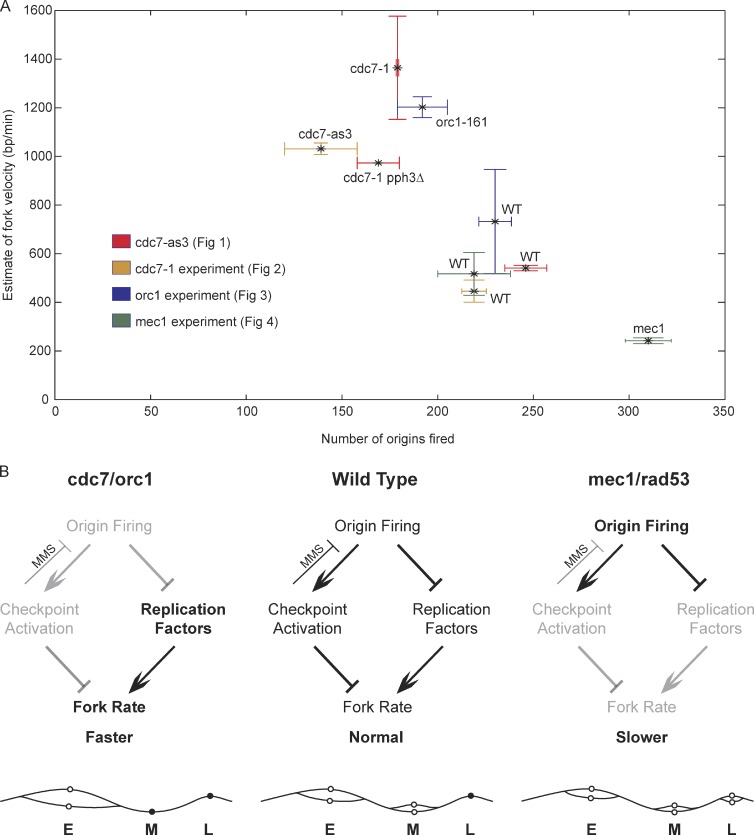
Cells with reduced origin firing have an increased rate of replication fork progression, whereas fork progression is slowed in cells with excess origins.
Abstract
DNA damage slows DNA synthesis at replication forks; however, the mechanisms remain unclear. Cdc7 kinase is required for replication origin activation, is a target of the intra-S checkpoint, and is implicated in the response to replication fork stress. Remarkably, we found that replication forks proceed more rapidly in cells lacking Cdc7 function than in wild-type cells. We traced this effect to reduced origin firing, which results in fewer replication forks and a consequent decrease in Rad53 checkpoint signaling. Depletion of Orc1, which acts in origin firing differently than Cdc7, had similar effects as Cdc7 depletion, consistent with decreased origin firing being the source of these defects. In contrast, mec1-100 cells, which initiate excess origins and also are deficient in checkpoint activation, showed slower fork progression, suggesting the number of active forks influences their rate, perhaps as a result of competition for limiting factors.
Introduction
The replication of eukaryotic chromosomes requires the cell cycle–regulated initiation of numerous replication origins on each chromosome. Coordinating much of this process are two highly conserved kinases, S-phase Cdk and Dbf4-dependent kinase (DDK), which become active at the G1–S transition (Labib, 2010). During early G1 phase, before S-phase Cdk and DDK activation, origin recognition complex, Cdc6, and Cdt1 load minichromosome maintenance (MCM) helicase complexes, in an inactive state, onto DNA at potential origin loci. A key step in replication initiation is the conversion of MCM into the active helicase, resulting in DNA unwinding, replisome assembly, and DNA synthesis. DDK plays an essential role in MCM activation by phosphorylating MCM, particularly the Mcm4 (and Mcm6) subunit. In fact, this is the only essential function of DDK in yeast, as mutations in MCM subunits that mimic the DDK-phosphorylated state or cause conformational changes that activate the helicase, obviate the normal requirement for DDK function for DNA replication and cell viability (Hardy et al., 1997; Fletcher et al., 2003; Sheu and Stillman, 2010).
As the name implies, DDK is composed of a catalytic kinase subunit, Cdc7, whose activity depends on Dbf4 (Masai and Arai, 2002). Dbf4 binds Cdc7, activating the kinase and targeting it to specific substrates, such as Mcm4. Dbf4 also negatively regulates DDK function as a target of the intra-S checkpoint pathway in response to replication stress or DNA damage (Duncker and Brown, 2003). Activated checkpoint kinase Rad53 phosphorylates Dbf4, inhibiting DDK-dependent activation of unfired origins (Lopez-Mosqueda et al., 2010; Zegerman and Diffley, 2010). There are conflicting reports as to whether this regulation directly inhibits DDK activity or affects its targeting to substrate, or both (Oshiro et al., 1999; Weinreich and Stillman, 1999; Sheu and Stillman, 2006). Rad53 activity also regulates the rate of replication fork progression through damaged DNA, suggesting that Rad53 might modulate replication fork progression by regulating DDK activity (Szyjka et al., 2008). In this study, we have examined replication fork dynamics in cells depleted of Cdc7 function and find that replication forks progress more rapidly than in wild-type (WT) cells. Together with analysis of Orc1- and checkpoint-defective cells, we show that replication fork rate is sensitive to the level of origin firing.
Results and discussion
Cdc7 activity regulates replication fork progression
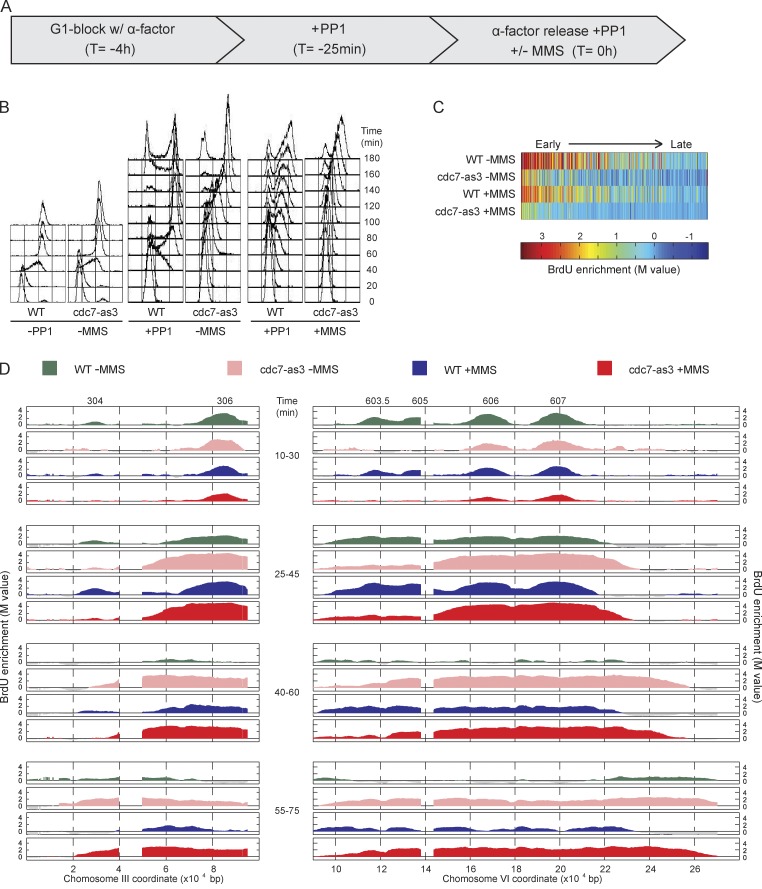
To address the potential function of DDK at replication forks, we analyzed the rate of DNA synthesis across two long replicons using BrdU immunoprecipitation (IP) analyzed by microarray (BrdU-IP-chip) in cells depleted of Cdc7 function. To deplete Cdc7 function, we used two well-characterized alleles: cdc7-as3 (L120A and V181A), the catalytic activity of which is directly inhibited by binding of ATP analogue PP1 within the ATP binding site (Wan et al., 2006), and cdc7-1, a temperature-sensitive kinase hypomorph, in the presence of the bob1 allele of MCM5, which enables reduced but sufficient origin firing for viability in the absence of Cdc7 kinase function (Hardy et al., 1997; Hoang et al., 2007).
WT and cdc7-as3 cells were synchronized in late G1 phase with α-factor and treated with PP1 25 min before release into S phase; upon release into S phase, aliquots of each culture were pulse labeled with BrdU for discrete intervals (Fig. 1 A). Analysis of bulk DNA content by FACScan showed rapid progression of WT cells through S phase, unaffected by the presence of PP1, whereas cdc7-as3 cells were delayed in bulk DNA synthesis, in a PP1-dependent manner (Fig. 1 B). Analysis of BrdU incorporation showed depletion of origin firing in PP1-treated cdc7-as3 cells, both in the number of origins that fired genome wide and in their levels of BrdU incorporation (see Materials and methods). We estimated that 234 origins fired in WT cells, and 157 fired in cdc7-as3 cells; these represent mainly earlier firing origins, as determination of later origins was precluded by possible BrdU signal from converging replication forks. In addition to fewer origins detected to fire, the level of BrdU incorporation was lower at these origins in cdc7-as3 cells, consistent with less efficient activation (Fig. 1 C). Arrangement of the origins’ BrdU incorporation levels according to their replication timing (see Materials and methods) showed that later origins were more diminished than earlier origins in cdc7-as3 cells (Fig. 1 C).
Figure 1.
Cdc7 function regulates replication fork progression. (A) WT and cdc7-as3 cells were synchronized with α-factor for 3 h and 35 min, treated with PP1 for 25 min, and released from α-factor with PP1 and with or without 0.033% MMS. (B) DNA content analysis by FACScan. Analysis of PP1 untreated cells is also shown. (C) Heat maps of BrdU incorporation levels at origins are arranged according to each origin’s published replication timing from early to late (left to right). (D) Aliquots were pulsed with BrdU for the indicated intervals and analyzed by BrdU-IP-chip. Results for segments of chromosomes III and VI are plotted, with origin locations indicated above. Data shown are from a single representative experiment out of two replicates, except data in C, and were calculated from both replicates.
This pattern of origin firing was observed along the chromosome III and VI regions that we analyzed in detail. In PP1-treated WT and cdc7-as3 cells, BrdU-IP-chip at 10–30 min showed similar levels of DNA synthesis occurring at the early origins, ARS306 and ARS607 (Fig. 1 D). However, at 10–30 and 25–45 min, BrdU incorporation at slightly later origins, ARS603.5 and ARS605, was diminished in cdc7-as3 cells, consistent with depletion of Cdc7 function. The activity of the earliest origins may reflect residual activity of Cdc7-as3 resistant to PP1 (perhaps bound to ATP) or instead may reflect the execution of Cdc7 function at these origins before Cdc7 depletion in late G1. We exploited this early origin firing to examine the consequences of Cdc7 inactivation on fork progression. During the 25–45- and 40–60-min intervals, the extent of BrdU incorporation along the ARS306 and ARS607 replicons was greater in cdc7-as3 than WT cells, suggesting a faster rate of replication fork progression in Cdc7-depleted cells. The distal BrdU incorporation apparent in WT cells during the 55–75-min pulse likely reflects subtelomeric origin activity. We also compared fork progression in WT and cdc7-as3 cells by analyzing the cumulative incorporation of BrdU over time (Fig. S1 B). This method yielded similar results as the pulse-labeling approach, showing more rapid progression of replication forks through the ARS306 and ARS607 replicons in cdc7-as3 than in WT cells. Together, these results indicate that Cdc7 is dispensable for replication fork progression and suggest that Cdc7 regulates the rate of replication fork progression along an undamaged DNA template.
Next, we analyzed whether Cdc7 function regulates the progression of replication forks traversing a damaged DNA template. G1-synchronized WT and cdc7-as3 cells were treated with PP1 and released into S phase in the presence of the DNA alkylating agent methyl-methane-sulfonate (MMS). Aliquots of each culture were pulsed with BrdU at defined intervals (Fig. 1 A). FACScan analysis showed slower progression of WT and cdc7-as3 cells through S phase as expected because of the presence of MMS, with somewhat slower bulk DNA replication in the cdc7-as3 cells, consistent with their reduced efficacy of origin firing (Fig. 1 B). We estimated that 219 origins fired in WT, and 134 fired in cdc7-as3 cells treated with MMS, and the efficiency of origin firing based on the level of BrdU incorporation was lower at most origins in cdc7-as3 cells (Fig. 1 C). The number of origins detected in MMS-treated versus untreated cells was only modestly decreased because the measurement in cells without MMS did not effectively detect later firing origins. However, comparison of WT and checkpoint-defective (mec1-100) cells indicates that >90 origins are detected as checkpoint inhibited by MMS treatment by our analysis (see Fig. 4).
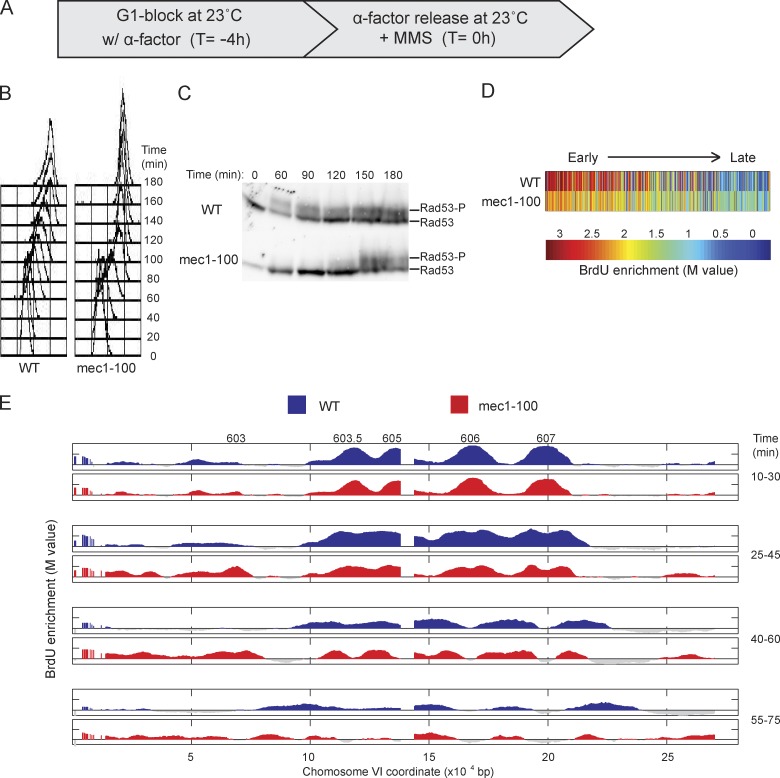
Figure 4.
Deregulated origin firing in mec1-100 slows replication forks. (A) WT and mec1-100 cells were synchronized with α-factor for 4 h at 23°C and released from α-factor at 23°C with 0.033% MMS. (B) DNA content analysis by FACScan. (C) Immunoblot analysis of phosphorylated Rad53 (Rad53-P); both rows are from the same blot and exposure. Molecular weight markers were not run on this gel; for the migration of size markers relative to the bands detected by this antibody, see Fig. 2 F. (D) Heat maps of BrdU incorporation levels at origins are arranged according to each origin’s published replication timing from early to late (left to right). (E) Aliquots were pulsed with BrdU for the indicated intervals and analyzed by BrdU-IP-chip. Results for entire chromosome VI are plotted, with origin locations indicated above. Data shown are from a single representative experiment out of two replicates, except data in D, and were calculated from both replicates.
In WT and cdc7-as3 cells, BrdU incorporation was similar for ARS306 and ARS607 at 10–30 min, whereas BrdU incorporation at the slightly later ARS603.5 and ARS605 was reduced in cdc7-as3 cells (Fig. 1 D). As in the absence of MMS, the rate of BrdU incorporation along the ARS306 and ARS607 replicons was greater in cdc7-as3 than in WT cells. We estimated replication fork rates in MMS using regression analysis based on the leading edge of BrdU incorporation across the ARS607 to chromosome VI right arm region (see Materials and methods). The firing of subtelomeric origins in WT cells precluded unambiguous determination of fork rate in the absence of MMS; however, the presence of MMS facilitated this analysis by inhibiting firing of subtelomeric origins through the intra-S checkpoint (Tercero et al., 2003). This analysis yielded a mean rate of 446 bp/min in MMS-treated WT cells and 1,031 bp/min in MMS-treated cdc7-as3 cells. The more rapid progression of BrdU incorporation in cdc7-as3 cells was dependent on PP1 (Fig. S1 E). Thus, Cdc7 is required for the normal rate of replication fork progression along undamaged and MMS-damaged templates.
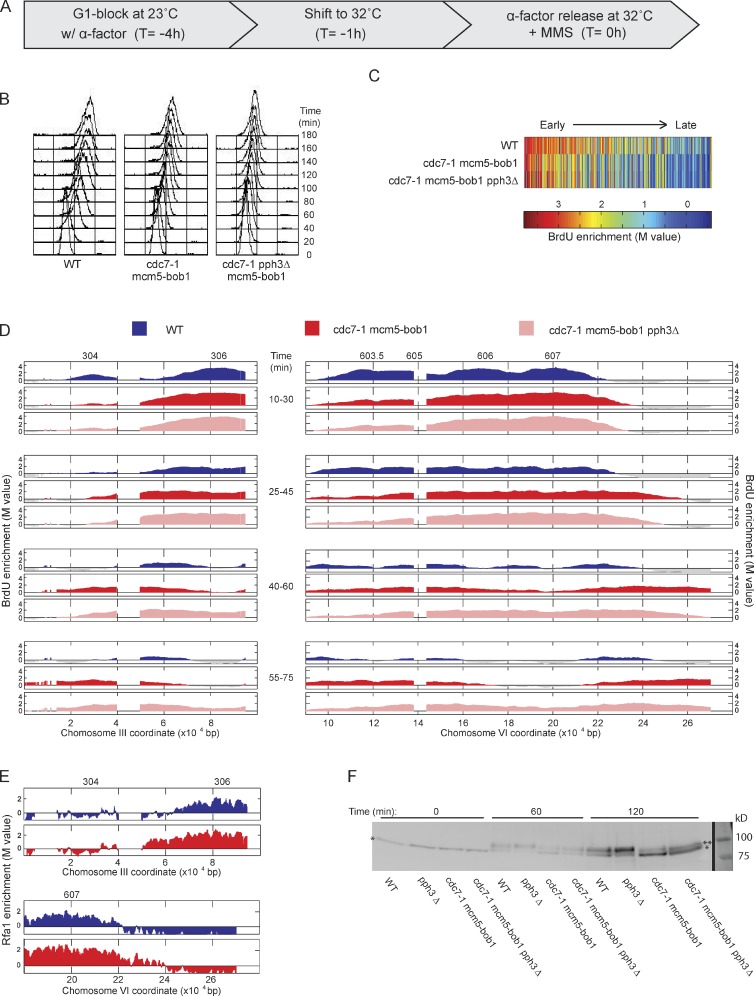
To corroborate these unexpected findings, we used the cdc7-1 allele in a similar analysis. G1-synchronized WT and cdc7-1 mcm5-bob1 cells were shifted to 32°C for 1 h before release into S phase in the presence of MMS and pulsed with BrdU (Fig. 2 A). Total DNA content analysis showed similar slow rates of DNA synthesis in WT and cdc7-1 mcm5-bob1 cells in the presence of MMS (Fig. 2 B), whereas cdc7-1 cells lacking the mcm5-bob1 suppressor allele showed tight arrest of DNA synthesis (Fig. S2 B), demonstrating the effective inhibition of Cdc7-1 function under these conditions, and at least partial restoration of origin firing by mcm5-bob1, as reported previously (Hoang et al., 2007). BrdU incorporation showed similar effects on origin firing of Cdc7 inhibition by cdc7-1 mcm5-bob1 as by cdc7-as3; we estimated that 221 origins fired in WT, and 175 fired in cdc7-1 mcm5-bob1 cells. As in cdc7-as3 cells, the level of BrdU incorporation at most origins was decreased; however, the earliest origins were least affected (Fig. 2 C). This result is consistent with the previous demonstration that the earliest origins are partially resistant to elimination of Cdc7 function in the presence of the mcm5-bob1 allele; the mcm5-bob1 allele alone does not affect origin firing (Hoang et al., 2007).
Figure 2.
Cdc7 functions upstream of Rad53 in fork regulation. (A) WT, cdc7-1 mcm5-bob1, and cdc7-1 mcm5-bob1 pph3Δ cells were synchronized with α-factor for 3 h at 23°C, shifted to 32°C for 1 h, and released from α-factor at 32°C with 0.033% MMS. (B) DNA content analysis by FACScan. (C) Heat maps of BrdU incorporation levels at origins are arranged according to each origin’s published replication timing from early to late (left to right). (D) Aliquots were pulsed with BrdU for the indicated intervals and analyzed by BrdU-IP-chip. Results for segments of chromosomes III and VI are plotted, with origin locations indicated above. (E) WT and cdc7-1 mcm5-bob1 cells expressing Rfa1-Myc18 were treated as in A and analyzed by ChIP-chip 35 min after release; plots are color coded as in D. (F) Immunoblot analysis of unphosphorylated (single asterisks) and phosphorylated Rad53 (double asterisk); molecular mass markers were visualized with Ponceau S. Black line indicates that intervening lanes have been spliced out. Data shown are from a single representative experiment out of two replicates, except data in C, and were calculated from both replicates.
BrdU incorporation at ARS306 and ARS607 was similar between WT and cdc7-1 mcm5-bob1 cells, whereas the firing of the slightly later origins was compromised specifically in the cdc7-1 mcm5-bob1 cells (Fig. 2 D). The incorporation of BrdU along the chromosome III and VI replicons progressed more rapidly in cdc7-1 mcm5-bob1 than WT cells, with rates of 1,364 and 541 bp/min, respectively (Fig. 2 D). The more rapid BrdU incorporation in the cdc7-1 mcm5-bob1 strain was accompanied by association of replication protein A, which binds single-stranded DNA at replication forks, consistent with more rapid progression of bona fide replication forks (Fig. 2 E). These observations support the results with the cdc7-as3 allele and indicate a role for DDK in regulating the rate of fork progression.
Cdc7 acts upstream of Rad53 in fork regulation
The aforementioned results show that Cdc7 function controls the rate of replication fork progression on undamaged and MMS-damaged DNA. We showed previously that deactivation of Rad53 promotes progression of forks slowed in response to MMS (Szyjka et al., 2008), and previous studies have shown differences in Rad53 activation in cells lacking Cdc7 activity (Tercero et al., 2003; Dohrmann and Sclafani, 2006; Ogi et al., 2008; Sheu and Stillman, 2010). Thus, reduced Cdc7 activity might permit rapid replication fork progression through damaged DNA by diminishing checkpoint signaling leading to Rad53 activation. To investigate whether Cdc7 depletion affects the level of checkpoint activation, we examined Rad53 activation as reflected in its altered electrophoretic mobility caused by its phosphorylation (Pellicioli et al., 1999). Under the conditions of the aforementioned experiments, Rad53 activation was reduced in cdc7-1 mcm5-bob1 compared with WT cells based on the smaller proportion of slower to faster migrating Rad53 (Fig. 2 F). This result is consistent with previous findings that Cdc7 is required for the normal level of checkpoint activity in cells undergoing replication stress and is consistent with the idea that reduced checkpoint activation resulting from reduced Cdc7 activity affects the rate of fork progression.
A previous study concluded that the requirement of Cdc7 for checkpoint activation in response to DNA damage reflects its function in initiation and the establishment of replication forks (Tercero et al., 2003). Cdc7 activity also is regulated as a direct target of the checkpoint via Rad53-dependent phosphorylation of Dbf4, which inhibits DDK function in origin firing. If Cdc7 is upstream of Rad53 in activation of the checkpoint, fork slowing should occur in response to increased Rad53 activity, even in the absence of Cdc7 function. Conversely, if Cdc7 activity is a downstream target or effector of the checkpoint required for regulation of fork rate, increased Rad53 activity should fail to slow forks in the absence of Cdc7 function. To increase Rad53 activity, we exploited our previous finding that deletion of the Rad53 phosphatase PPH3 results in Rad53 hyperactivity and slower replication fork progression in MMS (O’Neill et al., 2007; Szyjka et al., 2008). As predicted, pph3Δ resulted in increased Rad53 activity in WT and Cdc7-deficient, MMS-treated cells, although the level of Rad53 activity was lower in cells lacking Cdc7 function, consistent with reduced origin activation (Fig. 2 F). The increased Rad53 activity correlated with slower bulk DNA replication and slower fork progression (Fig. 2, B and D). Thus, enhanced Rad53 activity slows forks in the absence of Cdc7 activity, which is consistent with Cdc7 acting upstream of Rad53 in fork slowing in response to replication stress.
Decreased initiation from Orc1 depletion also deregulates fork progression
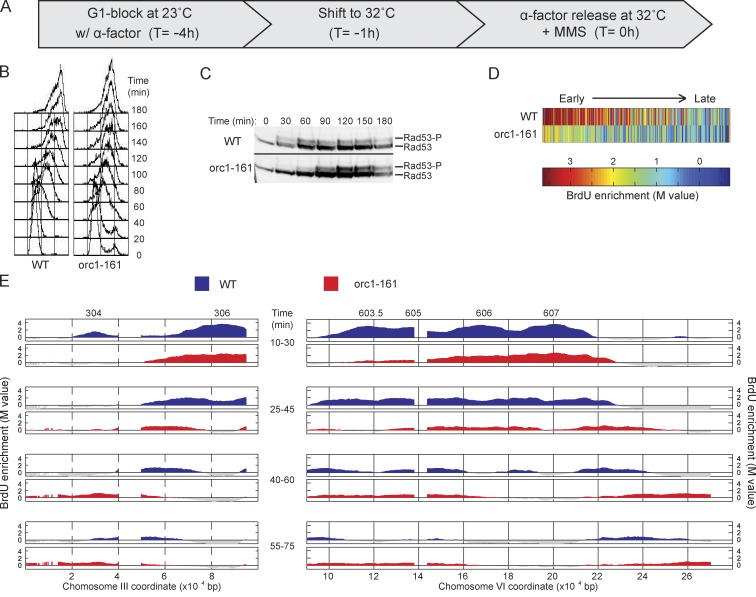
Our results suggest that the deregulated fork progression of Cdc7-depleted cells derives from Cdc7’s function in replication initiation. To address whether a decreased level of replication initiation is sufficient to deregulate fork progression, we examined fork progression in cells harboring orc1-161, a temperature-sensitive allele of ORC1, which is required for replication initiation; incubation of G1-arrested orc1-161 cells at the nonpermissive temperature reduces MCM occupancy at origins (Aparicio et al., 1997; Gibson et al., 2006). We performed the same temperature shift regimen and release into MMS as we did for the cdc7-1 cells (Fig. 3 A). Total DNA content analysis showed diminished progression through S phase of orc1-161 cells compared with WT, consistent with reduced origin firing in the mutant cells (Fig. 3 B). Rad53 activation also was reduced in orc1-161 cells, only reaching levels comparable to those of WT cells at ∼90 min (Fig. 3 C). Interestingly, these higher levels of Rad53 activation coincided with reduced progression of total DNA content in orc1-161 cells at these later times (Fig. 3 B), consistent with checkpoint regulation of fork rates (analysis of fork rates by BrdU-IP is not feasible at these later times). Analysis of BrdU incorporation showed a global reduction in the number of origins that fired and their BrdU incorporation levels in orc1-161 cells, consistent with depletion of Orc1 function. We estimated that 230 origins fired in WT, and 192 fired in orc1-161 cells. BrdU incorporation levels were also lower at most origins, including very early origins, which were only modestly affected in Cdc7-depleted cells (Fig. 3 D).
Figure 3.
Orc1 function regulates replication fork progression. (A) WT and orc1-161 cells were synchronized with α-factor for 3 h at 23°C, shifted to 32°C for 1 h, and released from α-factor at 32°C with 0.033% MMS. (B) DNA content analysis by FACScan. (C) Immunoblot analysis of phosphorylated Rad53 (Rad53-P); both rows are from the same blot and exposure. Molecular weight markers were not run on this gel; for the migration of size markers relative to the bands detected by this antibody, see Fig. 2 F. Black line indicates that intervening lanes have been spliced out. (D) Heat maps of BrdU incorporation levels at origins are arranged according to each origin’s published replication timing from early to late (left to right). (E) Aliquots were pulsed with BrdU for the indicated intervals and analyzed by BrdU-IP-chip. Results for segments of chromosomes III and VI are plotted, with origin locations indicated above. Data shown are from a single representative experiment out of two replicates, except data in D, and were calculated from both replicates.
Cells with diminished Orc1 activity exhibited initiation of ARS306 and ARS607 along with reduced initiation of the slightly later origins (ARS603.5 and ARS605; Fig. 3 E). Inactivation of Orc1 also affected the rate of fork progression, like Cdc7 inactivation, with a mean rate of 1,202 bp/min compared with 732 bp/min in WT cells (Fig. 3 E). Given the distinct roles of Cdc7 and Orc1 in replication initiation, we conclude that the common deficiency in origin activation best explains the diminished Rad53 activation and rapid fork rate.
Checkpoint elimination is not sufficient to deregulate fork rate
We have shown that decreased levels of initiation result in decreased Rad53 activation levels and faster fork rates. However, another feature of reduced Cdc7 and Orc1 activity that we hypothesized might contribute to faster fork rates is the reduced overall number of active forks, which might increase the availability of normally rate-limiting factors to the fewer active forks. To evaluate the effect of the number of active forks, we examined mec1-100 cells, which only weakly activate Rad53 in response to MMS (but sufficiently to maintain fork stability), while activating a larger than normal complement of origins, including late and normally dormant origins (Paciotti et al., 2001; Tercero et al., 2003). Therefore, these cells allow us to test the effect of higher numbers of active forks in combination with low levels of active Rad53. As shown previously, G1-synchronized mec1-100 cells released into MMS (Fig. 4 A) exhibit more rapid progression through S phase as measured by total DNA content (Fig. 4 B) and decreased Rad53 activation (Fig. 4 C; Paciotti et al., 2001). BrdU incorporation analysis showed increased numbers of active origins genomewide in mec1-100 cells, with 219 firing in WT and 310 in mec1-100 cells, the latter including many later origins (Fig. 4 D). Analysis of the chromosome III and VI regions showed similar levels of BrdU incorporation at earlier origins and higher levels at late origins like ARS603 in mec1-100 cells (Fig. 4 E). Replication forks progressed more slowly in mec1-100 cells than in WT cells (Fig. 4 E), with rates of 242 and 517 bp/min, respectively, despite lower levels of Rad53 activation in mec1-100 cells. We have observed similar BrdU incorporation profiles as in mec1-100 cells, including more origins firing and slower forks, in other intra-S checkpoint mutant strains, including rad53Δ and rad53Δ exo1Δ (EXO1 deletion suppresses the MMS sensitivity of rad53Δ cells; Fig. S3; Segurado and Diffley, 2008). A recent study in human cells reported slower fork progression in Ckh1-depleted cells, which was suppressed by additional depletion of Cdc7 activity (Petermann et al., 2010). These findings suggest that increased numbers of replication forks suppress more rapid fork progression, perhaps by depleting essential factors.
Replication fork and checkpoint levels regulate replication fork progression
Comparison of origin firing rates and replication fork rates across the experiments in MMS supports a model in which the rate of replication fork progression is inversely related to the number of active replication forks, which is determined by the level of origin firing (Fig. 5). We propose that the number of active forks influences overall fork rate in checkpoint-dependent and -independent ways. Robust checkpoint activation associated with substantial numbers of replication forks encountering DNA damage slows fork progression. Additionally, large numbers of forks deplete available replication factors or deoxynucleoside triphosphates, which limits fork rate even with a reduced or absent checkpoint. However, when fork numbers are reduced, as in cdc7 and orc1 mutant cells, reduced checkpoint activation and reduced competition from other forks for limiting factors allow more avid fork progression. In mec1-100 cells, where deficiency of Rad53 activation is associated with an excess of replication forks, replication factor depletion results in slower fork progression despite the lack of checkpoint activation. This model is based in part on our previous demonstration that suppression of Rad53 activity restores robust fork progression through MMS-damaged DNA (Szyjka et al., 2008). Further supporting the idea that fork rate is under checkpoint regulation, a recent study has shown that Ckh2 kinase (the metazoan equivalent of Rad53) inhibits the replicative helicase complex (Cdc45–MCM–GINS; Ilves et al., 2012). In addition, recent studies have shown that DDK and several other replication proteins, as well as deoxynucleoside triphosphates, are rate limiting for chromosomal DNA replication in yeast (Patel et al., 2008; Mantiero et al., 2011; Tanaka et al., 2011; Poli et al., 2012). Collectively, we conclude that replication fork rate is sensitive to levels of origin firing and checkpoint activity.
Figure 5.
Replication fork and checkpoint levels regulate replication fork progression. (A) Genome-wide origin firing and local fork rate for the experiments in MMS are plotted; mean and standard deviation are shown (n = 2). Data points are color coded for the experimental group represented. (B) The model depicts fork rate regulation in WT and mutant strains with different levels of origin firing and checkpoint functions. The font intensities and line/arrow thicknesses represent the relative strength of the corresponding pathway or signal. For example, gray fonts indicate a weak or defective function or pathway, and bold fonts indicate a hyperactive function or pathway. The chromosome graphic below each model depicts the levels of origin firing and fork rate in each condition. Open circles represent fired origins, and filled circles represent unfired origins; E, M, and L indicate early, middle, and late firing origins, respectively.
Materials and methods
Plasmid and strain constructions
All strains are derived from W303 and are described in Table 1. Gene disruptions were constructed by PCR-based methods (Güldener et al., 1996; Longtine et al., 1998). Plasmid p306-ars305Δ-BrdU-Inc was constructed by three-way ligation of 620-bp NotI–BglII PCR-amplified fragment 5′-flanking ARS305 and 560-bp BglII–SacI PCR-amplified fragment 3′-flanking ARS305 into NotI–SacI-digested p306-BrdU-Inc (Viggiani and Aparicio, 2006). Plasmid p306-ars305Δ-BrdU-Inc digested with BglII was used to simultaneously delete ARS305 and integrate the 10.3-kb plasmid with the BrdU-Inc cassette by gene replacement. Correct replacement was confirmed by PCR. The 1.6-kb HindIII–EcoRI fragment of cdc7-as3 containing the kinase-inactivating mutations L120A and V181A was isolated from pRS551-cdc7-as3 (L120A and V181A; a gift from N. Hollingsworth, Stony Brook University, Stony Brook, NY; Wan et al., 2006) and subcloned into EcoRI–HindIII-digested pRS306. The resulting plasmid, pRS306-cdc7-as3, was linearized with EcoRI and used to exchange CDC7 with cdc7-as3 by pop-in/pop-out replacement. pPP117, which contains a 3.6-kb EcoRI–SalI cdc7-1 fragment from pRH301 (Hollingsworth et al., 1992) in URA3 integrating vector pRS306, was linearized with ClaI and used to exchange CDC7 with cdc7-1 by pop-in/pop-out replacement followed by screening for temperature sensitivity at 37°C. The resultant cdc7-1 strain was then transformed with MluI-digested pRAS490 (Dohrmann and Sclafani, 2006), which contains mcm5-bob1-2 (CT to TC change at codon 83 to create the DdeI site and P83L mutation) in pRS306, to exchange MCM5 with mcm5-bob1-2 by pop-in/pop-out replacement followed by screening for suppression of temperature sensitivity at 37°C. The 7.5-kb SacI–SpeI fragment containing the mec1-100 allele was isolated from plasmid pML258.51 (a gift from M.P. Longhese, University of Milan, Milan, Italy; Paciotti et al., 2001) and subcloned into SacI–SpeI-digested pRS406. The resulting plasmid, pRS406-mec1-100, was linearized with BstEII and used to exchange MEC1 with mec1-100 by pop-in/pop-out replacement. All allele replacements were confirmed by DNA sequencing.
Table 1.
List of strains used in this study
| Strain | Genotype |
| JPy8 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) RFA1-18Myc (KanMX) |
| JPy9 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) RFA1-18Myc (KanMX) cdc7-1 mcm5-bob1 |
| JYy3 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) sml1Δ::HIS3 |
| JYy4 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) sml1Δ::HIS3 exo1Δ::TRP1 |
| RSy1298 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) cdc7-1 |
| RSy1307 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) cdc7-1 mcm5-bob1 |
| T2y41 | ars608Δ::HIS3 ars305Δ::BrdU-Inc (KanMX) orc1Δ::hisG leu2::ORC1 (LEU2) |
| T2y42 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (KanMX) orc1Δ::hisG leu2::orc1-161 (LEU2) |
| YZy2 | lys2Δ::hisG trp1::BrdU-Inc (TRP1) orc1Δ::hisG leu2::ORC1 (LEU2) |
| YZy3 | lys2Δ::hisG trp1::BrdU-Inc (TRP1) orc1Δ::hisG leu2::orc1-161 (LEU2) |
| YZy8 | ars608Δ::HIS3 ars609Δ::TRP1 leu2::BrdU-Inc (LEU2) |
| YZy10 | ars608Δ::HIS3 ars609Δ::TRP1 leu2::BrdU-Inc (LEU2) cdc7-as3 |
| YZy18 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (URA3) leu2::BrdU-Inc (LEU2) |
| YZy19 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (URA3) leu2::BrdU-Inc (LEU2) cdc7-as3 |
| YZy34 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) pph3Δ::KanMX |
| YZy35 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) cdc7-1 mcm5-bob1 pph3Δ::KanMX |
| YZy50 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) |
| YZy52 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) mec1-100 |
| YZy60 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) bar1Δ::LEU2 sml1Δ::HIS3 rad53Δ::KanMX exo1Δ::TRP1 |
| YZy61 | ars608Δ::HIS3 ars609Δ::TRP1 ars305Δ::BrdU-Inc (TRP1) bar1Δ::LEU2 sml1Δ::HIS3 rad53Δ::KanMX |
All strains share the W303a RAD5 genotype MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::hisG except where noted.
Yeast methods
Cells were grown in YEPD (yeast extract, peptone, and dextrose) for all experiments. Cell were synchronized in G1 phase by incubation with 5 nM α-factor (T6901; Sigma-Aldrich) for 4 h at 23°C and released by resuspension and gentle sonication in fresh YEPD lacking α-factor and containing 200 µg/ml Pronase E (P5147; Sigma-Aldrich). For DNA content analysis, cells were fixed with 70% ethanol overnight, washed, and resuspended in 50 mM sodium citrate, pH 7.4, and RNase A was added to 0.2 mg/ml and incubated for 3 h at 50°C. Proteinase K was added to 0.5 mg/ml and incubated 50°C for 2 h, after which SYTOX Green (Molecular Probes) was added to 1 µM for at least 30 min before analysis on a FACScan instrument (BD). PP1 (Tocris Biosciences) was used at 25 µM. For BrdU-IP-chip, 20 ml of culture (OD of ∼1) was pulse labeled with 800 µg/ml BrdU (B5002; Sigma-Aldrich) and harvested with addition of NaN3 to 0.1%, and genomic DNA was prepared by disruption with glass beads. 1 µg genomic DNA was sonicated to ∼500 bp, denatured, and immunoprecipitated with the anti-BrdU antibody (RPN202; GE Healthcare) at 1:1,000. For chromatin IP (ChIP) analyzed by microarray (ChIP-chip), 50 ml of culture (OD of ∼1) was fixed with formaldehyde, and chromatin was isolated by disruption with glass beads and sonicated to ∼500 bp. Chromatin was immunoprecipitated with the anti-Myc 9E10 antibody (MMS150; Covance) at 1:100. Immunoprecipitated and total DNA samples (from BrdU-IP-chip and ChIP-chip) were amplified using WGA2 (Sigma-Aldrich), labeled with Cy5 and Cy3, respectively, and hybridized to custom-designed oligonucleotide-based tiling microarrays (Roche) using the hybridization system (Maui; Roche) according to the manufacturer’s instructions; further details are provided in Knott et al. (2012) and Viggiani et al. (2009, 2010). Rad53 immunoblot analysis was performed with anti-Rad53 at 1:1,000 (SC6749; Santa Cruz Biotechnology, Inc.) as described previously (Gibson et al., 2004).
Microarray normalization
BrdU-IP-chip normalization from the Roche arrays was performed as previously described (Knott et al., 2009). In brief, probes from the most dense regions of the corresponding M-A plot were isolated, and principle component analysis was performed on their corresponding M (= log(IP/Total)) and A (= log(IP × Total)) values. The resultant first and second principle components were then taken to represent each probe’s normalized A and M values, respectively. After this, loess normalization was performed to remove any residual array artifacts (Smyth and Speed, 2003). Analysis of Rfa1 ChIP was performed using MA2C (Song et al., 2007).
Data filtering
We used the values of ϕ = exp(M) obtained from the previous step in the subsequent analysis. An enriched probe was defined as one with a ϕ value of >1. An enriched region was defined as a sequence of consecutive enriched probes. Each enriched region was given an enrichment score (E score), which was the sum of the ϕ values of the probes within the region. In most cases, there was a single, clearly enriched region of BrdU signal to the right of ARS607. In cases with more than one enriched region, for the purpose of estimating the fork speed, we chose the region with the maximum E score. The sixth column of Table S1 indicates which time intervals were included in the analysis.
Fork rate analysis
For each experiment, we examined the ϕ values in the single enriched region identified in the previous paragraph. We assumed that the probability of having a fork in the interval defined by the position of a single probe is proportional to the ϕ value of that probe. To estimate the leading edge of the replication fork, we used P, the 90th percentile of the resulting probability distribution. For each experiment, we write T for the mean time of the BrdU pulse (if the pulse occurred between time points a and b, T = 1/2(a + b)). For each strain and experimental condition, we calculate the values of P and T and fit a linear regression of the form P = u + vT to the data, in which u and v are coefficients in the linear regression model. We obtained estimated value of u as û and v as The estimate of the fork rate is given by We note an analysis along the same lines, but using the values of M in place of ϕ gives essentially the same results.
Origin firing analysis
Using the piecewise cubic Hermite interpolating polynomial, we interpolated the normalized and smoothed M value of the probes for every 10 bp of the genome. Under the null hypothesis of no enrichment around an origin, the sum of the N interpolated M values of the probes in this region will have approximately a Normal distribution with mean μ and variance σ2, in which μ and σ2 are the mean and variance of a typical interpolated M value. Using all the observed M values, we can estimate the μ and σ2.
For each origin, the sum S of signal from the N interpolated probes within a distance of 1,500 bp was calculated. An origin is determined to have significantly enriched signal if S lies in the tails of the null distribution, using a significance cutoff of 0.05. We use a Bonferroni correction for multiple testing. In all cases, we either used the earliest time point or the sum of two earliest time points as a representative early signal for determining whether the origin fired. The third column of Table S1 indicates which time intervals were included in this analysis. The color of individual cells in the heat maps in Figs. 1, 2, 3, and 4 represent this sum S assigned to each origin. The fourth column of Table S1 indicates which time intervals were included in calculating S for the heat map visualizations. We used the origin dataset from Knott et al. (2012) and timing data from Raghuraman et al. (2001).
Online supplemental material
Fig. S1 shows analysis of cumulative BrdU incorporation in cdc7-as3. Fig. S2 shows DNA content analysis of cdc7-1. Fig. S3 shows BrdU incorporation analysis of rad53Δ and rad53Δexo1Δ. Table S1 summarizes data used to calculate fork rates, origin firing, and BrdU signal at origins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201208060/DC1.
Supplementary Material
Acknowledgments
We thank N. Hollingsworth and M.P. Longhese for plasmids and S.L. Forsburg, M. Michael, and J. Bachant for critical reading of the manuscript.
This study was supported by National Institutes of Health grants 5R01-GM065494 and 3R01-GM065494-S1 (to O.M. Aparicio), P50-HG002790 (to T. Nellimoottil, S.R.V. Knott, and S. Tavaré), and 5R01-GM035078-22 (to R.A. Sclafani).
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin IP
- DDK
- Dbf4-dependent kinase
- IP
- immunoprecipitation
- MCM
- minichromosome maintenance
- MMS
- methyl-methane-sulfonate
- WT
- wild type
References
- Aparicio O.M., Weinstein D.M., Bell S.P. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 91:59–69 10.1016/S0092-8674(01)80009-X [DOI] [PubMed] [Google Scholar]
- Dohrmann P.R., Sclafani R.A. 2006. Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae. Genetics. 174:87–99 10.1534/genetics.106.060236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker B.P., Brown G.W. 2003. Cdc7 kinases (DDKs) and checkpoint responses: lessons from two yeasts. Mutat. Res. 532:21–27 10.1016/j.mrfmmm.2003.08.007 [DOI] [PubMed] [Google Scholar]
- Fletcher R.J., Bishop B.E., Leon R.P., Sclafani R.A., Ogata C.M., Chen X.S. 2003. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 10:160–167 10.1038/nsb893 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Aparicio J.G., Hu F., Aparicio O.M. 2004. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:10208–10222 10.1128/MCB.24.23.10208-10222.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Bell S.P., Aparicio O.M. 2006. Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes Cells. 11:557–573 10.1111/j.1365-2443.2006.00967.x [DOI] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524 10.1093/nar/24.13.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F., Dryga O., Seematter S., Pahl P.M., Sclafani R.A. 1997. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA. 94:3151–3155 10.1073/pnas.94.7.3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M.L., Leon R.P., Pessoa-Brandao L., Hunt S., Raghuraman M.K., Fangman W.L., Brewer B.J., Sclafani R.A. 2007. Structural changes in Mcm5 protein bypass Cdc7-Dbf4 function and reduce replication origin efficiency in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:7594–7602 10.1128/MCB.00997-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth R.E., Jr, Ostroff R.M., Klein M.B., Niswander L.A., Sclafani R.A. 1992. Molecular genetic studies of the Cdc7 protein kinase and induced mutagenesis in yeast. Genetics. 132:53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I., Tamberg N., Botchan M.R. 2012. Checkpoint kinase 2 (Chk2) inhibits the activity of the Cdc45/MCM2-7/GINS (CMG) replicative helicase complex. Proc. Natl. Acad. Sci. USA. 109:13163–13170 10.1073/pnas.1211525109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott S.R., Viggiani C.J., Aparicio O.M., Tavaré S. 2009. Strategies for analyzing highly enriched IP-chip datasets. BMC Bioinformatics. 10:305 10.1186/1471-2105-10-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott S.R., Peace J.M., Ostrow A.Z., Gan Y., Rex A.E., Viggiani C.J., Tavaré S., Aparicio O.M. 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 148:99–111 10.1016/j.cell.2011.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24:1208–1219 10.1101/gad.1933010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J., Maas N.L., Jonsson Z.O., Defazio-Eli L.G., Wohlschlegel J., Toczyski D.P. 2010. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature. 467:479–483 10.1038/nature09377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D., Mackenzie A., Donaldson A., Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 30:4805–4814 10.1038/emboj.2011.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Arai K. 2002. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell. Physiol. 190:287–296 10.1002/jcp.10070 [DOI] [PubMed] [Google Scholar]
- Ogi H., Wang C.Z., Nakai W., Kawasaki Y., Masumoto H. 2008. The role of the Saccharomyces cerevisiae Cdc7-Dbf4 complex in the replication checkpoint. Gene. 414:32–40 10.1016/j.gene.2008.02.010 [DOI] [PubMed] [Google Scholar]
- O’Neill B.M., Szyjka S.J., Lis E.T., Bailey A.O., Yates J.R., III, Aparicio O.M., Romesberg F.E. 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. USA. 104:9290–9295 10.1073/pnas.0703252104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro G., Owens J.C., Shellman Y., Sclafani R.A., Li J.J. 1999. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol. 19:4888–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Clerici M., Scotti M., Lucchini G., Longhese M.P. 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21:3913–3925 10.1128/MCB.21.12.3913-3925.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P.K., Kommajosyula N., Rosebrock A., Bensimon A., Leatherwood J., Bechhoefer J., Rhind N. 2008. The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol. Biol. Cell. 19:5550–5558 10.1091/mbc.E08-06-0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561–6572 10.1093/emboj/18.22.6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Woodcock M., Helleday T. 2010. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. USA. 107:16090–16095 10.1073/pnas.1005031107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J., Tsaponina O., Crabbé L., Keszthelyi A., Pantesco V., Chabes A., Lengronne A., Pasero P. 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 31:883–894 10.1038/emboj.2011.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L. 2001. Replication dynamics of the yeast genome. Science. 294:115–121 10.1126/science.294.5540.115 [DOI] [PubMed] [Google Scholar]
- Segurado M., Diffley J.F. 2008. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 22:1816–1827 10.1101/gad.477208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y.J., Stillman B. 2006. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 24:101–113 10.1016/j.molcel.2006.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y.J., Stillman B. 2010. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 463:113–117 10.1038/nature08647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K., Speed T. 2003. Normalization of cDNA microarray data. Methods. 31:265–273 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- Song J.S., Johnson W.E., Zhu X., Zhang X., Li W., Manrai A.K., Liu J.S., Chen R., Liu X.S. 2007. Model-based analysis of two-color arrays (MA2C). Genome Biol. 8:R178 10.1186/gb-2007-8-8-r178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjka S.J., Aparicio J.G., Viggiani C.J., Knott S., Xu W., Tavaré S., Aparicio O.M. 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 22:1906–1920 10.1101/gad.1660408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Nakato R., Katou Y., Shirahige K., Araki H. 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21:2055–2063 10.1016/j.cub.2011.11.038 [DOI] [PubMed] [Google Scholar]
- Tercero J.A., Longhese M.P., Diffley J.F. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 11:1323–1336 10.1016/S1097-2765(03)00169-2 [DOI] [PubMed] [Google Scholar]
- Viggiani C.J., Aparicio O.M. 2006. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast. 23:1045–1051 10.1002/yea.1406 [DOI] [PubMed] [Google Scholar]
- Viggiani C.J., Aparicio J.G., Aparicio O.M. 2009. ChIP-chip to analyze the binding of replication proteins to chromatin using oligonucleotide DNA microarrays. Methods Mol. Biol. 521:255–278 10.1007/978-1-60327-815-7_14 [DOI] [PubMed] [Google Scholar]
- Viggiani C.J., Knott S.R., Aparicio O.M. 2010. Genome-wide analysis of DNA synthesis by BrdU immunoprecipitation on tiling microarrays (BrdU-IP-chip) in Saccharomyces cerevisiae. Cold Spring Harb Protoc. 2010:pdb.prot5385 10.1101/pdb.prot5385 [DOI] [PubMed] [Google Scholar]
- Wan L., Zhang C., Shokat K.M., Hollingsworth N.M. 2006. Chemical inactivation of cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 174:1767–1774 10.1534/genetics.106.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Stillman B. 1999. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18:5334–5346 10.1093/emboj/18.19.5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J.F. 2010. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 467:474–478 10.1038/nature09373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.