Staufen1-mediated mRNA decay (SMD) is a mechanism to degrade mRNAs that contain Staufen1-binding sites (SBSs) in their 3′-untranslated regions (UTRs). Human SBSs can form by base-pairing between a 3′ UTR and a long noncoding RNA (lncRNA). Wang et al. now show that SMD occurs in mouse cells via mRNA–lncRNA base-pairing. They identify several lncRNAs that regulate myoblast cell differentiation. This study thus defines new roles for lncRNAs and 3′ UTR elements in development.
Keywords: B elements, ID elements, ½-sbsRNAs, lncRNA–mRNA base-pairing, Traf6 mRNA, Cdc6 mRNA, C2C12 myogenesis
Abstract
Staufen1-mediated mRNA decay (SMD) degrades mRNAs that harbor a Staufen1-binding site (SBS) in their 3′ untranslated regions (UTRs). Human SBSs can form by intermolecular base-pairing between a 3′ UTR Alu element and an Alu element within a long noncoding RNA (lncRNA) called a ½-sbsRNA. Since Alu elements are confined to primates, it was unclear how SMD occurs in rodents. Here we identify mouse mRNA 3′ UTRs and lncRNAs that contain a B1, B2, B4, or identifier (ID) element. We show that SMD occurs in mouse cells via mRNA–lncRNA base-pairing of partially complementary elements and that mouse ½-sbsRNA (m½-sbsRNA)-triggered SMD regulates C2C12 cell myogenesis. Our findings define new roles for lncRNAs as well as B and ID short interspersed elements (SINEs) in mice that undoubtedly influence many developmental and homeostatic pathways.
Staufen 1 (STAU1)-mediated mRNA decay (SMD) is a translation-dependent mechanism that is imperative for a number of mammalian cell processes, including adipogenesis (Cho et al. 2012), cell motility (Gong and Maquat 2011a), and myogenesis (Gong et al. 2009). According to the current model, SMD occurs when translation terminates sufficiently upstream of a STAU1-binding site (SBS) so that the terminating ribosome does not remove SBS-bound STAU1 (Maquat and Gong 2009). Recent data indicate that SMD is promoted by STAU1 dimerization, which augments the efficiency of SMD by increasing the interaction of STAU1 with the ATP-dependent RNA helicase UPF1 (Gleghorn et al. 2013). Additionally, SMD also involves the STAU1 paralog STAU2, and STAU2 can likewise form homodimers as well as heterodimers with STAU1 (Park et al. 2013). Both STAU paralogs increase UPF1 helicase activity, which is critical for SMD, without enhancing UPF1 ATPase activity, presumably by changing the conformation of UPF1 to mimic the ATP-bound activated configuration (Park et al. 2013).
In humans, SBSs can be formed by intramolecular base-pairing within an mRNA 3′ untranslated region (UTR), as exemplified by the 19-base-pair stem within mRNA encoding human ADP ribosylation factor 1 (ARF1) (Kim et al. 2005, 2007). Alternatively, human SBSs can be formed by intermolecular base-pairing between an Alu element within an mRNA 3′ UTR and a partially complementary Alu element within one or more long noncoding RNAs (lncRNAs) that we called ½-sbsRNAs (Gong and Maquat 2011a,b). Alu elements are a type of short interspersed element (SINE) of up to ∼300 nucleotides (nt) and are particular to primate genomes. Intermolecular SBSs are estimated to typify at least 13% of all human SMD targets (Gong and Maquat 2011a).
The role of Alu SINEs in primate SMD (Gong and Maquat 2011b) and the absence of Alu SINEs in mice led us to inquire whether the relatively shorter SINEs of mice could likewise function in SMD. The Mus musculus genome contains five major and unrelated SINE families of retrotransposons of up to ∼190 nt. These families consist of B1, B2, B4, identifier (ID), and mammalian-wide interspersed repeat (MIR) SINEs that are present at, respectively, ∼564,000, ∼348,000, ∼391,000, ∼79,000, and ∼115,000 copies per genome (Kass and Jamison 2007 and references therein). While all of the ∼1.4 million Alu SINEs in the human genome derive from 7SL RNA (Batzer and Deininger 2002), mouse SINEs derive from a broader range of RNA polymerase III (pol III) transcripts that include not only 7SL RNA, but also tRNAs and 5S rRNA (Kramerov and Vassetzky 2011).
Here we mined mouse transcriptome databases to identify lncRNAs that contain one or more SINEs and mRNAs that contain a single 3′ UTR SINE. We focused on single 3′ UTR SINEs, since they would not undergo intramolecular base-pairing. Base-pairing between partially complementary rodent SINEs that reside in cis could result in A-to-I editing and nuclear retention, although to a lesser extent than typifies inverted repeats of the longer and less diverged primate Alu SINEs (Neeman et al. 2006). Our data indicate that intermolecular base-pairing between SINE-containing lncRNAs, hereafter called mouse ½-sbsRNAs (m½-sbsRNAs), and SINE-containing mRNA 3′ UTRs in mice occurs and results in SMD. We previously demonstrated that the efficiency of SMD increases during the differentiation of mouse C2C12 myoblasts (MBs) to multinucleated myotubes (MTs) so as to augment the rate of myogenesis by, e.g., degrading mRNA encoding paired box 3 (mPax3), which is a protein that maintains C2C12 MBs in an undifferentiated state (Gong et al. 2009). Thus, here we used the C2C12 myogenic process to assay for the physiological relevance of m½-sbsRNA function in SMD. Remarkably, down-regulating the abundance of three out of the four m½-sbsRNAs tested altered the rate of myogenesis. In particular, down-regulating B2 SINE-containing m½-sbsRNA2 [m½-sbsRNA2(B2)], which we show triggers the SMD of mRNA encoding the E3 ubiquitin ligase called tumor necrosis factor receptor-associated factor 6 (mTraf6), promotes myogenesis. Consistent with this being attributable to a decrease in the efficiency of m½-sbsRNA2(B2)-mediated mTraf6 SMD, down-regulating mTraf6 mRNA was found to inhibit myogenesis.
Results
Identification of putative mouse lncRNA–mRNA duplexes that form via SINE base-pairing
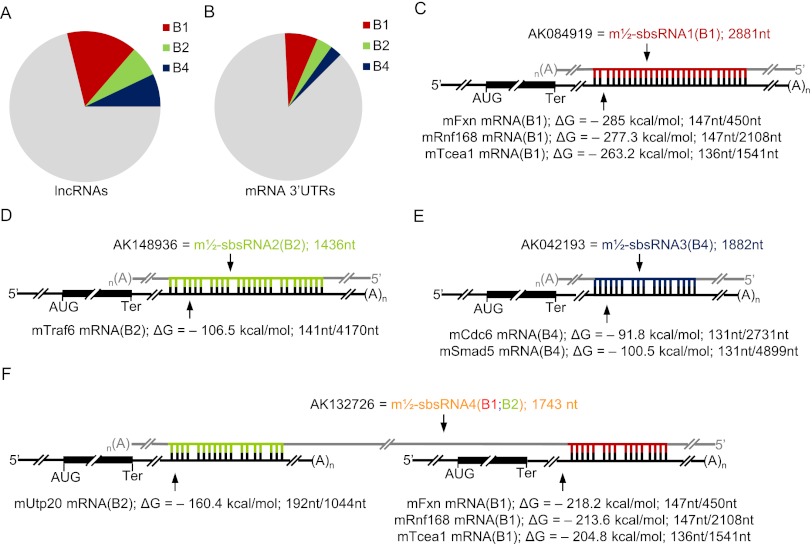
Using the Antisense ncRNA Pipeline data set (Engström et al. 2006; Pang et al. 2007) and RepeatMasker, we identified 578, 250, and 278 mouse lncRNAs that harbor, respectively, a B1, B2, and/or B4 SINE and in total account for a remarkable 28.4% of all annotated lncRNAs (Fig. 1A; Supplemental Table 1). Using the NCBI mouse mRNA database, 3_UTR_Finder (http://www.urmc.rochester.edu/labs/Maquat-Lab/software) and RepeatMasker, we identified 1232, 581, and 437 mouse mRNAs that contain, respectively, a single 3′ UTR B1, B2, or B4 element and comprise 13.2% of annotated mouse mRNAs (Fig. 1B; Supplemental Table 2).
Figure 1.
Bioinformatic characterization of SINE-containing lncRNAs and 3′ UTR SINE-containing mRNAs in mice and illustration of theoretical lncRNA–mRNA duplexes. (A) Pie chart showing the relative distribution of mouse lncRNAs containing a B1, B2, and/or B4 SINE (Supplemental Table 1). (B) As in A, but for mRNAs containing a single 3′ UTR SINE (Supplemental Table 2). (C–F) Diagrams of predicted base-pairing between the SINE of the denoted NCBI-named lncRNA (also called a m½-sbsRNA), whose length is provided in nucleotides, and the 3′ UTR SINE of one or more mRNAs. Computationally calculated values of the Gibbs free energy of formation (ΔG) (Supplemental Fig. 1) are also provided. Additionally, the length of each SINE is presented as a ratio of the length of each 3′ UTR in nucleotides, the latter of which was determined using the Ensembl database. (AUG) Translation initiation codon; (Ter) termination codon; (A)n, poly(A); (gray) lncRNA sequences flanking the differently colored SINEs.
RNA_RNA_anneal (Mathews et al. 1999, 2004; Turner and Mathews 2009; Gong and Maquat 2011a) was used to predict potential base-pairing between the full-length SINEs of each of the identified lncRNAs and mRNAs (provided they derived from the same family) and the associated Gibbs free energy of duplex formation (ΔG). This program allows no more than two consecutive mismatches. Eight duplexes that were predicted (1) to be among the most stable or to involve mRNAs known to be down-regulated during the differentiation of mouse C2C12 MBs to MTs (Chen et al. 2006) and (2) to involve lncRNAs that are largely cytoplasmic and were amenable to down-regulation using siRNA (see below) were chosen for further analysis. The B1 SINE-containing lncRNA that was assigned the NCBI accession number AK084919 [hereafter called m½-sbsRNA1(B1) for reasons shown below] was predicted to base-pair with three 3′ UTR B1 SINE-containing mRNAs. These mRNAs encode (1) frataxin (mFxn), which is a mitochondrial protein mutated in Friedreich ataxia; (2) E3 ubiquitin–protein ligase ring finger protein 168 (mRnf168); and (3) transcription elongation factor A protein 1 (mTcea1). The corresponding duplexes (Supplemental Fig. 1A–C) have computationally calculated ΔG values of, respectively, −285.0 kcal/mol, −263.2 kcal/mol, and −277.3 kcal/mol (Fig. 1C). Other lncRNA–mRNA duplexes, hereafter called m½-sbsRNA–mRNA duplexes (Supplemental Fig. 1D–I), were also predicted to form via intermolecular (1) B2–B2 base-pairing between m½-sbsRNA2(B2) and the 3′ UTR of mRNA encoding mTraf6, (2) B4–B4 base-pairing between m½-sbsRNA3(B4) and the 3′ UTR of mRNA encoding cell division cycle 6 homolog (mCdc6; Sacharoymyces cerevisiae) as well as the 3′ UTR of mRNA encoding mothers against decapentaplegic homolog 5 (mSmad5), and (3) B1–B1 and B2–B2 base-pairing between m½-sbsRNA4(B1;B2) and, respectively, the 3′ UTRs of mFxn mRNA, mRnf168 mRNA, mTcea1 mRNA and the 3′ UTR of mRNA encoding small subunit processome component 20 (mUtp20). The computationally calculated ΔG values for these complexes range from −218.2 kcal/mol to −91.8 kcal/mol (Fig. 1D–F). In contrast, ΔG values for alternative mRNA–lncRNA base-pairs between B elements of different families (e.g., B1–B2) range from −0 kcal/mol to −79 kcal/mol. Notably, it has been reported that B1 and B2 elements do not base-pair (Neeman et al. 2006).
Evidence that m½-sbsRNAs can mediate the SMD of their predicted target mRNAs
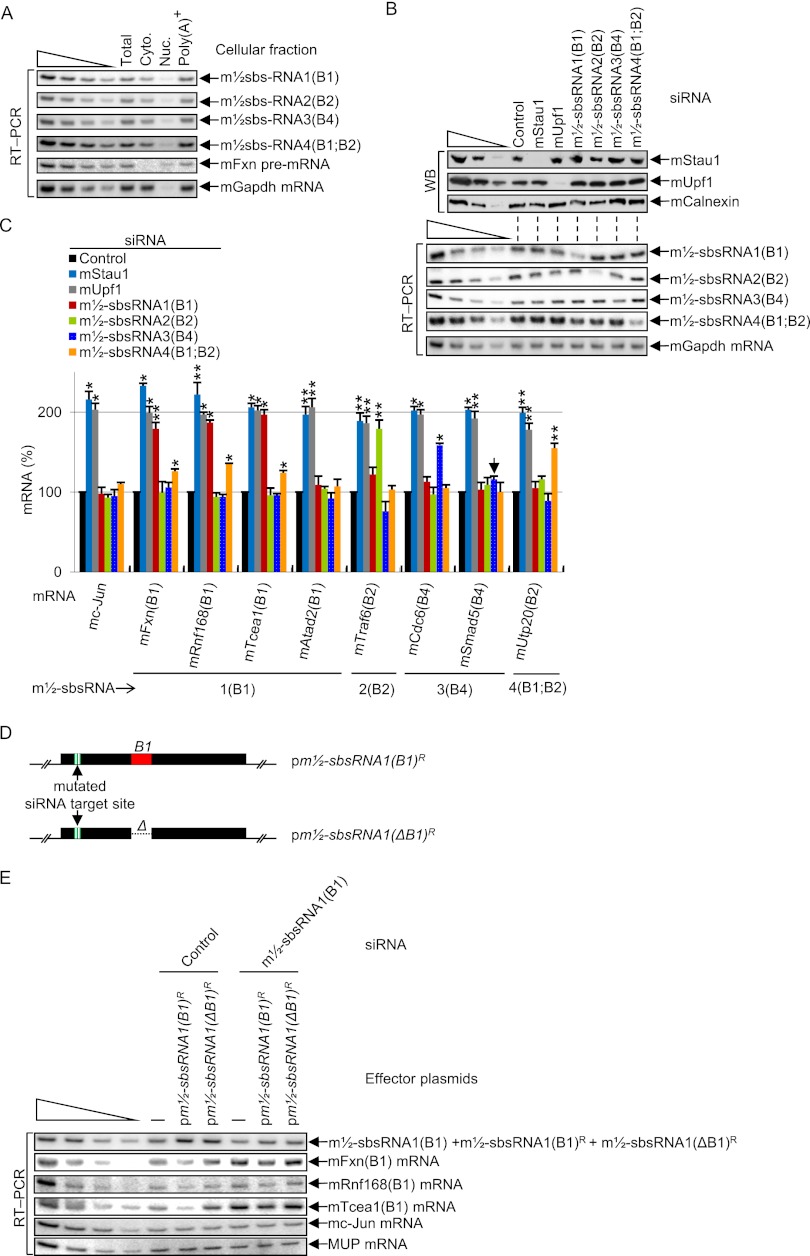
Reverse transcription (RT) coupled to semiquantitative PCR (RT–PCR) revealed that all four m½-sbsRNAs localize primarily to the cytoplasm of C2C12 MBs, thus reducing, if not precluding, the possibility of significant duplex formation and A-to-I editing within nuclei, and all are largely polyadenylated (Fig. 2A; Supplemental Fig. 2A). Western blotting of MB lysates demonstrated that the siRNA-mediated down-regulation of either mStau1 or mUpf1, a key SMD factor that is recruited to SBSs via SBS-bound mStau1 (Gong and Maquat 2011a), to <8% of its normal level (i.e., its level in the presence of control siRNA) had no effect on the abundance of any of the four m½-sbsRNAs but up-regulated the level of each 3′ UTR SINE-containing mRNA 1.5-fold to 2.3-fold above normal (Fig. 2B,C; data not shown for alternative mStau1 and mUpf1 siRNAs used in Kim et al. 2007). Down-regulating the abundance of each m½-sbsRNA to 25%–55% of its normal level increased each putative target mRNA 1.4-fold to 2.2-fold above normal with the exception of m½-sbsRNA3(B4), whose siRNA increased the level of mCdc6 mRNA but had no effect on the level of mSmad5 mRNA despite mSmad5 mRNA being up-regulated by both mStau1 siRNA and mUpf1 siRNA (Fig. 2C; Supplemental Fig. 2B,C, which shows corroborating RT coupled to quantitative PCR [qPCR] data). As expected based on computational predictions, only certain SINEs in a particular B-element family can base-pair, since not only their sequences, but also whether one is sense and the other is antisense relative to the RNA from which they derived matters. This is illustrated by our finding that m½-sbsRNA1(B1) siRNA failed to increase the level of the SMD target encoding mouse ATPase family AAA domain-containing 2 protein (mAtad2) (Fig. 2C): Even though the 3′ UTR of this mRNA contains a B1 SINE, it is in the same sense as the B1 SINE within m½-sbsRNA1(B1). An additional example includes the failure of m½-sbsRNA4(B1;B2) siRNA to down-regulate mTraf6(B2) mRNA: While this mRNA contains a 3′ UTR SINE that is antisense to that within m½-sbsRNA4(B1;B2), the predicted ΔG value of duplex formation is only −27.9 kcal/mol. That noted, the ΔG value of duplex formation is not the sole predictor of SMD (see the Discussion).
Figure 2.
Evidence that m½-sbsRNAs are largely cytoplasmic and polyadenylated, and trigger the SMD of mRNAs with which they are computationally predicted to base-pair. (A) RT–PCR of total cell (Total), cytoplasmic (Cyto.), nuclear (Nuc.), or total cell polyadenylated [Poly(A)+] RNA from the same number of mouse C2C12 MBs. RT was primed using oligo(dT)18. The analysis of mFxn pre-mRNA and mGapdh mRNA verified successful cell fractionation. (B) Western blotting (WB) using the designated antibody (α) (top) or RT–PCR (bottom) of lysates of C2C12 MBs (2 × 106 per 100-mm dish) that had been transiently transfected with 50 nM specified siRNA. mCalnexin protein and mGapdh mRNA served as loading controls. (C) As in B (bottom), but the specified mRNAs were analyzed, and the resulting quantitations (Supplemental Fig. 2B,C) are shown as histograms. The abundance of each mRNA was normalized to the abundance of its pre-mRNA, except that the level of mc-Jun mRNA, which derives from an intronless gene, was normalized to the level of mGapdh mRNA. Normalized levels were then presented as a percentage of the normalized level in the presence of control siRNA, which is defined as 100%. The downward-facing arrow marks the one m½-sbsRNA–mRNA duplex that is computationally predicted to occur but fails to trigger SMD of the constituent mRNA. (D) Schematic representations of siRNA-resistant pm½-sbsRNA(B1) plasmids, one of which lacks the B1 SINE (ΔB1). (E) RT–PCR of lysates of C2C12 MBs (2 × 106 per 100-mm dish) that had been transiently transfected with 50 nM specified siRNA and, 1 d later, with 1 μg of the specified plasmid and 1 μg of phCMV-MUP. All results derive from three independently performed experiments (Supplemental Fig. 2). (*) P < 0.05; (**) P < 0.01. P-values were calculated using the one-tailed student t-test.
To determine whether the m½-sbsRNA-mediated mRNA reduction is caused by the SINE within m½-sbsRNAs, C2C12 MBs were transiently transfected with control siRNA or m½-sbsRNA1(B1) siRNA and, 1 d later, with either pm½-sbsRNA1(ΔB1)R, which produces siRNA-resistant (R) m½-sbsRNA1(B1) that lacks the B1 element or, as a positive control, pm½-sbsRNA1(B1)R (Fig. 2D). In the presence of control siRNA, m½-sbsRNA1(B1)R promoted the SMD of its known targets relative to either m½-sbsRNA1(ΔB1)R or no exogenous nucleic acid (Fig. 2E; Supplemental Fig. 2D). Furthermore, in the presence of m½-sbsRNA1(B1) siRNA, m½-sbsRNA1(B1)R restored the SMD of its known targets, unlike either m½-sbsRNA1(ΔB1)R or no exogenous nucleic acid (Fig. 2E; Supplemental Fig. 2D).
We conclude that mStau1 and mUpf1 mediate a decrease in the abundance of mRNAs that contain a 3′ UTR B1, B2, or B4 SINE. Furthermore, m½-sbsRNAs that contain a B1, B2, or B4 SINE generally mediate a decrease in the abundance of mRNAs with which they are predicted to base-pair in a mechanism that does not alter m½-sbsRNA abundance. Thus, like human ½-sbsRNAs, m½-sbsRNAs are not SMD targets, presumably because they are not translated.
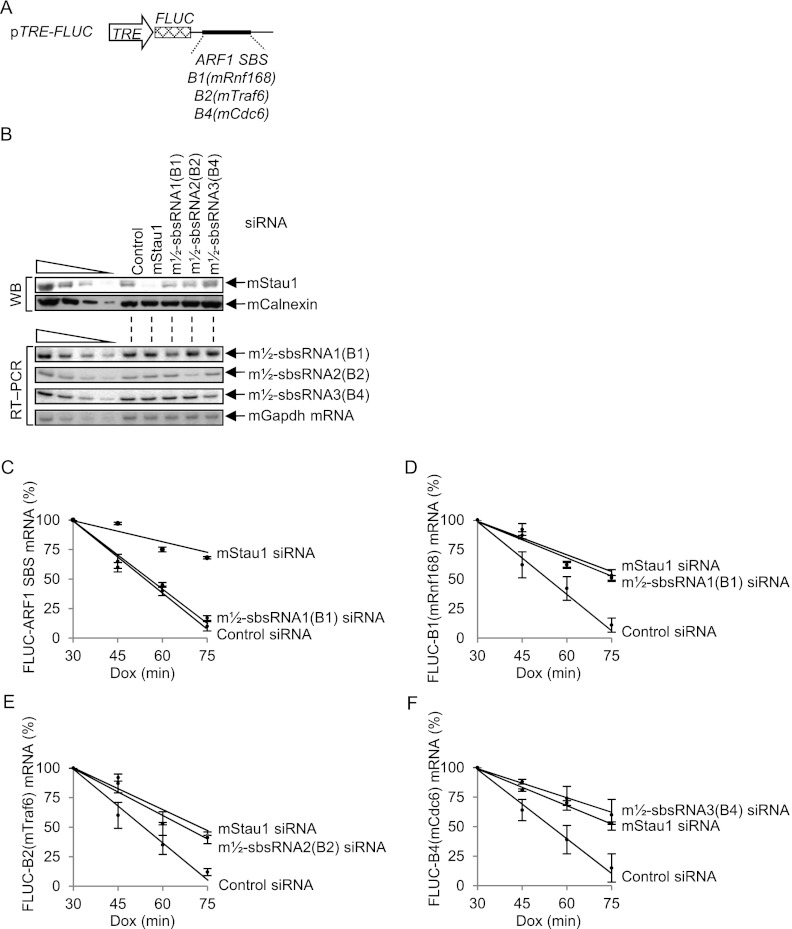
To determine whether the m½-sbsRNA-mediated reduction in mRNA abundance is due to a decrease in mRNA half-life via the predicted 3′ UTR SINE, mouse NIH3T3 B2A2 Tet-off cells (Chen et al. 2008) were transiently transfected in the presence of 0.5 μg/mL doxycycline (Dox) with control siRNA, mStau1 siRNA, m½-sbsRNA1(B1) siRNA, m½-sbsRNA2(B2) siRNA, or m½-sbsRNA3(B4) siRNA. Two days later, Dox was removed, and cells were transfected with (1) pTRE-FLUC-B1(mRnf168), pTRE-FLUC-B2(mTraf6), pTRE-FLUC-B4(mCdc6), and, as a positive control for SMD, pTRE-FLUC-ARF1 SBS reporter plasmids (Fig. 3A), and (2) the phCMV-MUP reference plasmid. Each reporter plasmid produces firefly luciferase (FLUC) mRNA that harbors the specified SINE downstream from the FLUC termination codon using a promoter that is repressed by Dox because it resides in cis to the tetracycline response element (TRE). Four hours later, an aliquot of cells was harvested at time 0; 0.5 μg/mL Dox was then added to the remaining cells to silence reporter gene transcription, and aliquots of cells were harvested as a function of time thereafter.
Figure 3.
Evidence that m½-sbsRNAs reduce mRNA abundance by reducing mRNA half-life. (A) Schematic representation of pTRE-FLUC reporter plasmids, the 3′ UTR of which contains the specified 3′ UTR SINE or, as a positive control, the ARF1 SBS. The cross-hatched box represents the FLUC open translational reading frame. (B–F) Mouse NIH3T3 B2A2 Tet-off cells (4 × 106 per 100-mm dish) that had been transiently transfected with 50 nM specified siRNA in the presence of 0.5 μg/mL Dox and, after removing Dox 48 h later, with a mixture of 1.5 μg of each pTRE-FLUC test plasmid and 1 μg of the phCMV-MUP reference plasmid. After an additional 4 h (time 0), 0.5 μg/mL Dox was added, and cells were harvested at the specified times thereafter. (B) Western blotting (top) or RT–PCR (bottom) of lysates at time 0. (C–F) Plot of RT–PCR quantitations (Supplemental Fig. 3) as a function of time after Dox addition. For each time point, the level of each pTRE-FLUC reporter mRNA was normalized to the level of MUP mRNA. Normalized levels were calculated as a percentage of the normalized level of each mRNA at 30 min in the presence of each siRNA, which is defined as 100%. All results derive from three independently performed experiments (Supplemental Fig. 3).
Western blotting or RT–PCR confirmed that each siRNA down-regulated its target protein or m½-sbsRNA (Fig. 3B). In control experiments, mStau1 siRNA prolonged the half-life of each reporter mRNA (Fig. 3C–F), consistent with each being targeted for SMD, and m½-sbsRNA1(B1) siRNA was of no consequence to the half-life of FLUC-ARF1 SBS mRNA, which lacks a 3′ UTR SINE but harbors an intramolecular SBS (Kim et al. 2007). m½-sbsRNA1(B1) siRNA, m½-sbsRNA2(B2) siRNA, and m½-sbsRNA3(B4) siRNA indeed increased the half-life of, respectively, FLUC-B1(mRnf168) mRNA, FLUC-B2(mTraf6) mRNA, and FLUC-B4(mCdc6) mRNA (Fig. 3C–F). We conclude that m½-sbsRNAs that are computationally predicted to base-pair with mRNA 3′ UTRs via partially complementary SINEs can trigger SMD.
Evidence for the existence of m½-sbsRNAs–mRNA duplexes that recruit mStau1 and mUpf1
To test the hypothesis that m½-sbsRNAs can create SBSs by base-pairing with 3′ UTR sequences, five reporter plasmids were constructed that harbor a 3′ UTR SINE downstream from the FLUC open translational reading frame (Fig. 4A). All five reporter plasmids together with pFLUC-No SBS, the 3′ UTR of which lacks an SBS (Kim et al. 2007), the phCMV-MUP reference plasmid, and either the pSTAU1-HA3 (Kim et al. 2005) or, as a negative control, pUC19 effector plasmid, were transiently introduced into C2C12 MBs. MBs were cross-linked using formaldehyde prior to lysis to ensure that detected interactions occurred in cells and not as an experimental artifact after cell lysis. Lysates were immunoprecipitated using either anti-HA or, as a negative control, rat IgG (rIgG).
Figure 4.
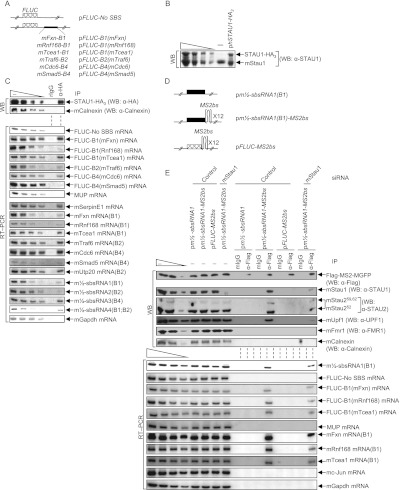
Confirmation that the predicted m½-sbsRNA–mRNA duplexes function as SBSs in C2C12 MBs. (A) Diagrams of pFLUC reporter plasmids, the 3′ UTR of which contains no SBS or the denoted mRNA 3′ UTR SINE situated 224 nt downstream from the FLUC termination codon. The cross-hatched box represents the FLUC open translational reading frame. (B) Western blotting using lysates of C2C12 MBs prior to immunoprecipitation. MBs (4 × 106 per 150-mm dish) had been transiently transfected with 10 μg of pSTAU1-HA3 or pUC19, 1 μg of each of the seven FLUC reporter plasmids, and 2 μg of phCMV-MUP and formaldehyde-cross-linked prior to lysis. (C) Western blotting (top) or RT–PCR (bottom) of lysates analyzed in B before (−) or after immunoprecipitation (IP) using anti-HA (α-HA) or, as a control for nonspecific immunoprecipitation, mIgG. (D) Diagrams of plasmids encoding m½-sbsRNA1(B1), m½-sbsRNA1(B1) harboring 12 copies of the MS2bs, or FLUC mRNA harboring 12 copies of the MS2bs. (E) Western blotting (top) or RT–PCR (bottom) before (−) or after immunoprecipitation of lysates of formaldehyde-cross-linked C2C12 MBs (4 × 106 per 150-mm dish) that had been transiently transfected with 50 nM specified siRNA and, 1 d later, with 5 μg of pFlag-MS2-hMGFP, 1 μg of each of the seven reporter plasmids shown in A, 2 μg of phCMV-MUP, and the denoted plasmid encoding m½-sbsRNA1(B1), m½-sbsRNA1(B1)-MS2bs, or FLUC-MS2bs. Immunoprecipitations were performed using anti-Flag or mIgG. All results are representative of at least two independently performed experiments (Supplemental Fig. 4; Supplemental Table 4).
STAU1-HA3 was expressed at ∼0.8-fold the level of cellular mStau1 (Fig. 4B). Anti-HA immunoprecipitated not only STAU1-HA3, but also each FLUC mRNA that contains the 3′ UTR B1 SINE from mFxn, mRnf168, or mTcea1 mRNA; the 3′ UTR B2 SINE from mTraf6 mRNA; or the 3′ UTR B4 SINE from mCdc6 mRNA but not the 3′ UTR B4 SINE of mSmad5 mRNA (Fig. 4C) as predicted from Figure 2C. As confirmation of immunoprecipitation specificity, anti-HA failed to detectably immunoprecipitate cellular mCalnexin protein or exogenous MUP mRNA but, as expected, did immunoprecipitate the cellular SMD target that encodes serpin peptidase inhibitor clade E, member 1 (mSerpinE1) (Fig. 4C) and that lacks a 3′ UTR SINE (Gong et al. 2009). Cellular mRNAs for mFxn, mRnf168, mTcea1, mTraf6, mCdc6, and mUtp20 and cellular m½-sbsRNAs1, m½-sbsRNA2, m½-sbsRNA3, and m½-sbsRNA4 were likewise immunoprecipitated by anti-HA. Our finding that cellular mSmad5 mRNA was also immunoprecipitated indicates that mStau1 binds to this mRNA. However, according to results obtained using FLUC-B4(mSmad5) mRNA, binding is not to the 3′ UTR SINE (Fig. 4C). No protein or RNA that was assayed was detected in the rIgG immunoprecipitation (Fig. 4C). We conclude that all tested SMD targets except mSmad5 mRNA bind mStau1 via base-pairing of their 3′ UTR B element with an m½-sbsRNA.
As additional evidence that m½-sbsRNA can create SBSs by base-pairing with complementary mRNAs, lysates of formaldehyde-cross-linked C2C12 MBs expressing (1) Flag-MS2-hMGFP (Gong and Maquat 2011a), which is a fusion of Flag, the viral MS2 coat protein (MS2), and monster green florescent protein (hMGFP); (2) m½-sbsRNA1(B1)-MS2bs (Fig. 4D), which harbors 12 copies of the MS2 coat protein-binding site (MS2bs); and (3) the reference MUP transcript were immunoprecipitated using anti-Flag or, as a negative control, mouse IgG (mIgG). The anti-Flag immunoprecipitation of Flag-MS2-hMGFP bound to m½-sbsRNA1(B1)-MS2bs coimmunoprecipitated endogenous mStau1, mStau2, mUpf1, mFxn mRNA, mRnf168 mRNA, and mTcea1 mRNA as well as the FLUC reporter mRNAs that harbored the 3′ UTR SINE of these mRNAs (Fig. 4E). Notably, human STAU2 functions in SMD (Park et al. 2013). Thus, mStau2 most likely also functions in SMD. Irrelevant proteins, such as mCalnexin, the ssRNA-binding protein mFmr1 (Ashley et al. 1993), and mRNAs that are not predicted to base-pair with m½-sbsRNA1(B1), such as the SMD target mc-Jun mRNA, mGapdh mRNA, or MUP mRNA, were not coimmunoprecipitated (Fig. 4E). Anti-Flag immunoprecipitations using lysates of MBs expressing either m½-sbsRNA1(B1), which lacks the MS2bs repeats, or FLUC-MS2bs mRNA, which harbors the MS2bs repeats, failed to detectably coimmunoprecipitate any of the SMD-relevant proteins or RNAs analyzed, which was also true of the mIgG immunoprecipitation using any lysate (Fig. 4E). mStau1 siRNA reduced the coimmunoprecipitation of mFxn mRNA, mRnf168 mRNA, and mTcea1mRNA as well as FLUC-B1(mFxn) mRNA, FLUC-B1(mRnf168) mRNA, and FLUC-B1(mTcea1) mRNA with m½-sbsRNA1(B1)-MS2bs (Fig. 4E; Supplemental Table 4), indicating that mStau1 stabilizes the duplex formed between m½-sbsRNA1 and each mRNA. Notably, experiments assaying m½-sbsRNA1(ΔB1)-MS2bs demonstrated that m½-sbsRNA1 lacking the B1 SINE failed to coimmunoprecipitate with mFxn mRNA, mRnf168 mRNA, or mTcea1 mRNA and thus mStau1, mStau2, or mUpf1 (Supplemental Fig. 4).
We conclude that duplexes of a m½-sbsRNA SINE and a sufficiently complementary SINE of an mRNA 3′ UTR form SBSs within cells. These duplexes are stabilized by the binding of mStau1.
m½-sbsRNA1(B1), m½-sbsRNA2(B2), and m½-sbsRNA3(B4) regulate the differentiation of MBs to MTs
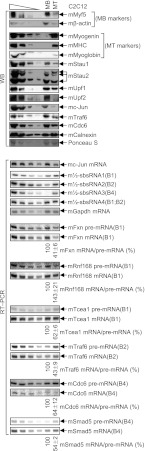
To investigate the physiological relevance of m½-sbsRNAs, the levels of m½-sbsRNAs and their target mRNAs were compared in C2C12 MBs relative to MTs given that the efficiency of SMD is known to increase during C2C12 myogenesis (Gong et al. 2009). As indicators of myogenesis and as expected (e.g., see Gong et al. 2009), the levels of the myogenic regulatory factor mMyf5 as well as mβ-actin, mStau1, mStau2, mUpf1, the NMD factor mUpf2, and mc-Jun, which derives from an mRNA that is an SMD target, decreased, while the levels of myogenin, myosin heavy chain (MHC), and myoglobin increased (Fig. 5; Supplemental Fig. 5B) where the levels of mCalnexin and Ponseau S-staining (Deato and Tjian 2007) controlled for variations in protein loading (Supplemental Fig. 5A). The levels of mTraf6 and mCdc6 proteins also decreased during myogenesis concomitantly with an increase in the levels of the m½-sbsRNA that is complementary to their mRNAs; i.e., m½-sbsRNA2(B2) and m½-sbsRNA3(B4), respectively. Attesting to the importance of SMD as a contributing, although not the sole, regulator of mTraf6 and mCdc6 protein abundance (Wu and Arron 2003; Yim and Erikson 2010), myogenesis was accompanied by a decrease in the levels of mTraf6 and mCdc6 mRNAs due to an increase in the efficiency of SMD. This was evidenced by a decrease in the ratio of mTraf6 mRNA/mTraf6 pre-mRNA and mCdc6 mRNA/mCdc6 pre-mRNA (Fig. 5; Supplemental Fig. 5B, where the level of pre-mRNA was used to control for any change in the level of product mRNA that was due to a change in gene transcription). Notably, the concomitant decrease in the ratio of mTraf6 protein to mTraf6 mRNA suggests that the translation of mTraf6 mRNA or stability of mTraf6 protein is decreased in MTs compared with MBs. Considering that mTraf6 protein promotes myogenesis (Mueck et al. 2011; Xiao et al. 2012), its decrease in abundance in MTs compared with MBs demonstrates the importance of measuring protein levels throughout the myogenic process rather than at only two time points. In fact, mTraf6 protein abundance increases before it decreases during the transition from MBs to MTs (see below), and this fluctuation undoubtedly contributes to muscle cell differentiation. In contrast to mTraf6 protein, mCdc6 protein promotes MB proliferation (Zhang et al. 2010) and thus would be predicted to inhibit myogenesis. While the level of mCdc6 protein also varies during the transition from MBs to MTs, a priori its decreased level in MTs compared with MBs as a result of the increased efficiency with which mCdc6 mRNA is targeted for SMD makes sense. The efficiency with which mFxn, mTcea1, and mSmad5 mRNA were targeted for SMD was also increased in MTs relative to MBs, as evidenced by a decrease in the ratio of each mRNA relative to its pre-mRNA (Fig. 5; Supplemental Fig. 5B). Our finding that the ratio of mRnf168 mRNA to mRnf168 pre-mRNA increased in MTs compared with MBs (Fig. 5; Supplemental Fig. 5B) even though mRnf168 mRNA is an SMD target (Figs. 2C, 3D, 4C,E) indicates that this mRNA is subject to regulatory mechanisms in addition to SMD.
Figure 5.
Evidence that three of the four m½-sbsRNAs are up-regulated during C2C12 cell myogenesis. C2C12 MBs (2 × 107 per 100-mm dish) were split between two dishes. Two days later, one dish was harvested as MBs, while the other was cultured in differentiation medium, and the resulting MTs were harvested 6 d later. (Top) Western blotting (WB) of MB and MT lysates using the specified antibody (α). The level of mCalnexin and PonseauS-staining (Supplemental Fig. 5A) served as loading controls. (mMHC) Mouse MHC. (Bottom) RT–PCR essentially as for Figure 2C, where the level of mGapdh mRNA served as a loading control, and the level of each mRNA was normalized to the level of its pre-mRNA. All results are representative of at least two independently performed experiments that did not vary by more than the amount shown (Supplemental Fig. 5B).
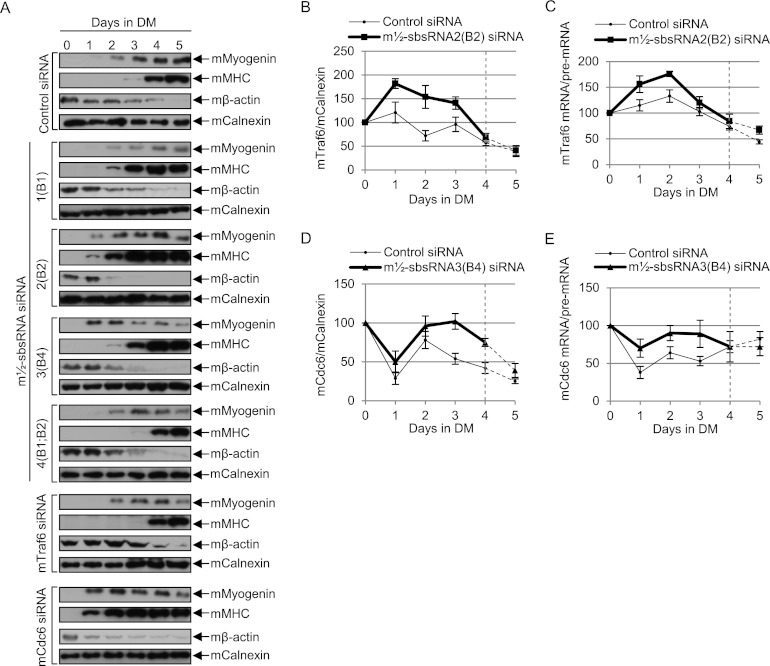
Given that the levels of three of the four m½-sbsRNAs increased in MTs compared with MBs and one [m½-sbsRNA4(B1;B2)] was only slightly reduced (Fig. 5; Supplemental Fig. 5B), we next tested whether down-regulating each individually affects the rate of myogenesis. C2C12 MBs were transfected on day −1 with each m½-sbsRNA siRNA, control siRNA, or, as additional controls, mTraf6 siRNA or mCdc6 siRNA and subsequently propagated in differentiation medium on days 0–5. Each m½-sbsRNA siRNA down-regulated its target to 15%–40% of its level in the presence of control siRNA (Supplemental Fig. 6A); the levels of Traf6 and mCdc6 proteins were down-regulated to, respectively, 22% and 17% of normal; and no siRNA affected the level of proteins or m½-sbsRNAs that were not predicted to be targets (Supplemental Fig. 6A). After using confocal microscopy to analyze DAPI-stained cells that had been transfected with control siRNA and finding that multinucleated MTs were apparent as early as day 3 (Supplemental Fig. 6B), the differentiation rates of all transfected cells were assessed using one or both of two methods: (1) transmitted light microscopy to visualize changes in cell morphology and (2) Western blotting to quantitate the level of three myogenic markers—namely, the induction of mMyogenin production and mMHC production (Sun et al. 2006) and the inhibition of mβ-actin production (Sun et al. 2006)—relative to the level of mCalnexin.
Remarkably, compared with control siRNA, all m½-sbsRNA siRNAs except m½-sbsRNA4(B1;B2) siRNA promoted the rate of myogenesis as assayed using either transmitted light microscopy (Supplemental Fig. 6C) or Western blotting (Fig. 6A; Supplemental Fig. 7A–G), and their relative promotional efficiencies were m½-sbsRNA2(B2) > m½-sbsRNA3(B4) > m½-sbsRNA1(B1). As expected, Traf6 siRNA slowed and Cdc6 siRNA increased the rate of myogenesis (Fig. 6A; Supplemental Fig. 7F,G). Thus, our finding that m½-sbsRNA2(B2) siRNA not only promoted myogenesis but also up-regulated the level of mTraf6 protein and the ratio of mTraf6 mRNA/mTraf6 pre-mRNA (Fig. 6B,C; Supplemental Fig. 7A,C,F,H) is consistent with the observed decrease in the efficiency of mTraf6 SMD promoting myogenesis. The simplest interpretation of our unexpected finding that m½-sbsRNA3(B4) siRNA also promoted myogenesis even though, relative to control siRNA, it increased the level of mCdc6 protein and the ratio of mCdc6 mRNA/mCdc6 pre-mRNA (Fig. 6D,E; Supplemental Fig. 7A,D,G,I) is that m½-sbsRNA3(B4) triggers the SMD of some mRNAs—either directly or indirectly—that encode proteins that promote myogenesis but have yet to be identified.
Figure 6.
Evidence that m½-sbsRNAs can influence C2C12 cell myogenesis by mediating SMD. (A–E) C2C12 MBs (5 × 106 per 100-mm dish) were transiently transfected with 50 nM specified siRNA on day 0 and induced to differentiate to MTs by propagating in differentiation medium (DM) 1-d post-transfection. (A) Western blotting of the specified protein (see Supplemental Fig. 6A for siRNA-mediated down-regulation of each target at day 0), where the level of mCalnexin controls for variations in protein loading. Importantly, the same dilution standards in the left-most four lanes were used in all panels to control for variations in blotting and exposure times, and 10 μg of total cell protein was analyzed in the right-most seven lanes (see Supplemental Fig. 7A–G for blots showing these standards). (B) Plot of Western blotting of mTraf6 protein in the presence of control siRNA or ½-sbsRNA2(B2) siRNA as a function of days in DM, where the normalized level of each on day 0 is defined as 100% (see Supplemental Fig. 7A,C,F, H for data). Dashed lines denote time points at which ½-sbsRNA2(B2) siRNA no longer down-regulated its target. (C) As in B but showing RT–PCR quantitations of mTraf6 mRNA/mTraf6 pre-mRNA (see Supplemental Fig. 7H for data). The dashed lines are as in B. (D) As in B except mCdc6 protein was analyzed (see Supplemental Fig. 7A,D,G,I for data). Dashed lines denote time points at which ½-sbsRNA3(B4) siRNA no longer down-regulated its target. (E) As in C but showing mCdc6 mRNA/mCdc6 pre-mRNA (see Supplemental Fig. 7I for data). The dashed lines are as in D. All results are representative of at least two independently performed experiments that did not vary by more than the amount shown.
An ID–ID duplex can constitute an SBS and trigger SMD, but those MIR–MIR duplexes that were tested do not
We also identified 71 mouse lncRNAs that harbor an ID SINE and 159 mouse mRNAs that contain a single 3′ UTR ID SINE (Supplemental Fig. 8A,B). Our studies of mRNA encoding cyclin-dependent protein kinase 23 (mCdc23) demonstrate that 3′ UTR ID-containing mRNAs can also be SMD targets: mCdc23 mRNA is up-regulated by mStau1 siRNA or mUpf1 siRNA and coimmunoprecipitates with STAU1-HA3, presumably due to its base-pairing with the largely cytoplasmic and polyadenylated AK076245(ID) lncRNA (Supplemental Fig. 8C–G).
Notably, 3.6% of mRNAs (i.e., 621 mRNAs) contain a single 3′ UTR MIR SINE and 6.1% of lncRNAs (i.e., 234 lncRNAs) contain a MIR SINE (J Wang and LE Maquat, unpubl.). The most stable two duplexes between a lncRNA(MIR) and a 3′ UTR MIR are computationally predicted to have ΔG values of −92.4 kcal/mol or −85.6 kcal/mol (Supplemental Fig. 8C,H,I). However, neither duplex appears to form even though the lncRNAs are largely cytoplasmic and polyadenylated (Supplemental Fig. 8D–F). MIR SINEs were amplified before the mammalian radiation (Smit and Riggs 1995) and therefore are likely to have accumulated more nucleotide changes per unit length than more recently evolved SINEs.
Discussion
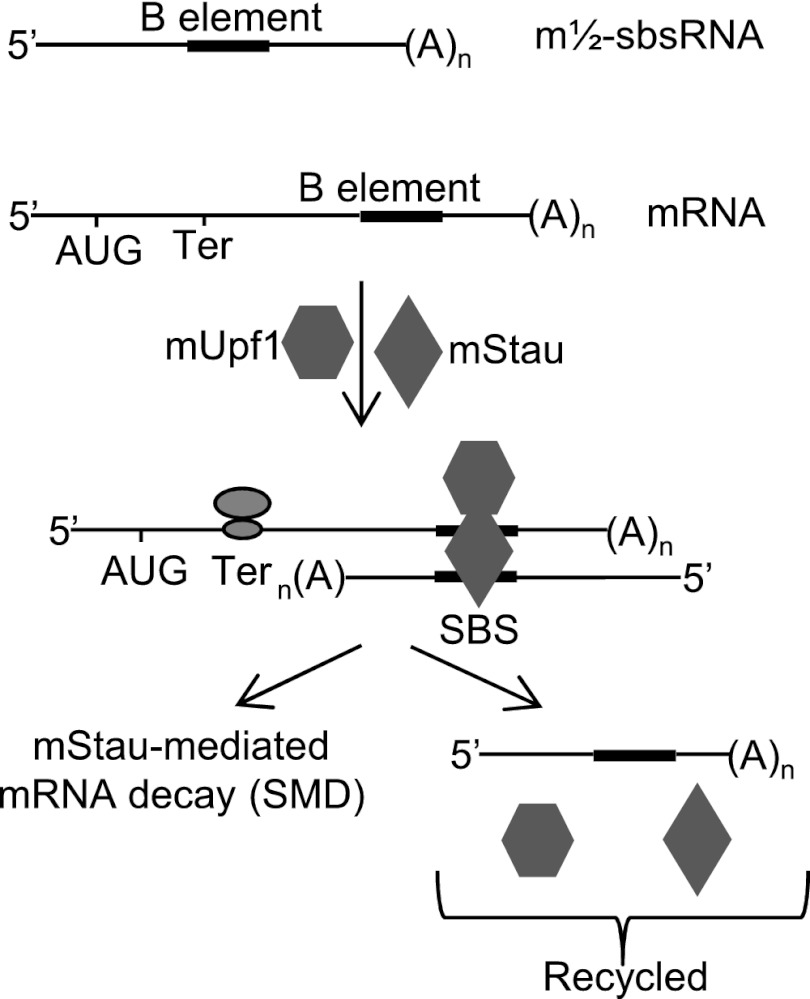
The integration of SINEs into mouse and other mammalian genomes can affect nuclear RNA metabolism by creating positive or negative regulators of transcription, splicing, or polyadenylation (Kramerov and Vassetzky 2011; Hancks and Kazazian 2012; Roy-Engel 2012). Here we provide the first report that the integration of B1, B2, B4, and ID SINEs into the mouse genome can also influence cytoplasmic RNA metabolism: Imperfect base-pairing between a SINE within a largely cytoplasmic and polyadenylated lncRNA, which we call an m½-sbsRNA, and a SINE from the same family within the 3′ UTR of an mRNA can generate an SBS and target the mRNA for degradation by SMD. We infer from data shown here and current models (Gong and Maquat 2011b; Park et al. 2013) that when translation terminates sufficiently upstream of an intermolecular SBS so that bound mStau is not removed by the terminating ribosome, mUpf1, which is recruited to SBSs via bound mStau, is activated and triggers decay of the constituent mRNA but, at least for those tested, not the constituent m½-sbsRNA (Fig. 7).
Figure 7.
Model for the m½-sbsRNA-mediated SMD of mRNAs. mRNAs harboring a 3′ UTR B1, B2, and/or B4 SINE base-pair with a m½-sbsRNA via a partially complementary SINE of the same family to form a B1–B1, B2–B2, or B4–B4 SBS. At least for human SBSs, STAU1 and/or STAU2 can associate with the SBS and recruit UPF1 (Park et al. 2013). When translation terminates upstream of the SBS, SBS-bound mStau, which recruits mUpf1, triggers SMD of the mRNA. Data indicate that m½-sbsRNAs are not degraded, presumably because they are not translated.
mStau1 binding to an SBS stabilizes the duplex (Fig. 4E). If the duplex is sufficiently long, multiple mStau1 and mStau2 molecules are envisioned to stabilize the SBS, considering that both human STAU1 and STAU2 (1) bind SBSs (Furic et al. 2008; Park et al. 2013), (2) form homodimers and heterodimers if not multimers (Martel et al. 2010; Park et al. 2013), and (3) bind UPF1 (Park et al. 2013). We show here that multiple m½-sbsRNAs can trigger the SMD of an individual mRNA, and an individual m½-sbsRNA can trigger the SMD of multiple mRNAs (Figs. 1–4). In fact, our finding that a single m½-sbsRNA can contain more than one SINE further illustrates the remarkable complexity of this post-transcriptional regulatory pathway.
Taking cues from our studies of 3′ UTR Alu elements within human mRNAs, not all mouse mRNAs harboring a 3′ UTR B element can be expected to be SMD targets, even in the presence of a complementary m½-sbsRNA that functions in the SMD of one or more other mRNAs. For example, the 3′ UTR Alu element of human BAG5 mRNA is predicted to base-pair with ½-sbsRNA2 with a ΔG of −416 kcal/mol (Gong and Maquat 2011a). For comparison, the 3′ UTR Alu element of human CDCP1 mRNA is predicted to base-pair with ½-sbsRNA2 with a ΔG of −153.7 kcal/mol (Gong and Maquat 2011a). Nevertheless, ½-sbsRNA2 coimmunoprecipitates with CDCP1 mRNA and triggers CDCP1 SMD but fails to coimmunoprecipitate with BAG5 mRNA, presumably because of an inhibitory BAG5 mRNP structure (Gong et al. 2012). As another example provided here, m½-sbsRNA3 triggers the SMD of mCdc6 mRNA but not mSmad5 mRNA despite predictions that the duplex of m½-sbsRNA3(B4) is stronger with mSmad5 mRNA (ΔG of −100.5 kcal/mol) than with mCdc6 mRNA (ΔG of −91.8 kcal/mol) (Figs. 1E, 2C, 3C; Supplemental Fig. 1E,F). While it remains to be determined how mStau1 and mUpf1 reduce mSmad5 mRNA abundance (Fig. 2C), the simplest explanation is that a 3′ UTR SBS in mSmad5 mRNA does not reside exclusively, if at all, in its B4 SINE. Alternatively, it may be that mSmad5 mRNA is an indirect target of SMD: While mSmad5 mRNA coimmunoprecipitates with mStau1 (Fig. 3C), it may not do so within its 3′ UTR.
Indeed, while it is possible to computationally predict those cellular SINE-containing lncRNAs that base-pair with a particular mRNA [e.g., see Supplemental Table 5 for mRnf168 mRNA(B1) predictions], computational predictions do not necessarily reflect reality. Thus, the lncRNA must be shown to be present in the cell type of interest, and base-pairing must be shown to occur in that cell type and to trigger SMD. In addition to the reasons stated above, we found that most mouse SINE-containing lncRNAs analyzed are not significantly cytoplasmic in C2C12 MBs and therefore are not expected to efficiently trigger SMD (J Wang and LE Maquat, unpubl.). Furthermore, our computational analyses allow for no more than two consecutive mismatches between SINEs. Considering that mStau1 binding to an SBS stabilizes the SBS (Fig. 4E) and that mStau1 undoubtedly dimerizes in a way that promotes SMD, as does its human ortholog (Gleghorn et al. 2013), it is likely that functional SBSs are tolerant of mismatches that are >2 nt, depending on the extent of base-pairing in the rest of the duplex. Thus, there are many variables to what defines a functional SBS that triggers SMD. SINEs have been identified in all mammals that have been examined and also in a number of fish, reptiles, and invertebrates. Considering that Stau and Upf1 proteins exist for members of all of these groups (Lynch et al. 2006; Imamachi et al. 2012; Tosar et al. 2012), it will be interesting to determine how widely ½-sbsRNA-mediated SMD has been distributed during evolution as a means of post-transcriptional gene control. The first integrations of B1, B2, B4, and ID SINES into the mouse genome occurred, respectively, ∼80 million, ∼55 million, <80 million, and ∼80 million years ago (Kass et al. 1996; Lee et al. 1998; Kass and Jamison 2007; Tsirigos and Rigoutsos 2009). We suggest that the SINE-mediated post-transcriptional network continues to build on existing and coevolving gene regulatory pathways.
Materials and methods
Computational analyses
A Perl program, 3_ UTR_Finder, was written to define mouse mRNA 3′ UTR sequences using the NCBI mouse mRNA database (http://www.urmc.rochester.edu/labs/Maquat-Lab/software). RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) was then used to identify B1, B2, B4, ID, and MIR SINEs in the 3′ UTR sequences and in mouse lncRNAs that are present in the Antisense ncRNA Pipeline (Engström et al. 2006; http://research.imb.uq.edu.au/rnadb/rnadb2_archive.htm). RNA_RNA_anneal (Gong and Maquat 2011a), which uses a recursive algorithm to predict the most stable duplexes and their folding free-energy change (ΔG) at 25°C (Mathews et al. 1999, 2004; Turner and Mathews 2009), was used to predict SINE base-pairing between lncRNAs and mRNA 3′ UTRs.
Cell cultures and transient transfections
Mouse skeletal muscle C2C12 MBs, a type of pluriopotent mesynchymal precursor cell, were propagated in DMEM (GIBCO-BRL) containing 15% fetal bovine serum and, where specified, induced to differentiate (at a concentration of 2 × 106 per 60-mm dish or 7.5 × 107 per 150-mm dish) to MTs using DMEM containing 5% horse serum (GIBCO-BRL). MBs or MTs were harvested as previously described (Gong et al. 2009) using 0.15% trypsin (Sigma) rather than scrapping to avoid harvesting undifferentiated reserve cells along with MTs. MBs were transiently transfected with siRNA and/or plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs consisted of mStau1 siRNA (Gong et al. 2009), mUpf1 siRNA (Gong et al. 2009), mTraf6 siRNA (Xiao et al. 2012), mCdc6 siRNA (Zhang et al. 2010), m½-sbsRNA1 siRNA (5′-GCUCUGGACUUGUCAGUUAdTdT-3′), m½-sbsRNA2 siRNA (5′-GCAACUAAUCAGUAACCGUUUdTdT-3′), m½-sbsRNA3 siRNA (5′-GAUGCUAGCUGUCUACUUAUUdTdT-3′), m½-sbsRNA4 siRNA (5′-GUGUAGGGCAUGAAUUUAAUUdTdT-3′), or the nonspecific Silencer Negative Control #1 (Ambion). No siRNA targeted SINE nucleotides.
For mRNA half-life studies using NIH3T3 B2A2 Tet-off cells, see Chen et al. (2008) and the legend for Figure 3.
Cell lysis, protein purification, immunoprecipitation, and Western blotting
Cells that were not subjected to immunoprecipitation were lysed using hypotonic buffer (Gong and Maquat 2011a). For all immunoprecipitations, cells were cross-linked using 0.75% formaldehyde for 10 min at room temperature, subsequently quenched using 0.25 M glycine for 5 min at room temperature, lysed using sonication, and immunoprecipitated as previously described (Gong and Maquat 2011a). Immunoprecipitations used anti-HA (Roche), anti-Flag (Sigma), rIgG (Sigma), or mIgG (Sigma). Cross-links were reversed after immunoprecipitation by heating for 60 min at 65°C. Western blotting was performed as described (Gong and Maquat 2011a) using antibodies described in the Supplemental Material. Total cell protein was quantitated after electrophoresis and transfer using Ponceau S-staining (Sigma) and/or anti-Calnexin. Each Western blot includes a series of threefold dilutions of protein that are shown under the wedge in the left-most part of each figure, demonstrating that data reside within the linear range of analysis.
Plasmid constructions, RNA purification, and RT coupled to either semiquantitative PCR or real-time (q)PCR, and microscopy
For plasmid constructions, RNA purification, and RT coupled to either semiquantitative PCR or real-time (q)PCR, and microscopy, see the Supplemental Material.
Acknowledgments
We thank Louise Baskin and Sarah Anderson at Dharmacon for help designing lncRNA siRNAs, Susanna de Lucas and Juan Ortín for anti-STAU1 antibodies, Ann-Bin Shyu for NIH3T3 B2A2 cells, Dave Mathews and Yinghan Fu for computational advice, Keith Nehrke for access to his inverted fluorescence microscope, Linda Callahan for confocal microscope assistance, and Max Popp and Riad Elbarbary for comments on the manuscript. This work was supported by the National Institutes of Health (GM074593 to L.E.M.) and a Messersmith Graduate Student Fellowship (C.G.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.212639.112.
References
- Ashley CT, Wilkinson KD, Reines D, Warren ST 1993. FMR1 protein: Conserved RNP family domains and selective RNA binding. Science 262: 563–566 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Chen IHB, Huber M, Guan T, Bubeck A, Gerace L 2006. Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis. BMC Cell Biol 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYA, Ezzeddine N, Shyu AB 2008. Messenger RNA half-life measurements in mammalian cells. Methods Enzymol 448: 335–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS, Kim YK 2012. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell 46: 495–506 [DOI] [PubMed] [Google Scholar]
- Deato MD, Tjian R 2007. Switching of the core transcription machinery during myogenesis. Genes Dev 21: 2137–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L 2006. Complex loci in human and mouse genomes. PLoS Genet 2: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Maher-Laporte M, DesGroseillers L 2008. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1-and Staufen2-containing ribonucleoprotein complexes. RNA 14: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn ML, Gong C, Kielkopf CL, Maquat LE 2013. Staufen1 dimerizes via a conserved motif and a degenerate double-stranded RNA-binding domain to promote mRNA decay. Nat Struct Mol Biol 20: 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011a. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011b. ‘Alu’strious long ncRNAs and their role in shortening mRNA half-lives. Cell Cycle 10: 1882–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE 2009. SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes Dev 23: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Popp M, Maquat LE 2012. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods 58: 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH 2012. Active human retrotransposons: Variation and disease. Curr Opin Genet Dev 22: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamachi N, Tani H, Akimitsu N 2012. Up-frameshift protein 1 (UPF1): Multitalented entertainer in RNA decay. Drug Discovery Ther 6: 55. [PubMed] [Google Scholar]
- Kass DH, Jamison N 2007. Identification of an active ID-like group of SINEs in the mouse. Genomics 90: 416–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass DH, Kim J, Deininger PL 1996. Sporadic amplification of ID elements in rodents. J Mol Evol 42: 7–14 [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, DesGroseillers L, Maquat LE 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′ UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 26: 2670–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS 2011. SINEs. Wiley interdisciplinary reviews RNA 2: 772–786 [DOI] [PubMed] [Google Scholar]
- Lee IY, Westaway D, Smit AFA, Wang K, Seto J, Chen L, Acharya C, Ankener M, Baskin D, Cooper C 1998. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res 8: 1022–1037 [DOI] [PubMed] [Google Scholar]
- Lynch M, Hong X, Scofield DG 2006. NMD and the evolution of eukaryotyic gene structure. In Nonsense-mediated mRNA decay. (ed LE Maquat) pp. 197–211, Landes Bioscience, Georgetown, TX [Google Scholar]
- Maquat LE, Gong C 2009. Gene expression networks: Competing mRNA decay pathways in mammalian cells. Biochem Soc Trans 37: 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Dugré-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trépanier V, Duchaîne T, Bouvier M, DesGroseillers L 2010. Multimerization of Staufen1 in live cells. RNA 16: 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci 101: 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueck T, Berger F, Buechsler I, Valchanova RS, Landuzzi L, Lollini PL, Klingel K, Munz B 2011. TRAF6 regulates proliferation and differentiation of skeletal myoblasts. Differentiation 81: 99–106 [DOI] [PubMed] [Google Scholar]
- Neeman Y, Levanon EY, Jantsch MF, Eisenberg E 2006. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA 12: 1802–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Dinger ME, Engström PG, Lenhard B, Mattick JS 2007. RNAdb 2.0—an expanded database of mammalian non-coding RNAs. Nucleic Acids Res 35: D178–D182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE 2013. Staufen 2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci 110: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel AM 2012. LINEs, SINEs and other retroelements: Do birds of a feather flock together? Front Biosci 17: 1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Riggs AD 1995. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res 23: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Trausch-Azar J, Ciechanover A, Schwartz A 2006. E2A protein degradation by the ubiquitin-proteasome system is stage-dependent during muscle differentiation. Oncogene 26: 441–448 [DOI] [PubMed] [Google Scholar]
- Tosar L, Thomas MG, Baez MV, Ibanez I, Chernomoretz A, Boccaccio GL 2012. Staufen: From embryo polarity to cellular stress and neurodegeneration. Front Biosci 4: 432. [DOI] [PubMed] [Google Scholar]
- Tsirigos A, Rigoutsos I 2009. Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Comput Biol 5: e1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DH, Mathews DH 2009. NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 38: D280–D282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Arron JR 2003. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays 25: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Xiao F, Wang H, Fu X, Li Y, Wu Z 2012. TRAF6 promotes myogenic differentiation via the TAK1/p38 mitogen-activated protein kinase and Akt pathways. PLoS ONE 7: e34081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H, Erikson RL 2010. Cell division cycle 6, a mitotic substrate of polo-like kinase 1, regulates chromosomal segregation mediated by cyclin-dependent kinase 1 and Separase. Proc Natl Acad Sci 107: 19742–19747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Sha J, Harter ML 2010. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol 188: 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]