In sensory organ precursor cells, Sanpodo can enhance or suppress Notch signaling by promoting interaction with Presenilin or driving receptor internalization, respectively.
Abstract
In Drosophila peripheral neurogenesis, Notch controls cell fates in sensory organ precursor (SOP) cells. SOPs undergo asymmetric cell division by segregating Numb, which inhibits Notch signaling, into the pIIb daughter cell after cytokinesis. In contrast, in the pIIa daughter cell, Notch is activated and requires Sanpodo, but its mechanism of action has not been elucidated. As Sanpodo is present in both pIIa and pIIb cells, a second role for Sanpodo in regulating Notch signaling in the low-Notch pIIb cell has been proposed. Here we demonstrate that Sanpodo regulates Notch signaling levels in both pIIa and pIIb cells via distinct mechanisms. The interaction of Sanpodo with Presenilin, a component of the γ-secretase complex, was required for Notch activation and pIIa cell fate. In contrast, Sanpodo suppresses Notch signaling in the pIIb cell by driving Notch receptor internalization. Together, these results demonstrate that a single protein can regulate Notch signaling through distinct mechanisms to either promote or suppress signaling depending on the local cellular context.
Introduction
Asymmetric cell division (ACD) is an evolutionarily conserved mechanism for generating cell fate diversity during development (Horvitz and Herskowitz, 1992; Knoblich, 2010). A striking example of ACD occurs during adult peripheral neurogenesis in Drosophila: the sensory organ precursor (SOP) cell, an epithelial cell–derived progenitor, undergoes four rounds of asymmetric cell division, giving rise to four mature cells that differentiate into a functional mechanosensory organ. As the SOP cell enters mitosis, the mitotic spindle aligns along the anterior–posterior axis and Numb localizes asymmetrically along the anterior membrane (Roegiers and Jan, 2004). Numb inhibits Notch signaling in the anterior SOP daughter cell (pIIb), while its absence in the adjacent pIIa cell permits Notch signaling activity (Rhyu et al., 1994).
Notch signaling in the pIIa cell is activated in trans via binding of the membrane-tethered ligand Delta, present on the surface of the pIIb cell, to the Notch receptor in the pIIa cell (Kandachar and Roegiers, 2012). Ligand binding induces sequential cleavage of the Notch receptor at the plasma membrane first by the ADAM metalloprotease (S2 cleavage) and then by the γ-secretase complex (S3 cleavage; Bray, 2006; Ilagan and Kopan, 2007). The S3 cleavage releases the Notch intracellular domain (NICD) from the membrane, and the NICD translocates to the pIIa cell nucleus and interacts with the DNA-binding protein CSL (CBF1/Suppressor of Hairless/Lag1) and the transcriptional coactivator Mastermind to regulate Notch target gene expression.
Sanpodo, a four-pass transmembrane protein, is expressed in SOP cells, and unlike Numb, is distributed to both pIIa and pIIb cells after mitosis (O’Connor-Giles and Skeath, 2003). In the pIIa cell, activation of Notch requires sanpodo, and Sanpodo protein localizes primarily at the plasma membrane (Salzberg et al., 1994; Park et al., 1998; Skeath and Doe, 1998; O’Connor-Giles and Skeath, 2003; Roegiers et al., 2005). However, the mechanism of sanpodo action in the pIIa cell has yet to be defined. In contrast, observations that Sanpodo localizes primarily to endosomes in pIIb cells in a Numb-dependent manner (Hutterer and Knoblich, 2005; Langevin et al., 2005; Roegiers et al., 2005) suggests a possible role for Sanpodo in modulating Notch activity in pIIb cells as well (O’Connor-Giles and Skeath, 2003; Babaoglan et al., 2009). In this study, we uncover the mechanistic role of Sanpodo in promoting Notch signaling in the pIIa cell, and determine the function of Sanpodo in regulating pIIb cell fate.
Results
Sanpodo binds the Presenilin subunit of the γ-secretase complex
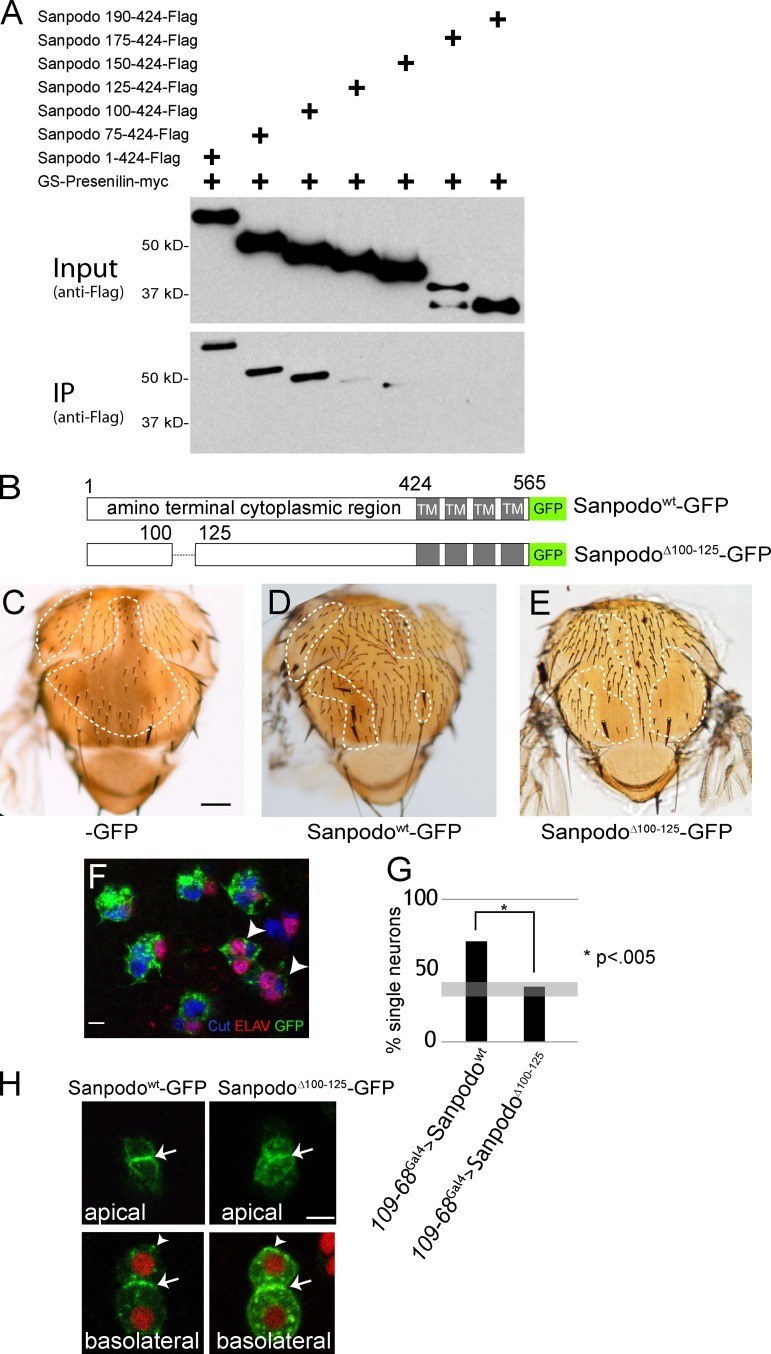
sanpodo loss-of-function mutants show an incompletely penetrant phenotype of failure to induce Notch signaling in pIIa cells, resulting in areas of balding on the pupal thorax (Jafar-Nejad et al., 2005; Roegiers et al., 2005). Release of the Notch intracellular domain from the plasma membrane after ligand binding requires the proteolytic activity of the γ-secretase complex, which is composed of four distinct transmembrane protein subunits: Pen-2, Aph-1, Nicastrin, and Presenilin. Genetic analysis suggests a requirement for sanpodo at the γ-secretase cleavage step of Notch activation (O’Connor-Giles and Skeath, 2003). We therefore tested whether Sanpodo physically associates with the γ-secretase complex in a coimmunoprecipitation assay. The four components of the γ-secretase complex must be present in roughly stoichiometric levels for the ordered assembly of the complex, and subsequent processing and trafficking to the plasma membrane (Hu and Fortini, 2003; Stempfle et al., 2010). We coexpressed all four γ-secretase complex components in S2 cells from a single plasmid together with the Sanpodo amino-terminal cytoplasmic region (ATCR; amino acids 1–424). We found that the Sanpodo ATCR coimmunoprecipitated with myc-tagged Presenilin, whereas a Sanpodo amino-terminal deletion mutant (SanpodoΔN190) did not (Fig. 1 A). We failed to detect an interaction between Sanpodo and overexpressed Presenilin alone in vivo, suggesting that assembly of the γ-secretase complex may be required for the Sanpodo interaction (unpublished data). To further narrow down the region of the Sanpodo cytoplasmic domain responsible for Presenilin binding, we generated a series of amino-terminal truncations of the Sanpodo ATCR and assessed their ability to bind Presenilin. We found that the region between 100 and 125 amino acids of the Sanpodo amino terminus is necessary for binding in CoIP assays in S2 cells (Fig. 1 A).
Figure 1.
Sanpodo’s interaction with Presenilin is required for pIIa cell fate. Coexpression of the γ-secretase complex with myc-tagged Presenilin and Flag-tagged Sanpodo amino-terminal fragments in Drosophila S2 cells. (A) Binding of the amino-terminal cytoplasmic portion of Sanpodo (amino acids 1–424) to Presenilin-myc requires amino acids 100–125. (B) Schematic of SanpodoΔ100–125 mutant, deleting amino acids 100–125 within the amino terminus. (C–E) Thoraces of flies with sanpodoc55Sb e mutant clones (dotted line: approximate clone boundaries, as determined by e/e and Sb/Sb) with 109-68-Gal4–driven expression of GFP, Sanpodowt-GFP, or SanpodoΔ100–125–GFP using the MARCM system. Sanpodowt-GFP expression largely restores the wild-type bristle pattern to the thorax in sanpodo mutant clones (C; bar, 200 µm), whereas SanpodoΔ100–125–GFP expression fails to suppress the characteristic balding (D). Supernumerary neurons (marked with both Cut [blue] and ELAV [red]) in pupal sanpodoC55 mutant sensory organs expressing SanpodoΔ100–125–GFP (green) using the MARCM system (F, arrowhead). Bar, 5 µm. (G) Quantification of pupal sanpodo mutant sensory organs with only one ELAV+ neuron, indicating suppression of the sanpodo mutant phenotype; when expressing either Sanpodowt-GFP or SanpodoΔ100–125–GFP in MARCM clones using 109-68-Gal4, expression of SanpodoΔ100–125–GFP does not suppress the incompletely penetrant sanpodo mutant phenotype (gray bar indicates previously reported range of single neuron sensory organs in sanpodo null mutant clones; Jafar-Nejad et al., 2005; Roegiers et al., 2005). (H) SanpodoΔ100–125–GFP localizes to the apical and basolateral plasma membrane (white arrows) and intracellular puncta (white arrowheads) in pIIb/pIIa cells, similarly to wild-type Sanpodo-GFP (Histone RFP [red], Sanpodowt-GFP and SanpodoΔ100–125–GFP [green]). Bar, 2 µm.
The 25–amino acid region that is required for Presenilin binding is not strongly conserved among other sanpodo orthologues in insects (Fig. S1 A); however, this region contains two six–amino acid sequences beginning with RY and enriched in hydrophobic residues. Interestingly, we observed variable numbers of these RYXXXX sequences are clearly present in Sanpodo orthologues from a broad range of insects (Fig. S1 A). Although we found that specific deletion of amino acids 100–125 in the Sanpodo ATCR abrogated Presenilin binding in vitro, mutation of the RY residues to alanines did not affect Presenilin binding in vitro (Fig. S2), suggesting other residues or overall secondary structure of this region may mediate the interaction.
To determine whether loss of the 25–amino acid region alters Sanpodo function in vivo, we generated flies carrying GFP-tagged full-length Sanpodo with a specific deletion of amino acids 100–125 (Fig. 1 B) under UAS control. sanpodo is required for Notch-dependent specification of the pIIa cell after asymmetric cell division, and sanpodo mutant clones on the adult fly thorax exhibit a significant loss of both hair and socket cells, which are the differentiated progeny of the pIIa cell, and a concomitant increase in neurons, which are progeny of the pIIb cell (Roegiers et al., 2005). We therefore tested whether SanpodoΔ100–125–GFP could rescue the loss of pIIa cell progeny in sanpodoC55 mutant clones and restore the wild-type bristle pattern on the thorax. Expression of SanpodoΔ100–125–GFP in sanpodoC55 mutant clones did not rescue, leaving large regions of the adult cuticle devoid of bristles, and failing to restrict neuronal differentiation in the lineage (Fig. 1, C–G). In contrast, wild-type Sanpodo-GFP or mutant versions of Sanpodo with deletions of similar size in nearby regions (amino acids 72–94 or 194–255) completely restore the wild-type bristle pattern (Fig. 1, C–G; Tong et al., 2010; and unpublished data). By live-cell imaging, we observed that SanpodoΔ100–125–GFP localization was grossly similar to Sanpodo-GFP in pIIa and pIIb cells (Fig. 1 H). From these observations we conclude that Sanpodo interacts with Presenilin via the region between 100 and 125 amino acids of the Sanpodo amino terminus, and that binding to Presenilin has an essential role in promoting Notch pathway activation in SOP lineage cells.
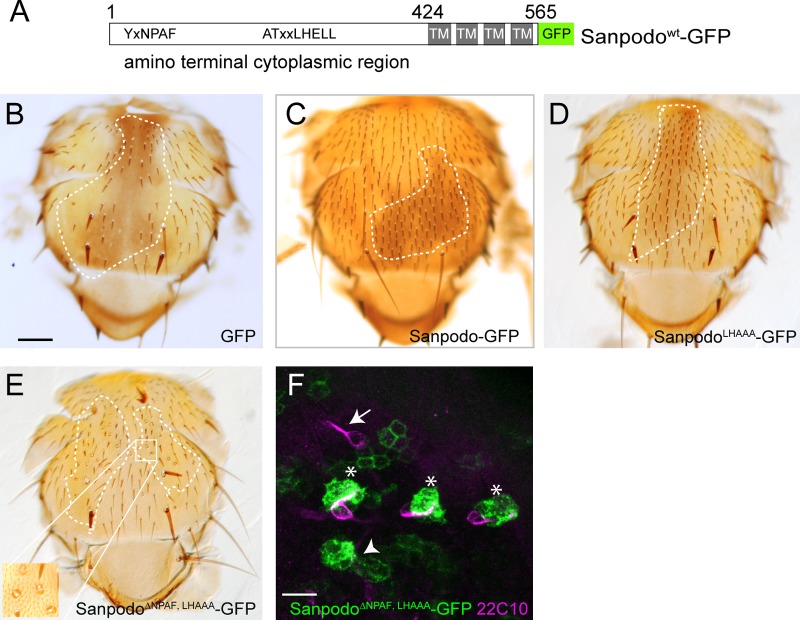
The Sanpodo NPAF and ELL motifs inhibit Notch signaling in the pIIb cell
Previously, we identified a conserved motif, NPAF, within the Sanpodo cytoplasmic domain that binds Numb and regulates Sanpodo targeting to endosomes in dividing SOP lineage cells (Tong et al., 2010). The NPAF motif is a variant of the NPxY motif that is a known endocytic trafficking signal associated with membrane proteins such as integrins and the LDL receptor. However, removal of this motif had no effect on Sanpodo’s ability to regulate Notch signaling in SOP lineage cells. We therefore searched for additional conserved motifs that could act together with the NPAF motif to control Sanpodo membrane targeting. Analysis of sanpodo homologues in other insect species revealed that in addition to the NPAF motif, there is a third conserved motif, ATxxLHELL (LHELL), at residues 293–301 in the amino-terminal cytoplasmic domain of Sanpodo (Fig. 2 A; Fig. S1 A). The LHELL motif contains a dileucine and an upstream acidic residue, characteristic of endocytic sorting signals (Bonifacino and Traub, 2003). To test the functional role of the LHELL motif, we generated a mutant form of Sanpodo-GFP substituting the ELL residues of the motif with alanines (LHAAA). To determine whether the ELL motif was required for controlling Notch signaling in pIIa and pIIb cells, we tested SanpodoLHAAA-GFP’s ability to rescue the sanpodoC55 mutant bristle loss phenotype in clones. We showed previously that Sanpodowt-GFP and SanpodoΔNPAF-GFP could restore the wild-type bristle pattern in sanpodoC55 clones when expressed using the scabrous-Gal4 driver (Fig. 2, B and C; Tong et al., 2010). Similarly, SanpodoLHAAA-GFP fully rescued sanpodoC55 clones, in contrast to GFP controls that exhibited extensive bristle loss (Fig. 2, B and D). From this result, we conclude that the ELL motif is dispensable for Sanpodo function in SOP lineage cells. We next tested whether disruption of both the NPAF and ELL motifs altered Sanpodo function. In the rescue assay, SanpodoΔNPAF, LHAAA-GFP expression reversed the strong balding phenotype of sanpodo mutant clones, indicating that the NPAF and ELL motifs together are not required for promoting Notch signaling in the pIIa cell (Fig. 2 E). However, closer examination revealed that while 70.1% of the rescued organs exhibited the wild-type single hair and socket cell (n = 578 organs), the remaining mutant organs contained supernumerary socket cells, a phenotype consistent with persistent Notch activation in the sensory organ lineage (Fig. 2 E, inset). We further characterized cell fate assignments of SanpodoΔNPAF, LHAAA-GFP rescued sanpodo mutant clones by labeling neurons in pupal sensory organ cell clusters. Expression of the SanpodoΔNPAF, LHAAA-GFP mutant completely suppressed supernumerary neuron phonotype observed in sanpodo mutant clones, limiting most organs to a single neuron (438/476 labeled cell clusters from 23 flies), but nearly 8% of rescued organs showed no neuronal marker expression (38/476, Fig. 2 F). These findings indicate that the Sanpodo NPAF and ELL motifs function to restrict socket cell fate and to promote neuronal cell fate in adult sensory organ cells.
Figure 2.
Sanpodo NPAF and ELL motif are required for Notch inhibition in pIIb cells. (A) Schematic of Sanpodowt-GFP transgene with NPAF and LHELL motifs shown. (B–E) scabrous-Gal4–driven expression of wild-type and mutant Sanpodo transgenes in sanpodoc55Sb e mutant clones using the MARCM system (approximate clone borders are marked with dotted line). Bar, 200 µm. (B) GFP expression fails to restore the wild-type bristle pattern to the thorax in sanpodoc55Sb e mutant clones, whereas both Sanpodowt-GFP and SanpodoLHAAA-GFP expression (C and D) fully rescue the sanpodo mutant phenotype. (E) SanpodoΔNPAF-LHAAA–GFP expression suppresses the bristle loss phenotype associated with the Sanpodo mutant, but a number of rescued bristles exhibit supernumerary socket cells (inset). (F) Sensory organs from pupae Expressing SanpodoΔNPAF-LHAAA-GFP (green) in sanpodoc55Sb e mutant clones using the MARCM system, labeled with the neuronal marker 22C10 (purple). Bar, 10 µm. Single 22C10-positive neurons are visible in most GFP+ sanpodo mutant sensory organs (*), as well as outside the clone region (white arrow), but are absent in others (white arrowhead).
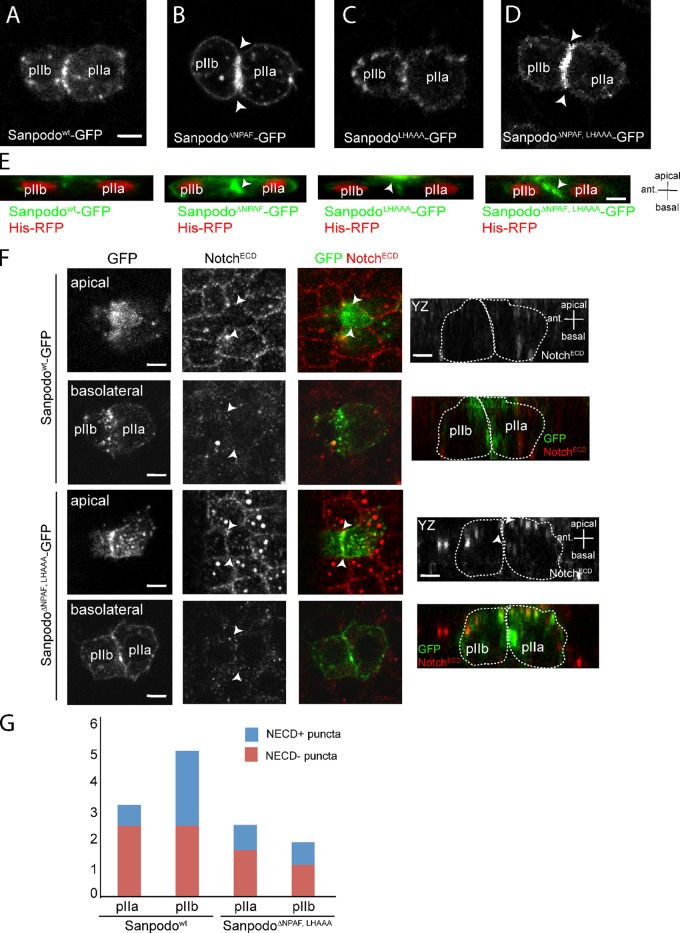
Deletion of the NPAF motif alone in Sanpodo decreases Sanpodo targeting to endosomes and increases Sanpodo levels at the plasma membrane in SOP lineage cells (Fig. 3, A and B; Tong et al., 2010). We used live-cell imaging to determine the subcellular localization of the SanpodoLHAAA-GFP mutant in SOP lineage cells. Like Sanpodowt-GFP, SanpodoLHAAA-GFP localized to both the basolateral plasma membrane and to endocytic vesicles (Fig. 3, A–C). However, SanpodoLHAAA-GFP exhibited enrichment at the apical region of the pIIa/pIIb daughter cells, as compared with Sanpodowt-GFP (Fig. 3 E). Interestingly, deletion of both the NPAF and ELL motifs suppressed the apical accumulation observed in the ELL mutant alone, and resulted in accumulation at the basolateral membrane in pIIa/pIIb cells (Fig. 3, D and E). This result indicates the apical accumulation of SanpodoLHAAA-GFP requires the NPAF motif and is potentially due to recycling of the protein from the endosomal compartment to the apical membrane.
Figure 3.
The NPAF and ELL motifs control Sanpodo trafficking in pIIa and pIIb cells. (A–D) Live-cell imaging of neuralized-Gal4–driven expression of GFP-tagged Sanpodo transgenes in pIIa/pIIb cell pairs; XY equatorial view. Bar, 2 µm. Sanpodowt-GFP (A) and SanpodoLHAAA-GFP (C) localize to the plasma membrane and in intracellular puncta, whereas SanpodoΔNPAF-GFP (B) and SanpodoΔNPAF, LHAAA–GFP (D) accumulate at the plasma membrane in both daughter cells, particularly at the membrane interface between pIIa/pIIb (B and D, white arrowheads). (E) Live-cell analysis of Sanpodo mutant transgenes (green) in pIIa and pIIb cells expressing His-RFP (red) 7 min after SOP mitotic exit; YZ view, apical at top, anterior to the left. Bar, 2 µm. SanpodoΔNPAF-GFP and SanpodoΔNPAF-LHAAA–GFP accumulate strongly at the basolateral interface (white arrowhead), whereas SanpodoLHAAA-GFP accumulates at the apical plasma membrane in pIIa and pIIb cells, compared with Sanpodowt-GFP. Notch (NotchECD, red) is depleted from apical (white arrowheads) pIIa/pIIb membrane interface in Sanpodo-GFP–expressing cells. (F) Apical membrane Notch (white arrowheads) is detected at low levels in cell rescued with SanpodoΔNPAF, LHAAA-GFP, whereas basolateral Notch levels are similar to wild type (YZ planes, apical is at the top, approximate cell outlines represented by dashed lines). Bar, 2 µm. (G) Quantification of colocalization of Sanpodo and NECD in intracellular vesicles in pIIa and pIIb cells. Wild-type Sanpodo-GFP accumulates in large intracellular (>0.5 µm) puncta, which partially colocalize with Notch, in higher numbers in pIIb cells. SanpodoΔNPAF, LHAAA-GFP puncta are less abundant and are equally distributed in pIIa and pIIb cells (standard deviation of 2.215–3.599 total Sanpodo + puncta per cell, n = 7 WT; n = 22 NPAF, LHAAA).
Because we observed inappropriate Notch activation in some cells when we expressed the SanpodoΔNPAF, LHAAA-GFP mutant in a rescue assay, we asked whether Notch subcellular localization was altered in SOP lineage cells. In wild-type pIIa/pIIb cells expressing Sanpodo-GFP, low levels of apical membrane Notch are present, and Notch levels in intracellular vesicles are higher in pIIb cells (Fig. 3, F and G). In contrast, we detected Notch at the apical region coincident with the plasma membrane interface between the pIIa and pIIb cell, whereas Notch was not enriched at the basolateral plasma membrane interface. Mutation of NPAF and ELL motifs also decreased the number of Sanpodo-colocalizing Notch intracellular puncta in pIIa/pIIb cell pairs. From these findings we concluded that the NPAF and ELL motifs are required for controlling Sanpodo membrane targeting and may contribute to regulating membrane Notch levels in pIIb/pIIa cells.
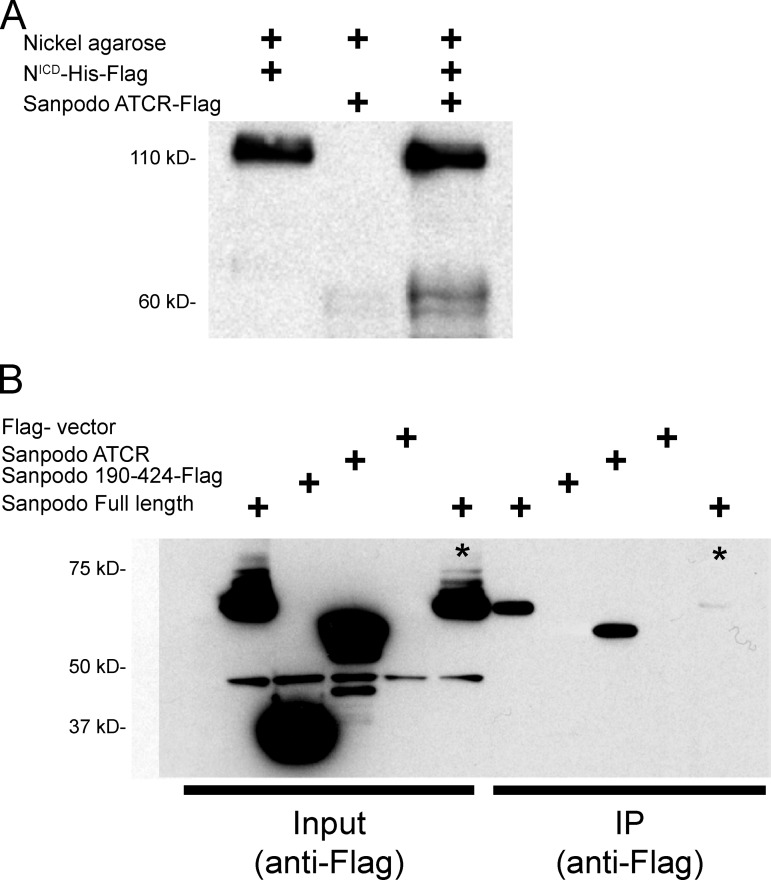
Sanpodo binds the Notch receptor directly
Although Sanpodo has been shown to associate with Notch in vivo and is required for robust Notch signaling activity in the pIIa cell (Roegiers et al., 2001, 2005; O’Connor-Giles and Skeath, 2003), it is unclear if the physical association between Sanpodo and Notch is direct, or whether it depends on other proteins such as Numb. We assessed the nature of the Sanpodo–Notch interaction using in vitro protein interaction assays and coimmunoprecipitation studies. We generated tagged versions of the Notch intracellular domain (NICD) and Sanpodo amino-terminal cytoplasmic domain in vitro and found that in binding assays, Notch and Sanpodo intracellular domains coprecipitated (Fig. 4 A). We next verified that Sanpodo’s interaction with Notch requires the amino-terminal portion of the Sanpodo cytoplasmic domain, which we have previously shown is required for Sanpodo function in Notch signaling in vivo (Tong et al., 2010). Whereas the NICD immunoprecipitates with both full-length Sanpodo and the Sanpodo amino-terminal cytoplasmic domain in lysates from S2 cells, a truncated version of the Sanpodo cytoplasmic domain, which lacks the amino-terminal 180 amino acids, fails to bind robustly in this assay (Fig. 4 B). We therefore conclude that Sanpodo binds directly to the NICD, and this binding interaction requires the same amino-terminal region of Sanpodo that is necessary for Notch regulation.
Figure 4.
Sanpodo binds the Notch intracellular domain. (A) Binding of Flag-tagged Sanpodo amino-terminal cytoplasmic domain (424 amino acids, detected at ∼60 kD) to the nickel Agarose beads is strongly increased when preincubated with the Flag- and His-tagged NICD, which binds to nickel Agarose beads robustly and is detected at 110 kD with the anti-Flag antibody. (B) Full-length Notch coimmunoprecipitates with either transfected full-length Sanpodo (all Sanpodo transgenes are Flag tagged) or the Sanpodo cytoplasmic domain in the induced S2-MT-Notch Drosophila cell line, whereas the amino-terminal truncated Sanpodo cytoplasmic domain (190–424) does not (asterisk indicates Notch uninduced).
Sanpodo controls Notch trafficking via conserved NPAF and ELL motifs
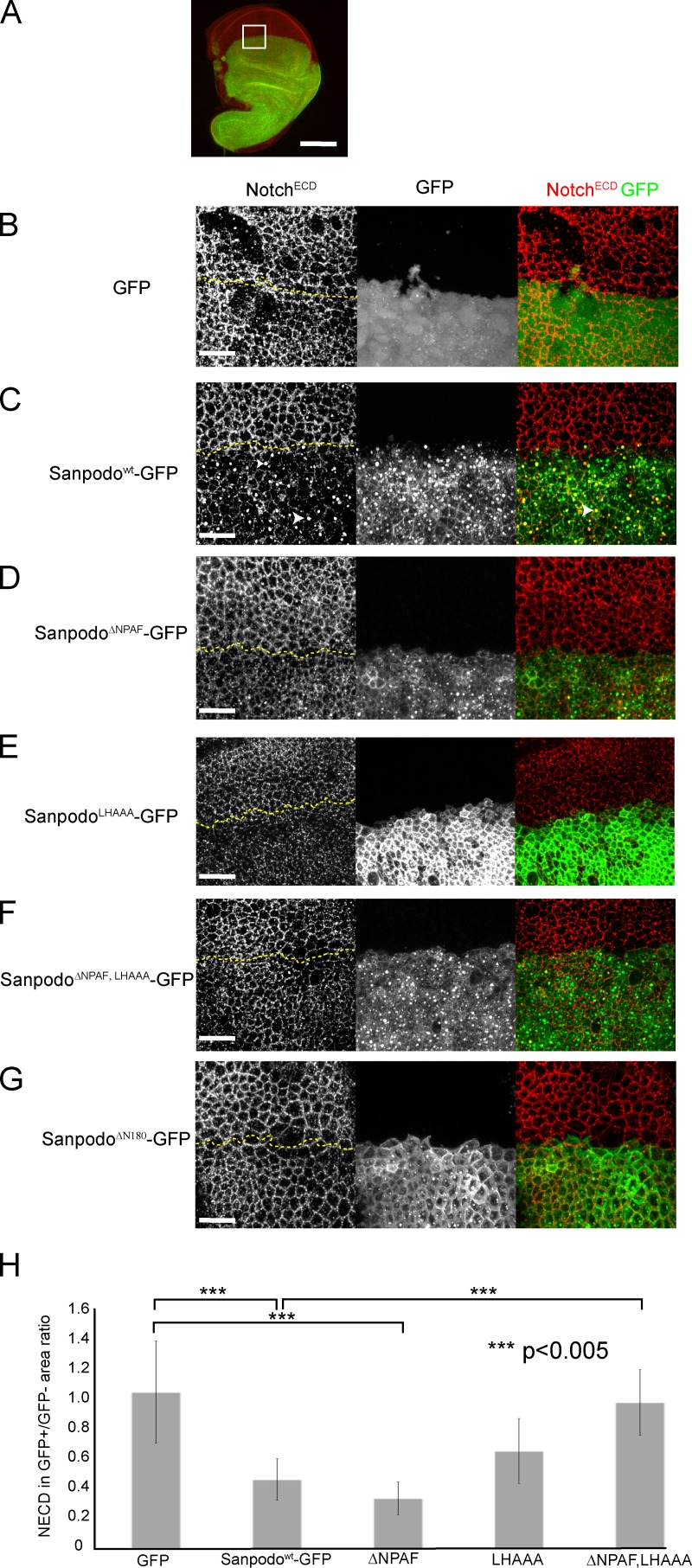
Based on our observations in SOP cells expressing wild-type and mutant Sanpodo transgenes, we hypothesized that if Sanpodo is being actively endocytosed and recycled, and binding to the Notch receptor directly, we would expect to see alterations in Notch receptor localization in the presence of Sanpodo. We tested this in wing disc epithelial cells in flies, which endogenously express Notch and Numb but not Sanpodo (Fig. 5 A). Ectopic expression of Sanpodowt-GFP, or a Sanpodo mutant lacking the region containing the Numb-binding amino-terminal NPAF motif (SanpodoΔNPAF-GFP) with the apterous-Gal4 driver, which drives expression in a subset of wing-disc epithelial cells, depleted Notch from the apical junctions of epithelial cells, and targeted Notch to intracellular puncta (Fig. 5, B–D and H), consistent with recently reported findings (Couturier et al., 2012). Mutation of ELL motif had at best a modest effect on Sanpodo’s ability to deplete Notch from the apical junction of epithelial cells (Fig. 5 E; Fig. S1 A). However, disruption of both the NPAF and ELL motifs from Sanpodo-GFP abrogated Notch depletion from the apical cell junctions (Fig. 5, F and H), suggesting that either motif is sufficient to regulate Notch levels at the membrane, but Notch trafficking regulation is lost in the absence of both. In contrast, the SanpodoΔ100–125 mutant, which does not bind Presenilin, depleted apical Notch similarly to wild-type Sanpodo-GFP (unpublished data), whereas the amino-terminal truncated SanpodoΔN180-GFP, which does not bind Notch in vitro, failed to deplete Notch from the apical membrane (Fig. 5 G). We conclude from this analysis that Sanpodo promotes Notch endocytosis from the apical junctions of epithelial cells through binding to the Notch receptor and driving Notch internalization through the redundant activity of the NPAF and ELL motifs.
Figure 5.
Sanpodo promotes Notch endocytosis. (A) apterous-Gal4–driven ectopic expression of GFP-tagged Sanpodo transgenes (green) in wing disc epithelial cells labeled with the NotchECD antibody (red). Bar, 100 µm. White box is the approximate region of disc shown in B–G. Merged apical XY confocal sections through wing disc epithelial cells (1.5-µm thickness, the border between GFP+ and GFP− regions are marked by a dashed yellow line). Expression of GFP alone has no effect on NECD localization to apical cell junctions in wing disc epithelial cells (B), whereas expression of Sanpodowt-GFP depletes Notch from the apical membrane in epithelial cells and causes Notch to accumulate in large apical vesicles, which in some cases colocalize with Sanpodo-GFP (C, white arrowheads). Both the ΔNPAF mutant (D) and the LHAAA mutant (E) retain the ability to deplete apical Notch, whereas the ΔNPAF, LHAAA mutant (F) or ΔN180 mutant of Sanpodo (G, ΔN180) abrogate Notch apical depletion. Quantification of apical NECD staining represented as a ratio of the area occupied by apical NECD staining in the GFP+/GFP− regions of merged apical XY planes from apterous-Gal4–driven ectopic expression of GFP-tagged Sanpodo transgenes and a GFP control 9H). Sanpodo-GFP and SanpodoΔNPAF-GFP significantly deplete membrane Notch levels, whereas Notch levels are decreased in the LHAAA; they do not attain statistical significance. In contrast, Notch levels in the SanpodoΔNPAF, LHAAA–GFP expression region were indistinguishable from GFP control expression, and significantly increased relative to Sanpodo-GFP. Bars, 25 µm.
Localization of Sanpodowt-GFP, SanpodoLHAAA-GFP, and SanpodoΔNPAF, LHAAA–GFP in epithelial cells was similar to localization of these proteins in SOP lineage cells, with SanpodoLHAAA-GFP mutant concentrated at the apical membrane and in large cytoplasmic vesicles whereas SanpodoΔNPAF-GFP, and SanpodoΔNPAF, LHAAA–GFP enriched along the basolateral membrane and sparsely at the apical membrane (Fig. 5, C–F). Previous studies have shown that ectopic expression of Sanpodo in epithelial cells interferes with Notch signaling during lateral inhibition (Fig. S3; and Babaoglan et al., 2009, Couturier et al., 2012). We find that overexpression of Numb enhances Sanpodo’s effect on lateral inhibition, and that deletion of the Sanpodo amino terminus (SanpodoΔN180) or mutation of the ELL motif abrogates the lateral inhibition phenotype (Fig. S3), suggesting that these regions are important for reducing Notch activity in this assay.
Discussion
Notch is a central and conserved regulator of neural cell fate. In asymmetrically dividing neural progenitors, activation of Notch in one daughter cell and suppression of the pathway in the other determines whether a cell will proliferate, differentiate, or die; or decide between neural versus glial cell fates. The mechanisms by which Notch is activated or inhibited after mitosis have been extensively studied, but our understanding remains incomplete (Kandachar and Roegiers, 2012). In contrast to well-studied proteins like Numb, we show that the four-pass transmembrane protein Sanpodo acts both to promote Notch signaling in one daughter cell by linking the Notch receptor to the γ-secretase complex, and to inhibit signaling by internalizing the Notch receptor in the paired daughter cell (Fig. 6).
Figure 6.
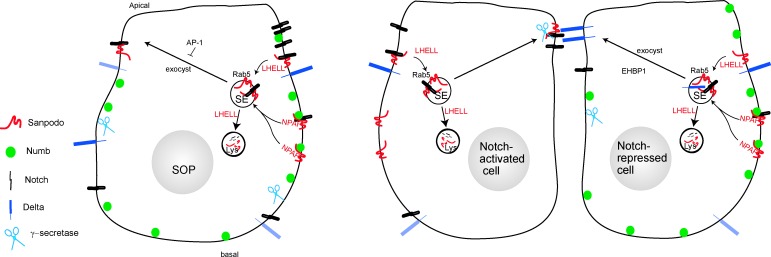
A potential model for Sanpodo trafficking in SOP lineage cells. Sanpodo expression is induced in SOP cells and the bulk of Sanpodo (red) enters the endocytic system, along with Notch, from the basolateral membrane via an interaction of Numb (green) and the Sanpodo NPAF motif. An alternative route for Sanpodo and Notch endocytosis requires the ELL motif. Endocytosed Sanpodo is directed to the lysosome and shunted away from the recycling to the apical membrane by the ELL motif. Sanpodo targeting to the apical membrane is regulated by the exocyst complex and the AP-1 complex of endocytic adaptor proteins.
An elaborate network controls endocytosis, recycling, and membrane targeting of the Notch ligand Delta in pIIb cells to activate signaling in the pIIa cell. Shortly after mitosis, Delta is enriched in an actin-rich structure proximal to the apical adherens junction in the pIIb cell, adjacent to the pIIa cell membrane (Kandachar and Roegiers, 2012). Defects in formation of the actin structure, or failure of Delta endocytosis or recycling, blocks Notch signaling in the pIIa cell (Ben-Yaacov et al., 2001; Le Borgne and Schweisguth, 2003; Rajan et al., 2009; Banks et al., 2011; Giagtzoglou et al., 2012). Similarly, genetic screens had identified sanpodo as an essential gene in activation of the Notch pathway for pIIa cell fate, but it has remained unclear how Sanpodo fulfills this function. The Sanpodo–Presenilin interaction we describe may provide a mechanism to ensure that low levels of ligand-bound Notch receptor in the pIIa cell are linked to the γ-secretase complex for prompt proteolytic processing upon ligand binding. Although our analysis suggests that Sanpodo could act as a linker between Notch and the γ-secretase complex, Sanpodo could also control either the activity of the enzyme or regulate its subcellular localization.
Control of membrane trafficking of transmembrane proteins relies on the interaction between tyrosine-based and dileucine-based motifs within the cytoplasmic domain of the membrane protein and specific endocytic adapter–protein complexes. In a previous study, our laboratory established that the Numb phosphotyrosine-binding domain binds to a tyrosine-based YxxNPAF motif at the Sanpodo amino terminus. Although disruption of this interaction clearly altered Numb-dependent bulk trafficking of Sanpodo from the basolateral plasma membrane to endosomes, the SanpodoΔNPAF mutant was fully able to restore sanpodo function in SOP lineage cells in vivo (Tong et al., 2010). Here, we show that the ELL motif limits Sanpodo accumulation to the apical membrane, both in epithelial cells and in SOP lineage cells. Dileucine signals regulate both internalization and lysosomal targeting of membrane proteins (Bonifacino and Traub, 2003). In Sanpodo, a dileucine signal could either function to promote endocytosis of Sanpodo from apical membrane, or to shunt endocytosed Sanpodo from the recycling pathway to the lysosome, or both. Our analysis of the trafficking of the SanpodoΔNPAF, LHAAA mutant shows that the NPAF-dependent sorting of Sanpodo from the basolateral membrane to endosomes is required for apical accumulation of the SanpodoLHAAA mutant. Therefore, the most likely scenario is that Sanpodo enters the endocytic system primarily from the basolateral membrane via the NPAF motif and the dileucine signal diverts most of the internalized Sanpodo to the lysosome. The dileucine may also play a role in endocytosis of Sanpodo–Notch complexes, as it is redundant with the NPAF motif for depletion of Notch from apical cell junctions in epithelial cells. This would be consistent with our observation that in the sanpodo clones rescued with the SanpodoΔNPAF, LHAAA mutant, Notch accumulates at the apical membrane in the pIIb cells, resulting in a cell fate change from pIIb to pIIa.
Regulation of membrane Notch levels in pIIa and pIIb cells correlate with changes in cell fate (Benhra et al., 2011; Couturier et al., 2012). For example, asymmetric segregation of Numb into the pIIb cell acts to inhibit Notch membrane localization in pIIb cells and promote pIIb cell fate. However, endogenous Numb has virtually no inhibitory effect on Notch signaling in cells where Notch membrane levels are high, such as epithelial cells undergoing Notch-mediated lateral inhibition. In contrast, Numb overexpression strongly enhances Sanpodo’s dominant-negative effect on lateral inhibition (Fig. S3). Numb interacts with endocytic proteins such as α-adaptin and EPS15 (Santolini et al., 2000; Berdnik et al., 2002; Tang et al., 2005), and a recent live-cell imaging study supports the notion that Numb suppresses Notch receptor accumulation at the membrane interface between pIIb and pIIa cells through endocytosis, thereby inhibiting Notch activation in the pIIb cell (Couturier et al., 2012). Our findings support the notion that Sanpodo-mediated endocytosis of apical Notch is essential for robust inhibition of Notch signaling in the pIIb cell. Sanpodo may therefore amplify the difference in Notch signaling in the pIIa cell and pIIb cell by first depleting membrane Notch levels before asymmetric division, then further reducing Notch levels together with Numb in the pIIb cell after SOP mitosis. The mutation of the NPAF and ELL motifs results in a fairly weak Notch activation phenotype in SOP lineage cells, which suggests either that Sanpodo activity is partially redundant with Numb in controlling membrane Notch levels, or that other sorting signals, such as putative tyrosine-based motifs (RYXXXX, which are conserved in sanpodo insect orthologues; Fig. S1 A), contribute to further Notch internalization.
Although sanpodo orthologues have yet to be identified in vertebrates, it is likely that similar mechanisms evolved to control Notch activation and Notch membrane levels, perhaps by more than one protein. Little is currently known about the regulation of membrane trafficking of the Notch receptor in mammalian cells, but both Numb and the ubiquitin ligase Itch/Su(dx) have been implicated in the control of membrane levels of vertebrate Notch1 (Chastagner et al., 2008; McGill et al., 2009). Future studies may therefore identify regulators of Notch trafficking and activation that act like Sanpodo in the vertebrate context and further enhance our understanding of the regulation of Notch signaling during development.
Materials and methods
Generation of mutant Sanpodo-GFP transgenes
The PfuII (Agilent Technologies) amplified coding region of Sanpodo was cloned into a pENTR/D-TOPO vector (Invitrogen), and swapped by LR recombination into the Drosophila Gateway pTWG destination vector containing the UAS–carboxy-terminal GFP (T. Murphy, Carnegie Institute of Washington, Washington, DC; Tong et al., 2010). Sanpodo deletion mutant constructs were made using primers containing targeted deletions. Site-specific mutants of Sanpodo were generated using the QuikChange II mutagenesis kit (Agilent Technologies). Transgenic fly lines were generated by Bestgene. Independent GFP-tagged transgene lines inserted in both the second and third chromosomes behaved similarly in our experiments.
Drosophila genetics, imaging, and immunohistochemistry
Crosses and fly stocks were maintained at 20 or 25°C. Stocks used for these studies were as follows: laboratory stocks yw; apterous-Gal4, yw; neuralized-Gal4/TM3, Sb, yw; scaberous-Gal4/CyO, yw; 109–68-Gal4 FRT40A/CyO, yw; UAS-eGFP, yw; UAS-Sanpodowt-GFP, yw; UAS-SanpodoΔN180-GFP, yw; UAS-SanpodoΔNPAF-GFP, yw; UAS-SanpodoLHAAA-GFP, yw; UAS-SanpodoΔNPAF, LHAAA-GFP, yw; UAS-SanpodoΔ100–125–GFP, yw;FRT82B spdoC55, Sb1, e1/TM6B, ubx. Stocks derived from Bloomington Drosophila Stock Center stocks: w; UAS-Sanpodo, yw, ubx-FLP; FRT82B tubulin-Gal80. Pupae were removed from the pupal case, dissected in PBS and fixed for immunohistochemistry, and mounted in Vectashield (Vector Laboratories; Roegiers et al., 2001; Zitserman and Roegiers, 2011). Antibodies used were as follows: mouse anti-NECD (1:100, 458.2H, Developmental Studies Hybridoma Bank [DSHB]), mouse anti-Cut (1:100, 2B10, DSHB), Rat anti-ELAV 7E8A10 (1:200, DSHB), and mouse anti-22C10/Futch (1:200, DSHB); secondary antibodies were conjugated to Alexa Fluor 555 or Alexa Fluor 633 Fluorochromes (Invitrogen). Pupae were dissected out of the pupal case and mounted between slide and coverslip for live-cell imaging (Roegiers et al., 2001; Zitserman and Roegiers, 2011). All images were acquired at room temperature (23°C) on an inverted microscope (model TE2000U; Nikon) equipped with a C1 confocal imaging system (488-, 543-, and 633-nm lasers; Nikon) or the SFC (swept field confocal) live-imaging system (488/514-nm laser; Nikon), using either 60 or 100× 1.49 NA objective lenses. All measurements were done using EZ-C1 software (Nikon). Wing disc apical Notch staining was quantified using MetaMorph imaging software (Molecular Devices) by acquiring XY images of the apical region of epithelial cells, merging the XY planes, and applying a threshold to both the NECD and the GFP images. The GFP threshold was used to establish GFP+ and GFP− regions. The NECD staining threshold was set to include the apical Notch staining in the GFP− (control) region. We then calculated the percentage of the total area of the region (GFP+ or GFP−) that contained apical NECD. These analyses were conducted at multiple regions of at least three wing discs for each genotype.
Protein biochemistry and coimmunoprecipitation
For γ-secretase coimmunoprecipitation assays, 7 × 106 Drosophila S2 cells seeded in a 10-cm plate and were cotransfected with 3 µg of pIZ-GS-PSN-Myc (kindly provided by M. Fortini, Thomas Jefferson University, Philadelphia, PA) and 3 µg of pAWF Sanpodo wild-type or mutant constructs. After 48 h of transfection, cells were harvested and lysed in 0.5 ml lysis buffer and the cell lysates were incubated with 40 µl of anti-Myc Agarose (Sigma-Aldrich) at 4°C overnight after being precleared in 40 µl of mouse IgG agarose (Sigma-Aldrich). The immunoprecipitates were washed five times in 1× phosphate-buffered saline (PBS) and resolved on NuPAGE gels (Invitrogen) along with input control. The blots were detected with anti–Flag-HRP (Sigma-Aldrich).
For Notch-binding assays, 3 µg of either pAWF Sanpodo wild-type or mutant constructs were transfected into S2 cells stably expressing NotchFULL under metallothion promoter or 3 µg of pUAS-NotchICD cotransfected along with 1 µg pActin-Gal4 in S2 cells. The expression of Notch, in S2 cells stably expressing Notch, was induced by incubation of the cells with 0.7 mM CuSO4 for 4 h before lysis. In both cases, cells were harvested 48 h after transfection and lysed for CoIP as described previously. The coimmunoprecipitation was performed at 4°C overnight with 4 µg of NotchICD (C17.9c6) preconjugated with Protein A/G (Thermo Fisher Scientific). After CoIP, antibody–antigen conjugates were washed, and protein bands were resolved via NuPAGE gels (Invitrogen) and visualized with anti-Flag-HRP (Sigma-Aldrich).
Online supplemental material
Fig. S1 shows the manual alignment of the Presenilin-binding region with other sanpodo homologues, and sequences of the amino-terminal region of these sanpodo homologues with the conserved NPAF, LHELL, and RYXXXX motifs highlighted. Fig. S2 shows that deletion of aa 100–125 of the Sanpodo ATCR abrogates Presenilin binding, but has no effect Notch ICD binding, and that mutation of the RY residues to alanine has no effect on Presenilin binding in vitro. Fig. S3 shows that Sanpodo misexpression in wing disc cells results in wing Notching and lateral inhibition defects on the scutellum. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201209023/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank M. Fortini, J. Skeath, the Developmental Studies Hybridoma Bank, and the Bloomington Drosophila Stock Center for generously providing fly stocks and reagents used in this study. We would also like to acknowledge the Fox Chase Cancer Center Molecular Modeling, Glasswashing and Drosophila Media, DNA sequencing, and Cell Culture core facilities. We would also like to thank D. Silverman, E. Cuikerman, and M. Andrake for assistance with experiments and analysis and A. O’Reilly, A. Bellacosa, D. Wiest, members of Fox Chase Fly group, and the anonymous reviewers for insightful input on this work.
This work was supported by National Institutes of Health grant RO1 NS059971 (to F. Roegiers).
Footnotes
Abbreviations used in this paper:
- ACD
- asymmetric cell division
- ATCR
- amino-terminal cytoplasmic region
- NECD
- Notch extracellular domain
- NICD
- Notch intracellular domain
- SOP
- sensory organ precursor cell
References
- Babaoglan A.B., O’Connor-Giles K.M., Mistry H., Schickedanz A., Wilson B.A., Skeath J.B. 2009. Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development. 136:4089–4098 10.1242/dev.040386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks S.M.L., Cho B., Eun S.H., Lee J.-H., Windler S.L., Xie X., Bilder D., Fischer J.A. 2011. The functions of auxilin and Rab11 in Drosophila suggest that the fundamental role of ligand endocytosis in notch signaling cells is not recycling. PLoS ONE. 6:e18259 10.1371/journal.pone.0018259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaacov S., Le Borgne R., Abramson I., Schweisguth F., Schejter E.D. 2001. Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J. Cell Biol. 152:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhra N., Lallet S., Cotton M., Le Bras S., Dussert A., Le Borgne R. 2011. AP-1 controls the trafficking of Notch and Sanpodo toward E-cadherin junctions in sensory organ precursors. Curr. Biol. 21:87–95 10.1016/j.cub.2010.12.010 [DOI] [PubMed] [Google Scholar]
- Berdnik D., Török T., González-Gaitán M., Knoblich J.A. 2002. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell. 3:221–231 10.1016/S1534-5807(02)00215-0 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- Bray S.J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Chastagner P., Israël A., Brou C. 2008. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE. 3:e2735 10.1371/journal.pone.0002735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier L., Vodovar N., Schweisguth F. 2012. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 14:131–139 10.1038/ncb2419 [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N., Yamamoto S., Zitserman D., Graves H.K., Schulze K.L., Wang H., Klein H., Roegiers F., Bellen H.J. 2012. dEHBP1 controls exocytosis and recycling of Delta during asymmetric divisions. J. Cell Biol. 196:65–83 10.1083/jcb.201106088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H.R., Herskowitz I. 1992. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 68:237–255 10.1016/0092-8674(92)90468-R [DOI] [PubMed] [Google Scholar]
- Hu Y., Fortini M.E. 2003. Different cofactor activities in γ-secretase assembly: evidence for a nicastrin-Aph-1 subcomplex. J. Cell Biol. 161:685–690 10.1083/jcb.200304014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A., Knoblich J.A. 2005. Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 6:836–842 10.1038/sj.embor.7400500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan M.X.G., Kopan R. 2007. SnapShot: notch signaling pathway. Cell. 128:1246 10.1016/j.cell.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H., Andrews H.K., Acar M., Bayat V., Wirtz-Peitz F., Mehta S.Q., Knoblich J.A., Bellen H.J. 2005. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev. Cell. 9:351–363 10.1016/j.devcel.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Kandachar V., Roegiers F. 2012. Endocytosis and control of Notch signaling. Curr. Opin. Cell Biol. 24:534–540 10.1016/j.ceb.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2010. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11:849–860 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin J., Le Borgne R., Rosenfeld F., Gho M., Schweisguth F., Bellaïche Y. 2005. Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr. Biol. 15:955–962 10.1016/j.cub.2005.04.054 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Schweisguth F. 2003. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell. 5:139–148 10.1016/S1534-5807(03)00187-4 [DOI] [PubMed] [Google Scholar]
- McGill M.A., Dho S.E., Weinmaster G., McGlade C.J. 2009. Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284:26427–26438 10.1074/jbc.M109.014845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor-Giles K.M., Skeath J.B. 2003. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell. 5:231–243 10.1016/S1534-5807(03)00226-0 [DOI] [PubMed] [Google Scholar]
- Park M., Yaich L.E., Bodmer R. 1998. Mesodermal cell fate decisions in Drosophila are under the control of the lineage genes numb, Notch, and sanpodo. Mech. Dev. 75:117–126 10.1016/S0925-4773(98)00098-7 [DOI] [PubMed] [Google Scholar]
- Rajan A., Tien A.-C., Haueter C.M., Schulze K.L., Bellen H.J. 2009. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat. Cell Biol. 11:815–824 10.1038/ncb1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu M.S., Jan L.Y., Jan Y.N. 1994. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 76:477–491 10.1016/0092-8674(94)90112-0 [DOI] [PubMed] [Google Scholar]
- Roegiers F., Jan Y.N. 2004. Asymmetric cell division. Curr. Opin. Cell Biol. 16:195–205 10.1016/j.ceb.2004.02.010 [DOI] [PubMed] [Google Scholar]
- Roegiers F., Younger-Shepherd S., Jan L.Y., Jan Y.N. 2001. Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat. Cell Biol. 3:58–67 10.1038/35050568 [DOI] [PubMed] [Google Scholar]
- Roegiers F., Jan L.Y., Jan Y.N. 2005. Regulation of membrane localization of Sanpodo by lethal giant larvae and neuralized in asymmetrically dividing cells of Drosophila sensory organs. Mol. Biol. Cell. 16:3480–3487 10.1091/mbc.E05-03-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg A., D’Evelyn D., Schulze K.L., Lee J.K., Strumpf D., Tsai L., Bellen H.J. 1994. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 13:269–287 10.1016/0896-6273(94)90346-8 [DOI] [PubMed] [Google Scholar]
- Santolini E., Puri C., Salcini A.E., Gagliani M.C., Pelicci P.G., Tacchetti C., Di Fiore P.P. 2000. Numb is an endocytic protein. J. Cell Biol. 151:1345–1352 10.1083/jcb.151.6.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath J.B., Doe C.Q. 1998. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 125:1857–1865 [DOI] [PubMed] [Google Scholar]
- Stempfle D., Kanwar R., Loewer A., Fortini M.E., Merdes G. 2010. In vivo reconstitution of gamma-secretase in Drosophila results in substrate specificity. Mol. Cell. Biol. 30:3165–3175 10.1128/MCB.00030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Rompani S.B., Atkins J.B., Zhou Y., Osterwalder T., Zhong W. 2005. Numb proteins specify asymmetric cell fates via an endocytosis- and proteasome-independent pathway. Mol. Cell. Biol. 25:2899–2909 10.1128/MCB.25.8.2899-2909.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Zitserman D., Serebriiskii I., Andrake M., Dunbrack R., Roegiers F. 2010. Numb independently antagonizes Sanpodo membrane targeting and Notch signaling in Drosophila sensory organ precursor cells. Mol. Biol. Cell. 21:802–810 10.1091/mbc.E09-09-0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitserman D., Roegiers F. 2011. Live-cell imaging of sensory organ precursor cells in intact Drosophila pupae. J. Vis. Exp. 51:e2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.