Abstract
For biosynthesis of bacillamide C by Bacillus atrophaeus C89 associated with South China sea sponge Dysidea avara, it is hypothesized that decarboxylation from L-tryptophan to tryptamine could be performed before amidation by the downstream aromatic L-amino acid decarboxylase (AADC) to the non-ribosomal peptide synthetases (NRPS) gene cluster for biosynthesizing bacillamide C. The structural analysis of decarboxylases' known substrates in KEGG database and alignment analysis of amino acid sequence of AADC have suggested that L-tryptophan and L-phenylalanine are the potential substrates of AADC. The enzymatic kinetic experiment of the recombinant AADC proved that L-tryptophan is a more reactive substrate of AADC than L-phenylalanine. Meanwhile, the AADC-catalyzed conversion of L-tryptophan into tryptamine was confirmed by means of HPLC and LC/MS. Thus during bacillamide C biosynthesis, the decarboxylation of L-tryptophan to tryptamine is likely conducted first under AADC catalysis, followed by the amidation of tryptamine with the carboxylic product of NRPS gene cluster.
Bacillamides, which have four members of bacillamide A, B, C and D1,2,3,4 (Fig. S1), contain a tryptamide thiazole, a common building block in potential bioactive cyclic peptides such as the protein synthesis inhibitors A21459 A and B from an Actinoplanes sp.5, antibiotic zelkovamycin produced by a Streptomyces sp. K96-06706 and immunosuppressive argyrins from myxobacteria7. Bacillamide A and its derivatives show antibiosis against dinoflagellates, raphidophytes2 and a particular species of cyanobacteria8. In our previous studies, bacillamide C and a new compound of neobacillamide A were isolated from Bacillus atrophaeus C89 associated with the South China Sea sponge Dysidea avara9. Since bacillamide C is obtained in higher yield than that of neobacillamide A9, the fermentation optimization of bacillamide C was carried out, and a 26-fold increase in bacillamide C production (71 mg/L) by B. atrophaeus C89 was achieved10. Though chemical synthesis of bacillamide C was carried out recently11,12, the biosynthetic pathway of bacillamide C remains unknown.
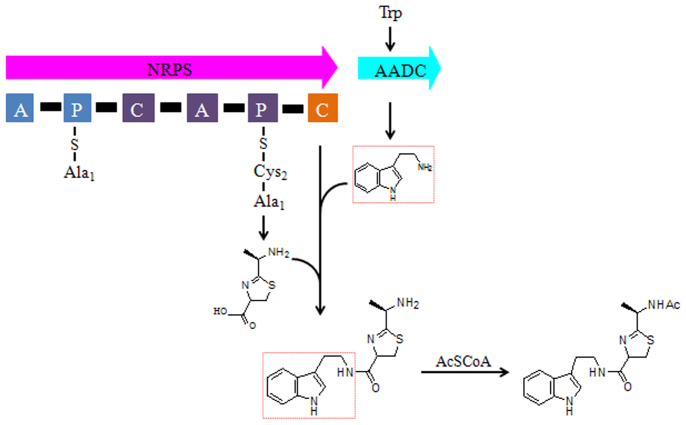
Nonribosomal peptides generated by nonribosomal peptide synthetases (NRPS) are extraordinary important for modern medicine because of their diverse biological activities, for example antibiotics tyrocidin A13, vancomycin14, immunosuppressive agents cyclosporine A15, cytostatics bleomycin A216 and toxins thaxtomin A17. Biogenetically, bacillamides including neobacillamide A and bacillamide C belong to nonribosomal peptides. Similarly as neobacillamide A9, bacillamide C could be derived from amino acids alanine, cysteine and tryptophan through NRPS biosynthesis strategy, which was supported by the finding of a NRPS gene cluster (7, 011 bp) by sequencing the genome of B. atrophaeus C89 (Genbank No. JQ 687535)18. Accordingly, the putative biosynthetic pathway of bacillamide C could be proposed based on the NRPSs domain organization (Fig. 1). Among the six domains of the NRPS gene cluster, the first module contains an adenylation domain and a peptidyl carrier protein domain (A-PCP), which selects and activates alanine. The peptide bond formation and heterocyclization is performed with cysteine by the adjacent elongation module containing a condensation domain, an adenylation domain and a peptidyl carrier protein domain (C-A-PCP). However, we do not know whether the decarboxylation of tryptophan to tryptamine is performed before amidation or after.
Figure 1. Supposed biosynthetic pathway of bacillamide C. Each square represents a NRPS enzymatic domain: C: condensation domain; A: adenylation domain; P: peptidyl carrier protein domain.
According to the genomic data of B. atrophaeus C89 (Genbank No. JQ 687535), there are fifteen putative decarboxylases in B. atrophaeus C89, so, which one is responsible for the decarboxylation needs to be defined. Here we put forward the hypothesis that the decarboxylase gene (AADC gene), which is at the nearest downstream after the NRPS gene cluster in B. atrophaeus C89, is probably involved in the decarboxylation of L-tryptophan to tryptamine, and then tryptamine is combined with the product of NRPS gene cluster by amidation (Fig. 1). In order to clarify the above hypothesis, under the guide of bioinformatics analysis of substrates' similarity and amino acid sequence, the AADC gene of B. atrophaeus C89 was cloned and expressed in Escherichia coli BL21. Subsequently, the isolated AADC enzyme was characterized as a highly efficient catalyst for the decarboxylation of tryptophan to tryptamine, suggesting that tryptamine rather than tryptophan was incorporated into the non-ribosomal peptide bacillamide C. Thus, this study provides an insight to elucidate the biosynthetic mechanism of bacillamide C in B. atrophaeus C89.
Results
Analysis on the potential substrate of AADC
According to the bioinformatic analysis18, there were 15 possible decarboxylases in the genome of B. atrophaeus C89 and all of them were exhibited high similarity (from 72% to 100%) to each other (Table 1). Usually, an enzyme is named based upon its substrate. For example, decarboxylyases are named based upon their substrates19. There are a quite few known substrate molecules in KEGG database. With the substrate-enzyme relationship in KEGG, a few of substrates of decarboxylases in B. atrophaeus C89 were predicted except for phenolic acid decarboxylase (id001001), phenylacrylic acid decarboxylase (id 003690) and pyridoxal-dependent decarboxylyase (id 003505) (Table 1). The alternative name of phenolic acid decarboxylase (id001001) is 4-hydroxybenzoate decarboxylase, thus 4-hydroxybenzoate could be the substrate of phenolic acid decarboxylase (id001001). Amino acid sequence of phenylacrylic acid decarboxylase (YP_004875917.1) showed 94% identity with phenylacrylic acid decarboxylase (id003690) from B. atrophaeus C89. YP_004875917.1 catalyzes the conversion of 4-coumarate (p-coumaric acid) to 4-4-vinylphenol20, so 4-coumarate could be a substrate of phenylacrylic acid decarboxylase (id003690). Similarly, the putative full length amino acid sequence of JQ400024 (id003505) showed 100% and 97% identity to the pyridoxal-dependent decarboxylyase of B. atrophaeus 1942 (YP_003975768.1) and aromatic-L-amino-acid decarboxylase of Muricauda ruestringensis DSM 13 (YP_004788433.1) respectively. Therefore, L-tryptophan and L-phenylalanine are the possible substrates for the functional gene JQ400024/id003505. As shown in Table 1, 14 different substrates corresponding to the 15 decarboxylases were searched, in which JQ608457 (id001001), JQ608467 (id003691) and JQ608468 (id003692) have the same substrate, 4-hydroxybenzoate. Whereas, JQ400024 (id003505) has two possible substrates L-tryptophan and L-phenylalanine.
Table 1. Putative substrates of 15 decarboxylases from B. atrophaeus C89.
| Gene No. | Enzyme (id) | Putative function homolog | Protein accession No | Identity (%) | Deduced enzyme substrate |
|---|---|---|---|---|---|
| JQ608455 | 000589 | alpha-acetolactate decarboxylase | NP_391481.1 | 86 | 2-Acetolactate |
| JQ608456 | 000869 | oxalate decarboxylase | NP_391204.1 | 85 | Oxalate |
| JQ608457 | 001001 | phenolic acid decarboxylase | YP_080504.1 | 90 | 4-Hydroxybenzoate |
| JQ608458 | 001372 | lysine decarboxylase | NP_387908.1 | 72 | L-Lysine |
| JQ608459 | 001860 | acetoacetate decarboxylase | YP_001422565.1 | 91 | Acetoacetate |
| JQ608460 | 002294 | aspartate alpha-decarboxylase | NP_390122.1 | 90 | L-Aspartate |
| JQ608461 | 002972 | uroporphyrinogen decarboxylase | NP_388893.1 | 88 | Uroporphyrinogen III |
| JQ608462 | 003331 | phosphopantothenoylcy steine decarboxylase | YP_004207623 | 87 | (R)-4′-Phosphopantothenoyl-L-cysteine |
| JQ608463 | 003347 | orotidine 5′-phosphate decarboxylase | NP_389438.1 | 85 | Orotidine 5′-phosphate |
| JQ608464 | 003445 | arginine decarboxylase | NP_389346.1 | 90 | L-Arginine |
| JQ400024 | 003505 | pyridoxal-dependent decarboxylase | YP_003975768.1 | 100 | L-Tryptophan |
| L-Phenylalanine | |||||
| JQ608465 | 003545 | phosphatidylserine decarboxylase | YP_001419902.1 | 74 | Phosphatidylserine |
| JQ608466 | 003690 | phenylacrylic acid decarboxylase | YP_004875917.1 | 94 | 4-Coumarate |
| JQ608467 | 003691 | 4-hydroxybenzoate decarboxylase | NP_388246.1 | 96 | 4-Hydroxybenzoate |
| JQ608468 | 003692 | 4-hydroxybenzoate decarboxylase | YP_003097675.1 | 93 | 4-Hydroxybenzoate |
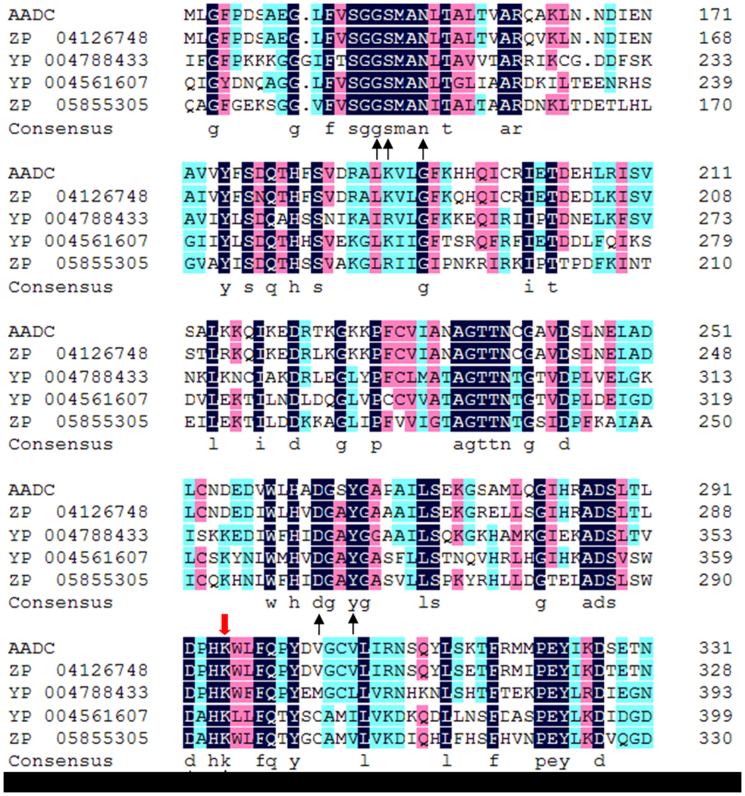
The amino acid sequence of the catalytic domain of functional gene JQ400024 (id003505) was aligned with certain reported decarboxylases in the GenBank (No: ZP 04126748, YP 004788433, YP 004561607 and ZP 05855305) (Fig. 2). AADC amino acid sequence (No: ZP 04126748) of Bacillus thuringiensis serovar sotto str.T04001 showed 88% similarity with JQ400024 (id003505). Both YP 004561607 and ZP 05855305 were the same type of decarboxylases, which belong to aromatic L-amino acid decarboxylases. Although the AADC sequence has lower similarity (less than 50%) with these aromatic L-amino acid decarboxylases, all of them have identical conserved pyridoxal 5′-phosphate (PLP) binding pockets and catalytic residue (Lys) (Fig. 2). The alignment result suggested that the JQ400024 (id003505) was a type of aromatic L-amino acid decarboxylase and it could catalyze the decarboxylation of aromatic L-amino acids.
Figure 2. Amino acid sequence multiple alignments of decarboxylases catalytic domains from B. atrophaeus C89 (AADC), Bacillus thuringiensis serovar sotto str. T04001 (ZP 04126748), Muricauda ruestringensis DSM13258 (YP 004788433), Erysipelothrix rhusiopathiae str. Fujisawa (YP 004561607) and Blautia hansenii DSM20583 (ZP 05855305).
The pyridoxal 5′-phosphate binding pocket was indicated by black arrow. The catalytic residue was indicated by red arrow.
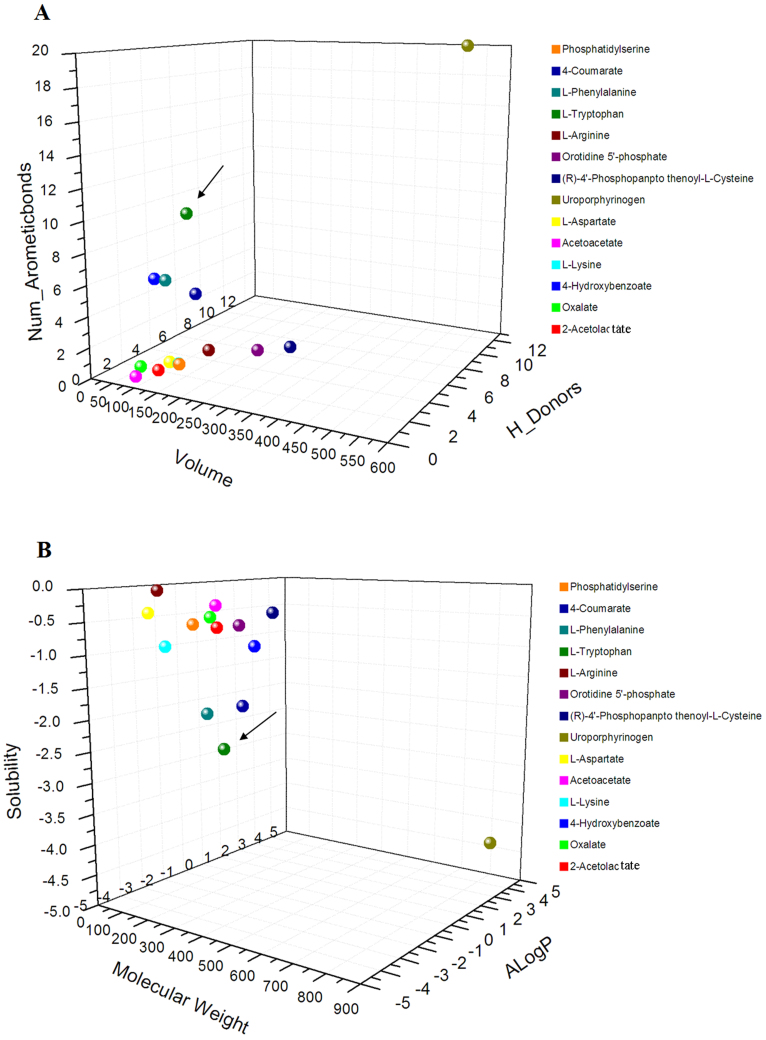
Owing to the relatively low sequence similarity for the same functional decarboxylases and the possible diversity in terms of substrates (as discussed above), a comparison was carried out between the observed substrate molecules. It is straightforward for us to characterize substrate properties based on their volume size, molecular weight, numbers of hydrogen bonding donor and acceptor, oil-water distribution, number of aromatic rings, and so on. As shown in Table 2, uroporphyrinogen III, L-tryptophan, L-phenylalanine, 4-hydroxybenzoate and 4-courmarate are aromatic carboxylates and others are not aromatic carboxylates. For these aromatic substrates, molecular size was in order of uroporphyrinogen III > L-tryptophen > L-phenylalanine > 4-courmarate > 4-hydroxybenzoate. Actually, molecular volume of L-tryptophan, L-phenylalanine and 4-courmarate were in a range of 100–130 Å. However, molecular weight (molar mass) and oil-water solubility of L-trytophan and L-phenylalanine were much lower than those of 4-courmarate and 4-hydroxybenzoate. As a result, we concluded that L-tryptophan and L-phenylalanine shared high similarity among the known substrates of decarboxylases (Fig. 3).
Table 2. Molecular characteristics of 14 decarboxylases' substrates.
| Deduced enzyme substrate | H Donor | Number of Aromatic bonds | Volume | Molecular Weight | Log P* | Solubility |
|---|---|---|---|---|---|---|
| 2-Acetolactate | 2 | 0 | 86.77 | 132.115 | −0.634 | −0.702 |
| Oxalate | 2 | 0 | 45.27 | 90.0349 | −0.429 | −0.553 |
| 4-Hydroxybenzoate | 2 | 6 | 84.37 | 138.121 | 1.217 | −1.092 |
| L-Lysine | 3 | 0 | 101.52 | 146.188 | −3.24 | −0.891 |
| Acetoacetate | 1 | 0 | 68.25 | 102.089 | −0.294 | −0.355 |
| L-Aspartate | 3 | 0 | 80.26 | 133.103 | −3.78 | −0.368 |
| Uroporphyrinogen III | 12 | 20 | 524.78 | 836.795 | 4.06 | −4.289 |
| (R)-4′-Phosphopantothenoyl-L-cysteine | 7 | 0 | 246.61 | 402.358 | −1.503 | −0.365 |
| Orotidine 5′-phosphate | 6 | 0 | 196.53 | 368.191 | −2.663 | −0.526 |
| L-Arginine | 5 | 0 | 110.1 | 174.201 | −3.831 | −0.018 |
| L-Tryptophan | 3 | 10 | 128.96 | 204.225 | −1.312 | −2.641 |
| L-Phenylalanine | 2 | 6 | 109.41 | 165.189 | −1.605 | −2.051 |
| Phosphatidylserine | 3 | 0 | 207.85 | 343.224 | −4.374 | −0.459 |
| 4-Coumarate | 1 | 6 | 102.89 | 163.15 | 0.211 | −2.054 |
Note: * LogP means logarithm of the octanol-water partition coefficient.
Figure 3. 3D scatterplot for the molecular properties of fourteen deduced substrates from fifteen decarboxylases.
A: based on molecular properties of volume, H: donor and aromatic bond; B: based on molecular properties of molecular weight, Alogp and solubility. Target substrate was indicated by black arrow.
Function of AADC in the transformation of L-tryptophan into tryptamine
The AADC gene was overexpressed in E.coli BL21 (DE3) as an N-termal His6-tagged fusion protein and purified by an affinity chromatography. The protein purity was judged by SDS-PAGE analysis which revealed AADC with a molecular mass of 58 kDa (Fig. 4). The MW and pI of the enzyme were calculated as 55.899 kDa and 5.96, respectively.
Figure 4. SDS-PAGE of purified recombinant decarboxylase.
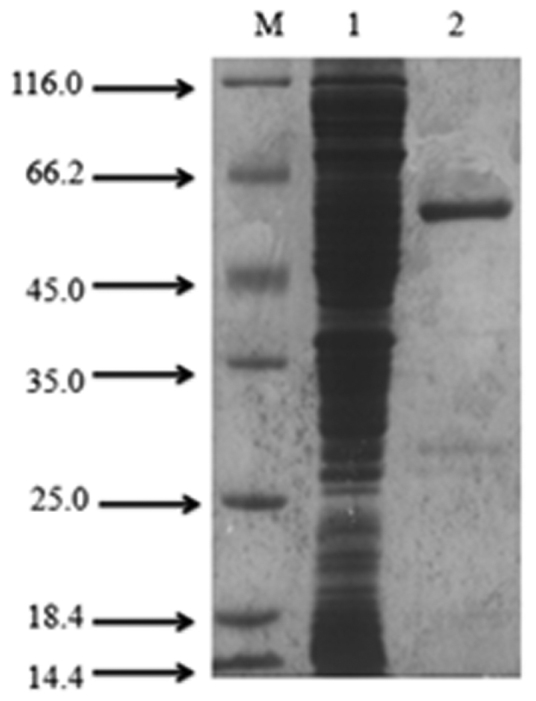
M: protein Marker; Lane 1: unpurified sample; Lane 2: purified enzyme.
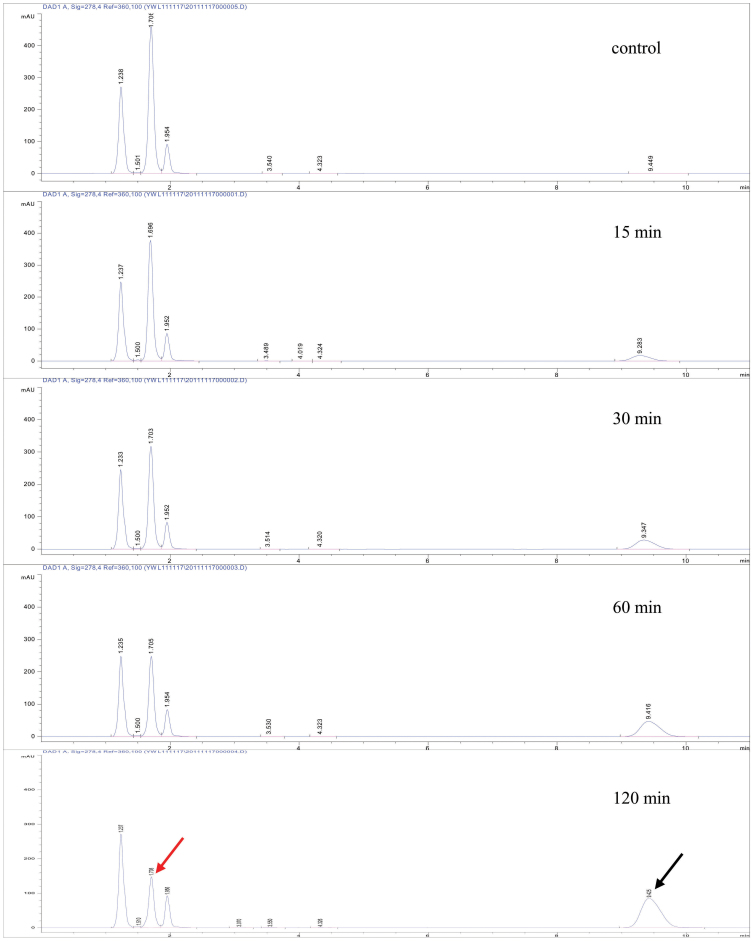
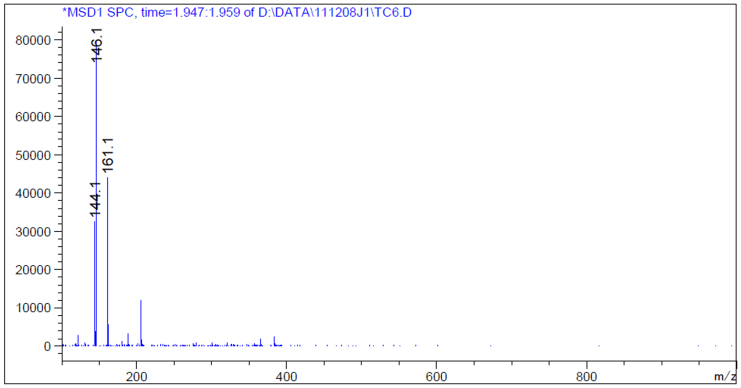
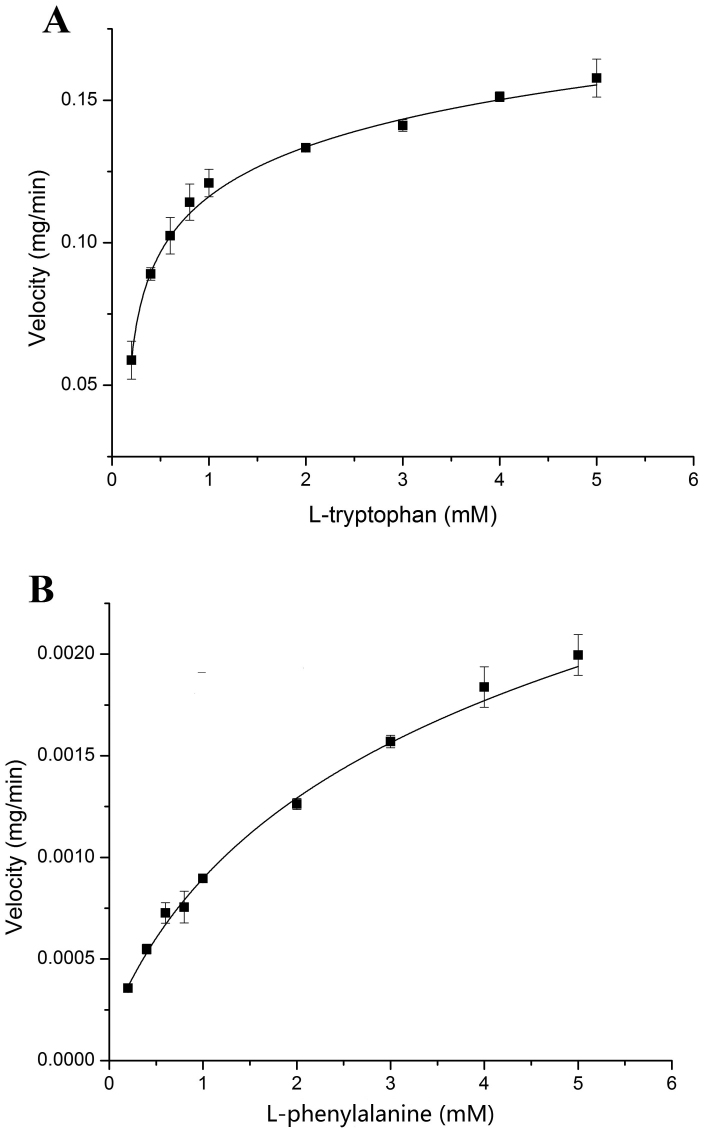
As proposed in Fig. 1, L-tryptophan was tested first as a putative substrate of AADC. Thin layer chromatography (TLC) and high pressure liquid chromatography (HPLC) were employed to monitor the production of tryptamine, the proposed product of AADC. After incubation with 1 mM of AADC and 1 mM of L-tryptophan for 15 min, a small peak occurred at 9.5 min (Fig. 5). The peak was increased with increase in incubation time, with decrease in L-tryptophan (rt = 2.5 min). The peak of 161.1 m/z ([M + H]+) in Fig. 6 proved the product of tryptamine because of its exact mass is 160.22. The fragmentation ion at 144.1 m/z is the product of the loss of ammonia from tryptamine, and the fragmentation ion at 146.1 m/z is [M + 2H]2+ (Fig. 6)21. Subsequently, the kinetic parameters were determined based on the tryptamine production. Km and Vmax values were calculated as 0.35 mmol/L and 0.163 mg product/min·mg protein, respectively (Fig. 7). Next, we used two other aromatic amino acids L-phenylalanine and L-tyrosine as substrates to assay the activity of AADC. Thin layer chromatography (TLC) (ninhydrin staining; mobile phase butanol/acetic acid/water: 4/1/1) showed that AADC could not use L-tyrosine as substrate (data not shown). However, AADC could recognize L-phenylalanine as a substrate and catalyze its decarboxylation. The kinetic parameters of AADC toward L-phenylalanine were determined as 0.872 mmol/L and 0.00184 mg product/min·mg protein, respectively (Fig. 7). But, L-tryptophan exhibited lower Km (0.35 mmol/L) and higher Vmax values (0.163 mg product/min mg protein) indicating that L-tryptophan was a better substrate for AADC than L-phenylalanine.
Figure 5. HPLC analysis of L-tryptophan and tryptamine in AADC enzyme reaction at different time points.
Red arrow: L-tryptophan, black arrow: tryptamine.
Figure 6. LC-MS result of AADC enzyme reaction with L-tryptophan as substrate.
Figure 7. Determination of Km and Vmax values for L-tryptophan (A) and L-phenylalanine (B) as substrate, respectively.
Values are means ± standard deviation.
Discussion
As shown in Fig. 1, bacillamide C was suggested to be derived from the amino acids alanine, cysteine and tryptophan through nonribosomal peptide synthetase (NRPS) biosynthesis strategy. Decarboxylation and amidation are two important steps in the biosynthesis of bacillamide C. Whether the decarboxylation of L-tryptophan to tryptamine is performed before amidation and which decarboxylase is responsible for the decarboxylaiton are two critical issues in the understanding of the biosynthesis mechanism of bacillamide C.
The 15 decarboxylases in B. atrophaeus C89 have 14 different substrates (Table 1). The suggested substrates and their molecular properties analysis might exclude the possibility of decarboxylation after amidation. According to the location of decarboxylases in the genome of B. atrophaeus C89, we deduce that the aromatic L-amino acid decarboxylase (AADC, JQ400024; id003505), which is the closest downstream decarboxylase to NRPS gene cluster, is most likely involved in the biosynthesis of bacillamide C. This was proved by bioinformatics analysis and AADC gene cloning, expression, enzymatic dynamics and activity tests.
The analysis of amino acid sequence alignment between the AADC and some reported decarboxylases showed that the AADC contained the high conserved identical PLP binding pockets and catalytic residue (lys) of aromatic L-amino acid decarboxylase (Fig. 2). Meanwhile, the molecular property analysis of 14 decarboxylases' substrates (Fig. 3; Table 1 and Table 2) suggested that L-tryptophan and L-phenylalanine are probably the substrates based on the substrates' similarity analysis, L-tryptophan and L-phenylalanine are the possible substrates of aromatic L-amino acid decarboxylase (AADC, JQ400024; id003505). This was supported by the production of bacillamide C and neobacillamide A by B. atrophaeus C89 synchronously9.
The analysis of enzymatic dynamics proved that L-tryptophan was a better substrate for AADC than L-phenylalanine because of its lower Km and higher Vmax values (Fig. 7). This result was consistent with our previous study that the yield of bacillamide C was higher than that of neobacillamide A isolated from B. atrophaeus C899. In contrast, if L-phenylalanine is provided as substrate instead of L-tryptophan, the product obtained will be neobacillamide A. Neobacillamide A may share the similar biosynthetic pathway as bacillamide C, only the substrates for AADC are different.
Lots of literatures have reported that aromatic L-amino acid decarboxylases are involved in the biosynthesis of secondary metabolites including terpenoid indole alkaloids, hydroxycinnamic acid amides, serotonin and phenylacetaldehyde22,23,24. AADCs L-tryptophan decarboxylases have been intensively studied because of their importance in the biosynthesis of pharmaceutical secondary metabolites23,24,25,26. When L-tryptophan was used as the substrate, the Km of AADC in B. atrophaeus C89 was 0.35 mM which is four-fold higher than that of the C. roseus (0.075 mM)25. Whereas, the decarboxylases from Ophiorrhiza pumila26 and rice24 showed two-fold higher Km values (0.72 mM, 0.69 mM, respectively) than that of B. atrophaeus C89 AADC. It was suggested that the affinity of L-tryptophan for AADC in B. atrophaeus C89 is different from that of other microorganisms.
On conclusion, the bioinformatics analysis and in vitro dynamics and activity of AADC indicated that AADC is probably involved in the biosynthesis of bacillamide C through decarboxylation of L-tryptophan to tryptamine. In NRPS modular organization (Fig. 1), the last module is only composed of C domain without A domain. This C domain is responsible for peptide bond formation between tryptamine and the growing peptide chain intermediates, which is catalyzed by the other two modules (A-PCP and C-A-PCP). The knockout of AADC gene from B. atrophaeus C89 will provide the direct evidence in future. Even so, the results from this study suggested that the bacillamide C production by B. atrophaeus C89 could be improved with the addition of L-tryptophan. Most importantly, based on the characterization of AADC, manipulation of AADC's specificity by point mutation of the substrate-coordinating amino acid residues mighty be an approach to generate novel bacillamide compounds in further research.
Methods
Reagents
L-tryptophan and L-phenylalanine were purchased from Sigma Chemical Company (St. Louis, MO, USA). Trans Taq™ DNA polymerase and pEASY™-E1 Expression Kit were purchased from TransGen Biotech (Beijing, China). Plasmid Mini Kit I was purchased from Omega (USA). Ni-NTA agarose was purchased from Qiagen (Qiagen Co., Hilden, Germany). All other chemicals used were analytical grade reagents unless stated.
Strains and plasmids
B. atrophaeus C89 isolated from South China Sea sponge Dysidea avara was identified according to the 16S rRNA gene27 (Genbank No. DQ091007). E. coli Trans1-T1 and BL21 (DE3) strains were used for propagation of plasmids and as host for expression of the decarboxylase gene, respectively. B. atrophaeus C89 was incubated in a medium containing 5 g of beef extract and 10 g of peptone in a liter of artificial seawater28. The culture of B. atrophaeus C89 was maintained at 28°C. E. coli Trans1-T1 and BL21 were grown in Luria-Bertani (LB) medium at 37°C.
Analysis of molecular properties of decarboxylases' substrates
The GenBank accession numbers of 15 decarboxylases' sequences from B. atrophaeus C89 were JQ608455 (id 000589), JQ608456 (id 000869), JQ608457 (id 001001), JQ608458 (id 001372), JQ608459 (id 001860), JQ608460 (id 002294), JQ608461 (id 002972), JQ608462 (id 003331), JQ608463 (id 003347), JQ608464 (id 003445), JQ400024 (id 003505), JQ608465 (id 003545), JQ608466 (id 003690), JQ608467 (id 003691) and JQ608468 (id 003692) (Table 1). Amino acid sequences of the 15 decarboxylases and related homolog protein sequences were analyzed based on the Orthologous Genes (COG) and KEGG databases18. Six different characteristics (H Donor, Num_Aromatic bonds, Volume, Molecular Weight, ALogP29 and Solubility, Table 2) of different deduced substrates were selected to construct molecular structure models using Discovery Studio and the energy minimization of molecular structure models through CHARMM (Chemistry at HARvard Macromolecular Mechanics, http://www.charmm.org/)30,31. ALogP indicated the log of the octanol-water partition coefficient using Ghose and Crippen's method. Molecular solubility was expressed as LogS, where S was the solubility in mol/L. Volume is the quantity of three-dimensional space enclosed by some closed boundary. Molecular weight was calculated as the sum of the individual isotopic masses of all the atoms in any molecule. Hydrogen (H) bonds occur when a “donor” atom donates its covalently bonded hydrogen atom to an electronegative “acceptor” atom. An aromatic bond type is used for the ring bonds of benzenoid systems, 6-membered aromatic heterocycles and cyclopentadienyl systems.
Alignment analysis of AADC
The conserved catalytic domain of AADC (JQ400024, id003505) was analyzed in Conserved Domain Database at the NCBI. Database homology searches of amino acid sequences were carried out using BLASTp in NCBI protein database. Amino acid sequence alignment between B. atrophaeus C89 AADC and reference sequences was carried out by DNAMAN.
Genomic DNA extraction
B. atrophaeus C89 was grown at 28°C for 24 h. Genomic DNA was isolated by the modification of Marmur method32; cells were collected by centrifugation at 12,000 × g for 5 min. The pellets were washed twice with 1 mL of TE buffer (10 mM of Tris, 1 mM of EDTA, pH 8.0) and centrifuged at 12,000 × g for 5 min. The pellets were suspended in 500 μL of TE buffer. The suspended cells were treated with lysozyme (2 mg/mL) at 37°C for 1 h followed by 20% sodium dodecyl sulfate (SDS) and proteinase K (20 mg/mL) at 55°C for 1 h. The lysate was extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, V:V:V) twice and washed with an equal volume of chloroform/isoamyl alcohol (24:1, V:V). DNA was precipitated with an equal volume of isopropanol at −20°C for 1 h and harvested by centrifugation at 12,000 × g for 15 min. Finally, DNA pellets were washed with 70% ethanol and resuspended in 30 μL of TE Buffer. The extracted DNA was analyzed on 1.0% agarose gel.
PCR amplification of AADC gene
Forward primer decF0403 (5′-GTGAAACAAGTGTC GGAAAACCTG-3′) and reverse primer decR0403 (5′-TTATTCAGCAACGCATGGATACG-3′) used for amplification of AADC genes were designed using Primer Premier 5.0 based on the AADC sequence in the genome of B. atrophaeus C89. In PCR reaction, two hundred nanogram of template DNA was used in 50 μL of reaction system. The PCR mixture (50 μL) contained 0.2 mM of dNTPs, 0.4 μM of each primer, 5 U of Trans Taq™-T DNA Polymerase (Transgen, China) and 1 × PCR buffer (20 mM Tris-HCl , pH 9.0, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4). PCR was carried out as follows: initial denaturation at 94°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 90 s; and a final extension of 10 min at 72°C. PCR products were purified with Cycle Pure kit and Gel Extraction kit according to the manufacturer's instructions (Axygen, USA). The purified PCR products were ligated into the pEASY™-E1 Expression Vectors (Transgen, China) and the ligated mixture was transformed into competent E. coli Trans1-T1. Positive recombinants were identified by DNA sequencing with T7 promoter and T7 terminator primer. The recombinant plasmids containing AADC gene were purified using the Plasmid Mini Kit I (Omega, China) and transformed into competent cells of E. coli BL21. Transformed cells were plated on LB-agar plates containing 50 μg/mL of ampicillin and incubated at 37°C overnight.
Expression and purification of recombinant AADC
Five milliliter of LB broth containing 100 μg/mL of ampicillin were inoculated with a single transformed E. coli colony and incubated at 37°C overnight. The 5 mL of cells was inoculated into 500 mL of LB broth containing antibiotic and incubated at 37°C. IPTG and PLP (pyridoxal-5′-phosphate) at final concentration of 0.5 mM and 0.1 mM were then added to the culture medium when OD600 of the culture medium was 0.4–0.6, respectively. The incubation was continued at 28°C for 16 h. After centrifugation at 12000 × g, 4°C for 20 min, the cells pellets were suspended in lysis buffer at 2–5 mL per gram wet weight and incubated on ice with lysozyme (1 mg/mL), PMSF (1 mM) and PLP (0.5 mM) and disrupted by sonication. The clear supernatant was collected by centrifugation at 12000 × g and applied to Ni-NTA agarose resin column equilibrated with lysis buffer (pH 8.0) containing 50 mM of NaH2PO4, 300 mM of NaCl and 10 mM of imidazole. The enzyme was eluted with 20, 150 and 250 mM of imidazole in 50 mM of phosphate buffer (pH 8.0) containing 300 mM of NaCl. Recombinant decarboxylse was eluted with the above buffer by gravity. The purified enzyme was monitored by Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 12.0% (w/v) polyacrylamide gel. Protein molecular weight standards in the range of 14.4–116.0 kDa (Fermentas, USA) were used as marker. Protein bands were visualized by Coomassie brilliant blue R-250 staining33.
Activity and dynamics analyses of AADC
AADC activity was assayed according to the slightly modified method of Pennings34. The standard mixture (0.2 mL; final pH 8.0 at 35°C) contained 0.1 M of Tris-HCl buffer, 1 mM of L-tryptophan or 1 mM of L-phenylalanine, 0.4 mM of pyridoxal-5′-phosphate, 3 mM of dithiothreitol and 0.05 mL of the suitably diluted enzyme solution. A blank was prepared by placing the mixture in boiling water for 5 min. The incubation was started by placing the tubes in a water bath at 35°C for 1 h and terminated by adding an equal volume of methanol. The amount of products formed was measured by HPLC and calculated from relative calibration curves. One unit of decarboxylase activity was defined as the amount of enzyme that produced 1 μM of tryptamine or phenylethylamine equivalent per minute34. Protein concentrations were measured by Nano Vue using BSA (bovine serum albumin) as the standard.
The purified decarboxylase was incubated with nine different concentrations (from 0.2 mM to 5.0 mM) of each substrate (tryptophan and phenylalanine) respectively in 0.1 M Tris-Hcl buffer (pH 8.0) at 35°C for 30 min. The Km and Vmax values were calculated by the relative Lineweaver-Burk plot.
For the detection of changes from L-tryptophan to tryptamine during the reaction process, the amount of tryptamine in reaction sample was assayed at different reaction times (0 min, 15 min, 30 min, 60 min and 120 min) by HPLC (Aglient 1100 Series, ZORBAX Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm), mobile phase: methanol/H2O (70/30,V/V, 7 mM SDS,10 mM KH2PO4), 25°C, velocity of flow: 1 mL/min, detection time: 0–11 min, sample size: 20 μL, detection wavelength: 278 nm). The reaction samples were analyzed by LC/MS using an Aglient 1200 Series LC system and 6140 quadrupole MS. Mobile phase A was water with 0.0375% TFA and mobile phase B was ACN with 0.01875% TFA. The detailed mobile phase gradient was shown as follows: 0–0.4 min, 0% B; 0.4–3.4 min, 0–80% B; 3.4–4.0 min, 80%–100% and 4.0–4.5 min, 0% B, while the flow rates were 0.6 mL/min, 0.6 mL/min, 0.8 mL/min and 0.8 mL/min, respectively. The mass spectrometer was operated in a positive ion electrospray ionization mode (capillary potential 3.5 kV, desolvation temperature 350°C, source temperature 100°C).
Author Contributions
L.Y., F.Z., Z.L. and S.L. conceived and designed the experiments. L.Y. performed the experiments. L.Y., Q.C., Z.L., S.L. and Y.Z. analyzed the data. F.Z. contributed reagents/materials/analysis tools. L.Y., F.Z. and Z.L. wrote the paper. All authors reviewed the manuscript.
Supplementary Material
Fig. S1
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) (30821005) and the National High-tech R & D Program of China “863” (2013AA092901; 2012AA020403).
References
- Ivanova D. V. et al. Microbiaeratin, a new natural indole alkaloid from a Microbispora aerata strain, isolated from Livingston Island, Antarctica. Prep. Biochem. Biotechnol. 37, 161–168 (2007). [DOI] [PubMed] [Google Scholar]
- Jeong S. Y., Ishida K., Ito Y., Okada S. & Murakami M. Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides. Tetrahedron Lett. 44, 8005–8007 (2003). [Google Scholar]
- Omura S., Suzuki Y., Kitao C., Takahashi Y. & Konda Y. Isolation of a new sulfur-containing basic substance from a thermo actinomyces species. J. Antibiot. 28, 609–610 (1975). [DOI] [PubMed] [Google Scholar]
- Socha A. M., Long R. A. & Rowley D. C. Bacillamides from a hypersaline microbial mat bacterium. J. Nat. Prod. 70, 1793–1795 (2007). [DOI] [PubMed] [Google Scholar]
- Ferrari P. et al. Antibiotics A21459 A and B, new inhibitors of bacterial protein synthesis. II. Structure elucidation. J. Antibiot. 49, 150–154 (1996). [DOI] [PubMed] [Google Scholar]
- Tabata N., Tomoda H., Zhang H., Uchida R. & Omura S. Zelkovamycin, a new cyclic peptide antibiotic from Streptomyces sp. K96-0670: II. Structure elucidation. J. Antibiot. 52, 34–39 (1999). [DOI] [PubMed] [Google Scholar]
- Vollbrecht L., Steinmetz H. & Hofle G. Argyrins, immunosuppressive cyclic peptides from myxobacteria II. Structure elucidation and stereochemistry. J. Antibiot. 55, 715–721 (2002). [DOI] [PubMed] [Google Scholar]
- Churro C. et al. Effects of bacillamide and newly synthesized derivatives on the growth of cyanobacteria and microalgae cultures. J. Appl. Phycol. 21, 429–442 (2009). [Google Scholar]
- Yu L. L., Li Z. Y., Peng C. S. & Guo Y. W. Neobacillamide A, a novel thiazole-containing alkaloid from the marine bacterium Bacillus vallismortis C89, associated with South China Sea sponge Dysidea avara. Helv. Chim. Acta. 92, 607–612 (2009). [Google Scholar]
- Jin L. et al. Bacillamide C production by the optimized cultivation of the Bacillus atrophaeus strain C89 associated with the South China Sea sponge Dysidea avara. Proc. Biochem. 46, 1153–1159 (2011). [Google Scholar]
- Li D., Yang H. S., Cui Q., Mao S. J. & Xu X. H. Synthesis of bacillamide 3 and its analogue. Chin. Chem. Lett. 20, 1195–1197 (2009). [Google Scholar]
- Wang W., Joyner S., Khoury K. A. S. & Dömling A. (−)-Bacillamide C: the convergent approach. Org. Biomol. Chem. 8, 529–532 (2009). [DOI] [PubMed] [Google Scholar]
- Mootz H. D. & Marahiel M. A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179, 6843–6850 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wageningen A. M. A. et al. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5, 155–162 (1998). [DOI] [PubMed] [Google Scholar]
- Weber G., Schorgendorfer K., Schneider-Scherzer E. & Leitner E. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 26, 120–125 (1994). [DOI] [PubMed] [Google Scholar]
- Du L., Sánchez C., Chen M., Edwards D. J. & Shen B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7, 623–642 (2000). [DOI] [PubMed] [Google Scholar]
- Healy F. G., Wach M., Krasnoff S. B., Gibson D. M. & Loria R. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38, 794–804 (2000). [DOI] [PubMed] [Google Scholar]
- Liu F., Sun W., Su F., Zhou K. & Li Z. Draft genome sequence of the sponge-associated Bacillus atrophaeus C89, a potential producer of marine drugs. J. Bacteriol. 194, 4454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson F. R. Prokaryotic and eukaryotic pyridoxal-dependent decarboxylases are homologous. J. Mol. Evol. 31, 325–329 (1990). [DOI] [PubMed] [Google Scholar]
- Jiang H., Wood K. V. & Morgan J. A. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 2962–2969 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C. P. B. et al. Fingerprint analysis of thermolytic decarboxylation of tryptophan to tryptamine catalyzed by natural oils. J. Chromatogr. A. 1210, 115–120 (2008). [DOI] [PubMed] [Google Scholar]
- Facchini P. J., Huber-Allanach K. L. & Tari L. W. Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochem. 54, 121–138 (2000). [DOI] [PubMed] [Google Scholar]
- Kaminaga Y. et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylase and oxidation. J. Biol. Chem. 281, 23357–23366 (2006). [DOI] [PubMed] [Google Scholar]
- Kang S., Kang K., Lee K. & Back K. Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta. 227, 263–272 (2007). [DOI] [PubMed] [Google Scholar]
- Noé W., Mollenschott C. & Berlin J. Tryptophan decarboxylase from Catharanthus roseus cell suspension cultures: purification, molecular and kinetic data of the homogenous protein. Plant Mol. Biol. 3, 281–288 (1984). [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Sudo H., Yamazaki M., Aimi N. & Saito K. Camptothecin biosynthetic genes in hairy roots of Ophiorrhiza pumila: cloning, characterization and differential expression in tissues and by stress compounds. Plant Cell Physiol. 44, 395–403 (2003). [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Hu Y., Huang Y. Q. & Huang Y. Isolation and phylogenetic analysis of the biologically active bacteria associated with three South China Sea sponges. Microbiol. 76, 494–499 (2007). [PubMed] [Google Scholar]
- Li Z. Y. & Liu Y. Marine sponge Craniella austrialiensis-associated bacterial diversity revelation based on 16S rDNA library and biologically active actinomycetes screening, phylogenetic analysis. Lett. Appl. Microbiol. 43, 410–416 (2006). [DOI] [PubMed] [Google Scholar]
- Ghose A. K., Viswanadhan V. N. & Wendoloski J. J. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: an analysis of ALOGP and CLOGP Methods. J. Phys. Chem. A 102, 3762–3772 (1998). [Google Scholar]
- Brooks B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J. & Swaminathan S. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983). [Google Scholar]
- Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3, 208–218 (1961). [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Pennings E. J. M., Hegger I., Heijden R., Duine J. A. & Verpoorte R. Assay of tryptophan decarboxylase from Catharanthus roseus plant cell cultures by high-performance liquid chromatography. Anal. Biochem. 165, 133–136 (1987). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1