Metalloproteinase-mediated cleavage of EphA2 induces breast tumor cells to shift from collective invasion to single-cell invasion.
Abstract
Changes in EphA2 signaling can affect cancer cell–cell communication and motility through effects on actomyosin contractility. However, the underlying cell–surface interactions and molecular mechanisms of how EphA2 mediates these effects have remained unclear. We demonstrate here that EphA2 and membrane-anchored membrane type-1 matrix metalloproteinase (MT1-MMP) were selectively up-regulated and coexpressed in invasive breast carcinoma cells, where, upon physical interaction in same cell–surface complexes, MT1-MMP cleaved EphA2 at its Fibronectin type-III domain 1. This cleavage, coupled with EphA2-dependent Src activation, triggered intracellular EphA2 translocation, as well as an increase in RhoA activity and cell junction disassembly, which suggests an overall repulsive effect between cells. Consistent with this, cleavage-prone EphA2-D359I mutant shifted breast carcinoma cell invasion from collective to rounded single-cell invasion within collagen and in vivo. Up-regulated MT1-MMP also codistributed with intracellular EphA2 in invasive cells within human breast carcinomas. These results reveal a new proteolytic regulatory mechanism of cell–cell signaling in cancer invasion.
Introduction
Cancer metastasis involves tumor cell invasion across basement membranes and interstitial tissues. The invasion can occur by collective cell groups and by individual cells displaying either an elongated mesenchymal morphology or a less polarized rounded morphology and amoeboid movement (Friedl and Wolf, 2010; Sanz-Moreno and Marshall, 2010). Collective and mesenchymal invasion depend on the ECM proteolysis, whereas compromised proteolytic activity has been linked to a switch to amoeboid-type invasion (Sabeh et al., 2009; Sanz-Moreno and Marshall, 2010; Wolf and Friedl, 2011). Extensive evidence supports the importance of such plasticity for tumor spread and anti-cancer drug resistance (Alexander and Friedl, 2012). However, it is unclear how the ECM microenvironment or cell-surface and soluble cell migration and segregation cues regulate switches between the interchangeable modes of invasion (Giampieri et al., 2010; Friedl and Wolf, 2010; Sanz-Moreno and Marshall, 2010; Yilmaz and Christofori, 2010).
Eph receptors have emerged as important regulators of cancer cell migration and segregation through cell–cell and cell–ECM interactions (Nievergall et al., 2012). Eph binding to membrane-bound ephrin ligand induces tyrosine-kinase activation, clustering, and trans-phosphorylation of the receptors, creating docking sites for cytoplasmic signaling proteins (Himanen et al., 2007, 2010; Seiradake et al., 2010; Janes et al., 2012). This triggers bidirectional signaling in receptor- and ligand-expressing cells (Himanen et al., 2007; Pasquale, 2008). At cell–cell contacts, Eph signaling is regulated by receptor interactions and cross-talk with transmembrane cofactors including adhesion and growth factor receptors, other Eph receptors, and proteases with a disintegrin and metalloprotease domain (ADAMs; Pasquale, 2005; Himanen et al., 2007, 2010; Janes et al., 2012; Miao and Wang, 2012). However, the consequences and context-dependent effectors of Eph signaling remain unclear.
EphA2 has been linked to aggressive progression of breast, prostate, pancreatic, colon, and lung carcinoma as well as melanoma (Wykosky and Debinski, 2008; Margaryan et al., 2009; Brantley-Sieders, 2012). In breast cancer and glioblastoma, EphA2 overexpression is often coupled with low ephrinA1 expression (Macrae et al., 2005; Wykosky et al., 2005). Although this can be reflected by low receptor tyrosine phosphorylation, alternative ligand-independent signaling has also been implicated (Macrae et al., 2005; Miao et al., 2009; Hiramoto-Yamaki et al., 2010). Upon cancer cell–cell contacts, EphA2-Rho signaling regulates contact inhibition of locomotion by enhanced contractility and rounding, and EphA2 has also been linked to amoeboid movement (Parri et al., 2009; Astin et al., 2010; Taddei et al., 2011). Although EphA2 cooperates with E-cadherin in epithelial cell junctions, its interactions in cancer cell–cell contact regulation have remained unclear (Zantek et al., 1999; Miura et al., 2009). We describe here a unique protein interaction between EphA2 and membrane type-1 matrix metalloproteinase (MT1-MMP). This protease is induced at tumor edges and upon tumor cell transition to an invasive mesenchymal phenotype in multiple types of cancer including breast carcinoma (Ota et al., 2009; Sugiyama et al., 2010b). Although MT1-MMP has been reported to drive invasion of these cells largely by degrading ECM barriers, current results identify a novel activity whereby MT1-MMP regulates cell junctional dynamics and dissemination of single cells via repulsive responses triggered by EphA2 cleavage (Ota et al., 2009; Sabeh et al., 2009; Sugiyama et al., 2010b).
Results
EphA2 and MT1-MMP are coexpressed and regulate collagen invasion in breast carcinoma cells
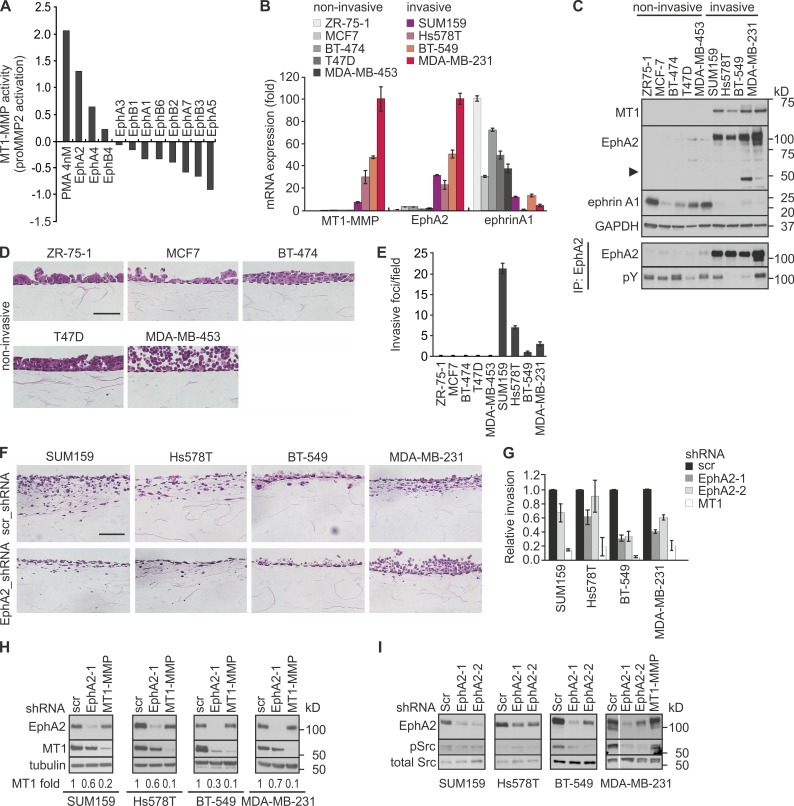
Using a human kinome cDNA library, we have identified EphA2 as an MT1-MMP regulator (Sugiyama et al., 2010a). This library contained 11 Eph receptors, of which only EphA2 increased MT1-MMP–mediated MMP-2 activation more than twofold (Fig. 1 A). To investigate the potential relevance of such regulation in cell invasion, we first analyzed the mRNA and protein expression of EphA2 and MT1-MMP in nine breast carcinoma cell lines. ZR-75-1, MCF-7, BT-474, T47D, and MDA-MB-453 cells expressed ephrinA1 along with negligible MT1-MMP and EphA2 (Fig. 1, B and C; and Fig. S1 A). Conversely, high MT1-MMP and EphA2 (relatively poorly phosphorylated) coupled with low ephrinA1 expression was observed in SUM159, Hs578T, BT-549, and MDA-MB-231 cells (Fig. 1, B and C; and Fig. S1 A). Interestingly, the coexpression of EphA2 and MT1-MMP was associated with different levels of truncated C-terminal EphA2 fragments in BT-549 and MDA-MB-231 cells (Fig. 1 C).
Figure 1.
EphA2 and MT1-MMP are coexpressed in invasive breast carcinoma cells and regulate collagen invasion. (A) EphA2 is a positive regulator of MT1-MMP activity. Charts show relative pro-MMP2 activation in cells overexpressing 11 Eph receptors as quantified by gelatin zymography in our primary genome-wide gain-of-function kinome screen for MT1-MMP activity (for each kinase, n = 1; Sugiyama et al., 2010a). The relative value of mock control has been set to zero. PMA treatment served as a positive control. (B) Relative expression of MT1-MMP, EphA2, and ephrinA1 mRNAs was assessed by qPCR in the indicated human breast carcinoma cell lines (error bars indicate mean ± SEM, n = 3). (C) The corresponding protein expression and EphA2 tyrosine phosphorylation were assessed as indicated (n = 3). The arrowhead indicates truncated EphA2. GAPDH served as a loading control. (D) Light micrographs of collagen cross sections visualize the H&E-stained noninvasive breast carcinoma cells. Broken lines indicate the surface of collagen gels. Bar, 20 µm. (E) Quantification of collagen invasion of the breast carcinoma cell lines (error bars indicate mean ± SEM, n = 3). (F) Light micrographs of collagen cross sections visualize the H&E-stained invasive cells after EphA2 knockdown (EphA2-1 shRNA). Bar, 20 µm. (G) Relative invasion of EphA2 and MT1-MMP knockdown cells (error bars indicate mean ± SEM, n = 3). Invasion of the control cells (scr shRNA) was set to one. (H) Immunoblotting for MT1-MMP and EphA2 in the stable knockdown cells (n = 3). Mean values of MT1-MMP normalized with tubulin (loading control) are indicated below each blot. (I) EphA2 silencing reduces Src activation in BT-549 and MDA-MB-231 cells. Immunoblotting for the phosphorylated (pSrc; Tyr416) and total Src in the invasive cells (n = 3) is shown.
To correlate the coexpression results to cell invasion, the cells were seeded atop 3D cross-linked collagen matrix that typifies the ECM of collagen-rich stroma (Sabeh et al., 2009; Egeblad et al., 2010). All ephrin-expressing cell lines grew in multilayer colonies but displayed negligible invasive activities into the underneath matrix (Fig. 1, D and E). In contrast, the MT1-MMP– and EphA2-coexpressing cells invaded into collagen, a result consistent with their reported invasiveness also across a laminin-rich hydrogel (Fig. 1 E; Neve et al., 2006). In our collagen assay, SUM159 and Hs578T cells invaded with elongated morphologies more efficiently than MDA-MB-231 and BT-549 cells (Fig. 1 E).
EphA2 enhances breast carcinoma cell invasion and MT1-MMP expression
To determine the effects of EphA2 on cell invasion, EphA2 and MT1-MMP were silenced by lentiviral shRNAs. EphA2 knockdown impaired SUM159 and Hs578T cell invasion into collagen, whereas BT-549 and MDA-MB-231 cell invasion was more dramatically inhibited (Fig. 1, F and G; and Fig. S1, B and C). The invasion of all these cells was also inhibited by MT1-MMP depletion, which indicates that EphA2 and MT1-MMP promoted invasion cooperatively (Fig. 1 G and S1 C). Moreover, EphA2 silencing decreased MT1-MMP protein and mRNA, thus providing one explanation for the regulation of pro-invasive MT1-MMP activity through a transcriptional response (Fig. 1 H and S1 C). Interestingly, in BT-549 and MDA-MB-231 cells, but not in SUM-159 and Hs578T cells, EphA2 knockdown decreased the activation of Src, which can regulate overall cell motility as well as both transcriptional activation and phosphorylation of MT1-MMP (Fig. 1 I; Barbolina et al., 2007; Nyalendo et al., 2007; Sugiyama et al., 2010a). Consistently, Src inhibitor prevented MDA-MB-231 cell invasion, and MT1-MMP overexpression partially rescued the invasion of EphA2 knockdown cells (Fig. S1, D–G). These results establish EphA2 as a multifaceted regulator of MT1-MMP–dependent breast carcinoma cell invasion.
MT1-MMP interacts with EphA2 to modulate receptor localization and cell invasive growth
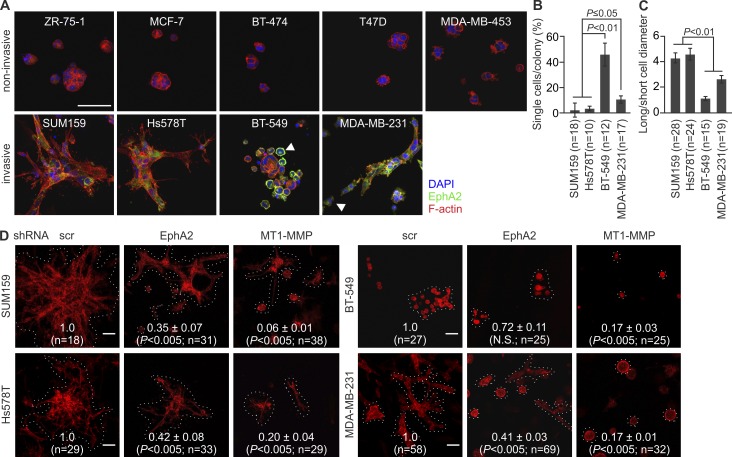
In addition to invasion, MT1-MMP enhances invasive cell growth within collagen (Hotary et al., 2003). To assess such invasive growth properties, single cells were implanted inside 3D collagen. ZR-75-1, MCF-7, BT-474, T47D, and MDA-MB-453 cells with minor MT1-MMP and EphA2 formed small noninvasive colonies, whereas SUM159, Hs578T, BT-549, and MDA-MB-231 cells grew and invaded efficiently within collagen (Fig. 2 A). SUM159 and Hs578T colonies formed multicellular outgrowths rich in actin stress fibers (Fig. 2, A–C; and Fig. S1 H). In contrast, BT-549 cells grew in sphere-like colonies surrounded by rounded singly invasive cells with cortical actin. MDA-MB-231 cells had cortical actin coupled to more elongated morphology (Fig. 2, A–C; and S1 H). EphA2 and MT1-MMP knockdown reduced the invasive growth of all these cells (Fig. 2 D).
Figure 2.
EphA2 silencing reduces cell invasive growth in 3D collagen. (A) The breast carcinoma cells were embedded in 3D collagen as a single cell suspension followed by a 4-d culture. Confocal micrographs show EphA2 and filamentous actin (phalloidin) in representative cell colonies. Arrowheads indicate round single cells with prominent surface and intracellular EphA2. See Fig. S1 G for details. (B and C) Quantitative assessment of single-cell invasion (B) and cell shape (C; error bars indicate mean ± SEM; three collagen preparations per cell line). (D) Confocal micrographs show representative colonies of the phalloidin-stained (red) invasive cells after MT1-MMP and EphA2 knockdown. Relative colony size is indicated in each micrograph (mean ± SEM; six collagen preparations per cell line). Broken lines define individual colonies. P-values were determined with a Mann–Whitney U test (B–D). Bars, 50 µm.
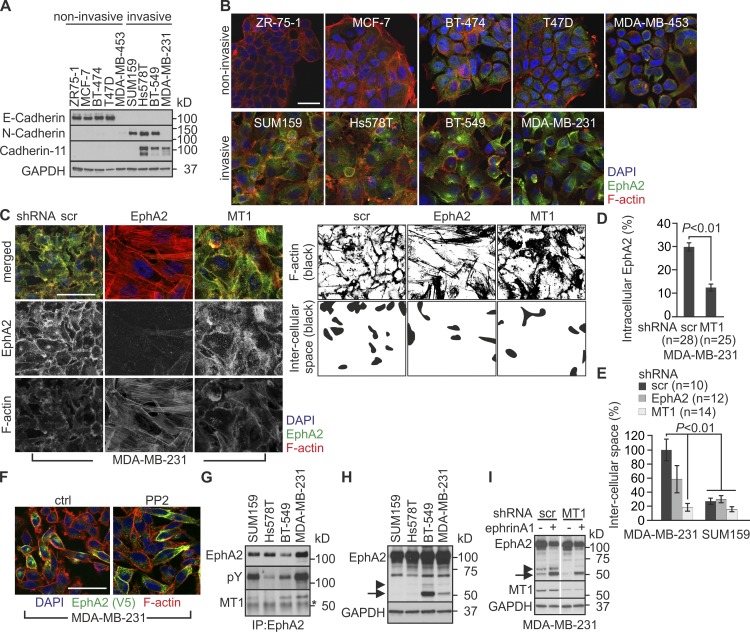
In 2D culture, EphA2 and MT1-MMP knockdown had less of an effect on cell growth (Fig. S1, I and J). In the absence of the MT1-MMP–dependent constraint of 3D collagen, the noninvasive, E-cadherin–expressing ZR-75-1, MCF-7, BT-474, and T47D cells formed tight epithelial-type colonies (Fig. 3, A and B). However, MDA-MB-453 cells with negligible expression of the tested cadherins showed a less adhesive rounded morphology (Fig. 3, A and B). Of the invasive cells, all displaying signs of EMT, e.g., expression switch from E-cadherin to N-cadherin and cadherin-11, SUM159 and Hs578T cells grew as compact mesenchymal cultures with prominent actin stress fibers, whereas BT-549 and MDA-MB-231 cells displayed more cortical actin and intercellular spaces (Fig. 3 B). Differentially from a dispersed EphA2 localization in SUM159 and Hs578T cells, the receptor was distinctly distributed to perinuclear compartments and cell surface in BT-549 and MDA-MB-231 cells (Figs. 2 A, 3 B, and S1 K). Importantly, MT1-MMP knockdown prevented intracellular EphA2 accumulation, resulting in more dispersed localization and reduced intercellular spaces in MDA-MB-231 cells (Fig. 3, C–E; and Fig. S1 J). Similar changes occurred after Src inhibition by PP2, which suggests that functional EphA2–MT1-MMP interaction coupled with EphA2-dependent Src activity regulates cell junctional properties (Figs. 3 F and S2 A). Although EphA2 knockdown resulted in an additional increase in actin stress fibers in 2D, the EphA2–MT1-MMP cooperation was strongly supported by their physical interaction concomitant with EphA2 processing in BT-549 and MDA-MB-231 cells (Fig. 3, G and H).
Figure 3.
The MT1-MMP– EphA2 axis modulates cell morphology. (A) The expression of indicated cadherins was assessed in the breast carcinoma cell lines (n = 3). (B) Representative confocal micrographs show EphA2 and filamentous actin (phalloidin) in the 2D cell cultures. (C, left) Representative confocal micrographs show EphA2 and filamentous actin (phalloidin) in control (scr), EphA2, and MT1-MMP knockdown MDA-MB-231 cells. (C, right) Black-and-white images visualize actin cytoskeleton (top) and intercellular spaces (bottom). (D and E) Quantitative assessment of intracellular EphA2 localization (D) and intercellular spaces within the indicated MDA-MB-231 and SUM159 knockdown cells (E; error bars indicate mean ± SEM; n = 3). P-values were determined with a Mann–Whitney U test. (F) The Src kinase inhibitor PP2 impairs intracellular EphA2 localization. MDA-MB-231 cells were incubated with 5 µM PP2 for 2 h. See Fig. S2 A for details. (G and H) Soluble lysates of the invasive cells were subjected to immunoprecipitation and immunoblotting as indicated (n = 3). The asterisk indicates IgG. Note the physical interaction of MT1-MMP and EphA2 (G) and EphA2 processing in MDA-MB-231 and BT-549 cells (H). (I) MT1-MMP is required for constitutive processing of endogenous EphA2 in MDA-MB-231 cells. Control and MT1-MMP knockdown cells were incubated with soluble ephrinA1 (1 µg/ml) for 2 h followed by immunoblotting as indicated (n = 2). Arrowheads and arrows indicate the truncated ∼60- and ∼50-kD C-terminal EphA2 fragments, respectively. GAPDH served as loading control. Bars, 50 µm.
MT1-MMP is required for constitutive EphA2 processing in MDA-MB-231 cells
To determine if MT1-MMP mediates EphA2 processing, we assessed the effects of MT1-MMP knockdown on EphA2 protein forms. The full-length receptor and ∼60- and ∼50-kD fragments were detected in control cells (Fig. 3 I). The ∼50-kD fragments were increased by soluble recombinant ephrinA1. Importantly, MT1-MMP knockdown prevented the processing to the ∼60 kD fragments, whereas some ∼50-kD fragments were also detected in the ephrinA1-stimulated knockdown cells (Fig. 3 I).
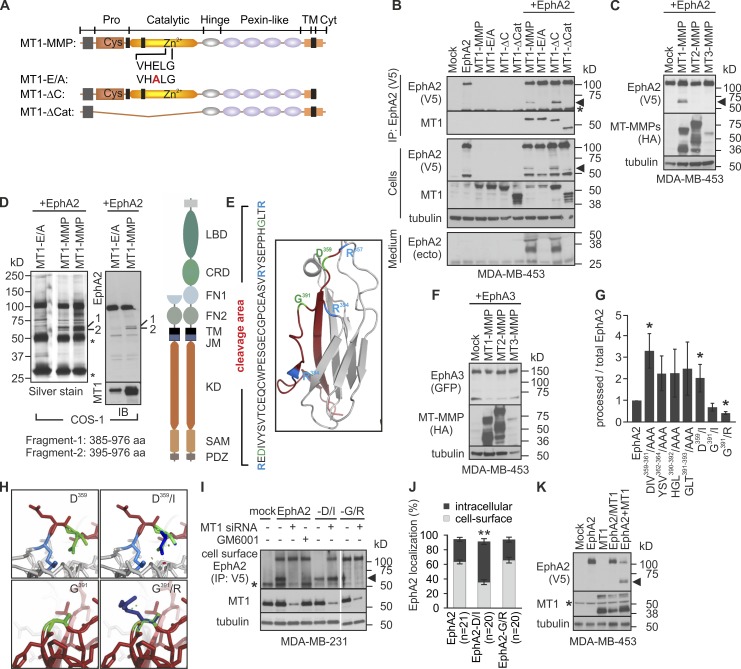
MT1-MMP interacts with and cleaves EphA2 in same membrane complexes
To examine the molecular basis of EphA2 processing, V5 epitope-tagged EphA2 and MT1-MMP or the MT1-MMP mutant proteins with inactivating point mutation (MT1-E/A), or deletion of either cytoplasmic (MT1-ΔC) or catalytic (MT1-ΔCat) domains were expressed alone or in combination in MDA-MB-453 cells (Fig. 4 A). Consistent with the endogenous interaction in invasive cells, ectopically expressed EphA2 and MT1-MMP were coprecipitated (Fig. 4 B). Both proteolytically active MT1-MMP and MT1-ΔC induced EphA2 cleavage, as indicated by the ∼60-kD fragments in the same complexes. Coincidentally, soluble ∼35-kD N-terminal fragments were detected in the conditioned medium (Fig. 4 B). The cleavage was prevented by the MT1-E/A mutation (Fig. 4 B) and was specific for MT1-MMP, as MT2- and MT3-MMP did not cleave EphA2 (Fig. 4 C). However, MT1-ΔCat was also detected in the EphA2 complexes, which indicates that, rather than being just an enzyme–substrate interaction, the MT1-MMP catalytic domain is not required for the physical EphA2–MT1-MMP interaction (Fig. 4 B).
Figure 4.
MT1-MMP cleaves EphA2 at the Fibronectin type-III domain on the cell surface in cis. (A) Schematic representation of MT1-MMP and mutant proteins encoded by the used cDNA constructs. (B and C) MDA-MB-453 cells were cotransfected with EphA2 and MT1-MMP mutants or HA-tagged MT-MMPs followed by immunoprecipitation and immunoblotting of cell lysates and conditioned media as indicated (n = 3). Arrowheads indicate the truncated ∼60-kD C-terminal EphA2 fragments. The asterisk indicates IgG. (D) Determination of the MT1-MMP cleavage sites in EphA2. Lysates of COS-1 cells coexpressing EphA2 and MT1-MMP or MT1-E/A were subjected to immunoprecipitation, immunoblotting, and silver staining. Approximately 60-kD EphA2 fragments (1 and 2) were subjected to mass spectrometry (see Tables S1 and S2). (D, right) Schematic picture of EphA2. LBD, ligand-binding domain; CRD, cysteine-rich domain; FN, Fibronectin domain; TM, transmembrane region; JM, juxtamembrane region; KD, kinase domain; SAM, sterile α motif; PDZ, Psd-95, Dlg, and ZO1 domain. (E) Crystal structure of FN1 and amino acid sequence of the cleavage area. Red, cleavage area; blue, R357, R384, and R394; green, D359 and G391 (green). (F) Lysates of MDA-MB-453 cells expressing EphA3 alone or in combination with the HA-tagged MT-MMPs were subjected to immunoblotting as indicated (n = 3). (G) Relative processing of EphA2 mutants was quantified and normalized with tubulin (error bars indicate mean ± SEM, n = 3). *, P < 0.05, Student’s t test. (H) Structural changes of EphA2-D/I and EphA2-G/R. Green, D359 or G391; dark blue, newly generated side chains. Short green lines and disks indicate atoms in contact or slightly overlapping, and red disks indicate significant van der Waals overlap. All possible rotamers at the mutation points are shown Video 1. (I) Cell-surface EphA2, EphA2-D/I, and EphA2-G/R in control or MT1-MMP knockdown MDA-MB-231 cells were detected by cell surface biotinylation followed by immunoprecipitation. Cell lysates were subjected to immunoblotting as indicated (n = 2). Where indicated, EphA2-expressing cells were treated with 1 µM GM6001 for 16 h. The arrowhead indicates ∼60-kD EphA2 fragments. The asterisk indicates IgG. (J) Quantitative assessment of EphA2 and the mutant protein localization by immunofluorescence (error bars indicate mean ± SEM; n = 3). **, P < 0.01, Mann–Whitney U test. (K) EphA2 and MT1-MMP were expressed either by cotransfection or co-culture of single transfected MDA-MB-453 cells. Cell lysates were subjected to immunoblotting as indicated (n = 3). The arrowhead indicates ∼60-kD EphA2 fragments. The asterisk indicates IgG. Tubulin served as loading control.
MT1-MMP cleaves EphA2 at Fibronectin type-III domain
The EphA2 cleavage site was next identified by mass spectrometry (LC-MS/MS) after V5-tag antibody immunoprecipitation. As assessed by silver staining and immunoblotting, ∼110-kD full-length EphA2 and a doublet of ∼60 kD fragments were precipitated from COS-1 cells after coexpression of MT1-MMP and C-terminally V5-tagged EphA2 (Fig. 4 D). These fragments were absent from parallel control precipitates from EphA2- and MT1-E/A–coexpressing cells (Fig. 4 D). Importantly, peptide sequences corresponding to the C-terminal Y385-I976 and T395-I976 EphA2 fragments were identified by mass spectrometry, after in-gel trypsin digestion of the two ∼60-kD protein bands (Fig. 4 D and Tables S1 and S2). The cleavage area was in Fibronectin type-III domain 1 (FN1) within tryptic peptides flanking amino acids E358-R384 and Y385-R394 (Fig. 4, D and E). Interestingly, P389HGL392 is a consensus P3-P1′ sequence for MT1-MMP substrates (P-X-G/P-L; http://merops.sanger.ac.uk/index.shtml). EphA3, with the closest homology within the identified cleavage area, lacks the consensus sequence and was not cleaved by MT1-MMP (Fig. 4 F). The MT1-MMP consensus and other plausible substrate sequences based on MEROPS were selected for mutation analysis. Although none of the tested triple mutations (DIV/AAA, YSV/AAA, EAS/AAA, RYS/AAA, HGL/AAA, and GLT/AAA) reduced the cleavage, G391/I point mutation at the MT1-MMP consensus site notably decreased the processing (Fig. 4 G). Moreover, G391/R mutation previously found in human lung cancer cells rendered EphA2 essentially resistant to the cleavage (Fig. 4 G; Faoro et al., 2010). Consistently, structure analysis suggested that the G391/R mutation stabilizes the C369-S396 loop, whereas D359/I mutation increased the processing by rendering the loop W348-V361 apparently more accessible for the protease (Fig. 4 G and H; and Video 1).
MT1-MMP–dependent EphA2 cleavage occurs in cis and is associated with intracellular receptor translocation
Because the EphA2–MT1-MMP interaction and EphA2 processing were coupled with intracellular EphA2 localization, the processing could either occur on the cell surface, triggering rapid receptor internalization, or within intracellular protein complexes. To define if MT1-MMP cleaved EphA2 on the cell surface, EphA2 immunoprecipitation after cell-surface biotinylation was performed in control and MT1-MMP knockdown MDA-MB-231 cells expressing EphA2, EphA2-D/I, or EphA2-G/R. Both full-length and truncated ∼60-kD EphA2 were exposed on the cell surface. The truncated cell-surface fragments were lost after MT1-MMP silencing (Fig. 4 I). Likewise, the cell membrane–impermeable MMP inhibitor GM6001 inhibited EphA2 processing, which indicates that the cleavage by MT1-MMP occurred on the cell surface (Fig. 4 I). Consistently, mostly cleaved cell-surface EphA2-D/I was detected unless the full-length protein was stabilized in the presence of only low levels of MT1-MMP after siRNA knockdown (Fig. 4 I). In contrast, only full-length EphA2-G/R was detected on the cell surface with or without MT1-MMP silencing (Fig. 4 I). Notably, the EphA2-D/I processing was associated with its increased intracellular localization and cell rounding (Figs. 4 J and S2 B). EphA2-D/I and MT1-MMP colocalized intracellularly and on the cell surface, whereas the cleavage-resistant EphA2-G/R colocalized more with cell-surface MT1-MMP (Figs. 4 J and S2 B). Although it remains possible that some EphA2-D/I is also cleaved intracellularly, e.g., during export to the cell membrane, these results altogether suggest that EphA2 processing on cell surface is associated with its internalization.
To define next if MT1-MMP cleaved EphA2 in cis on the same cell membrane or in trans on adjacent cells, EphA2 and MT1-MMP were expressed either by cotransfection or co-culture of single-transfected MDA-MB-453 cells. Despite comparable MT1-MMP expression, only cotransfection enabled EphA2 processing (Fig. 4 K), which indicates that MT1-MMP cleaves EphA2 in cis opposite to the reported ephrinA cleavage by ADAM (Janes et al., 2005).
MT1-MMP–dependent EphA2 processing triggers cell–cell repulsion
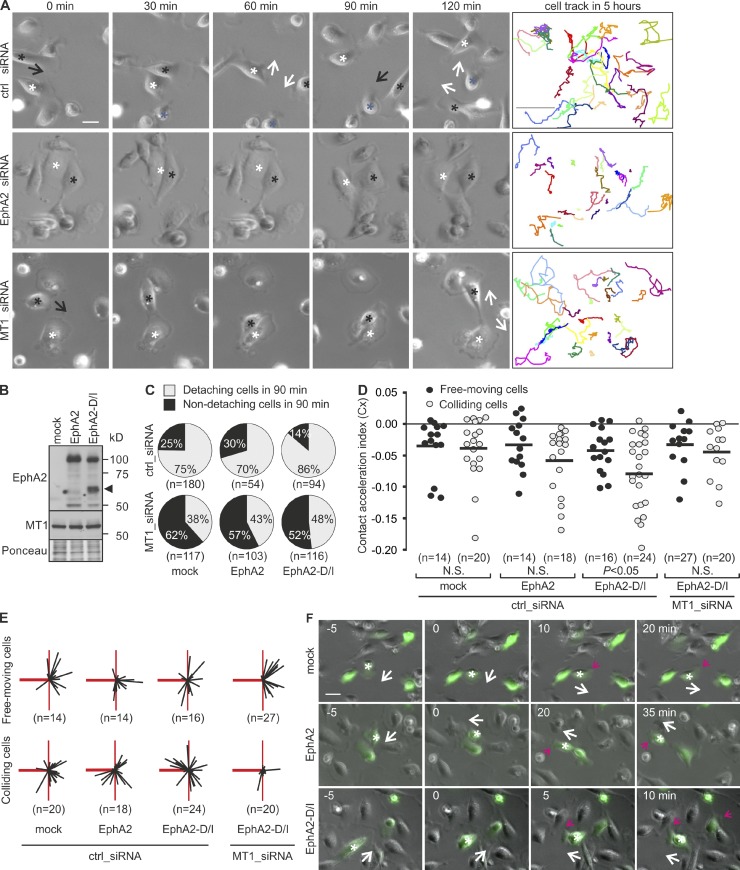
To test if the EphA2 cleavage functionally contributes to the observed cell junction disassembly, the effects of EphA2 and MT1-MMP knockdown were first assessed on MDA-MB-231 cells by live cell imaging. Control cells moved freely for long distances and underwent numerous adhesions and detachments (Fig. 5 A and Video 2). EphA2 knockdown reduced detachments and overall cell movement, whereas MT1-MMP silencing specifically hindered the detachment of colliding cells, thus reducing their movement (Fig. 5 A and Videos 3 and 4). To define if the detachment failure was caused by impaired EphA2 cleavage, detachment responses upon collision were followed in cells expressing EphA2 or EphA2-D/I (Fig. 5 B). In control cells, 25% of collisions were not followed by detachment (Fig. 5 C). EphA2 overexpression did not markedly affect this frequency, whereas EphA2-D/I notably decreased the number of non-detaching cells by 1.8-fold (Fig. 5 C and Videos 5–7). MT1-MMP knockdown rescued the detachments to a more comparable frequency in EphA2 and EphA2-D/I cells (Fig. 5 C and Videos 8–10), which suggests that EphA2 cleavage by MT1-MMP promotes cell–cell detachment.
Figure 5.
EphA2 processing by MT1-MMP enhances cell repulsion. (A) Representative time-lapse microscopy images of control, EphA2, or MT1-MMP knockdown MDA-MB-231 cells at the indicated time points. Black arrows indicate the moving direction of cells marked with black asterisks toward the cells marked with white or blue. White arrows indicate the moving direction of both cells after collision. Representative track plots are shown as colored lines (right; n = 25 cells, 5 h). See Videos 2–4. Bars: (time-lapse panels) 10 µm; (tracking panels) 300 µm. (B) Control, EphA2-expressing, and EphA2-D/I–expressing cells were subjected to immunoblotting as indicated (n = 3). Ponceau red staining served as a loading control. The arrowhead indicates ∼60-kD EphA2 fragments. (C) Control or MT1-MMP knockdown cells were followed by time-lapse microscopy for 12 h. The cells coexpressing GFP and EphA2 or EphA2-D/I were tracked. Pie charts show the percentages of non-detaching and detaching cells after collision (90 min). (D) Contact acceleration indices (Cx) of free-moving and colliding cells (see Fig. S2 C). P-values were determined with a Mann–Whitney U test; (E) Representation of velocity vectors of free-moving and colliding cells in compass plots. The heavy red line represents the scaled displacement of all cells before collision, and thin black lines show the scaled displacement of each cell after collision. The thin red line is a reference line that marks the angle of 90° relative to the displacement before contact (heavy red line). (F) Representative time-lapse microscopy images at the indicated time points. White arrows indicate the moving direction of cells marked with white asterisks, and red arrows indicate a newly formed leading edge. See Videos 5–7. Bar, 10 µm.
To further investigate the repulsive responses, the cells were tracked before and after collision. These migration paths (colliding) were compared with those of noncolliding (free-moving) cells (Fig. S2 C; Paddock and Dunn, 1986). EphA2 overexpression only slightly increased directional changes upon collision, as indicated by nonsignificant difference in contact acceleration indices and comparable scaled cell displacement vectors of free-moving and colliding cells (Fig. 5, D–F; and Fig. S2, D and E). In contrast, upon collision, EphA2-D/I triggered significantly greater directional switches, which were reverted to comparable levels with free-moving cells by MT1-MMP knockdown (Fig. 5, D–F). These results indicate that MT1-MMP is responsible for the contact-mediated cell-repulsive responses upon EphA2 cleavage.
Prominent EphA2 cleavages promote RhoA activation and single-cell invasion
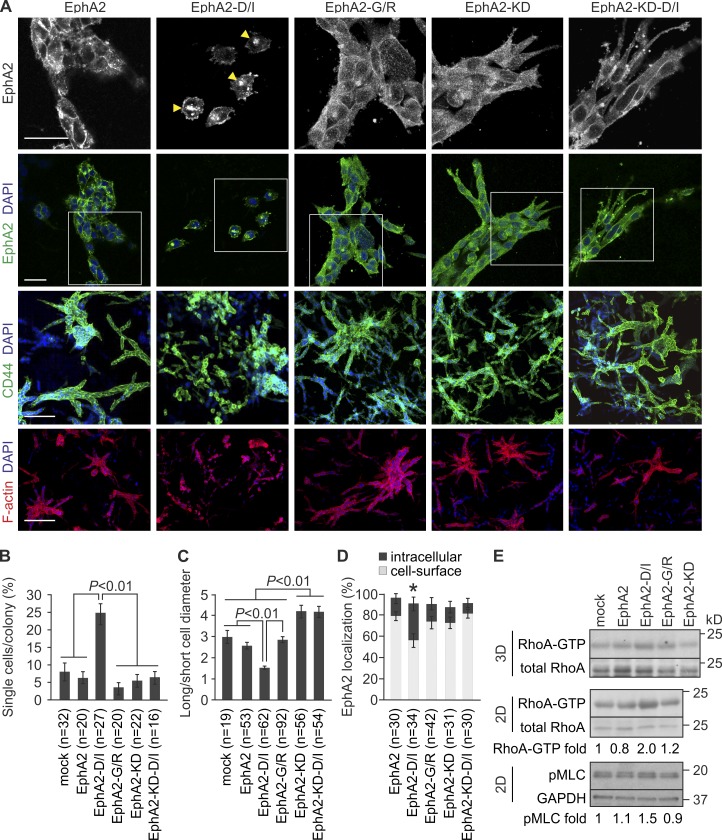
To test if the cell repulsion triggered by EphA2 cleavage was related to cell invasion, MDA-MB-231 cells expressing EphA2, EphA2-D/I, EphA2-G/R, kinase activity–deficient EphA2-KD, and EphA2-KD-D/I were implanted in collagen. Similar to control cells, EphA2 cells formed coherent colonies with mainly multicellular sprouts (Fig. 6, A–C; and Fig. S3, A and B). In these colonies, EphA2 localized intracellularly and on the cell surface (Fig. 6, A and D). Consistent with 2D results, EphA2-D/I accumulated in intracellular compartments coincidentally with loss of cell contacts and rounded single-cell invasion (Fig. 6, A–D). In contrast, cells expressing cleavage-resistant EphA2-G/R with more dispersed EphA2 distribution grew as large collectively invading colonies (Fig. 6, A–D). Moreover, cells expressing either EphA2-KD or the efficiently cleaved EphA2-KD-D/I formed coherent colonies with multicellular sprouts of elongated cells (Fig. 6, A–D; and Fig. S3, A and B). Thus, cleavage of catalytically active EphA2 by MT1-MMP triggered cell transition to rounded morphology and single-cell invasion.
Figure 6.
EphA2 cleavage promotes single-cell invasion. (A) MDA-MB-231 cells stably overexpressing EphA2, EphA2-D/I, EphA2-G/R, and the kinase activity–deficient EphA2-KD and EphA2-KD-D/I proteins were embedded in 3D collagen and allowed to grow for 4 d. Confocal micrographs show EphA2 and filamentous actin (phalloidin) in representative cell colonies. Boxes indicate the areas included as magnified insets on the top panel. CD44 was used as a cell-surface marker. Arrowheads indicate intracellular EphA2. Bars: (top two rows) 50 µm; (bottom two rows) 200 µm. (B–D) Quantitative assessment of single-cell invasion (B), cell shape (C), and EphA2 localization (D; error bars indicate mean ± SEM; three collagen preparations per cell). *, P < 0.05, Mann–Whitney U test. (E) EphA2-D/I increases RhoA activity within 3D collagen and in 2D. Soluble lysates from 3D and 2D cultures of EphA2 or the mutant protein–expressing cells were incubated with Rhotekin RBD-GST–conjugated agarose beads followed by immunoblotting as indicated (n = 4). Phosphorylated myosin light chain (pMLC) was detected from total cell extracts (n = 3). Normalized mean values of RhoA-GTP with total RhoA and pMLC with GAPDH are indicated below each blot.
EphA2 has been associated with RhoA activation and myosin light chain (MLC)-dependent actomyosin contractility leading to cell body retraction (Parri et al., 2009; Taddei et al., 2011). To assess if the EphA2 processing was linked to Rho signaling, we examined RhoA and MLC activation in cells expressing EphA2 or the mutant proteins. Although EphA2 and EphA2-G/R had minor effects on the RhoA activity, EphA2-D/I notably increased and EphA2-KD appeared to decrease active RhoA (Figs. 6 E and S3 C). MLC phosphorylation showed a similar trend (Fig. 6 E). These results suggest a mechanism for repulsion and cell junction disassembly via RhoA-mediated contractility upon cleavage and intracellular translocation of the active EphA2.
Ectopic assembly of the MT1-MMP–EphA2 axis induces repulsion and single-cell invasion
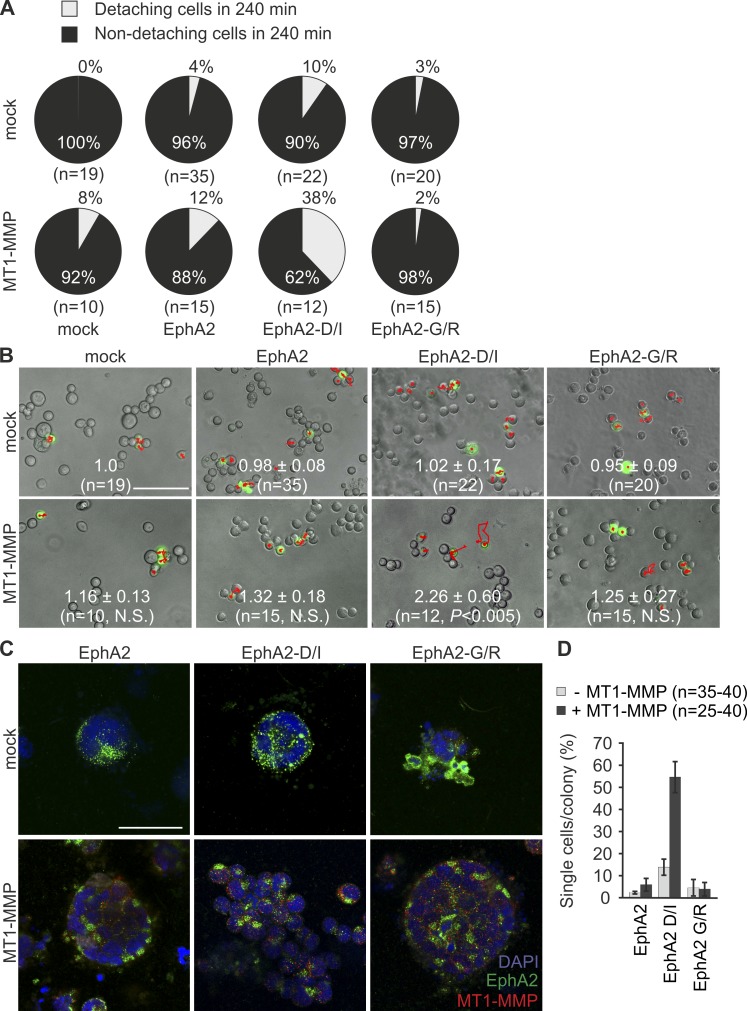
To confirm the cooperation of EphA2 and MT1-MMP on cell junction disassembly, MDA-MB-453 cells essentially devoid of these proteins were transfected to express EphA2, EphA2-D/I, or EphA2-G/R alone or in combination with MT1-MMP. Although EphA2, EphA2-D/I, and MT1-MMP alone or EphA2 with MT1-MMP only slightly affected cell detachment, the coexpression of EphA2-D/I and MT1-MMP specifically increased cell detachments and moving distances as assessed by live cell imaging (Fig. 7, A and B). In 3D collagen, EphA2- and EphA2-G/R–expressing cells formed small colonies, whose growth was enhanced by MT1-MMP (Fig. 7 C). Importantly, although EphA2-D/I expression alone did not alter invasive growth of these cells, the ectopic coexpression with MT1-MMP resulted in single-cell invasion (Fig. 7, C and D).
Figure 7.
Reconstituting MT1-MMP–dependent EphA2 processing by ectopic expression induces repulsion and single-cell invasion of MDA-MB-453 cells. (A and B) The cells transiently coexpressing GFP and EphA2, EphA2-D/I, or EphA2-G/R with or without MT1-MMP were followed by time-lapse microscopy for 4 h; GFP- and EphA2-coexpressing cells were tracked. Pie charts show the percentage of non-detaching and detaching cells after collision (A; mean ± SEM). Representative time-lapse microscopy images visualizing track plots (shown in red); relative cell moving distances are indicated in each micrograph (B; mean ± SEM; five time-lapse analyses per cell line). Broken lines in EphA2-D/I– and MT1-MMP–coexpressing cells define the position of the tracked cells at the 0 time point. P-values were determined with a Mann–Whitney U test. Bar, 300 µm. (C) The cells were cultured within 3D collagen for 6 d. Representative confocal micrographs show EphA2 and MT1-MMP in representative cell colonies. Bar, 50 µm. (D) Quantitative assessment of single-cell invasion (error bars indicate mean ± SEM; three collagen preparations per cell).
EphA2 cleavage promotes single cell dissemination in vivo
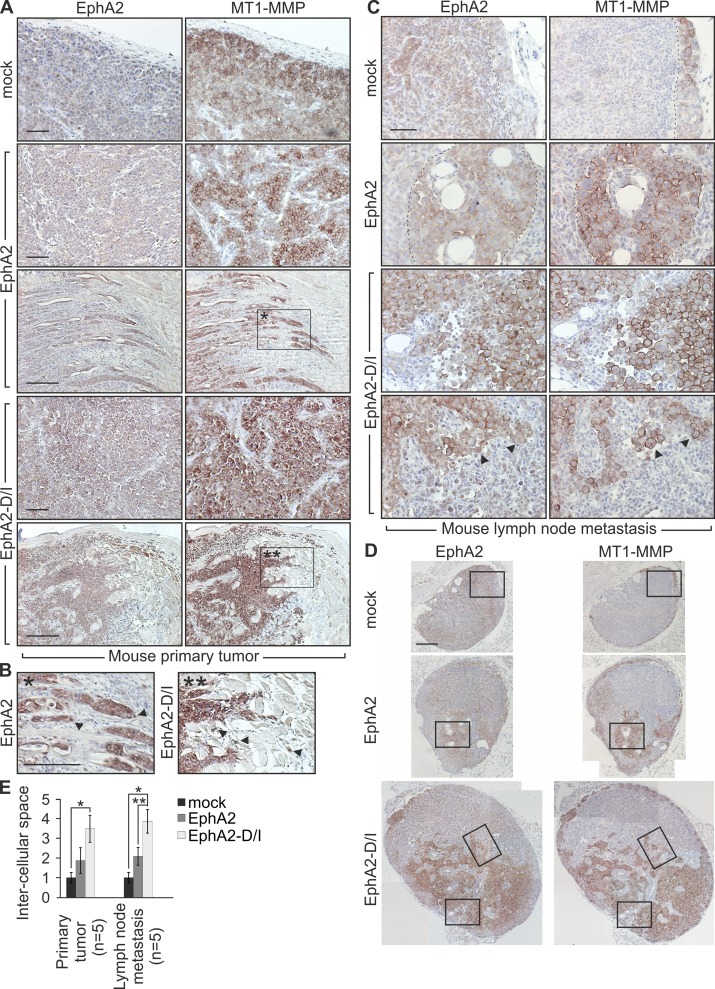
To assess if the cellular events triggered by EphA2 processing regulated the tumor cell phenotype in vivo, lentiviral EphA2 and EphA2-D/I were expressed in MDA-MB-231 cells (Fig. S3 D). After orthotopic injection into the mammary fat pad of severe combined immunodeficient (SCID) female mice, all xenografts formed tumors within 4 wk. In control and EphA2 tumors, the cells grew cohesively (Fig. 8, A and E; and Fig. S3 G). EphA2 and to a higher extent EphA2-D/I increased MT1-MMP throughout the tumors. Moreover, EphA2-D/I promoted dissociation of cell junctions and single-cell invasion into stroma (Fig. 8, A, B, and E; and Fig. S3 G). In lymph nodes of control tumor-bearing mice, carcinoma cells with less MT1-MMP relative to the primary tumors remained mainly within the cortex (Fig. 8, C and D; and Fig. S3, E and F; metastasis in sentinel lymph nodes; 2/5). EphA2 enhanced growth of coherent metastatic colonies with increased MT1-MMP (Fig. 8, C and D; and Fig. S3, E and F; metastasis in sentinel lymph nodes; 2/5). Even more than in primary tumors, the metastatic EphA2-D/I cells displayed poor cell–cell contacts (Fig. 8, C–E; and Fig. S3, E–G; metastasis in sentinel lymph nodes; 4/5).
Figure 8.
EphA2 cleavage promotes single cell phenotype of MDA-MB-231 cells in vivo. Control, EphA2, or EphA2-D/I stably expressing MDA-MB-231 cells were orthotopically injected into the mammary fat pad of SCID mice. Tumor growth was followed for 4 wk. Tumor sizes were not significantly different (mock, 567 ± 96.25 mm3; EphA2, 320 ± 145.75 mm3; EphA2-D/I, 601 ± 81.43 mm3). (A and B) EphA2 and MT1-MMP immunohistochemistry of paraffin sections of the primary xenograft tumors. Corresponding areas (A, boxed regions) and the magnified insets (B) are marked with single and double asterisks. Arrowheads indicate invasive front cells. (C and D) Immunohistochemistry of representative metastatic tumor cell colonies in lymph nodes. The broken lines demarcate the border between tumor cells and host tissue. Arrowheads indicate singly invading cells with high MT1-MMP and intracellular EphA2. Boxes (D) indicate the areas magnified in the insets (C). Bars: (A–C) 50 µm; (D) 200 µm. (E) Quantitative assessment of intercellular spaces within tumor cell colonies in mouse primary tumors and lymph nodes (error bars indicate mean ± SEM). The measured example areas are shown in Fig. S3 G. Relative intercellular space of the mock tumors is set to one. *, P < 0.01, **, P < 0.05; Mann–Whitney U test.
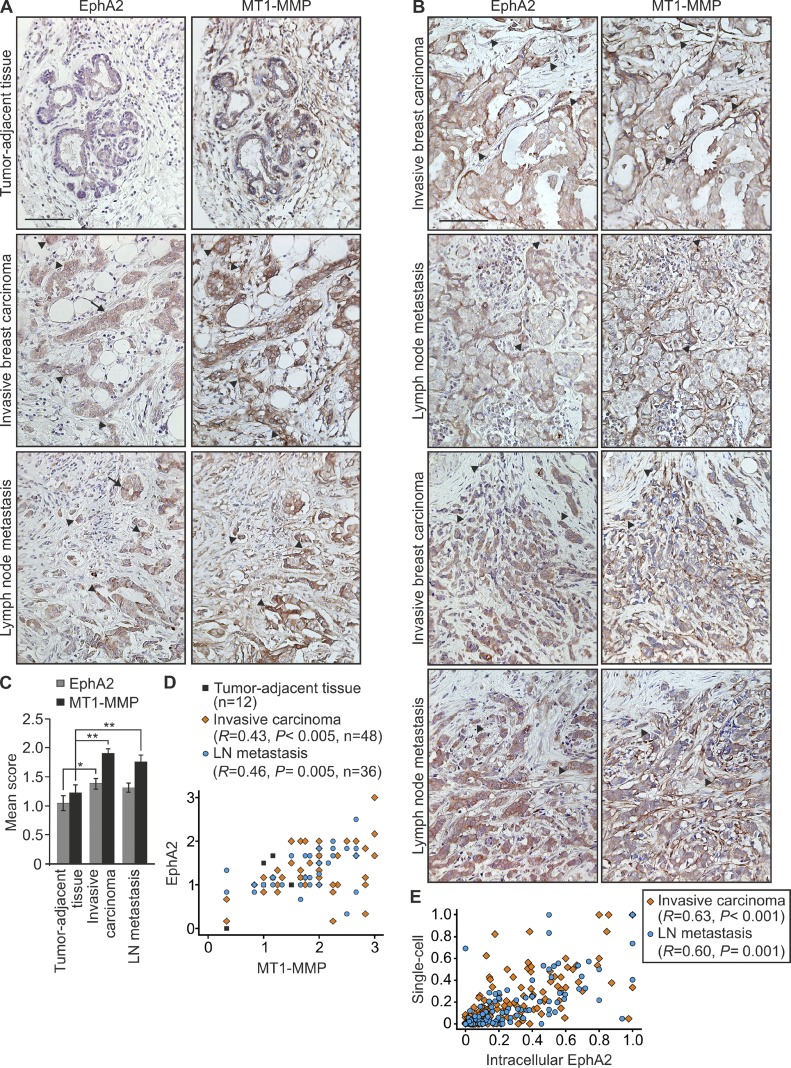
To further assess the relevance of these results for human breast carcinoma, we examined EphA2 and MT1-MMP coexpression by immunohistochemistry of a human tissue array containing 48 cases of invasive breast cancer with matched normal tumor-adjacent tissue (n = 12) or lymph node metastasis (n = 36). Variable membranous and cytoplasmic staining for both proteins was observed. EphA2 was detected in luminal epithelium and carcinoma cells, with minimal stromal staining (Fig. 9, A and B; and Fig. S4). MT1-MMP was detected in both the carcinoma compartment and stroma (Fig. 9, A and B; and Fig. S4). EphA2 and MT1-MMP levels were higher in primary tumors and metastases than in tumor-adjacent tissues, and showed a positive correlation (Fig. 9, A–D; and Fig. S4; primary tumors: EphA2 33/48, MT1-MMP 41/48; metastases: EphA2 21/36, MT1-MMP 27/36). In single tumor cells and small or loose cell groups within stroma, increased MT1-MMP was often coupled with intracellular EphA2, which resembles the receptor localization associated with EphA2 cleavage and single-cell phenotype in vitro (Fig. 9, A, B, and E; and Fig. S4). In cohesive cancer cell colonies, mainly low MT1-MMP was instead coupled with cell-surface EphA2 (Fig. 9, A and B; and Fig. S4). These results suggest that the MT1-MMP–EphA2 axis functions as a biologically relevant regulatory mechanism of cancer cell invasion.
Figure 9.
EphA2 and MT1-MMP are coexpressed in invasive human breast carcinomas. (A) EphA2 and MT1-MMP immunohistochemistry of tumor-adjacent tissue, invasive breast carcinoma, and lymph node metastasis from the same patient. (B) Immunohistochemistry of invasive breast carcinomas and matched lymph node metastases. Arrows indicate junctional EphA2, and arrowheads indicate intracellular EphA2 in highly MT1-MMP–expressing tumor cells. Bars, 100 µm. (C) Quantitative analysis of the EphA2 and MT1-MMP staining (error bars indicate mean ± SEM). *, P = 0.05; **, P < 0.01; Mann–Whitney U test. (D and E) Scatter plot of EphA2 versus MT1-MMP staining (D; mean score), and of single-cell phenotype versus intracellular EphA2 (E; invasive carcinoma, n = 48; LN metastasis, n = 36). R, Pearson correlation coefficient; p-values were determined with a Student’s t test.
Discussion
Metastasizing carcinoma cells infiltrate surrounding tissues by different invasion modes depending on microenvironmental cues (Friedl and Wolf, 2010). Collective and mesenchymal invasion within collagen-rich tissues involve ECM degradation by MT1-MMP (Sabeh et al., 2009; Friedl and Wolf, 2010; Sugiyama et al., 2010a). In addition, cancer cells can invade in an amoeboid manner by squeezing through small spaces (Giampieri et al., 2010; Sanz-Moreno and Marshall, 2010; Wolf and Friedl, 2011). Aberrant EphA2 expression has been associated with this type of movement characterized by rapid changes in cell shape and direction (Parri et al., 2009; Taddei et al., 2011). These processes and contact inhibition of locomotion upon cancer cell–cell contact can be induced by EphA2-dependent generation of actomyosin contractility, whereas amoeboid invasion is less dependent on ECM proteolysis (Parri et al., 2009; Astin et al., 2010; Taddei et al., 2011). However, we observed coexpressed EphA2 and MT1-MMP in all tested invasive breast carcinoma cells, which displayed either mesenchymal or rounded morphologies and used MT1-MMP for invasion into cross-linked collagen. Intrigued by further observation of EphA2–MT1-MMP interaction and EphA2-dependent Src activation, as well as EphA2 processing and intracellular translocation, we conducted mass spectrometry and mutation analyses that revealed a specific MT1-MMP cleavage in the EphA2 FN1 domain. This cleavage triggered dissociation of breast carcinoma cell junctions, cell repulsive responses, and rounded single-cell invasion in vitro and in vivo.
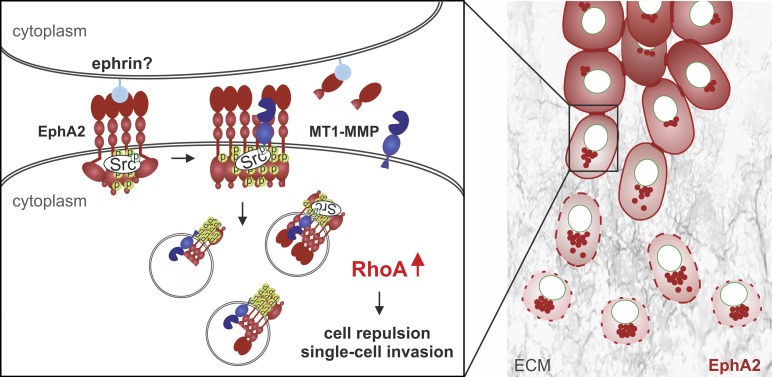
Pericellular proteolysis has emerged as a key mechanism to regulate cell functions (Blobel, 2005; Itoh and Seiki, 2006). As such, ADAMs regulate cell–cell signaling by releasing ephrinA–EphA complexes via in trans ephrin cleavage from the adjacent cell (Janes et al., 2012). Although endocytosis of the ephrinA–EphA complexes is associated with receptor degradation and signal termination, the molecular mechanisms of aberrant pro-migratory signaling via EphA2 in cancer cells have remained undefined (Janes et al., 2012; Miao and Wang, 2012). Current results identify a novel mechanism, whereby MT1-MMP cleaves EphA2 in cis in the same cancer cell–surface complexes (Fig. 10). Although this cleavage had minor effects on free cell movement in 2D, it increased directional switches and detachments upon cell collision. Therefore, the identified in cis cleavage also operates in cell–cell signaling, which was further supported by concurrent junctional disassembly in 2D and 3D cell colonies. Because the cleavage occurred constitutively in MDA-MB-231 cells, their low expression of ephrinA1 and other ligands, typically also seen in aggressive cancers, can be sufficient to trigger limited receptor activation and clustering upon cell–cell contact (Macrae et al., 2005). EphA2-dependent Src activation, which was required for the MT1-MMP cleavage-induced intracellular EphA2 translocation, also occurred without exogenous ligand. However, soluble recombinant ephrinA1 enhanced the accumulation of a smaller EphA2 fragment, which was less dependent on MT1-MMP and similar in size to the major constitutive EphA2 fragment in BT-549 cells. Further considering somewhat higher ephrinA1 expression in BT-549 cells, different types or magnitudes of receptor activation may initiate more complex proteolytic processes. These mechanisms and the differences in the MT1-MMP–EphA2 interrelationship in the most mesenchymally invasive cells (Hs578T and SUM159), relative to those in cells with physical MT1-MMP–EphA2 interactions and constitutive EphA2 cleavages (MDA-MB-231 and BT-549), will be of future interest. Nevertheless, current results support a model where physical EphA2–MT1-MMP interaction and EphA2 processing regulate signaling compartmentalization, cytoskeletal migratory responses, and invasion outcome (Fig. 10). This MT1-MMP–EphA2 axis is unique among other previously described physical MT1-MMP–RTK interactions, where the signaling is indirectly modulated by cleavage of a cofactor rather than the RTK itself by MT1-MMP (Lehti et al., 2005, 2009; Chan et al., 2012).
Figure 10.
Model of MT1-MMP and EphA2 interaction in breast carcinoma cells. In invasive cells, where MT1-MMP and EphA2 are coexpressed concomitantly with low ephrinA1, EphA2 promotes Src signaling and further increases MT1-MMP expression. At cell–cell junctions, the few ephrin ligands can be sufficient for limited receptor activation, thus triggering further receptor clustering and activation, which could also involve other ligand-independent mechanisms (Miao and Wang, 2012). Upon MT1-MMP–EphA2 interaction on the cell surface, cleavage of active EphA2 by MT1-MMP triggers Src activity-dependent intracellular translocation of the receptor parallel with increased actomyosin contractility through RhoA activation. These signaling events promote cell–cell repulsion, junctional disassembly, and dissemination of motile single cells within collagen and in vivo. Consistent with the model, prominent intracellular localization of EphA2-D/I can reflect increased cleavages and subsequent internalization events, although intracellular processing of this cleavage-prone receptor also remains a possibility.
Increasing evidence indicates that cell cytoskeleton and motility are differentially regulated in 2D and 3D via cadherin junctions, integrin-mediated ECM adhesion, and ECM degradation (Sabeh et al., 2009; Friedl and Wolf, 2010; Harunaga and Yamada, 2011). Consistently, EphA2 knockdown enhanced actin stress fibers only during MDA-MB-231 cell adhesion onto a rigid 2D substrate, and MT1-MMP knockdown inhibited cell invasion and growth within 3D collagen while having minor effects on free cell movement and growth in 2D. In spite of such differentially governed cell functions, the identified MT1-MMP–EphA2–RhoA axis more uniformly regulated cell junctional dynamics in both 2D and 3D. Notably, similar repulsive responses and invasion outcome as characterized in MDA-MB-231 cells were also activated upon ectopic reassembly of the MT1-MMP–EphA2 complexes in MDA-MB-453 cells. In these cells, lack of cadherins was associated with poor cell–cell adhesion in 2D. Yet, efficient repulsive responses of these otherwise poorly motile cells were activated only by coexpression of MT1-MMP and cleavage-prone EphA2-D/I. Within constrains of 3D collagen, these cells instead remained in collective colonies even when MT1-MMP overexpression allowed collagen degradation and colony growth. Although the nature of these cell–cell contacts remains to be elucidated, coexpression of MT1-MMP and EphA2-D/I to trigger both repulsion and ECM degradation was required for activation of efficient single-cell invasion in 3D. This suggests that MT1-MMP–EphA2 interaction and EphA2 cleavage can function as more general yet potentially context-dependent regulatory mechanisms of cancer cell invasion.
In human cancers, MT1-MMP induction is often restricted to invasive edges and reactive stroma, whereas EphA2 is typically expressed in proliferating breast carcinoma cells (Okada et al., 1995; Szabova et al., 2005; Sabeh et al., 2009; Sugiyama et al., 2010b; Brantley-Sieders et al., 2011). Considering the differential expression of multiple Eph receptors and their ligands in stromal cells, the signaling networks are likely to be more complicated in vivo. Nevertheless, analogously to the MT1-MMP cleavage-dependent intracellular EphA2 localization in singly invading cells in 3D cultures and mouse xenografts, intracellular EphA2 colocalized with the up-regulated MT1-MMP in invasive edges and single tumor cells within stroma of human breast carcinomas. Depending on the host tissue and target organs, tumor growth and metastasis may instead depend on cell–cell adhesion and collective invasion (Giampieri et al., 2009). Therefore, our results of the cleavage-resistant EphA2-G/R will help to explain the invasive growth-promoting function of this mutant protein earlier described in lung cancer (Faoro et al., 2010). By providing these new mechanisms of context-dependent cancer cell–cell communication, current results increase the understanding of cell invasion plasticity that critically contributes to both cancer metastasis and drug responses.
Materials and methods
Cell lines
Human breast carcinoma cells ZR75-1, MCF7, BT-474, T47D, MDA-MB-453, Hs578T, BT-549, MDA-MB-231 (American Type Culture Collection), and SUM159 (obtained from O. Kallioniemi, Institute for Molecular Medicine Finland, Helsinki, and VTT Technical Research Centre of Finland, Turku, Finland; originally established by S. Ethier, http://www.asterand.com/Asterand/human_tissues/hubrcelllines.htm; Neve et al., 2006), as well as COS-1 monkey kidney cells (American Type Culture Collection), were cultured according to the manufacturer’s instructions.
Antibodies and chemicals
Rabbit polyclonal antibodies against the C-terminal domain of EphA2, ephrinA1, total Src (Santa-Cruz Biotechnology, Inc.), phospho-Src (Cell Signaling Technology), the hinge domain of MT1-MMP (EMD Millipore), phospho-MLC (Abcam), goat polyclonal antibodies against the N-terminal domain of EphA2 (R&D Systems), and mouse monoclonal antibodies against the catalytic domain of MT1-MMP, HA, cadherin-11 (EMD Millipore), E-cadherin, N-cadherin (BD), V5 (Invitrogen), CD44, RhoA, Rac1, CDC42 (Santa Cruz Biotechnology, Inc.), tubulin, GAPDH (Sigma-Aldrich), phospho-tyrosine (EMD Millipore), and GFP (a gift from E. Verschuren, University of Helsinki, Helsinki, Finland) were used. MMP inhibitor GM6001, Src inhibitor PP2 (EMD Millipore), ephrinA1-Fc Chimera (R&D Systems), and Puromycin (Sigma-Aldrich) were also used.
cDNAs and mutation analyses
Cells were transfected with expression vectors encoding human EphA2 (Varjosalo et al., 2008), MT1-MMP (Lehti et al., 2002), MT2-MMP, MT3-MMP (HA epitope–tagged MT-MMP constructs were obtained from S.J. Weiss, University of Michigan, Ann Arbor, MI; Li et al., 2008), EphA3 (a gift from E. Verschuren), and EGFP (Invitrogen) using FuGENE HD (Roche) according to the manufacturer’s instructions. Based on a MEROPS database analysis (http://merops.sanger.ac.uk/index.shtml), the predicted MT1-MMP cleavage sites in EphA2 were mutated. The PCR-mediated overlap extension method was used for site-directed-mutagenesis (Agilent Technologies) using the following primers: EphA2-D/I forward, 5′-CCGGGGCCGCGAGATCATTGTCTACAGCG-3′, and reverse, 5′-CGCTGTAGACAATGATCTCGCGGCCCCGG-3′; EphA2-G/I forward, 5′-GGAGCCTCCTCACATACTGACCCGCACCAG-3′, and reverse, 5′-CTGGTGCGGGTCAGTATGTGAGGAGGCTCC-3′; EphA2-G/R forward, 5′-GAGCCTCCTCACCGACTGACCCGCACCAG-3′, and reverse, 5′-CTGGTGCGGGTCAGTCGGTGAGGAGGCTC-3′; EphA2-DIV/AAA forward, 5′-GGGGCCGCGAGGCCGCTGCCTACAGCGTCACC-3′, and reverse, 5′-GGTGACGCTGTAGGCAGCGGCCTCGCGGCCCC-3′; EphA2-YSV/AAA forward, 5′-CGTCACCTGCGAACAGGCCGCGGCCGAGTCTGGGG-3′, and reverse, 5′-CCCCAGACTCGGCCGCGGCCTGTTCGCAGGTGACG-3′; EphA2-EAS/AAA forward, 5′-GGGCCGTGTGCGGCCGCTGTGCGCTACTC-3′, and reverse, 5′-GAGTAGCGCACAGCGGCCGCACACGGCCC-3′; EphA2-RYS/AAA forward, 5′-GAGGCCAGTGTGGCCGCCGCGGAGCCTCCTCACG-3′, and reverse, 5′-CGTGAGGAGGCTCCGCGGCGGCCACACTGGCCTC-3′; EphA2-HGL/AAA forward, 5′-CTCGGAGCCTCCTGCCGCAGCGACCCGCACCAGTG-3′, and reverse, 5′-CACTGGTGCGGGTCGCTGCGGCAGGAGGCTCCGAG-3′; and EphA2-GLT/AAA forward, 5′-GGAGCCTCCTCACGCAGCGGCCCGCACCAGTGTGAC-3′, and reverse, 5′-GTCACACTGGTGCGGGCCGCTGCGTGAGGAGGCTCC-3′. Molecular structure analysis and visualization of the mutated sequences were done using PyMOL software.
Real-time quantitative PCR (qPCR)
RNA was extracted with an RNeasy Mini kit (QIAGEN) followed by reverse transcription with iScript reverse transcription (Life Technologies). mRNA expression was quantified using TaqMan Universal PCR Master Mix and validated primers (MT1-MMP; Hs 01037006_gH, EphA2; Hs00171656_m1, ephrinA1; Hs00358886_m1; Applied Biosystems). The expression was normalized with TATA-binding protein (TBP) mRNA expression.
shRNA and siRNA
shRNAs targeted against MT1-MMP (TRCN0000050855) and EphA2 (TRCN0000006403, TRCN0000006404, TRCN0000006405, TRCN0000006406, and TRCN0000006407; Thermo Fisher Scientific) or nontargeting scrambled shRNA were used. The packaging plasmid (pCMVdr8.74), envelope plasmid (pMD2-VSVG), and MT1-MMP, EphA2, or scrambled shRNA plasmid (hairpin-pLKO.1 vector) were cotransfected into 293FT producer cells using Lipofectamine 2000 reagent (Invitrogen). Complete breast carcinoma cell growth medium was changed on 293FT cells 24 h after transfection. The viral supernatants were collected after 48 h, passed through a 0.4-µm filter, and incubated with human breast carcinoma cells. After 16 h of infection, the supernatants were replaced with complete media followed by Puromycin selection (2 µg/ml; Tatti et al., 2008) of the transduced cells. siRNA against human MT1-MMP (SI03648841) and nonsilencing control siRNA (QIAGEN) were transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Knockdown efficiency of all shRNAs was monitored by qPCR 48 h after transfection. The two most efficient EphA2 shRNAs (TRCN0000006403 and TRCN0000006405) were used for this study.
Immunoblotting, immunoprecipitation, cell-surface biotinylation, and immunofluorescence
Immunoblotting, immunoprecipitation, cell-surface biotinylation, and immunofluorescence staining were performed as described previously (Lehti et al., 2002). Soluble cell lysates of cells embedded in type I collagen were obtained by mechanical homogenization in RIPA lysis buffer and clarification by centrifugation. When indicated, the soluble cell lysates were subjected to SDS-PAGE and immunoblotting (Lehti et al., 2002) or immunoprecipitation using anti-V5, anti-HA antibody conjugated agarose (Sigma-Aldrich), or EphA2 polyclonal antibodies (Santa Cruz Biotechnology, Inc.). For cell-surface biotinylation, cells were rinsed at 4°C with ice-cold PBS and incubated in PBS containing 0.5 mg/ml of Sulfo-NHS-biotin (Thermo Fisher Scientific) on ice for 1 h. The reaction was terminated by washing with 10 mM Tris/HCl buffer, pH 7.4, containing 150 mM glycine for 10 min. The soluble cell lysates were subjected to immunoprecipitation with EphA2 polyclonal antibodies (Santa Cruz Biotechnology, Inc.). The immune complexes were detected with horseradish peroxidase–conjugated streptavidin (Dako).
Cells grown on glass coverslips were washed with PBS and fixed with 4% PFA at 4°C for 10 min. After washing with Dulbecco’s PBS, the cells were incubated in Dulbecco’s PBS containing 5% BSA to prevent nonspecific binding of the antibodies. The cells were then incubated with the primary antibody in 5% BSA in Dulbecco’s PBS for 1 h. The bound antibodies were detected using Alexa Fluor–conjugated secondary antibodies (Invitrogen). The coverslips were mounted on glass slides using Vectashield anti-fading reagent (Vector Laboratories). Confocal micrographs of immunofluorescence stainings were obtained using a confocal microscope (LSM 5 DUO) with a Plan-Apochromat 40×, 1.3 NA oil objective lens and with a Plan-Apochromat 10×, 0.45 NA objective lens, or using a confocal microscope (LSM 780) with a Plan-Neofluar 40×, 1.3 NA oil objective lens (all from Carl Zeiss). Brightness and contrast were linearly adjusted using Photo-Paint X5 (Corel). Single optical sections or a combination of two serial optical sections were used for image display.
Rho-GTPase activity assay
Soluble cell lysates were incubated with Rhotekin RBD-GST (for the RhoA activity assay) or PAK-1 PDB-conjugated agarose beads (for Rac1 and CDC42 activity assay) for 4 h at +4°C. The resulting bound Rho–GTPase complexes were analyzed by immunoblotting. Total RhoA, Rac1, and Cdc42 were detected from soluble cell lysates. For detection of phospho-MLC, cells were lysed in Laemmli sample buffer and passed through a 20-gauge needle 15 times before SDS-PAGE.
Immunohistochemistry
Mouse tumors and lymph nodes and human breast carcinoma tissue arrays containing paraffin-embedded tissue biopsies (BRM961; US Biomax, Inc.) were stained with pretitrated dilutions of rabbit antibodies against the C-terminal domain of EphA2 and mouse antibodies against the hinge domain of MT1-MMP. Mouse tumors and lymph nodes were fixed with 4% paraformaldehyde, dehydrated, and embedded in paraffin. Tissue sections and human breast carcinoma tissue arrays were de-paraffinized in TissueClear (Tissue-Tek) and rehydrated in a graded ethanol series. After antigen retrieval by boiling the tissue sections in sodium citrate buffer (10 mM sodium citrate and 0.05% Tween 20, pH 6.0), sections were incubated for 10 min in 0.6% (vol/vol) hydrogen peroxide. Tissue sections were subsequently rinsed and incubated with 2.5% normal horse serum (Vector Laboratories) for 30 min. Primary antibodies were incubated for 2 h in blocking buffer, followed by incubation with peroxidase-conjugated anti–mouse Ig and anti–rabbit Ig for 30 min. Detection was performed using a Vectastain ABC kit (Vector Laboratories) according to the manufacturer’s instructions. After washes, slides were dehydrated and mounted using Cytoseal mounting medium (Electron Microscopy Sciences). Light micrographs were obtained using a microscope (DM LB; Leica) with N-Plan 10×, bright field (BF), 0.25 NA and N-Plan, 20×, BF, 0.4 NA objective lenses. Brightness and contrast were linearly adjusted using Photo-Paint X5. Immunohistochemical stainings of human breast carcinoma tissue array for EphA2 and MT1-MMP were performed twice and scored by three independent observers (N. Sugiyama, E. Gucciardo, and K. Lehti; scores: 0, negligible; 1+, weak positive; 2+, moderate; and 3+, high). Single cells and cells with intracellular EphA2 staining were counted from four nonoverlapping areas in equal relative positions within the core biopsy images.
3D type I collagen growth and invasion assays
Collagen invasion and growth were assessed essentially as described previously (Sugiyama et al., 2010b). Rat tail collagen I (Sigma-Aldrich) was dissolved in 0.2% acetic acid, neutralized with NaOH, and diluted to a final concentration of 2.2 mg/ml in MEM. For the 3D collagen cell growth assay, single cell suspension (3 × 103 cells/ml) was prepared in collagen I solution followed by casting 50-µl gels onto 24-well plates and culturing them for 4 d in complete media. The cell colony growth was followed by phase-contrast imaging using an inverted epifluorescence microscope (Axiovert 200; Carl Zeiss) with a Plan-Neofluar 10×, 0.3 NA, Ph1, DIC objective lens (Carl Zeiss), and stained after 4% PFA fixation with TRITC-conjugated phalloidin (Sigma-Aldrich) or antibodies against CD44, MT1-MMP, or EphA2. Confocal imaging was performed using a confocal microscope (LSM 5 DUO; Carl Zeiss).
For the 3D collagen invasion assay, 2.2 mg/ml of rat tail collagen I (Sigma-Aldrich) solution was cast 150 µl into the upper chamber of Falcon cell culture inserts in 24-well cell culture plates and allowed to form a gel at +37°C for 1 h. Tumor cells (3 × 105/ insert) in RPMI containing 10% fetal bovine serum were added on top of the gel. The medium supplemented with recombinant human HGF (25 ng/ml; R&D Systems) as a chemoattractant was added to the lower chamber. The cells were cultured at 37°C for 5 d, replenishing them with complete medium every second or third day. The gels were then fixed in 4% PFA, dehydrated, and embedded into paraffin. Hematoxylin and eosin (H&E)-stained sections were photographed using a microscope (DM LB; Leica).
Mass spectrometry analysis for identification of MT1-MMP cleavage sites in EphA2
C-terminal V5-epitope tagged EphA2 was transfected together with active MT1-MMP or inactive MT1-E/A in COS-1 cells. The soluble cell lysates were incubated with anti-V5 epitope antibody-conjugated agarose beads, and the protein complexes were eluted with Laemmli SDS-PAGE sample buffer. Immunoprecipitated proteins were resolved by SDS-PAGE with a linear 4–20% gradient gel under reducing conditions followed by immunoblot analysis using anti-V5 antibody or by silver staining using Proteosilver Plus Stain kit (Sigma-Aldrich). Two separate bands of ∼60 kD were cut out from the silver-stained gels and subjected to mass-spectrometry analysis (LC-MS/MS) after trypsin digestion.
Time-lapse imaging
Time-lapse microscopy imaging was performed on a Stallion HIS microscope (Intelligent Imaging Innovations, Inc.) with an EC Plan-Neofluar 20×, 0.50 Ph2 objective lens (Carl Zeiss), a camera (AxioCam MRm; Carl Zeiss), and SlideBook 4.1 software (Intelligent Imaging Innovations, Inc.). The cells were seeded on chambers (BD) coated with monomeric collagen I (1 × 105 cells/cm2). An image was taken every 5 min at 37°C with 5% CO2 for up to 5 or 12 h. All movies were compiled and cell tracking was done using ImageJ MTrackJ and Manual Tracking Plug-In.
Cell detachment and cell–cell repulsion analyses
The cell detachment or attachment responses were determined by counting the number of detaching or attaching cells followed for 90 min after cell collision. Quantification of cell repulsion was performed as explained in Fig. S2 C (Paddock and Dunn, 1986). In brief, the displacements 35 min before collision (A) and for 35 min after collision (B) were measured. The contact acceleration index (Cx) of vector b-a represents the difference between how far the cell has progressed in the direction of A and how far it would have gone had there been no collision. Cx values were also calculated for the same population of free-moving cells tracked for the same time period (70 min). The statistical significance of the Cx values between free-moving cells and colliding cells was determined using a Mann–Whitney U test. The velocity of each cell was determined by tracking the position of the nucleus in the first and last images divided by the total time and represented in a compass graph using MATLAB software (MathWorks).
Tumor growth in mice
Experiments were approved by the State Provincial Office of Southern Finland. MDA-MB-231 cells were lentivirally transduced with a Renilla luciferase–GFP fusion reporter protein. Stable cell pools expressing EphA2 or EphA2-D/I proteins were selected by puromycin (Sigma-Aldrich). Cell pools (2 × 106 cells/mouse) were orthotopically injected into the mammary fat pad of SCID female mice (5–7 wk of age; Taconic) and the tumor growth was followed for 4 wk. Tumor growth and metastasis to local lymph nodes were followed by noninvasive bioluminescence measurement visualized after i.p. injection of coelenterazine (35 µg in 100 µl PBS; Synchem) using an in vivo imaging system (Xenogen IVIS System).
Image analysis
All image quantifications were performed by processing all obtained micrographs using ImageJ software. For relative collagen invasion, light micrographs of H&E-stained collagen cross sections were converted into 8-bit (black-and-white) images, a constant threshold was applied, and cells (displayed as saturated black areas) located below the collagen surface were counted by performing the “Analyze Particles” function (see Fig. 1, D and F). For quantification of single-cell invasion, cells within each colony in 3D collagen and surrounding invading single cells were manually counted from epifluorescence micrographs (Figs. 2 B and 6 B). Colony size was calculated by measuring the area occupied by each colony within the 3D matrix (Fig. 2 D). Cell morphology was quantitatively assessed from confocal micrographs by measuring the longest and shortest cell diameter crossing nuclei and calculating their ratio (Figs. 2 C and 6 C). Quantitative assessment of EphA2 localization was performed by calculating the percentage of EphA2 staining localized to perinuclear compartments (defined by the trans-Golgi network, Golgin97; Invitrogen) of total receptor staining (Fig. 3 D). Alternatively, EphA2 localization was quantified by calculating the percentage of EphA2 staining displayed on cell surface and intracellular compartments. Confocal micrographs visualizing EphA2 staining were converted into 8-bit images, a constant threshold level was set to exclude the background, and EphA2 signals, displayed as saturated black areas, on the cell surface, perinuclear compartments, or cytoplasmic region were measured by using the “Analyze Particles” function (Figs. 4 J and 6 D). Intercellular space on the monolayer was drawn and measured from a fixed area within confocal micrographs (Fig. 3 E). For visualization of the actin cytoskeleton, confocal micrographs visualizing phalloidin staining were converted into black-and-white images (Figs. 3 C and S1 G). For quantitative assessment of intercellular spaces within tumor cell colonies in mouse primary tumors and lymph nodes, light micrographs visualizing MT1-MMP staining were converted into 8-bit images, a constant threshold level was set to select only the intercellular spaces, and the “Analyze Particles” function was used to measure their total area (Fig. 8 E and S3 G).
Statistical analysis
All numerical values represent mean ± SEM. Statistical significance was determined using two-tail Student´s t tests or Mann–Whitney U test.
Online supplemental material
Fig. S1 shows relative EphA2 and MT1-MMP protein expression in human breast carcinoma cells and validation of EphA2 and MT1-MMP shRNAs, as well as the activities of Src and MT1-MMP in collagen invasion, EphA2 activities in MT1-MMP regulation, 2D cell proliferation analysis after EphA2 and MT1-MMP silencing, and the effects of EphA2 and MT1-MMP knockdown on cell morphology. Fig. S2 shows the effects of EphA2 processing on the receptor subcellular distribution, schematic explanation of the quantification method for cell repulsive movement, and quantitative assessment of the effects of MT1-MMP silencing on repulsive cell movement. Fig. S3 shows the cleavage of EphA2 proteins, activities of cleavage on Rac1 and Cdc42, source and magnified images of the representative light micrographs of EphA2 and MT1-MMP immunohistochemistry of metastatic tumors in lymph nodes from mouse xenografts, and example images of the representative light micrographs of MT1-MMP immunohistochemical staining of mouse primary tumors for intercellular space analysis. Fig. S4 shows the representative light micrographs of EphA2 and MT1-MMP immunohistochemistry of tissue biopsies from human breast tissue arrays. Video 1 shows the possible rotamers of EphA2-D/I and G/R mutant proteins. Videos 2–4 show time-lapse imaging of control EphA2 knockdown or MT1-MMP knockdown MDA-MB-231 cells. Videos 5–7 show time-lapse imaging of control, EphA2-expressing, or EphA2-D/I–expressing MDA-MB-231 cells. Videos 8–10 show time-lapse imaging of MT1-MMP knockdown MDA-MB-231 cells expressing control, EphA2, and EphA2-D/I proteins. Tables S1 and S2 show Mascot search engine output for Eph2 fragments 1 and 2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205176/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201205176.dv.
Supplementary Material
Acknowledgments
We thank Anne Aarnio and Beatriz Martins for excellent technical assistance; S.J. Weiss (University of Michigan), E. Verschuren, and O. Kallioniemi for cDNAs and cells; J. Keski-Oja, T. Mäkelä, V.-M. Leppänen, and K. Alitalo (University of Helsinki, Helsinki, Finland) for advise on reagents and software; the Biomedicum Molecular Imaging Unit for imaging facilities; the Meilahti Animal Center for animal husbandry; and the Biomedicum Tissue Preparation and Histochemistry Unit for paraffin sections.
This work was funded by University of Helsinki Foundations, the Academy of Finland, the Association for International Cancer Research, Finnish Cancer Foundations, the Sigrid Juselius Foundation, the Graduate School of Musculoskeletal Disorders and Biomaterials (to N. Sugiyama), the Helsinki Biomedical Graduate Program (to E. Gucciardo), the Paulo Foundation, the Emil Aaltonen Foundation, and the Iida Montinin Foundation.
Footnotes
Abbreviations used in this paper:
- ADAM
- a disintegrin and metalloprotease
- H&E
- hematoxylin and eosin
- MLC
- myosin light chain
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type-1 MMP
- qPCR
- quantitative PCR
- SCID
- severe combined immunodeficient
References
- Alexander S., Friedl P. 2012. Cancer invasion and resistance: interconnected processes of disease progression and therapy failure. Trends Mol. Med. 18:13–26 10.1016/j.molmed.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Astin J.W., Batson J., Kadir S., Charlet J., Persad R.A., Gillatt D., Oxley J.D., Nobes C.D. 2010. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat. Cell Biol. 12:1194–1204 10.1038/ncb2122 [DOI] [PubMed] [Google Scholar]
- Barbolina M.V., Adley B.P., Ariztia E.V., Liu Y., Stack M.S. 2007. Microenvironmental regulation of membrane type 1 matrix metalloproteinase activity in ovarian carcinoma cells via collagen-induced EGR1 expression. J. Biol. Chem. 282:4924–4931 10.1074/jbc.M608428200 [DOI] [PubMed] [Google Scholar]
- Blobel C.P. 2005. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 6:32–43 10.1038/nrm1548 [DOI] [PubMed] [Google Scholar]
- Brantley-Sieders D.M. 2012. Clinical relevance of Ephs and ephrins in cancer: lessons from breast, colorectal, and lung cancer profiling. Semin. Cell Dev. Biol. 23:102–108 10.1016/j.semcdb.2011.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantley-Sieders D.M., Jiang A., Sarma K., Badu-Nkansah A., Walter D.L., Shyr Y., Chen J. 2011. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PLoS ONE. 6:e24426 10.1371/journal.pone.0024426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.M., Wong H.L., Jin G., Liu B., Cao R., Cao Y., Lehti K., Tryggvason K., Zhou Z. 2012. MT1-MMP inactivates ADAM9 to regulate FGFR2 signaling and calvarial osteogenesis. Dev. Cell. 22:1176–1190 10.1016/j.devcel.2012.04.014 [DOI] [PubMed] [Google Scholar]
- Egeblad M., Rasch M.G., Weaver V.M. 2010. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 22:697–706 10.1016/j.ceb.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faoro L., Singleton P.A., Cervantes G.M., Lennon F.E., Choong N.W., Kanteti R., Ferguson B.D., Husain A.N., Tretiakova M.S., Ramnath N., et al. 2010. EphA2 mutation in lung squamous cell carcinoma promotes increased cell survival, cell invasion, focal adhesions, and mammalian target of rapamycin activation. J. Biol. Chem. 285:18575–18585 10.1074/jbc.M109.075085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Wolf K. 2010. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188:11–19 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri S., Manning C., Hooper S., Jones L., Hill C.S., Sahai E. 2009. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 11:1287–1296 10.1038/ncb1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri S., Pinner S., Sahai E. 2010. Intravital imaging illuminates transforming growth factor beta signaling switches during metastasis. Cancer Res. 70:3435–3439 10.1158/0008-5472.CAN-10-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harunaga J.S., Yamada K.M. 2011. Cell-matrix adhesions in 3D. Matrix Biol. 30:363–368 10.1016/j.matbio.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen J.P., Saha N., Nikolov D.B. 2007. Cell-cell signaling via Eph receptors and ephrins. Curr. Opin. Cell Biol. 19:534–542 10.1016/j.ceb.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen J.P., Yermekbayeva L., Janes P.W., Walker J.R., Xu K., Atapattu L., Rajashankar K.R., Mensinga A., Lackmann M., Nikolov D.B., Dhe-Paganon S. 2010. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. USA. 107:10860–10865 10.1073/pnas.1004148107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto-Yamaki N., Takeuchi S., Ueda S., Harada K., Fujimoto S., Negishi M., Katoh H. 2010. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J. Cell Biol. 190:461–477 10.1083/jcb.201005141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K.B., Allen E.D., Brooks P.C., Datta N.S., Long M.W., Weiss S.J. 2003. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 114:33–45 10.1016/S0092-8674(03)00513-0 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Seiki M. 2006. MT1-MMP: a potent modifier of pericellular microenvironment. J. Cell. Physiol. 206:1–8 10.1002/jcp.20431 [DOI] [PubMed] [Google Scholar]
- Janes P.W., Saha N., Barton W.A., Kolev M.V., Wimmer-Kleikamp S.H., Nievergall E., Blobel C.P., Himanen J.P., Lackmann M., Nikolov D.B. 2005. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 123:291–304 10.1016/j.cell.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Janes P.W., Nievergall E., Lackmann M. 2012. Concepts and consequences of Eph receptor clustering. Semin. Cell Dev. Biol. 23:43–50 10.1016/j.semcdb.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Lehti K., Lohi J., Juntunen M.M., Pei D., Keski-Oja J. 2002. Oligomerization through hemopexin and cytoplasmic domains regulates the activity and turnover of membrane-type 1 matrix metalloproteinase. J. Biol. Chem. 277:8440–8448 10.1074/jbc.M109128200 [DOI] [PubMed] [Google Scholar]
- Lehti K., Allen E., Birkedal-Hansen H., Holmbeck K., Miyake Y., Chun T.H., Weiss S.J. 2005. An MT1-MMP-PDGF receptor-beta axis regulates mural cell investment of the microvasculature. Genes Dev. 19:979–991 10.1101/gad.1294605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti K., Rose N.F., Valavaara S., Weiss S.J., Keski-Oja J. 2009. MT1-MMP promotes vascular smooth muscle dedifferentiation through LRP1 processing. J. Cell Sci. 122:126–135 10.1242/jcs.035279 [DOI] [PubMed] [Google Scholar]
- Li X.Y., Ota I., Yana I., Sabeh F., Weiss S.J. 2008. Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol. Biol. Cell. 19:3221–3233 10.1091/mbc.E08-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae M., Neve R.M., Rodriguez-Viciana P., Haqq C., Yeh J., Chen C., Gray J.W., McCormick F. 2005. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 8:111–118 10.1016/j.ccr.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Margaryan N.V., Strizzi L., Abbott D.E., Seftor E.A., Rao M.S., Hendrix M.J., Hess A.R. 2009. EphA2 as a promoter of melanoma tumorigenicity. Cancer Biol. Ther. 8:279–288 10.4161/cbt.8.3.7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Wang B. 2012. EphA receptor signaling—complexity and emerging themes. Semin. Cell Dev. Biol. 23:16–25 10.1016/j.semcdb.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Li D.Q., Mukherjee A., Guo H., Petty A., Cutter J., Basilion J.P., Sedor J., Wu J., Danielpour D., et al. 2009. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 16:9–20 10.1016/j.ccr.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Nam J.M., Kojima C., Mochizuki N., Sabe H. 2009. EphA2 engages Git1 to suppress Arf6 activity modulating epithelial cell-cell contacts. Mol. Biol. Cell. 20:1949–1959 10.1091/mbc.E08-06-0549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10:515–527 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergall E., Lackmann M., Janes P.W. 2012. Eph-dependent cell-cell adhesion and segregation in development and cancer. Cell. Mol. Life Sci. 69:1813–1842 10.1007/s00018-011-0900-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyalendo C., Michaud M., Beaulieu E., Roghi C., Murphy G., Gingras D., Béliveau R. 2007. Src-dependent phosphorylation of membrane type I matrix metalloproteinase on cytoplasmic tyrosine 573: role in endothelial and tumor cell migration. J. Biol. Chem. 282:15690–15699 10.1074/jbc.M608045200 [DOI] [PubMed] [Google Scholar]
- Okada A., Bellocq J.P., Rouyer N., Chenard M.P., Rio M.C., Chambon P., Basset P. 1995. Membrane-type matrix metalloproteinase (MT-MMP) gene is expressed in stromal cells of human colon, breast, and head and neck carcinomas. Proc. Natl. Acad. Sci. USA. 92:2730–2734 10.1073/pnas.92.7.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota I., Li X.Y., Hu Y., Weiss S.J. 2009. Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc. Natl. Acad. Sci. USA. 106:20318–20323 10.1073/pnas.0910962106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock S.W., Dunn G.A. 1986. Analysing collisions between fibroblasts and fibrosarcoma cells: fibrosarcoma cells show an active invasionary response. J. Cell Sci. 81:163–187 [DOI] [PubMed] [Google Scholar]
- Parri M., Taddei M.L., Bianchini F., Calorini L., Chiarugi P. 2009. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 69:2072–2081 10.1158/0008-5472.CAN-08-1845 [DOI] [PubMed] [Google Scholar]
- Pasquale E.B. 2005. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 6:462–475 10.1038/nrm1662 [DOI] [PubMed] [Google Scholar]
- Pasquale E.B. 2008. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 133:38–52 10.1016/j.cell.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Sabeh F., Shimizu-Hirota R., Weiss S.J. 2009. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185:11–19 10.1083/jcb.200807195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V., Marshall C.J. 2010. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 22:690–696 10.1016/j.ceb.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Seiradake E., Harlos K., Sutton G., Aricescu A.R., Jones E.Y. 2010. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat. Struct. Mol. Biol. 17:398–402 10.1038/nsmb.1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N., Varjosalo M., Meller P., Lohi J., Chan K.M., Zhou Z., Alitalo K., Taipale J., Keski-Oja J., Lehti K. 2010a. FGF receptor-4 (FGFR4) polymorphism acts as an activity switch of a membrane type 1 matrix metalloproteinase-FGFR4 complex. Proc. Natl. Acad. Sci. USA. 107:15786–15791 10.1073/pnas.0914459107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N., Varjosalo M., Meller P., Lohi J., Hyytiäinen M., Kilpinen S., Kallioniemi O., Ingvarsen S., Engelholm L.H., Taipale J., et al. 2010b. Fibroblast growth factor receptor 4 regulates tumor invasion by coupling fibroblast growth factor signaling to extracellular matrix degradation. Cancer Res. 70:7851–7861 10.1158/0008-5472.CAN-10-1223 [DOI] [PubMed] [Google Scholar]
- Szabova L., Yamada S.S., Birkedal-Hansen H., Holmbeck K. 2005. Expression pattern of four membrane-type matrix metalloproteinases in the normal and diseased mouse mammary gland. J. Cell. Physiol. 205:123–132 10.1002/jcp.20385 [DOI] [PubMed] [Google Scholar]
- Taddei M.L., Parri M., Angelucci A., Bianchini F., Marconi C., Giannoni E., Raugei G., Bologna M., Calorini L., Chiarugi P. 2011. EphA2 induces metastatic growth regulating amoeboid motility and clonogenic potential in prostate carcinoma cells. Mol. Cancer Res. 9:149–160 10.1158/1541-7786.MCR-10-0298 [DOI] [PubMed] [Google Scholar]
- Tatti O., Vehviläinen P., Lehti K., Keski-Oja J. 2008. MT1-MMP releases latent TGF-beta1 from endothelial cell extracellular matrix via proteolytic processing of LTBP-1. Exp. Cell Res. 314:2501–2514 10.1016/j.yexcr.2008.05.018 [DOI] [PubMed] [Google Scholar]
- Varjosalo M., Björklund M., Cheng F., Syvänen H., Kivioja T., Kilpinen S., Sun Z., Kallioniemi O., Stunnenberg H.G., He W.W., et al. 2008. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 133:537–548 10.1016/j.cell.2008.02.047 [DOI] [PubMed] [Google Scholar]
- Wolf K., Friedl P. 2011. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 21:736–744 10.1016/j.tcb.2011.09.006 [DOI] [PubMed] [Google Scholar]
- Wykosky J., Debinski W. 2008. The EphA2 receptor and ephrinA1 ligand in solid tumors: function and therapeutic targeting. Mol. Cancer Res. 6:1795–1806 10.1158/1541-7786.MCR-08-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykosky J., Gibo D.M., Stanton C., Debinski W. 2005. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol. Cancer Res. 3:541–551 10.1158/1541-7786.MCR-05-0056 [DOI] [PubMed] [Google Scholar]
- Yilmaz M., Christofori G. 2010. Mechanisms of motility in metastasizing cells. Mol. Cancer Res. 8:629–642 10.1158/1541-7786.MCR-10-0139 [DOI] [PubMed] [Google Scholar]
- Zantek N.D., Azimi M., Fedor-Chaiken M., Wang B., Brackenbury R., Kinch M.S. 1999. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 10:629–638 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.