is known about the role of HP1 homologues in stem cells. Extensive BLAST (Basic Local Alignment Search Tool) searches revealed that the planarian genome harbors two HP1 isoforms, Smed–HP1-1 (HP1-1) and Smed–HP1-2 (HP1-2). The characteristic domains (Eissenberg and Elgin, 2000) are highly conserved in these two HP1 proteins (Fig. S2 A) and are closely related to their homologues from plants to human (Fig. 2 A).
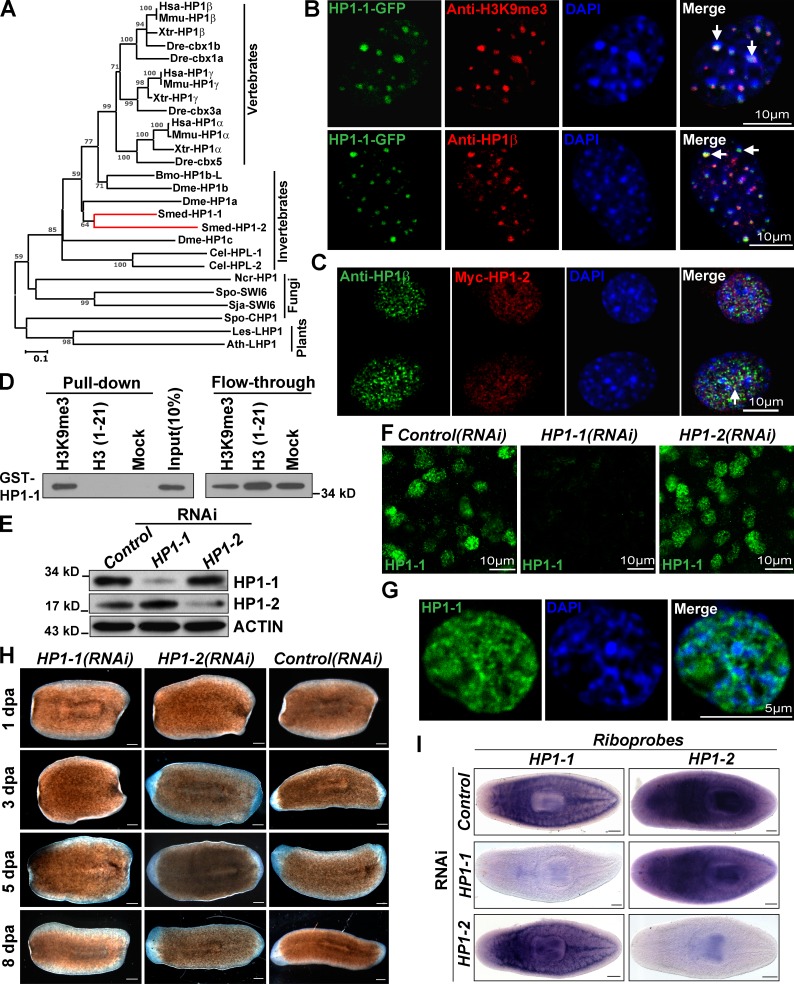
Figure 2.
Knockdown of HP1-1 causes regeneration defects. (A) Phylogenetic tree of HP1 family proteins. Bar, 10% amino acid substitution. C. elegans (Cel), D. melanogaster (Dme), D. virilis (Dv), D. rerio (Dre), B. mori (Bmo), H. sapiens (Hsa), M. musculus (Mmu), X. tropicalis (Xtr), N. crassa (Ncr), S. japonicus (Sja), S. pombe (Spo), L. esculentum (Les), A. thaliana (Ath), and S. mediterranea (Smed) are shown. (B and C) IF of NIH3T3 cells transfected either with EGFP-tagged HP1-1 (B) or Myc–HP1-2 (C). Antibodies against mouse HP1-β or H3K9me3 were used for examining colocalization. Shown are the nuclear regions stained by DAPI. White arrows indicate overlapping dots. (D) GST–HP1-1 was tested for binding to Lys 9 methylated peptide (first lane), unmethylated peptide (second lane), or mock (third lane). (E) Western blot analysis showing the specificity of HP1 antibodies. (F and G) IF analysis showing HP1-1 localization in whole-mount animals (F) or in a single cell (G). Nuclear regions were counterstained with DAPI. (H) Phenotypic analysis of regenerating animals (n = 60 for each gene). (I) WISH showing the efficiency and specificity of gene knockdown by D10 (12/12 animals per condition showed similar results). Bars, 0.1 mm, unless otherwise indicated.
When ectopically overexpressed in NIH3T3 cells, both HP1-1 and HP1-2 exhibited nuclear localization with a pattern similar to endogenous HP1 protein; they also localized to H3K9me3 (lysine 9 of histone H3)-dense foci (Fig. 2, B and C). However, in HeLa cells, only a small fraction of HP1-1 colocalized with H3K9me3 (Fig. S2 B), suggesting that the subnuclear localization of HP1-1 may be cell type dependent. Furthermore, far Western–type overlay assays showed that bacterially expressed GST–HP1-1, but not GST alone, strongly bound to a single band corresponding to the position of core histones (Fig. S2 C). Peptide pull-down assays further revealed that HP1-1 specifically bound to the methylated Smed-H3 peptide but not to the unmethylated peptide (Fig. 2 D and Fig. S2 D). These data demonstrated the evolutionarily conserved recognition of methylated H3 tails by HP1-1 (Bannister et al., 2001; Lachner et al., 2001), suggesting that these two proteins are bona fide HP1 homologues. We further generated two antibodies, which specifically detect HP1-1 and HP1-2 (Fig. 2 E). Interestingly, immunofluorescence (IF) with HP1-1 antibody labeled the nuclei of mesenchymal cells with a diffuse staining pattern and showed little overlap with DAPI-dense heterochromatin regions (Fig. 2, F and G), suggesting a euchromatin distribution for HP1-1.
We then evaluated the effects of the two HP1 genes on regeneration. By 1 d postamputation (dpa), both HP1-1(RNAi) and HP1-2(RNAi) worms initiated blastema formation. However, from then on, blastema growth appeared to cease completely, and the wound epithelium began to show signs of regression by 5 dpa in HP1-1(RNAi) worms. Further defects, such as curling and decreased motility, appeared by 8 dpa (Fig. 2 H), displaying a phenotype reminiscent of neoblast loss. In contrast, neither Control nor HP1-2(RNAi) worms showed any defects in regeneration (Fig. 2 H). WISH validated the efficacy and specificity of RNAi knockdown and showed that whereas HP1-2 displays a ubiquitous expression pattern, HP1-1 resides between the gut branches and is absent from the pharynx and the region anterior to photoreceptors (Fig. 2 I). Thus, of the two evolutionarily conserved HP1-like proteins, only loss of HP1-1 function leads to a defect in blastema development.
HP1-1 marks ASCs of planarian
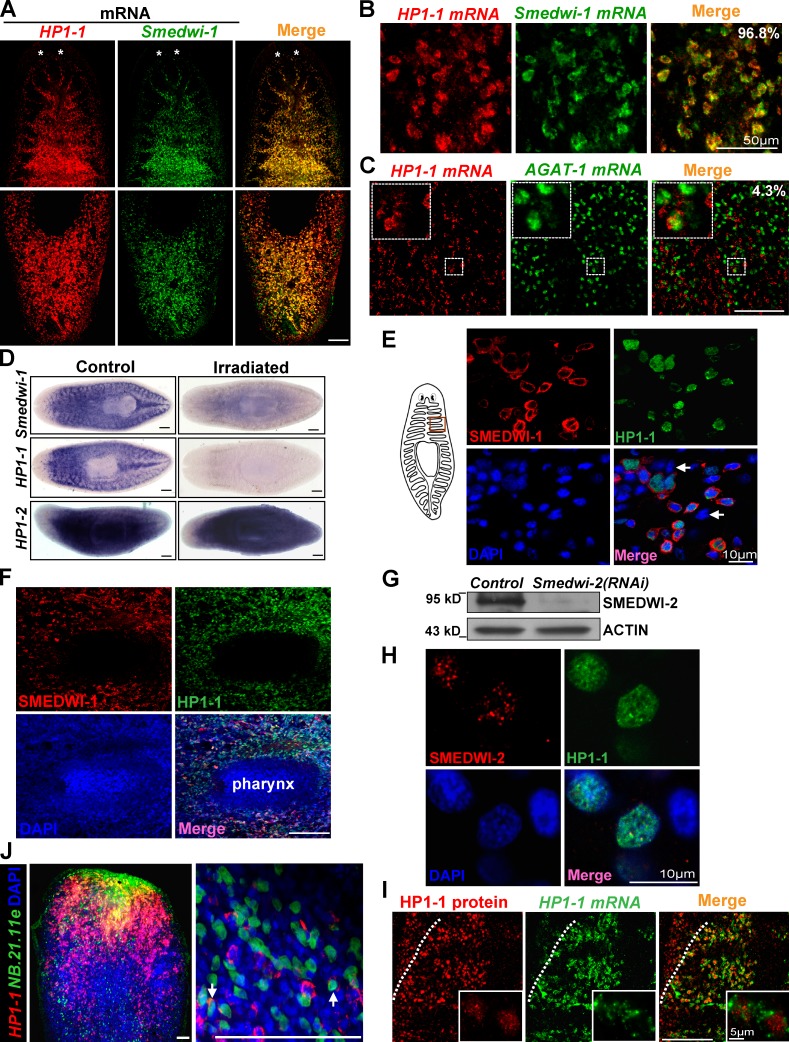
To more precisely define the cells marked by HP1-1, we compared the localization of HP1-1 with other markers. Quantification of double FISH showed that 96.8% of HP1-1–expressing cells (n = 970) are smedwi-1 positive (Fig. 3, A and B), whereas only 4.3% (n = 670) express a late-progeny marker AGAT-1 (Fig. 3 C). Quantitative real-time PCR (qRT-PCR) of FACS-purified cells revealed an enrichment of HP1-1 in dividing neoblasts (Fig. 1 F, greater than sevenfold in X1 cells compared with Xins cells), and WISH confirmed that HP1-1 expression was eliminated after irradiation in a similar fashion to smedwi-1 loss, albeit HP1-2 showed no discernible changes (Fig. 3 D). Upon amputation, strong expression of HP1-1 was observed beneath the anterior stumps where proliferative neoblasts localize; irradiation also effectively diminished this expression (Fig. S2 E). Furthermore, IF showed that the HP1-1 protein was detected in a portion of the nuclei of mesenchymal cells, which are mostly surrounded by the SMEDWI-1 ringlike signal (91%, n = 540; Fig. 3 E and Fig. S2 F) and is absent from the pharynx (Fig. 3 F); irradiation abrogated this expression (Fig. S2, G and H). To further validate the subcellular localization of HP1-1, SMEDWI-2 antibody was generated (Fig. 3 G) to examine nuclear labeling. HP1-1 also overlapped substantially with SMEDWI-2 within the same nuclei (95%, n = 610; Fig. 3 H and Fig. S2 I). These data together provide compelling evidence that expression of the HP1-1 gene is largely specific for planarian ASCs.
Figure 3.
HP1-1 is a novel marker of planarian ASCs. (A–C) Double FISH for HP1-1 and the neoblast marker smedwi-1 (A and B) or the late-progeny marker AGAT-1 (C). (A) The top and bottom rows show the anterior and posterior region, respectively. Asterisks show photoreceptors. (B) Magnified views of coexpression. (C) Insets show a higher magnification of the small boxed regions. The number is the percentage of overlapping cells. n ≥ 6 animals for each condition. (D) WISH analysis of control and irradiated animals (1 d postirradiation). The smedwi-1 gene was used as a positive control. n = 5 for each condition. (E and F) Double IF on vibrating sections (E) or whole-mount animals (F) revealed that HP1-1–labeled nuclei (green) were surrounded by the cytoplasmic SMEDWI-1 (red) signal, and both proteins are absent from the pharyngeal areas. White arrows indicate the DAPI-stained nuclei that lack HP1-1. (E, left) Boxed inset represents the body parts shown in the right-side images. (G) Western blot analysis showing the specificity of SMEDWI-2 antibody. (H) Double IF with HP1-1 and SMEDWI-2 antibodies on vibrating sections. Nuclear regions were counterstained with DAPI. (I) Coexpression of HP1-1 mRNA and HP1-1 protein along gut branches. White dotted lines indicate the distribution boundary of mRNA. Insets represent a magnified view of the coexpression. Single confocal optical section. (J) Double FISH for HP1-1 and the early progeny marker NB.21.11e in a regenerating animal (2 dpa). Shown are anterior-facing wounds from a single confocal optical section. White arrows indicate coexpressing cells. Bars, 0.1 mm, unless otherwise indicated.
Similar to SMEDWI-1, a portion of HP1-1 protein is closer to the animal margin than its transcripts (Fig. 3 I). This suggests that HP1-1 also labels neoblasts undergoing differentiation caused by sustained protein levels (Guo et al., 2006; Scimone et al., 2010). HP1-1 is also localized to spermatogonial cells (Fig. S2 J), as demonstrated by localization in testes lobules (Wang et al., 2007, 2010). During regeneration, HP1-1–positive cells accumulated beneath the amputation boundary, and ∼6% of them coexpressed the early progeny marker NB.21.11e (Fig. 3 J), suggesting that HP1-1–expressing cells have the potential to differentiate into several cell types. In addition, all the H3S10P-labeled cells, representing mitotic neoblasts (Newmark and Sánchez Alvarado, 2000), were also HP1-1 positive (n = 420; Fig. S2 K). This indicates that HP1-1–expressing cells possess the ability to undergo mitosis in tissues. Together, these data suggest that HP1-1 marks planarian ASCs capable of both differentiation and self-renewal.
HP1-1 knockdown impairs ASC self-renewal
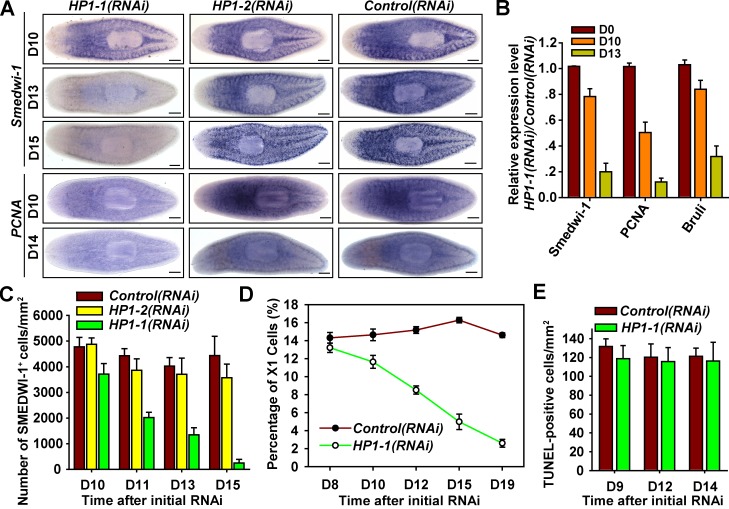
We next asked whether HP1-1 is required for neoblast regulation by monitoring the expression level of neoblast markers. When HP1-1 is depleted, smedwi-1 gradually decreases from 13 d after first dsRNA feeding (D13) and is severely diminished by D15 (Fig. 4, A and B). Moreover, the decrease of PCNA, a neoblast-specific gene involved in proliferation, is even more pronounced than that of smedwi-1. The SMEDWI-1 signal also progressively declined from D11 and was maximally diminished by D15 in HP1-1(RNAi) animals (Fig. 4 C and Fig. S3 A). These results suggest that HP1-1 knockdown reduced expression of neoblast markers. Furthermore, using FACS analyses, we observed a 40% reduction of the X1 population beginning on D12 and that was almost eliminated by D19 in HP1-1(RNAi) worms (Fig. 4 D and Fig. S3 B). This suggests that loss of HP1-1 leads to failure of maintaining neoblasts. The reduction of the stem cell population could be caused by induction of either cell death or differentiation or deprivation of proliferative capacity. However, apoptotic levels were indistinguishable between control and HP1-1(RNAi) worms (Fig. 4 E and Fig. S3 C), suggesting that the decrease of neoblasts was not caused by increased apoptosis.
Figure 4.
HP1-1 maintains the ASC population. (A) WISH showing the expression of neoblast markers smedwi-1 and PCNA, n = 14 for each condition. Bars, 0.1 mm. (B) qRT-PCR to analyze relative mRNA levels of neoblast-enriched genes in HP1-1(RNAi) worms. n = 3. (C) Quantitative analysis of SMEDWI-1–positive cell numbers in intact dsRNA-fed animals. n = 6 animals from three independent experiments. (D) Relative percentages of the X1 population in dsRNA-fed animals, from three independent experiments. (E) Quantification of TUNEL-positive nuclei over time. n = 5 animals. Error bars show SDs.
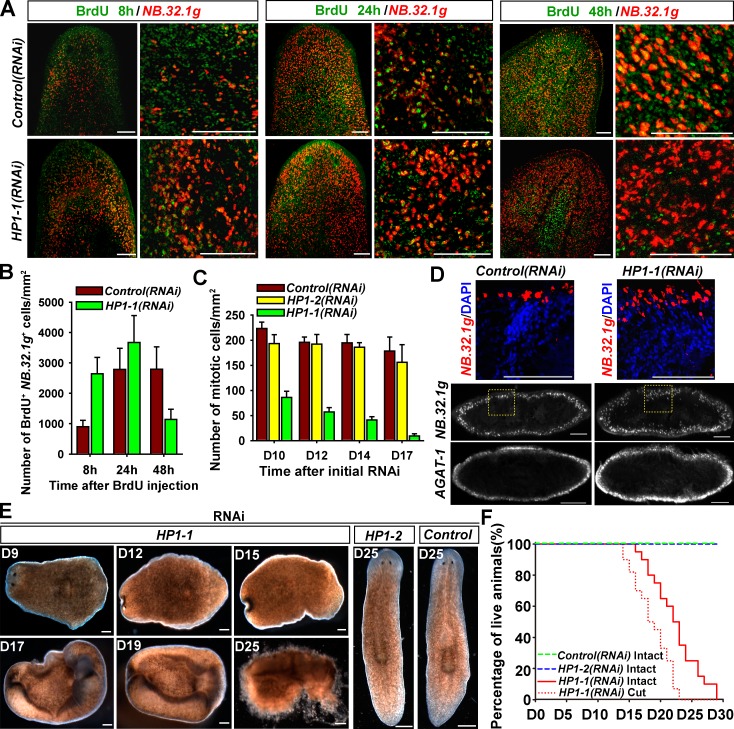
We then determined the cell fate of neoblasts with BrdU-mediated lineage tracing experiments. 8 h after administration of BrdU, labeled cells that lacked expression of early progeny marker NB.32.1g were found deep in the mesenchyme, representing proliferating neoblasts (Eisenhoffer et al., 2008). In contrast, HP1-1(RNAi) animals displayed reduced BrdU incorporation and enhanced costaining of NB.32.1g in the normally undifferentiated deep layer (Fig. 5, A and B, BrdU 8 h), indicating that premature differentiation occurs. Furthermore, BrdU-labeled postmitotic cells had migrated farther toward the peripheral margin with an increase in the number of cells that colabeled NB.32.1g at 24 h in control animals. Upon HP1-1 knockdown, the double-positive cells persisted for 24 h after BrdU administration but declined by 48 h, although NB.32.1g expression showed no discernible changes (Fig. 5, A and B, 48 h). These data suggest that failure of neoblast maintenance results from loss of proliferative potential and subsequent premature differentiation. Consistent with premature entry into the postmitotic differentiation pathway, HP1-1 depletion reduced H3S10P-positive cells to <50% of those in the control as early as D10 (Fig. 5 C) and led to expansion of NB.32.1g-expressing regions (Fig. 5 D). Mitotic cells further declined dramatically at week 2 and nearly disappeared by D17 upon HP1-1 knockdown (Fig. 5 C and Fig. S3 D). Single BrdU pulse experiments further validated impaired proliferative activity (Fig. S3 E). Thus, HP1-1 knockdown abolished proliferation and led to premature, continuous differentiation, indicating that HP1-1 controls ASC self-renewal rather than survival.
Figure 5.
HP1-1 is required for stem cell self-renewal and tissue homeostasis. (A) BrdU chase labeling (D10) combined with FISH of NB.32.1g. Shown are representative confocal images (single slice) after 8-h, 24-h, or 48-h BrdU incorporation. Shown are representative results from three independent biological replicates (more than four animals per time point). (B) Quantification of cells double labeled with BrdU and NB.32.1g. Error bars show SDs, n = 5 animals. (C) Quantitative analysis of H3S10P-positive cell numbers in intact dsRNA-fed worms. Error bars show SDs, n = 6 animals. (D) FISH of lineage markers on transverse sections after RNAi knockdown (D11), n = 5 animals for each condition. The top row shows a higher magnification of the boxed regions in the middle row. (E) Phenotypic analysis of intact RNAi worms; >20 animals per condition were similar. (F) Survival curve for dsRNA-fed worms. Control worms survived for >30 d. n = 100 for each condition. Bars, 0.1 mm.
Self-renewing neoblasts are critical for replacing cells lost during the course of physiological cell turnover. HP1-1 depletion led to impaired motility and sedentary action on D9 as well as head and tail tip regression on D13; HP1-1(RNAi) worms further curled ventrally and eventually lysed by D25, presenting a phenotype reminiscent of neoblast loss (Fig. 5, E and F). These results together suggest that HP1-1 maintains ASC self-renewal to contribute to tissue homeostasis.
HP1-1 sustains regenerative proliferation of ASCs
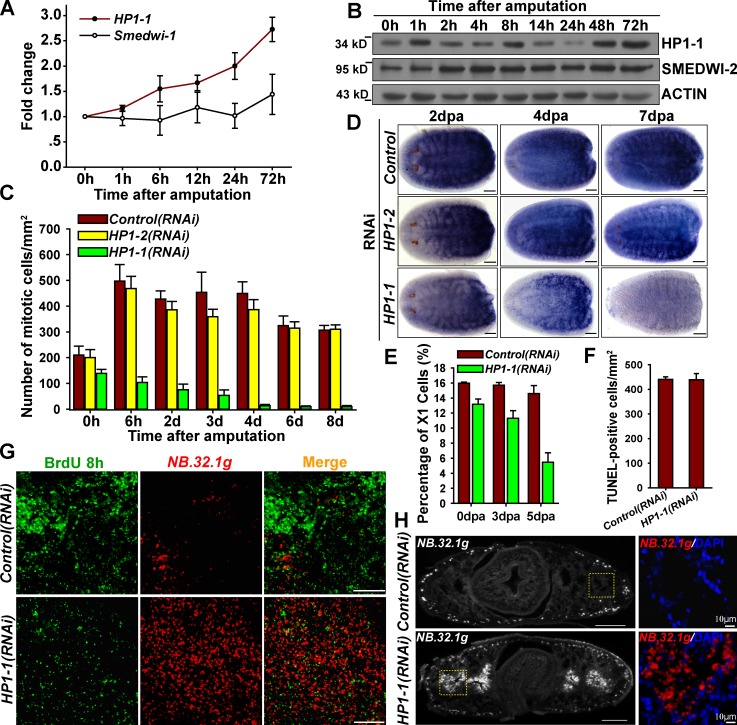
Interestingly, upon injury, HP1-1–expressing cells increased and accumulated beneath the blastema, where proliferating neoblasts usually localize (Fig. S4 A), and HP1-1 mRNA was up-regulated (Fig. 6 A). Western blot analyses further revealed two increases in HP1-1 protein within 8 h (Fig. 6 B, 8 h) after amputation and a third increase 48–72 h after wounding (Fig. 6 B). These changes in HP1-1 correspond well to the mitotic peaks of the neoblast wound response that induces formation of the regenerating blastema (Wenemoser and Reddien, 2010). As such, we examined the proliferation response by staining the mitotic marker H3S10P. As expected, amputation triggered a burst of neoblast proliferation in wild-type worms. Conversely, the proliferation peak induced by amputation was reduced by 80% at 3 dpa and almost completely eliminated by 6 dpa in HP1-1(RNAi) animals but not in control or HP1-2(RNAi) animals (Fig. 6 C). These data demonstrate that HP1-1 is indispensable for regenerative proliferation of ASCs in response to amputation.
Figure 6.
HP1-1 is essential for regenerative proliferation of ASCs in planarian. (A) qRT-PCR showing relative expression levels of HP1-1 and Smedwi-1. n = 3. (B) Western Blot analyses of HP1-1 levels during the course of regeneration. Shown are representative results from three independent biological replicates. (C) Quantitative analysis of H3S10P-positive cell numbers in regenerating worms. n = 6 animals. (D) WISH showing the expression levels of smedwi-2 in head fragments; anterior is to the left. n = 14 animals for each condition. (E) Relative percentages of the X1 population in regenerating animals. n = 3. (F) Quantification of TUNEL-positive nuclei at 4 dpa. n = 5 for each condition. (G) A single 8-h pulse of BrdU delivered by injection at 2 dpa combined with FISH of NB.32.1g. Shown are representative, single-slice confocal images from three independent biological replicates (more than three animals per time point). (H) FISH of NB.32.1g on transverse sections of regenerating worms (2 dpa). n = 4 animals for each condition. The right-side images show higher magnifications of the boxed regions in the left images. Bars, 0.1 mm, unless otherwise indicated. Error bars show SDs.
Neoblast markers smedwi-2 (Fig. 6 D) and SMEDWI-1 (Fig. S4 B) accumulated in a manner similar to that of controls by 2 dpa. FACS analysis showed that 75% of neoblasts are present by 3 dpa (Fig. 6 E). This excluded the possibility that loss of mitotic response results from absence of the neoblast population. However, SMEDWI-1 and smedwi-2 decreased dramatically by 4 dpa and almost diminished by 7 dpa (Fig. 6 D and Fig. S4 B). qRT-PCR confirmed reduced expression of several neoblast-specific genes (Fig. S4 C), and FACS analysis indicated that neoblasts are reduced to 30% by 5 dpa (Fig. 6 E). Thus, the failure of regenerative proliferation finally led to neoblast loss. Although amputation increased apoptosis as compared with intact worms (Fig. 4 E), HP1-1 knockdown did not alter apoptosis levels relative to controls (Fig. 6 F).
We next examined the process of neoblast commitment with lineage tracing experiments. By 2 dpa, BrdU labeling increased rapidly in the deep mesenchymal layer (Fig. 6 G, top) when compared with intact worms (Fig. 5 A). In contrast, neoblast proliferation induced by amputation was nearly absent in HP1-1(RNAi) worms at the 48 h peak of wild-type proliferation. Concomitantly, there was a significant increase in NB.32.1g expression in the deep layer with ≥36% of the remaining BrdU-positive cells (65–73 cells/180 counted, n = 3) displaying ectopic expression of NB.32.1g (Fig. 6 G, bottom). These data suggest that HP1-1 acts to support stem cell proliferation in response to injury, whereas loss of HP1-1 leads to a neoblast-intrinsic failure of the proliferative response and subsequent premature differentiation. Consistent with this notion, NB.32.1g is ectopically expressed around the gut branches (Fig. 6 H), where neoblasts localized, in HP1-1(RNAi) animals. Additionally, expression levels of several late-progeny markers were indistinguishable between control and HP1-1(RNAi) animals by 3 dpa (Fig. S4, D and E), even though smedwi-2 depletion apparently blocks differentiation (Fig. S4 F). This indicates that tissues continuously execute differentiation programs in the early stages of regeneration when HP1-1 is absent. However, AGAT-1 expression decreased dramatically by 7 dpa (Fig. S4 G), suggesting that the neoblast pool, lacking in proliferative potential, is progressively exhausted. Together, the defect in blastema formation may result from a combination of the failure to quench an ever-increasing demand for propagation, accelerated differentiation, and finally, exhaustion of the neoblast population.
Microarray analysis of genes downstream of HP1-1 during regeneration
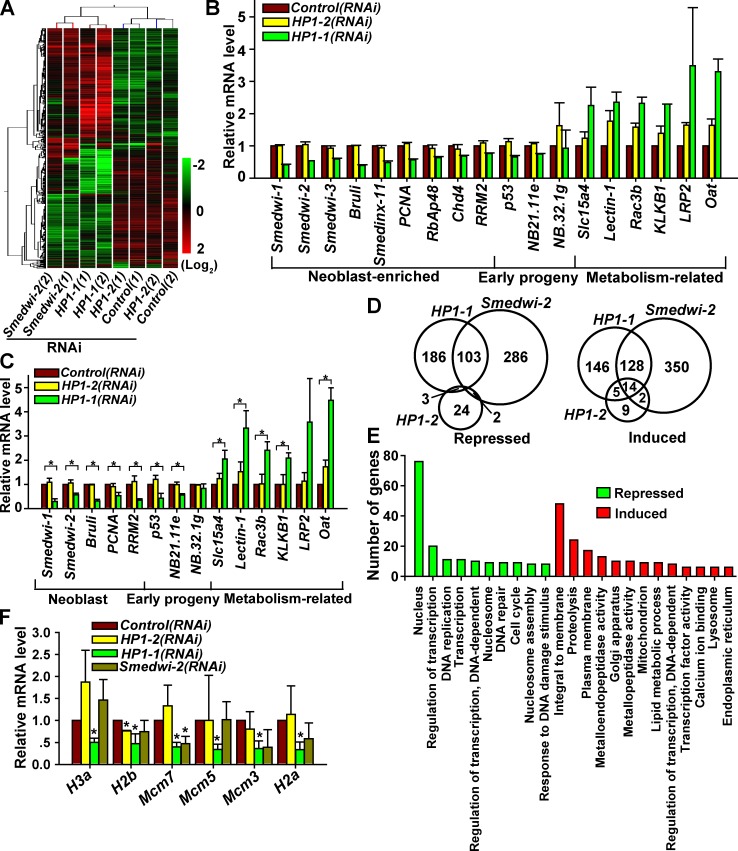
We next performed global gene expression profiling to analyze genes affected when regenerative proliferation is abolished (Fig. 7 A). qRT-PCR verified 33 of 34 selected genes, including nine neoblast marker genes that specifically decreased by 60% (Fig. 7, B and C; and not depicted). These data are consistent with the aforementioned failure in maintaining self-renewal and also validate the accuracy of the array. Notably, unsupervised hierarchical clustering clearly distinguished HP1-1 from HP1-2 (Fig. 7 A), and knockdown of HP1-1 affected a larger set of genes (293 induced and 292 repressed, >1.5-fold) than HP1-2 (30 induced and 29 repressed, >1.5-fold; Fig. 7 D).
Figure 7.
Microarray analysis of genes affected by HP1-1 knockdown. (A) Heat map of altered genes (>1.5-fold) shared among the four profiles of Control, Smedwi-2, HP1-1, and HP1-2(RNAi) regenerating worms at 3 dpa; log2-based scale. Numbers in parentheses represent replicate samples. (B and C) Microarray (B) and qRT-PCR (C) showing the relative expression levels of lineage markers. Error bars show SDs, n = 2 (B) or 3 (C). *, P < 0.01. (D) Venn diagram representation of differentially expressed genes in the three RNAi groups. (E) Gene ontology enrichment analysis of up- and down-regulated genes (>1.5-fold in both replicates) in HP1-1(RNAi) animals. (F) qRT-PCR showing expression levels of proliferation-related genes at 3 dpa. Error bars show SDs, n = 3. *, P < 0.05.
HP1 is known to contribute to heterochromatic gene silencing (Eissenberg et al., 1990). Unexpectedly, upon HP1-1 knockdown, the number of repressed genes was comparable with the number induced (Fig. 7 D). Gene ontology analysis showed that the most significantly reduced genes were those associated with the nucleus, especially genes involved in DNA replication and proliferation (Fig. 7 E). The top hits were the Mcm family and histone-related genes (Fig. 7 F); some of them are known to be expressed specifically in proliferating neoblasts (Salvetti et al., 2000; Solana et al., 2012). Considering that >70% dividing neoblasts are present at 3 dpa (Fig. 6 E), HP1-1 may act to sustain expression of proliferation-related genes in neoblasts during regeneration. In contrast, the induced genes mainly belong to the categories of membrane protein, proteolysis, and metabolic processes (Fig. 7, C and E), most of which are irradiation insensitive and are expressed in differentiated cells (Eisenhoffer et al., 2008). These data support the notion that HP1-1 controls a neoblast expression program compatible with its role in promoting proliferation and repressing differentiation. In addition, gene expression profiles revealed only a partial overlap between affected genes of SMEDWI-2 and HP1-1 (Fig. 7, A and D). Smedwi-2 knockdown primarily abrogated expression of neoblast progeny genes (Fig. S4 F) without affecting genes involved in proliferation (Fig. 7 F). Thus, the mechanism by which HP1-1 regulates ASCs may be distinctly different from that by SMEDWI-2.
HP1-1–induced expression of Mcm5 mediates regenerative proliferation
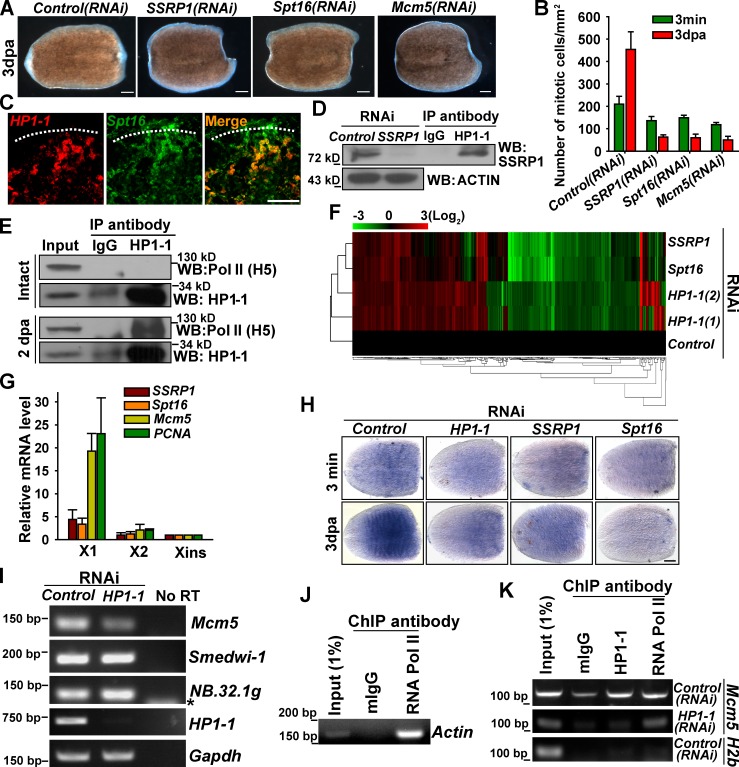
Because the multifaceted functions of HP1 isoforms depend on its different set of binding partners, we performed gene knockdown to search for genes showing similar phenotypes with HP1-1 in the list of known binding proteins (Maison and Almouzni, 2004; Fanti and Pimpinelli, 2008). Interestingly, individual depletion of 13 genes associated with gene silencing (Craig, 2005; Grewal and Jia, 2007) did not obviously abolish regeneration (Fig. S5 A). Moreover, knockdown of either HP1-1 or HP1-2 impaired H3K9me3 levels (Fig. S5 B), suggesting that the defects upon HP1-1 knockdown are not likely caused by general defects in heterochromatin formation or gene silencing. Interestingly, knockdown of FACT complex genes, SSRP1and Spt16, abolished regeneration capacity and proliferation response (Fig. 8, A and B) and resulted in defects reminiscent of HP1-1 depletion (Fig. S5, C–F). Both SSRP1 and Spt16 colocalized with HP1-1 in neoblasts (Fig. S5 G). Upon amputation, HP1-1–expressing cells were enriched beneath the amputation boundary, where proliferating neoblasts are localized, and they coexpressed Spt16 (Fig. 8 C). To further test whether HP1-1 interacts with the FACT complex, an antibody against SSRP-1 was generated (Fig. 8 D, left). Immunoprecipitation experiments showed that HP1-1 coimmunoprecipitated with SSRP1 in regenerating worms (Fig. 8 D, right). Given that the FACT complex is involved in transcription initiation and elongation (Orphanides et al., 1998), HP1-1 may function cooperatively with the FACT complex to activate gene transcription during regeneration. Consistent with this speculation, HP1-1 coimmunoprecipitated with the transcriptionally active form of RNA polymerase II (phospho-Ser2) in regenerating worms (Fig. 8 E). This result suggests that, upon injury, HP1-1 may associate with phosphorylated RNA polymerase II to bind their target genes.
Figure 8.
HP1-1–induced expression of Mcm5 mediates regenerative proliferation. (A) Phenotypic analysis of regenerating worms (n = 10). (B) Quantitative analysis of H3S10P-positive cell numbers in regenerating worms. Error bars show SDs, n = 6 animals. (C) Double FISH for HP1-1 and Spt16 in regenerating worms. White dotted lines indicate the amputation site. (D, right) HP1-1 immunoprecipitates from regenerating worms were immunoblotted with an antibody against SSRP1. (left) RNAi was used to validate the specificity of SSRP1 antibody. (E) HP1-1 immunoprecipitates from intact or regenerating worms (2 dpa) were immunoblotted with an antibody against the phosphorylated RNA polymerase II, H5. (F) Heat map of the altered genes shared between the four profiles (≥1.5-fold) at 3 dpa; red shows induced and green shows repressed, log2-based scale. Numbers in parentheses represent replicate samples. (G) qRT-PCR of isolated X1, X2, and Xins cells. Expression levels in Xins cells were set as 1. Error bars show SDs, n = 3. (H) Both SSRP1 and Spt16 are essential for induced Mcm5 expression during regeneration. n = 8 for each condition. Bar, 0.1 mm. (I) Semiquantitative PCR analysis of FACS-purified X1 cells. Shown are representative results from two independent biological replicates. No RT, no reverse transcriptase. Asterisk shows a primer dimer. (J) RNA polymerase II (RNA Pol II) is associated with the Actin promoter as demonstrated by ChIP followed by gene-specific PCR. mIgG, mouse IgG. (K) In vivo HP1-1 binding to the proximal promoter of the Mcm5 gene. Analysis of chromatin extracted from HP1-1(RNAi) worms (middle) was used as a control. Shown are representative results from three independent biological replicates. IP, immunoprecipitation; WB, Western blot.
We then performed additional microarray analysis of SSRP1- or Spt16-depleted worms and compared the gene expression profiles. Cluster analysis revealed that HP1-1 and FACT complex subunits behave strikingly similarly (Fig. 8 F), and transcripts associated with proliferation were likewise decreased by nearly 70% in worms depleted of SSRP1 or Spt16 (Fig. S5 E) when neoblasts are only moderately reduced (Fig. S5 H). Considering that decreased proliferation potential is the primary effect of HP1-1 loss (Fig. 5), genes whose expression was reduced may account for the phenotype. Thus, 85 decreased genes from the overlapping hits were selected for RNAi assay (Fig. S5 I), and five genes exhibiting phenotypes identical to that of HP1-1 knockdown were identified (Fig. S5 J). Among them, Mcm5 was one of the significantly down-regulated genes in the overlapping gene list. Interestingly, Mcm5 was highly enriched in proliferating neoblasts resembling the expression of a proliferation marker, PCNA (Fig. 8 G). Amputation led to an increase in Mcm5 expression, whereas injury-induced Mcm5 induction was severely compromised by knockdown of HP1-1 or FACT subunits (Fig. 8 H). Furthermore, X1 populations from control and HP1-1(RNAi) worms were isolated by FACS, and the expression of Mcm5 was assessed. The result shows that Mcm5 was indeed decreased upon HP1-1 knockdown, when smedwi-1 showed little change (Fig. 8 I). These results suggest that HP1-1 is required for expression of Mcm5.
To further ascertain whether Mcm5 is a direct target of HP1-1, we developed a whole-animal chromatin immunoprecipitation (ChIP; W-ChIP) assay (Fig. 8 J). Chromatin fragments were immunoprecipitated with HP1-1 antibody and extracted from regenerating worms at 3 dpa, when specific induction of Mcm5 transcripts is readily observed. The results showed that HP1-1 interacts with the proximal promoter region adjacent to the transcription start site of Mcm5, whereas HP1-1 knockdown abolishes this interaction (Fig. 8 K). This result suggests that Mcm5 is a direct target of HP1-1. Additionally, attenuation of Mcm5 led to failure in the injury-induced proliferative response (Fig. 8 B) and in regeneration as well (Fig. 8 A). Mcm5(RNAi) animals precisely phenocopy defects seen in HP1-1–depleted worms (Fig. S5, C–F), which suggests that Mcm5 is a downstream effector of HP1-1. This accounts, at least in part, for the HP1-1 phenotype in regenerative proliferation. Thus, our data indicate that HP1-1 and the FACT complex function together, through activating Mcm5 expression during transcription elongation, to support regenerative proliferation of ASCs and, consequently, to promote regeneration.
Discussion
Using an RNAi screen and gene expression profiling to initiate a survey of chromatin factors maintaining neoblast identity, we identified 12 chromatin genes with functions in neoblast-driven regeneration and established the first chromatin network underlying planarian neoblast regulation. Surprisingly, the key component of this network is an HP1-like protein, HP1-1, which is a novel marker for pluripotent neoblasts. HP1-1 maintains neoblast self-renewal in homeostatic tissues through promoting proliferation and repressing differentiation and, upon injury, elicits a proliferation burst. Mechanistically, our data support a model whereby an HP1 protein collaborates with SSRP1, a component of the FACT complex, to activate Mcm5 in ASCs and initiate regenerative proliferation. These data expand the inventory of genes regulating stem cell function and reveal an unexpected role for an HP1 gene in stem cell–mediated regeneration.
The plasticity of the cellular epigenome has been implicated in inducing regeneration (Barrero and Izpisua Belmonte, 2011). Planarian ASCs respond to injury and regenerate missing parts rapidly (Newmark and Sánchez Alvarado, 2000; Wenemoser et al., 2012), providing a promising system for studying epigenetic regulation of regeneration. In this study, we applied an RNAi screen to functionally test regeneration requirements for a total of 205 potential chromatin genes and introduced the use of qRT-PCR with FACS-sorted cells to test whether identified genes were enriched in neoblasts. We identified 12 genes representing at least six chromatin complexes (CAF1, BAF, NuRD, FACT, Cdk-activating kinase, and Mcm2–7 complex) essential for neoblast function and regeneration. Interestingly, several complexes have an analogous role in murine ES cell regulation (Fazzio et al., 2008). For instance, p150 is required for ES viability (Houlard et al., 2006), NuRD complex controls pluripotency (Kaji et al., 2006; Dovey et al., 2010), and the BAF complex regulates self-renewal and pluripotency (Ho et al., 2009). These data suggest that planarian ASCs are controlled by key chromatin regulators similar to those operating in ES cells and unveil an unexpected extent of deep conservation in epigenetic regulation between neoblasts and mammalian ES cells. Given that several genes we identified are highly expressed but not well understood in murine stem cells, individual analysis of their functions should provide valuable insights into mammalian stem cell biology and regenerative medicine. Additionally, recent advances have provided important insight into the gene expression programs operating in neoblasts, including some chromatin complexes (Rossi et al., 2007; Eisenhoffer et al., 2008; Labbé et al., 2012; Onal et al., 2012; Solana et al., 2012; Wagner et al., 2012), though detailed mechanistic studies have been hindered by less availability of reagents and assays. The antibodies validated and the assays developed in this study provide important tools for analyzing the function and mechanism of a particular gene in this emerging field.
Most species encode two or three HP1 isoforms that are ubiquitously expressed and believed to be general factors of heterochromatin formation and gene silencing (Li et al., 2002). Although HP1 homologues are involved in cell differentiation (Cammas et al., 2004; Panteleeva et al., 2007), the precise roles of HP1 proteins in stem cells are still elusive. Here, we focused on HP1-1 because of its specific expression pattern and the potential discovery of a novel mechanism and function of a general chromatin protein. We found unexpectedly that HP1-1, but not HP1-2, is exclusively expressed in ASCs and functions in balancing proliferation and differentiation, which together suggest key roles for HP1 in maintaining a stem cell gene expression program. In addition, our observations revealed a novel role for HP1 in promoting a proliferative response, possibly by inducing expression of Mcm5. Because the observation that expression of both HP1 and FACT genes is strongly correlated with the proliferation state of human cells (Ritou et al., 2007; Garcia et al., 2011), it will be interesting to evaluate whether this signaling axis plays general roles in regulating mammalian stem cells and regeneration capacity.
In addition, although there has been extensive efforts to investigate roles of HP1s in maintenance of heterochromatin (Maison and Almouzni, 2004; Fanti and Pimpinelli, 2008), some evidence has revealed a surprising role for HP1 in euchromatic gene expression (Piacentini et al., 2003; Vermaak and Malik, 2009). For instance, an HP1 isoform recruits the FACT complex to RNA polymerase II during heat shock stress in Drosophila melanogaster (Kwon et al., 2010) or maintains transcription of cell cycle regulators (De Lucia et al., 2005). However, it remains unclear whether HP1s regulate stem cells and, if so, whether it is dependent on gene activation. Here, we demonstrate that HP1-1 displays primarily euchromatic localization in neoblasts and interacts with SSRP1 and active RNA polymerase II after injury, suggesting that interaction of HP1-1 with SSRP1 is critical for inducing gene expression in stem cells. Because the pausing of RNA polymerase II at a promoter-proximal site early in transcription elongation is a general rate-limiting step in transcription (Core and Lis, 2008) and the FACT complex is involved in transcription elongation (Orphanides et al., 1998), HP1-1 may function in facilitating RNA polymerase II release from promoter-proximal pausing. The observed induction of HP1-1 expression and elevated interaction of HP1-1 with RNA polymerase II further supports this notion.
MCM proteins play essential roles in DNA replication and cell division. Although Mcm5 has been implicated in zebrafish retinal development (Ryu et al., 2005), in vivo analysis of gene regulation and function of Mcm5 in multicellular organisms and stem cells has been scarce. Our data identified Mcm5 as a downstream target of HP1-1 to support regenerative proliferation, suggesting a critical role for Mcm5 in stem cell mobilization. Nevertheless, our data do not exclude the possibilities that there are other target genes, and the function of HP1-1 for silencing genes may also be important for preventing premature differentiation. Once annotation of the planarian genome is completed, it will be important to define the binding sites of HP1-1 on a genome-wide scale.
Collectively, our results indicate that neoblasts are controlled by key chromatin regulators similar to those operating in murine ES cells and prompt consideration of a model whereby an HP1-like protein initiates regeneration through transcriptional elongation in ASCs. Identification of the HP1-1–Mcm5 cascade as the trigger of regenerative proliferation, and as an important regulator for maintaining the regenerative potential of adult tissues, provides new insights into how chromatin factors orchestrate stem cell activation and regeneration through transcriptional regulation.
Materials and methods
Animals
Clonal asexual (CIW4) and sexual strains of Schmidtea mediterranea were maintained in Montjuïch salts (1.6 mM NaCl, 1.0 mM CaCl2, 1.0 mM MgSO4, 0.1 mM MgCl2, 0.1 mM KCl, and 1.2 mM NaHCO3 prepared in autoclaved Milli-Q water) and 0.75× Montjuïch salts, respectively, at 20°C in the dark (Cebrià and Newmark, 2005). Animals were fed weekly with homogenized calf liver. All animals, 4–6 mm in length, were starved 1 wk before any experiments. For irradiation, planarians were exposed to 100 Gray of γ irradiation using a sealed source of Cesium 137 (Gammacell 3000). The animals were kindly provided by P. Newmark (University of Illinois at Urbana-Champaign/Howard Hughes Medical Institute, Urbana, IL), P. Reddien (Massachusetts Institute of Technology/Howard Hughes Medical Institute, Cambridge, MA), and N. Oviedo (University of California, Merced, Merced, CA).
Identification and cloning of planarian chromatin-related genes
A BLAST-based reciprocal best-hit method, in combination with protein sequence alignment and phylogenetic analysis, was used to identify orthologous genes in planarian. In brief, known chromatin proteins from the human, mouse, and fly genomes were retrieved by searching the NCBI database for an array of keywords consisting of protein domains commonly found in chromatin genes, such as “Chromo,” “Set,” “PhD,” and “Tudor.” Using these sequences as search queries, TBLASTN analysis was performed against the planarian genome database SmedGD (S. mediterranea Genome Database) and the hermaphroditic strain EST database (Zayas et al., 2005; Robb et al., 2008). The resulting ESTs and unigenes were used to deduce the putatively encoded protein sequences. Planarian genes or proteins were named after their closest human homologues after extensive comparison with BLASTP according to the standard nomenclature system (Reddien et al., 2008). The final list included 210 unique unigenes, 205 of which were successfully cloned from a cDNA library prepared from adult worms using PCR. The full-length sequences of HP1-1, HP1-2, Mcm5, SSRP1, Spt16, and H3 were obtained with the RNA ligase–mediated rapid amplification of cDNA ends kit (Ambion) and deposited in GenBank (accession nos. JN216838, JN216839, JX070079, JX070080, JX070081, and JX070082, respectively). Protein sequence prediction was performed using six-frame translation (Baylor College of Medicine, Houston, TX), and alignments were performed using ClustalW2 and the online version of MAFFT (K. Katoh, Osaka University, Osaka, Japan). A phylogenetic tree was built using the neighbor-joining algorithm in ClustalX. The species shown in the phylogenetic tree (Fig. 2 A) are Caenorhabditis elegans, D. melanogaster, Drosophila virilis, Danio rerio, Bombyx mori, Homo sapiens, Mus musculus, Xenopus tropicalis, Neurospora crassa, Schizosaccharomyces japonicus, Schizosaccharomyces pombe, Lycopersicon esculentum, Arabidopsis thaliana, and S. mediterranea.
RNAi experiments
The RNAi vector was constructed by inserting multiple cloning sites into the region between two T7 promoters in the pPR244 vector (a gift from P. Reddien; Reddien et al., 2005a). cDNAs of individual genes (∼1,000 bp) were cloned into the pPR244 vector using KpnI or BamHI and BglII (Thermo Fisher Scientific), and the coding sequence (∼1,000 bp) of the GFP gene was cloned as a control. All RNAi vectors were confirmed by sequencing before induction with 1 mM IPTG (at an OD600 of 0.4) in the HT115 strain (a gift from Z. Zhang, Changhai Hospital, Shanghai, China). For the RNAi knockdown, worms were fed three times over 8 d (first, fifth, and eighth) and were amputated into three fragments pre- and postpharyngeally at 24 h after the last feeding. For the RNAi screen, two rounds of feeding and amputation were used to minimize issues of protein perdurance. The effectiveness of RNAi was confirmed by in situ hybridization or qRT-PCR. All screens were repeated twice, and >10 worms were used for each treatment.
Antibodies and immunostaining
Mouse monoclonal antibodies against HP1-1 (9B11 and 13E6) and polyclonal antibodies for HP1-2, SSRP1, and SMEDWI-2 (1T1B1) were raised and affinity purified in our laboratory according to standard protocols. The antibodies used in IF or Western blotting are as follows: SMEDWI-1 (a gift from P. Newmark and Y. Wang, University of Illinois at Urbana-Champaign, Urbana, IL), H3S10P (06–570; EMD Millipore), BrdU (555746; BD), β-actin (M20010; Abmart), H3K9me3 (ab8898; Abcam), HP1-β (ab10478; Abcam), and RNA polymerase II (MMS-126R and 129R [Covance]; 05–623 [EMD Millipore]). Secondary antibodies were Alexa Fluor 488 and 555 obtained from Molecular Probes (Invitrogen). IF was performed as previously reported (Guo et al., 2006). In brief, fixed and bleached worms were rehydrated in graded PBSTx (PBS + 0.3% Triton X-100)/methanol solutions, blocked in PBSTx containing 0.25% IgG-free BSA (Sigma-Aldrich), and incubated with primary antibodies overnight at 4°C. After extensive washing with PBSTx, samples were incubated with Alexa Fluor 488– or Alexa Fluor 555–conjugated secondary antibody and mounted using Mowiol mounting medium or fluorescent mounting medium (Dako).
BrdU incorporation
For immunostaining alone, animals were fed a food mixture containing 5 mg/ml BrdU (Sigma-Aldrich) for half an hour. At 6 h after feeding, animals were sacrificed in 2% HCl, and IF was performed as previously described (Newmark and Sánchez Alvarado, 2000; Guo et al., 2006). For BrdU combined with FISH, BrdU (two to three injections of ∼30 nl of 10 mg/ml) in planarian water was injected prepharyngeally to animals. At appropriate times after microinjection (8, 24, or 48 h), animals were sacrificed, fixed, and bleached. After rehydration, FISH was performed first. Then, samples were incubated with 2 N HCl for 15–20 min at room temperature, neutralized for 2 min in 0.1 M borax (Sigma-Aldrich), washed twice for 5 min in PBSTx, and blocked with PBSTx + 0.6% BSA at room temperature for 4 h. Samples were incubated with mouse monoclonal anti-BrdU (1:30; BD), and the signal was amplified with Alexa Fluor secondary antibody at 4°C overnight.
TUNEL assay and transmission electron microscopy
Whole-mount TUNEL assay was performed as previously described (Pellettieri et al., 2010). In brief, worms were sacrificed in 10% n-acetyl cysteine (diluted in PBS), fixed in 4% formaldehyde (diluted in PBSTx), and permeabilized in 1% SDS (diluted in PBS) for 20 min. Fixed animals were bleached overnight in 6% H2O2 (diluted in PBST). Bleached animals were washed in PBST and further rinsed in PBS and equilibration buffer before incubating in terminal transferase enzyme (90418; EMD Millipore) for 4 h. Enzyme treatment was stopped by washing in stop/wash buffer (90419; EMD Millipore), and animals were rinsed in PBSTB (PBST with 0.25% BSA) and then incubated for 4 h in anti–digoxigenin-rhodamine (90429; EMD Millipore), which was diluted in blocking solution (90425; EMD Millipore). Stained animals were rinsed on a platform shaker at room temperature in PBSTB for 4 × 10 min and mounted under coverslips on glass slides with Mowiol mounting medium.
Transmission electron microscopy was performed as previously described (Salvetti et al., 2005; Bonuccelli et al., 2010). In brief, after fixation with 2.5% glutaraldehyde solution in 0.1 M cacodylate buffer, animals were postfixed in 2% osmium tetroxide. Ultrathin sections were stained with uranyl acetate and lead citrate and observed with a transmission electron microscope (H-7650; Hitachi).
WISH and FISH
WISH and FISH were performed as previously described (Pearson et al., 2009; Collins et al., 2010). In brief, worms were killed in 5% n-acetyl cysteine (Sigma-Aldrich), fixed in 4% PFA, permeabilized using reduction buffer, and dehydrated in a graded series of methanol in PBSTx before bleaching. After rehydration, hybridizations were performed with ∼0.5 ng/µl riboprobes. After proper washing and antibody incubation (anti–digoxigenin-AP, 1:4,000; Roche), signal was developed using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate substrate (1:50; Roche), and samples were mounted with 80% glycerol. For FISH, after blocking, samples were incubated with anti–digoxigenin-peroxidase (1:1,000; Roche) overnight and subsequently developed with FITC-tyramide generated by using fluorescein mono-N-hydroxysuccinimide-ester (46100; Thermo Fisher Scientific) and tyramide (T-2879; Sigma-Aldrich) in the presence of 0.0015% H2O2 in PBST (PBS + 0.01% Tween 20). For double FISH, digoxigenin-labeled probes were first developed with Cy3-tyramide (1:500). After inactivation of antibody-conjugated HRP, the second antibody against the FITC-labeled probe (1:500; Roche) was added followed by development with FITC-tyramide. Within a given experiment, all samples were developed in the fluorescent substrate for the same length of time and imaged using identical exposure conditions. FISH on cross sections was performed as previously described (Tu et al., 2012) with modifications. In brief, FISH-stained animals were transferred to a graded series of sucrose in PBS before embedding in optimum cutting temperature compound (Sakura). Specimens were brought to −80°C and were sectioned in a cryostat (Microm HM550; Thermo Fisher Scientific) at 14 µm at −20°C. The sections were placed on charged slides (Premiere) and mounted with Mowiol mounting medium before imaging.
Image acquisition, processing, and quantification
Live animals, WISH, and TUNEL samples mounted with 80% glycerol or Mowiol mounting medium were photographed using a microscope (SteREO Discovery.V20; Carl Zeiss) equipped with a Plan Apochromat 1.0× objective and a digital microscope camera (AxioCam HRc; Carl Zeiss) automated by AxioVision Rel.4.8 software (Carl Zeiss). IF and FISH specimens were mounted with fluorescence mounting medium (Dako) or Mowiol mounting medium, and images were captured with a laser-scanning confocal microscope (True Confocal Scanner SP5; Leica; HCX Plan Apochromat confocal scanning 10×/0.4 NA, 20×/0.7 NA, 40×/0.85 NA, or 63×/1.40 NA oil immersion objective lens) by LAS AF software (Leica). Images were processed with LAS AF Lite software and Photoshop software (Adobe) and were quantified using QWin software (Leica) or ImageJ software (National Institutes of Health). All IF and in situ hybridization experiments were performed, imaged, and processed identically (at room temperature, 22°C) to allow direct comparison between experimental animals and controls.
RNA extraction and gene expression profiling
Gene arrays, covering 9,981 of the 10,173 unigenes deposited in the NCBI (Entrez records, 2009), were designed with the platform at the eArray website (Agilent Technologies) and printed in a 4 × 44,000 slide format for oligonucleotide arrays (Agilent Technologies). The RNAi worms (three feedings total) were amputated pre- and postpharyngeally and allowed to regenerate for 3 d. Total RNA was isolated using TRIZOL (Invitrogen) and an RNeasy mini kit (QIAGEN). RNA quality was assessed using a NanoDrop 1000 (Thermo Fisher Scientific) and a 2100 Bioanalyzer (Agilent Technologies). RNA was amplified and labeled with Cy3-CTP using a low RNA input fluorescent linear amplification kit (Agilent Technologies). Labeled cRNA was assessed, and equal masses of the sample were hybridized to arrays for 14 h. Arrays were scanned with a microarray scanner (G2565BA; Agilent Technologies), and raw signal intensities were extracted with Feature Extraction v8.9 software (Agilent Technologies). All raw data were normalized and analyzed with GeneSpring GX 10.0 (Agilent Technologies), and hierarchical clustering and heat map generation were performed using GeneSpring GX software, Cluster 3.0 (University of Tokyo, Human Genome Center), and Java TreeView 1.1.6r2 software (Saldanha, 2004). All array experiments were performed in duplicate using independently prepared RNA and separate gene chips. We also restricted the analysis to genes with expression level changes of ≥1.5-fold in duplicate samples and an mean raw expression intensity of ≥1,000 in any group. For gene ontology analyses, probe IDs were converted to unigene IDs, and unigenes were assigned gene ontology terms from the gene ontology database based on the closest gene ontology–annotated BLASTX homologue.
Flow cytometry and qRT-PCR
Sorting by flow cytometry was performed as previously described (Hayashi et al., 2006; Peiris et al., 2012). In brief, planarians were diced into small pieces on a cold plate and incubated in 1 mg/ml collagenase (diluted in calcium- and magnesium-free medium plus 1% BSA) as previously described (Scimone et al., 2010). Dissociated cells were filtered with a cell strainer (BD) and stained with 0.2 µg/ml calcein acetoxymethyl ester and 18 µg/ml Hoechst 33342 for an appropriate time. After incubating with 5 µg/ml propidium iodide, analyses and sorts were performed using the FACSAria II (BD) or MoFlo XDP (Beckman Coulter). Data were processed by FlowJo V7.6.5 (Tree Star, Inc.).
qRT-PCR was performed as previously described (Li et al., 2011b). In brief, total RNA was isolated using TRIZOL (Invitrogen). cDNAs were generated from 300–500 ng of total RNA with moloney murine leukaemia virus reverse transcription (Promega). Gene-specific primers were designed with the Universal Probe library (Roche) or OligoPerfect designer (Invitrogen). qPCRs were performed with SYBR Green quantitative PCR master mix (Toyobo Co.) on a quantitative PCR system (LightCycler 480; Roche). At least three biological replicates were performed for each group, and each experiment was performed with triplicate or quadruplicate PCR reactions. Data are expressed using the comparative cycle threshold method. Relative expression levels were normalized to the levels of Gapdh (AY068133) mRNA and plotted with SigmaPlot 11.0 (Systat Software, Inc.).
Western blot analysis
A homogenate of planarians, generally two to three animals, was prepared quickly in 200 µl radioimmunoprecipitation assay buffer. After fractionation by SDS-PAGE, proteins in the gel were transferred to nitrocellulose membranes (Pall Corporation), and the membranes were incubated with primary antibodies in blocking buffer overnight at 4°C. After incubation with HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Inc.), bands were detected using an ECL Western blotting detection kit (Thermo Fisher Scientific). Western blot analysis was performed in at least three independent experiments, and representative data were shown. For assessing HP1-1 expression during regeneration, animals were cut into five fragments, and equal amounts of protein were loaded at each time point. For histone peptide pull-down assay, GST-tagged HP1-1 was overexpressed in Escherichia coli (BL21). Cells were grown at 37°C with shaking, and protein expression was induced with 0.1 mM IPTG. The GST–HP1-1 protein was purified using glutathione–Sepharose 4B (GE Healthcare) in accordance with the manufacturer’s instructions. Histone peptide pull-down assay was performed as previously described (Li et al., 2011a). In brief, 1 µg of purified GST–HP1-1 protein was incubated with 1 µg of synthesized biotinylated histone peptides (a gift from B. Li, Institute of Biochemistry and Cell Biology, Shanghai, China) in binding buffer at 4°C overnight. Then, 20 µl streptavidin beads (GE Healthcare) was added for another 2 h. After extensive washing, the bound proteins were eluted in 2× SDS loading buffer and were subjected to Western blot analysis.
Immunoprecipitation and W-ChIP assay
Immunoprecipitation was performed as previously described (Nielsen et al., 1999). In brief, dissociated cells were suspended in nuclei isolation buffer, and nuclei were pelleted by centrifugation. After addition of nuclei extraction buffer, appropriate antibodies were added for 2 h followed by incubation with protein A/G beads (Santa Cruz Biotechnology, Inc.) overnight at 4°C with rotation. Western blot was performed as described in the previous paragraph.
The W-ChIP assay was established according to the standard ChIP protocol (Lee et al., 2006) with modifications. In brief, dissociated planarian cells were fixed in 1% formaldehyde (10 min) and quenched with glycine (5 min). Cells were then homogenized in nuclei lysis buffer (50 mM Tris-HCl, pH 8/10 mM EDTA/1% SDS/protease inhibitor), and sonication conditions were optimized using a Bioruptor (Diagenode) to yield fragments of ∼300–800 bp. The lysate was centrifuged, and the supernatant was collected and further diluted with ChIP dilution buffer. After performing a preclearing step, the supernatant was incubated with appropriate antibodies overnight at 4°C with rotation. After incubation with 60 µl protein G beads (EMD Millipore) for an additional hour, bound complexes were extensively washed and were released from the beads with elution buffer (0.1 M NaHCO3/1% SDS). Cross-links were reversed, and chromatin was purified by treatment with RNase A (Takara Bio Inc.) followed by proteinase K (Invitrogen) digestion and DNA purification. PCR analysis was performed to confirm the enrichment of RNA polymerase II or HP1-1 on target promoters (Mcm5, Actin, and Gapdh). ChIP with normal murine IgG was used as a negative control.
Cell culture
HeLa and NIH3T3 cells were cultured in standard commercial DMEM medium (Hyclone) supplemented with 5% FBS. Full-length cDNAs of planarian HP1-1 and HP1-2 were cloned into the pEGFP-N1 or pCMV-Myc vector (Takara Bio Inc.) and were confirmed by sequencing. Purified HP1-1–EGFP and Myc–HP1-2 vectors were transfected into HeLa cells with Lipofectamine 2000 (Invitrogen). Cells were fixed in 4% PFA, permeabilized with PBSTx, blocked with 5% goat serum, and then incubated with the primary antibody at 4°C. After washing with PBS, samples were immunostained with the Alexa Fluor secondary antibody (Invitrogen) and counterstained with DAPI (Roche) at room temperature.
Statistical analysis
Results are presented as means ± SD, and statistical analyses were performed in SigmaPlot 11.0 using the Student’s t test for two groups or one-way analysis of variance for three or more groups. P < 0.05 was considered significant.
Online supplemental material
Fig. S1 shows identification of chromatin genes essential for regeneration. Fig. S2 shows that HP1-1 is localized in planarian neoblasts. Fig. S3 shows that HP1-1 is required for neoblast self-renewal. Fig. S4 shows that HP1-1 is essential for a proliferative response. Fig. S5 shows that knockdown of the FACT complex genes and Mcm5 leads to a phenotype resembling that of HP1-1 depletion. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207172/DC1.
Supplementary Material
Acknowledgments
We thank P. Newmark, P. Reddien, and N. Oviedo for kindly providing worms; P. Newmark and Y. Wang for the SMEDWI-1 antibody; P. Reddien for the pPR244 construct; Z. Zhang for the HT115 strain; B. Li for the histone peptide; and Y. Wang, N. Oviedo, J. Pellettieri, T. Guo, L. Cheng, Z. Zhang, C. Mao, and staffs in the core facility (Institute of Health Sciences) for technical assistance. We thank A. Sánchez Alvarado, Y. Wang, B. Li, X. Li, and R. Lu for critical reading of the manuscript and all Jing laboratory members for comments.
This work was supported in part by the National Natural Science Foundation of China (81130005 and 30971615), the Ministry of Science and Technology of China (2010CB945600, 2011CB811304, and 2007CB947002), and the Chinese Academy of Sciences (XDA01040306). G. Brewer was supported by a visiting professorship at the Institute of Health Sciences (Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences).
Footnotes
Abbreviations used in this paper:
- ASC
- adult stem cell
- BAF
- BRG1/Brm-associated factor
- ChIP
- chromatin immunoprecipitation
- dpa
- day postamputation
- dsRNA
- double-stranded RNA
- ES
- embryonic stem
- FACT
- facilitates chromatin transcription
- IF
- immunofluorescence
- MCM
- minichromosome maintenance
- NuRD
- nucleosome remodeling and deacetylase
- qRT-PCR
- quantitative real-time PCR
- W-ChIP
- whole-animal ChIP
- WISH
- whole-mount in situ hybridization
References
- Agata K. 2003. Regeneration and gene regulation in planarians. Curr. Opin. Genet. Dev. 13:492–496 10.1016/j.gde.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A.G. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8:532–538 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 410:120–124 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Barrero M.J., Izpisua Belmonte J.C. 2011. Regenerating the epigenome. EMBO Rep. 12:208–215 10.1038/embor.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125:315–326 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bonuccelli L., Rossi L., Lena A., Scarcelli V., Rainaldi G., Evangelista M., Iacopetti P., Gremigni V., Salvetti A. 2010. An RbAp48-like gene regulates adult stem cells in planarians. J. Cell Sci. 123:690–698 10.1242/jcs.053900 [DOI] [PubMed] [Google Scholar]
- Cammas F., Herzog M., Lerouge T., Chambon P., Losson R. 2004. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 18:2147–2160 10.1101/gad.302904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F., Newmark P.A. 2005. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 132:3691–3703 10.1242/dev.01941 [DOI] [PubMed] [Google Scholar]
- Collins J.J., III, Hou X., Romanova E.V., Lambrus B.G., Miller C.M., Saberi A., Sweedler J.V., Newmark P.A. 2010. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 8:e1000509 10.1371/journal.pbio.1000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Lis J.T. 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 319:1791–1792 10.1126/science.1150843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J.M. 2005. Heterochromatin—many flavours, common themes. Bioessays. 27:17–28 10.1002/bies.20145 [DOI] [PubMed] [Google Scholar]
- Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K. 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 4:80–93 10.1016/j.stem.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F., Ni J.Q., Vaillant C., Sun F.L. 2005. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 33:2852–2858 10.1093/nar/gki584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey O.M., Foster C.T., Cowley S.M. 2010. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA. 107:8242–8247 10.1073/pnas.1000478107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer G.T., Kang H., Sánchez Alvarado A. 2008. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 3:327–339 10.1016/j.stem.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., Elgin S.C. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204–210 10.1016/S0959-437X(00)00058-7 [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., James T.C., Foster-Hartnett D.M., Hartnett T., Ngan V., Elgin S.C. 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 87:9923–9927 10.1073/pnas.87.24.9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L., Pimpinelli S. 2008. HP1: a functionally multifaceted protein. Curr. Opin. Genet. Dev. 18:169–174 10.1016/j.gde.2008.01.009 [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Huff J.T., Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 134:162–174 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia H., Fleyshman D., Kolesnikova K., Safina A., Commane M., Paszkiewicz G., Omelian A., Morrison C., Gurova K. 2011. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget. 2:783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. 2011. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12:36–47 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Jia S. 2007. Heterochromatin revisited. Nat. Rev. Genet. 8:35–46 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- Guo T., Peters A.H., Newmark P.A. 2006. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell. 11:159–169 10.1016/j.devcel.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Asami M., Higuchi S., Shibata N., Agata K. 2006. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48:371–380 10.1111/j.1440-169X.2006.00876.x [DOI] [PubMed] [Google Scholar]
- Ho L., Ronan J.L., Wu J., Staahl B.T., Chen L., Kuo A., Lessard J., Nesvizhskii A.I., Ranish J., Crabtree G.R. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA. 106:5181–5186 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M., Berlivet S., Probst A.V., Quivy J.P., Héry P., Almouzni G., Gérard M. 2006. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2:e181 10.1371/journal.pgen.0020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B. 2006. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8:285–292 10.1038/ncb1372 [DOI] [PubMed] [Google Scholar]
- Kwon S.H., Florens L., Swanson S.K., Washburn M.P., Abmayr S.M., Workman J.L. 2010. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 24:2133–2145 10.1101/gad.1959110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé R.M., Irimia M., Currie K.W., Lin A., Zhu S.J., Brown D.D., Ross E.J., Voisin V., Bader G.D., Blencowe B.J., Pearson B.J. 2012. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells. 30:1734–1745 10.1002/stem.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 410:116–120 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., Young R.A. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729–748 10.1038/nprot.2006.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.Z., Huang Z., Cui Q.Y., Song X.H., Du L., Jeltsch A., Chen P., Li G., Li E., Xu G.L. 2011a. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 21:1172–1181 10.1038/cr.2011.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Clevers H. 2010. Coexistence of quiescent and active adult stem cells in mammals. Science. 327:542–545 10.1126/science.1180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kirschmann D.A., Wallrath L.L. 2002. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA. 99(Suppl. 4):16462–16469 10.1073/pnas.162371699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Q., Zeng A., Han X.S., Wang C., Li G., Zhang Z.C., Wang J.Y., Qin Y.W., Jing Q. 2011b. Argonaute-2 regulates the proliferation of adult stem cells in planarian. Cell Res. 21:1750–1754 10.1038/cr.2011.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Almouzni G. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296–304 10.1038/nrm1355 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 448:553–560 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P.A., Sánchez Alvarado A. 2000. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220:142–153 10.1006/dbio.2000.9645 [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Sánchez Alvarado A. 2002. Not your father’s planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 3:210–219 10.1038/nrg759 [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Reddien P.W., Cebrià F., Sánchez Alvarado A. 2003. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA. 100(Suppl. 1):11861–11865 10.1073/pnas.1834205100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18:6385–6395 10.1093/emboj/18.22.6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onal P., Grün D., Adamidi C., Rybak A., Solana J., Mastrobuoni G., Wang Y., Rahn H.P., Chen W., Kempa S., et al. 2012. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 31:2755–2769 10.1038/emboj.2012.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., Hochedlinger K. 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell. 145:835–850 10.1016/j.cell.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 92:105–116 10.1016/S0092-8674(00)80903-4 [DOI] [PubMed] [Google Scholar]
- Panteleeva I., Boutillier S., See V., Spiller D.G., Rouaux C., Almouzni G., Bailly D., Maison C., Lai H.C., Loeffler J.P., Boutillier A.L. 2007. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 26:3616–3628 10.1038/sj.emboj.7601789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B.J., Eisenhoffer G.T., Gurley K.A., Rink J.C., Miller D.E., Sánchez Alvarado A. 2009. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238:443–450 10.1002/dvdy.21849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris T.H., Weckerle F., Ozamoto E., Ramirez D., Davidian D., García-Ojeda M.E., Oviedo N.J. 2012. TOR signaling regulates planarian stem cells and controls localized and organismal growth. J. Cell Sci. 125:1657–1665 10.1242/jcs.104711 [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Fitzgerald P., Watanabe S., Mancuso J., Green D.R., Sánchez Alvarado A. 2010. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338:76–85 10.1016/j.ydbio.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L., Fanti L., Berloco M., Perrini B., Pimpinelli S. 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161:707–714 10.1083/jcb.200303012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P.W., Sánchez Alvarado A. 2004. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20:725–757 10.1146/annurev.cellbio.20.010403.095114 [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Bermange A.L., Murfitt K.J., Jennings J.R., Sánchez Alvarado A. 2005a. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 8:635–649 10.1016/j.devcel.2005.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sánchez Alvarado A. 2005b. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 310:1327–1330 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Newmark P.A., Sánchez Alvarado A. 2008. Gene nomenclature guidelines for the planarian Schmidtea mediterranea. Dev. Dyn. 237:3099–3101 10.1002/dvdy.21623 [DOI] [PubMed] [Google Scholar]
- Ritou E., Bai M., Georgatos S.D. 2007. Variant-specific patterns and humoral regulation of HP1 proteins in human cells and tissues. J. Cell Sci. 120:3425–3435 10.1242/jcs.012955 [DOI] [PubMed] [Google Scholar]
- Robb S.M., Ross E., Sánchez Alvarado A. 2008. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 36(Suppl. 1):D599–D606 10.1093/nar/gkm684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Salvetti A., Marincola F.M., Lena A., Deri P., Mannini L., Batistoni R., Wang E., Gremigni V. 2007. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol. 8:R62 10.1186/gb-2007-8-4-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L., Shibata N., Nishimura O., Agata K. 2010. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341:429–443 10.1016/j.ydbio.2010.02.037 [DOI] [PubMed] [Google Scholar]
- Ryu S., Holzschuh J., Erhardt S., Ettl A.K., Driever W. 2005. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc. Natl. Acad. Sci. USA. 102:18467–18472 10.1073/pnas.0506187102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics. 20:3246–3248 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Saló E., Baguñà J. 1984. Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J. Embryol. Exp. Morphol. 83:63–80 [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Deri P., Batistoni R. 2000. An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev. Dyn. 218:603–614 [DOI] [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Lena A., Batistoni R., Deri P., Rainaldi G., Locci M.T., Evangelista M., Gremigni V. 2005. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 132:1863–1874 10.1242/dev.01785 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A. 2006. Planarian regeneration: its end is its beginning. Cell. 124:241–245 10.1016/j.cell.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Scimone M.L., Meisel J., Reddien P.W. 2010. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development. 137:1231–1241 10.1242/dev.042051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J., Kao D., Mihaylova Y., Jaber-Hijazi F., Malla S., Wilson R., Aboobaker A. 2012. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol. 13:R19 10.1186/gb-2012-13-3-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu K.C., Pearson B.J., Sánchez Alvarado A. 2012. TORC1 is required to balance cell proliferation and cell death in planarians. Dev. Biol. 365:458–469 10.1016/j.ydbio.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D., Malik H.S. 2009. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 43:467–492 10.1146/annurev-genet-102108-134802 [DOI] [PubMed] [Google Scholar]
- Wagner D.E., Wang I.E., Reddien P.W. 2011. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 332:811–816 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D.E., Ho J.J., Reddien P.W. 2012. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 10:299–311 10.1016/j.stem.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zayas R.M., Guo T., Newmark P.A. 2007. nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. USA. 104:5901–5906 10.1073/pnas.0609708104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Stary J.M., Wilhelm J.E., Newmark P.A. 2010. A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev. 24:2081–2092 10.1101/gad.1951010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman I.L. 2000. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 100:157–168 10.1016/S0092-8674(00)81692-X [DOI] [PubMed] [Google Scholar]
- Wenemoser D., Reddien P.W. 2010. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344:979–991 10.1016/j.ydbio.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Lapan S.W., Wilkinson A.W., Bell G.W., Reddien P.W. 2012. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26:988–1002 10.1101/gad.187377.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas R.M., Hernández A., Habermann B., Wang Y., Stary J.M., Newmark P.A. 2005. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. USA. 102:18491–18496 10.1073/pnas.0509507102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.