Abstract
Cytochrome P450 family 1 (CYP1) includes four subfamilies of enzymes: CYP1A, CYP1B, CYP1C, and CYP1D. In many vertebrates, CYP1A, 1B, and 1C expression is induced by agonists of the aryl hydrocarbon receptor, including toxic contaminants such as chlorinated dioxins, coplanar chlorinated biphenyls, and polynuclear aromatic hydrocarbons. Assessed at the level of mRNA, protein, or enzyme activity, CYP1s (especially CYP1As) represent potent and popular biomarkers of contaminant exposure in aquatic vertebrates. Alkylated resorufins are synthetic substrates used to detect, quantify, and describe catalytic activities of cytochrome P450s. The ability to oxidize specific resorufin-based substrates can distinguish the catalytic activities of individual CYP1s. Xenopus laevis, the African clawed frog, is the most widely employed amphibian model in aquatic toxicology, yet the number, inducibility, and activities of CYP1s have not been systematically characterized in this species. Here we report the cloning of cDNAs encoding two new CYP1 family members, X. laevis CYP1B and CYP1C, along with an integrated assessment of the induction of alkyloxyuresorufin-O-dealkylase (AROD) activities and mRNA expression of four known X. laevis CYP1s: CYP1A6, CYP1A7, CYP1B, and CYP1C. Using XLK-WG, an X. laevis kidney epithelial cell line, we determined that EROD (ethoxyresorufin substrate) and MROD (methoxyresorufin) were both induced 3000- to 5000-fold following 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) exposure up to 250 nM, while BROD (benzyloxyresorufin) and PROD (pentyloxyresorufin) activity was not detectable regardless of TCDD treatment. TCDD induced CYP1A6 and CYP1A7 mRNAs by 2–3 orders of magnitude, while CYP1B and CYP1C were unchanged. The more potent AHR agonist, FICZ (6-formylindolo[3,2-b]carbazole), induced CYP1B up to 10-fold at concentrations between 0.1 and 250 nM, while CYP1C induction was less than 3-fold. CYP1B mRNA showed the highest constitutive mRNA expression, 5- to 75-fold greater than the other CYP1 transcripts. Taken together, these results suggest that CYP1A6 and CYP1A7 perform the bulk of EROD and MROD activities we observed in these cells. The ability of each X. laevis CYP1 to catalyze oxidation of individual resorufin substrates remains to be determined. Correlating CYP1 mRNA and induced AROD activity is a significant step toward clarifying the biochemical meaning of these biomarkers and the roles of CYP1 enzymes in X. laevis. The cell culture approach represents an important complement to the long standing use of frog embryos and tadpoles in toxicological studies, providing a well suited model system for determining the molecular mechanisms underlying the regulation of these important biomarkers of contaminant exposure.
Keywords: Cytochrome P450, CYP1 family, Xenopus laevis, FETAX, Ortholog, Aryl hydrocarbon receptor
1. Introduction
Enzymes comprising cytochrome P450 family I (CYP1s) bio-transform a wide range of xenobiotics and endogenous compounds. The CYP1 family is comprised of four sub-families, CYP1A, CYP1B, CYP1C and CYP1D, paralog groups arising from a common ancestral gene (Goldstone et al., 2009). While CYP1s seemingly exist in all vertebrate groups, gene duplication and loss have resulted in distinct complements of functional CYP1 genes in individual taxa. CYP1As and CYP1Bs exhibit wide taxonomical representation, but CYP1Cs have not been detected in mammals, and CYP1Ds are absent from the bird lineage (Jönsson et al., 2011b) and from many mammals, including humans (Uno et al., 2011). Taxonomic groups also differ in the number and evolutionary origin of individual paralogs within each CYP1 subfamily. Mammals have two CYP1A paralogs, CYP1A1 and 1A2, the orthologs of bird CYP1A4 and 1A5, respectively (Goldstone and Stegeman, 2006). Most fish harbor a single CYP1A (Morrison et al., 1995, 1998), although salmonids (Rabergh et al., 2000) and eels (Mahata et al., 2003) have multiple CYP1As that result from relatively recent, taxon-specific gene duplications. Multiple CYP1C paralogs have also resulted from teleost-specific gene duplications (Godard et al., 2005; Goldstone et al., 2007).
CYP1A, 1B, and 1C expression is induced by agonists of the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor (reviewed in Okey, 2007). Potent xenobiotic inducers include planar halogenated aromatic hydrocarbons (PHAH; e.g. TCDD, PCBs) and polynuclear aromatic hydrocarbons (PAH; e.g. benzo(a)pyrene), many of which are also CYP1 substrates. Expression of CYP1s, especially CYP1As, is a popular biomarker of aquatic animal exposure to these classes of toxic contaminants, and convenient measures exist to detect mRNA, immunoreactive protein, and enzyme activity (reviewed in Hahn, 2002).
CYP1s exhibit distinct but overlapping catalytic specificities for alkyl resorufin fluorescent substrates used in biomarker and kinetic studies (Burke and Mayer, 1983). Studies in mammals established 7-ethoxyresorufin dealkylation (EROD) as a predominantly CYP1A1 activity, although CYP1A2 and CYP1B1 can also utilize this substrate (Burke et al., 1994; Crespi et al., 1997). The MROD assay (7-methoxyresorufin dealkylation) is associated with CYP1A2 (Burke et al., 1994). Catalytic preferences of the orthologous enzymes in birds and fish suggest that extrapolating these canonical relationships to other animal groups is not necessarily straightforward. While EROD is a specific biomarker of CYP1A4 activity in chickens (Rifkind et al., 1994; Sinclair et al., 1997), in the common cormorant (Phalacrocorax carbo) both CYP1A4 and 1A5 exhibit substantial EROD activity, and catalytic specificity is instead defined by the high BROD (7-benzyloxyresorufin dealkylation) activity in CYP1A4 and high MROD activity of CYP1A5. (Kubota et al., 2009). In zebrafish CYP1A exhibits the greatest rate of EROD activity, although CYP1B1, 1C1 and 1C2 are substantially active as well. All four enzymes have roughly comparable MROD and BROD activity, while PROD (7-pentoxyresorufin dealkylation) is lacking only in CYP1B1 (Scornaienchi et al., 2010). CYP1D1 performs all four AROD reactions poorly (Scornaienchi et al., 2010). Overall, it is clear that differences in catalytic preferences between CYP1 isoforms in different animal groups can present complications in the interpretation and comparison of biomarker expression data in toxicological studies.
In contrast to mammals, birds, and fishes, relatively little is known about the activities and expression of CYP1s in amphibians. In this study, we sought to characterize important features of the CYP1 gene family in Xenopus laevis, the African clawed frog. X. laevis is a long standing model of vertebrate development in which morphological changes from fertilization through metamorphosis are well documented (Nieuwkoop and Faber, 1994). It is also a widely used aquatic model of developmental toxicity, including teratogenesis (e.g. the frog embryo teratogenesis assay – FETAX; American Society of Testing and Materials, 1998) and endocrine disruption (e.g. Degitz et al., 2005). Although X. laevis is insensitive to lethality induced by TCDD (2,3,7,8 tetrachlorodibenzo-p-dioxin) and other AHR agonists (Gutleb et al., 1999; Jung and Walker, 1997) and X. laevis AHRs have relatively low affinity for TCDD (Lavine et al., 2005), this species nonetheless expresses two CYP1A mRNAs, CYP1A6 and CYP1A7, which are inducible by 3-methylcholanthrene (Fujita et al., 1999), TCDD (Lavine et al., 2005) and FICZ (6-formylindolo[3,2-b]carbazole), a potent candidate endogenous AHR ligand (Laub et al., 2010). Like the numerous other closely related paralogs in X. laevis, CYP1A6 and 1A7 likely arose from a species-specific genome duplication event approximately 30 mya (Hughes and Hughes, 1993). Xenopus tropicalis, a diploid congeneric, harbors only one CYP1A gene. One gene each of CYP1B1 and CYP1C1 were recently shown to be up-regulated in X. tropicalis tadpoles exposed to PCB126 (Jönsson et al., 2011a). As in other vertebrates, expression of the single X. tropicalis CYP1D1 was not affected (Jönsson et al., 2011a). Taken together, observations in frogs raise questions about the number, orthology, and inducibility of additional CYP1s in X. laevis, the much more widely used toxicological model, as well as the correlation between mRNA expression and the inducibility of AROD activities commonly used as biomarkers in aquatic toxicology. The objective of this study was to resolve these questions. Here we report the cloning of CYP1B and CYP1C cDNAs from this species, as well as induction profiles of AROD activity biomarkers in XLK-WG, a convenient X. laevis cell line derived from kidney epithelium.
2. Materials and methods
2.1. Cell culture
XLK-WG and Hepa1c1c7 cells were obtained from the ATCC (Manassas, VA; Martin et al., 1998). XLK-WG cells were maintained at 30 °C with 5% CO2 in RPMI-1640 medium (ATCC) plus 20% fetal bovine serum (Invitrogen). Hepa1c1c7 cells were maintained at 37 °C with 5% CO2 in α-MEM medium (Sigma, St. Louis, MO) with 10% fetal bovine serum. Cells were routinely cultured in 75 cm2 plastic flasks (Greiner). To prevent formation of FICZ from exposure to fluorescent light, media were stored and warmed in dark conditions, including amber and/or foil-wrapped vessels, and culture hood lights remained off during all procedures (Diani-Moore et al., 2006; Oberg et al., 2005).
FICZ (95% purity) was obtained from Biomol (Plymouth Meeting, PA). TCDD (purity 97–99% purity) was obtained from Ultra Scientific (Kingstown, RI).
2.2. Enzyme activity assays
CYP1 activity was measured in whole cells using alkylresorufin-O-deethylase (AROD) assays (Kennedy and Jones, 1994). Substrates used in these studies were 7-ethoxyresorufin (the EROD assay), 7-methoxyresorufin (MROD), 7-benzyloxyresorufin (BROD), and 7-pentyloxyresorufin (PROD). Conditions were similar to those described in Hestermann et al. (2000). XLK-WG and Hepa1c1c7 cells were seeded in 96-well plates (Greiner, black wall/clear bottom and lid) with 250 μl medium (40,000 cells/well) and grown for 24 or 45 h before being dosed with TCDD, FICZ, or DMSO vehicle (0.25%). For XLK-WG cells, exposures ranged from 0.1 nM to 500 nM; for Hepa1c1c7 cells, concentrations ranged from 1 pM to 10 nM. Treatment lasted 3 or 24 h so that the total time for cell growth was 48 h. Cell viability was verified in representative wells by trypan blue staining. Cells were washed with 250 μl/well 1x PBS and subsequently treated with 100 μl/well 2 μM 7-ethoxyresorufin for 30 min. Resorufin was detected using a Gemini EM multi-well fluorescence plate reader (Molecular Devices, Sunnyvale, CA) with excitation and emission wavelengths of 530 and 586 nm, respectively. The addition of 75 μl/well fluorescamine (175 mg/ml) in acetonitrile was used to stop the reaction and measure protein content using the excitation and emission wavelengths of 400 and 460 nm, respectively (Kennedy and Jones, 1994). A standard curve used to determine resorufin and protein concentrations was constructed using known concentrations of resorufin and bovine serum albumin (Promega). Alkylresorufin substrates and resorufin were purchased from Anaspec (San Jose, CA).
Statistical analysis of all dose-response studies was completed using Prism 4.0b (GraphPad, San Diego, CA). EC50 values were determined via non-linear regression of the fractional response (Poland and Glover, 1975), constraining the background response to 0 and the maximal response to 1. All assays in XLK-WG cells were replicated three to four times, and Hepa1c1c7 experiments were replicated at least twice.
2.3. RNA extraction
Prior to exposure, cells were seeded in 25 cm2 flasks (Greiner) and grown to near confluence. Cells were exposed to graded concentrations of TCDD or FICZ dissolved in dimethylsulfoxide (DMSO) for 24 h or 3 h, respectively. For XLK-WG cells, exposures ranged from 0.1 nM to 500 nM; for Hepa1c1c7 cells, concentrations ranged from 1 pM to 10 nM. Control cells were exposed to an equal volume of DMSO (0.25% final concentration). Total RNA from each flask was extracted using QIAshredder spin columns and RNeasy kits (Qiagen, Valencia, CA) as directed by the manufacturer.
2.4. Amplification and cloning of X. laevis CYP1B and CYP1C
Partial cDNAs encoding CYP1B and CYP1C were amplified by RT-PCR using the GeneAmp Gold RNA PCR Reagent Kit (Applied Biosystems) with degenerate primers. Degenerate primers were designed based on conserved sequences of several vertebrate CYP1s (Table S1). Predicted sequences of X. tropicalis CYP1B and CYP1C were used to reduce degeneracy where possible, and inosine was inserted in select positions where 4-fold degeneracy remained (Wilkie and Simon, 1991). Reverse transcription of 1 μg total RNA was primed with random hexamers as directed by the manufacturer. PCR conditions were: 95°/10′; 43 cycles of [95°/15″; 50°/30″; 72°/60″]; 72°/7′. RT-PCR products were cloned into pGEM-T Easy (Promega) and sequenced. Blastx searches revealed that partial cDNAs for both CYP1B and CYP1C had been isolated.
Partial cDNA sequences were used to design gene-specific primers (Table S2) for the amplification of 5′ and 3′ ends of each cDNA using the SMART RACE cDNA amplification kit (Clontech, Mountain View, CA) under the following cycling conditions: 94 °C/2 min; 30 cycles of [94°/30″; annealing/30″; 72°/3′]; 72 °C/7′. Annealing temperatures varied with primer sequence.
Full length contiguous sequences were determined for each cDNA with the phred/phrap/cross-match algorithms in MacVector Assembler (MacVector, Cary, NC) using 3–10 clones of each overlapping PCR product. The existence of each contiguous DNA sequence was verified by amplifying each as a single PCR product encompassing the entire open reading frame using primers in Table S3.
Sequencing was performed by Retrogen (San Diego, CA) or by the University of Maine DNA Sequencing Facility (Orono, ME).
2.5. Sequence alignment and phylogenetic analysis
The translated CYP1B and CYP1C amino acid sequences were aligned with amino acid sequences for other CYP1 family proteins using CLUSTAL-W (Thompson et al., 1997) within MacVector 10.0.2. The Neighbor Joining algorithm (Saitou and Nei, 1987) with 100 bootstrap samplings was used to construct a phylogenetic tree. Sequences used in the phylogenetic analysis are indicated in Table S4.
2.6. Quantitative PCR
Total RNA was extracted as described above and treated with DNase using the Turbo DNAfree kit (Ambion). DNase-treated RNA (10 ng/PCR reaction) was converted to cDNA using random hexamers and Taqman Reverse Transcription Reagents (rMoMuLV reverse transcriptase; Applied Biosystems, Foster City, CA) before being quantified using Power SYBR Green PCR Master Mix (Applied Biosystems) with specific primers (Operon Biotechnologies). PCR conditions were the following: 95° C/10′; 50 cycles of [95° C/15 s, 60 °C/1 min]. Relative expression was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001) in SDS v1.4 software (Applied Biosystems). Error is expressed as the range of possible relative expression values determined by the standard error of ΔΔCt. β-Actin was used as the endogenous control. Primer sequences for each transcript are indicated in Table S5.
3. Results
3.1. Inducibility of AROD biomarkers in XLK-WG cells
The existence of X. laevis CYP1A6 and 1A7 genes was reported by Fujita et al. (1999), and the inducibility of their mRNAs by several AHR agonists has been amply demonstrated (e.g. Jelaso et al., 2003; Laub et al., 2010; Lavine et al., 2005). Less well examined have been the AROD activities associated with these or other frog CYP1s. Here we used XLK-WG, a kidney epithelial cell line (Martin et al., 1998), to demonstrate that EROD activity is induced by TCDD. Consistent with our previous observations (Laub et al., 2010), EROD induction in frog cells was three orders of magnitude less responsive than in Hepa1c1c7 (Fig. 1A; Table 1), a widely studied mouse hepatoma cell line. This difference is consistent with the low affinity of X. laevis AHRs for TCDD (Lavine et al., 2005). The mouse cells also exhibited around 5-fold greater maximal reaction rate than XLK-WG (Fig. 1B and C). We next examined the TCDD-inducibility of MROD activity in the same cell lines. TCDD was a dramatically less potent MROD inducer in the frog cells than in the mouse cells (more than 5000-fold; Fig. 2A, Table 1). The two cell lines exhibited less than 3-fold difference in the maximum reaction rates (Fig. 2B and C). Induction of BROD and PROD, activities observed previously in zebrafish CYP1 enzymes (Scornaienchi et al., 2010), was not detected in our assays with TCDD-treated XLK-WG cells (detection limit <0.5 pmol/mg protein/min; data not shown).
Fig. 1.
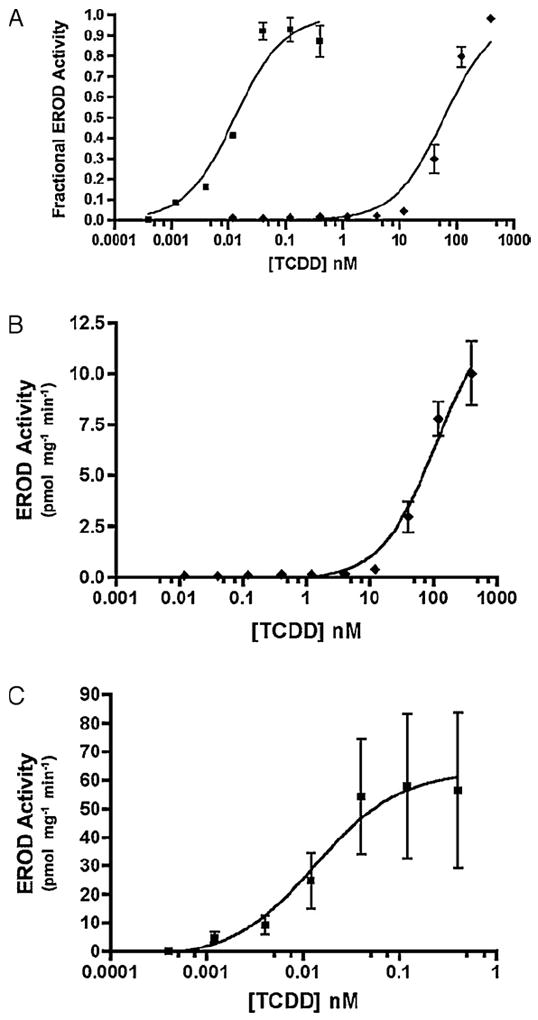
TCDD-induced EROD activity in XLK-WG (frog) and Hepa1c1c7 (mouse) cells. (A) Fractional induction of EROD activity in Hepa1c1c7 (mouse; squares) and XLK-WG (frog; diamonds). Fractional activity is normalized to the maximum value in each dose response, which is defined as 1.0. (B) Actual EROD activity in XLK-WG cells. (C) Actual EROD activity in Hepa1c1c7 cells. Error bars indicate standard error. n = 2–4.
Table 1.
Twenty-four hour TCDD treatments on amphibian and mammalian cell lines exhibit distinct EC50 values in EROD and MROD assays (half-maximal induction of each activity). Variability expressed as standard error. n = 2–4 per assay.
| XLK-WG | Hepa1c1c7 | |
|---|---|---|
| EROD EC50 (nM) | 73.7 ± 8.3 | 0.019 ± 0.006 |
| MROD EC50 (nM) | 96.1 ± 25.3 | 0.017 ± 0.002 |
Fig. 2.
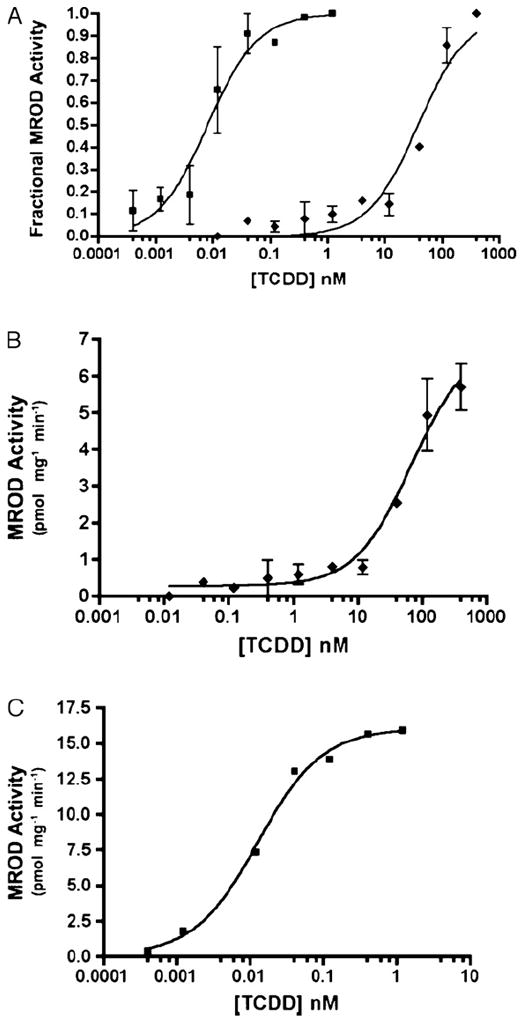
TCDD-induced MROD activity in XLK-WG (frog) and Hepa1c1c7 (mouse) cells. (A) Fractional induction of MROD activity in Hepa1c1c7 (mouse; squares) and XLK-WG (frog; diamonds). Fractional activity is normalized to the maximum value in each dose response, which is defined as 1.0. (B) Actual MROD activity in XLK-WG cells. (C) Actual MROD activity in Hepa1c1c7 cells. Error bars indicate standard error. n = 2–4.
3.2. Cloning and phylogenetic analysis of CYP1B and CYP1C from X. laevis
To begin understanding the relationship between AROD activities and expression of X. laevis CYP1 mRNAs, we sought to clone cDNAs encoding additional CYP1 family members expressed in XLK-WG cells. We designed degenerate primers using an alignment of multiple CYP1B and 1C amino acid sequences, carefully considering the nucleotide sequences of X. tropicalis CYP1s to reduce primer degeneracy. We successfully isolated partial cDNAs encoding single orthologs of CYP1B and CYP1C. Remaining sequence of each cDNA was determined by 5′ and 3′ RACE. To verify its existence in the cells, each open reading frame was amplified as a single RT-PCR product (data not shown) and sequenced in full. The orthology of the encoded proteins was confirmed by phylogenetic analysis. CYP1A6, CYP1A7, CYP1B, and CYP1C appear in appropriate monophyletic groups with orthologs from other species (Fig. 3). Perhaps surprisingly, the frog CYP1B and 1C amino acid sequences bore greater resemblance to their respective orthologs in zebrafish than in chicken, another tetrapod (Tables 2 and 3). This divergence may reflect different selective pressures in the adaptation to terrestrial vs. aquatic habitats. Although zebrafish CYP1C1 and 1C2 are co-orthologs of X. laevis CYP1C (Fig. 3), the frog protein shares a slightly greater degree of identity with CYP1C1 (Table 3). CYP1B and CYP1C cDNA sequences have been deposited in GenBank with accession numbers JN089388 and JN089389.
Fig. 3.
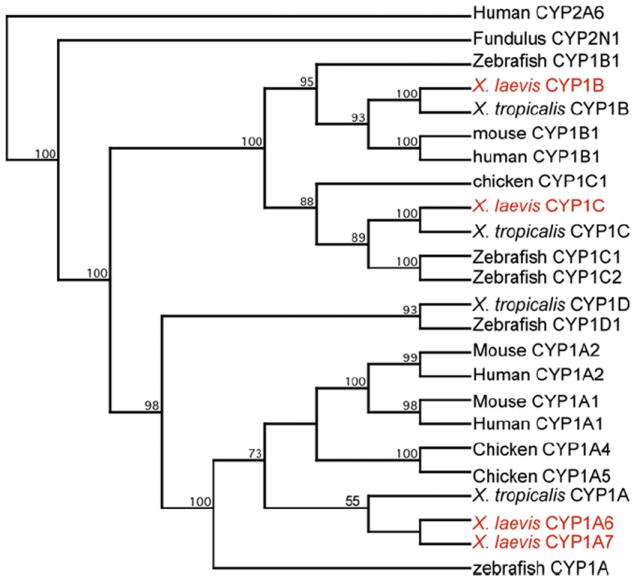
Phylogenetic analysis of X. laevis CYP1 sequences. Amino acid sequences of the indicated proteins were aligned, and a neighbor-joining tree was inferred using Clustal W. Numbers at the branch points represent the bootstrap values based on 100 samplings. Accession numbers of the sequences are found in Table S4 (online supplemental material).
Table 2.
Percent amino acid identity between Xenopus spp., chicken (Gallus gallus), and zebrafish (Danio rerio) CYP1B proteins.
| X. tropicalis 1B1 | G. gallus 1B1 | D. rerio 1B1 | |
|---|---|---|---|
| X. laevis 1B | 81 | 36 | 50 |
| X. tropicalis 1B1 | 38 | 52 | |
| G. gallus 1B1 | 35 |
Table 3.
Percent amino acid identity between Xenopus spp., chicken (Gallus gallus), and zebrafish (Danio rerio) CYP1C proteins.
| X. tropicalis 1C1 | G. gallus 1C1 | D. rerio 1C1 | D. rerio 1C2 | |
|---|---|---|---|---|
| X. laevis 1C | 91 | 54 | 60 | 52 |
| X. tropicalis 1C1 | 54 | 59 | 53 | |
| G. gallus 1C1 | 50 | 44 | ||
| D. rerio 1C1 | 69 |
3.3. CYP1 mRNA expression and inducibility by AHR agonists
We next used qRT-PCR to examine the inducibility of each X. laevis CYP1 mRNA. After 24 h of exposure to TCDD, there was no evidence of CYP1B or CYP1C expression beyond the constitutive level (data not shown), while the substantial induction of CYP1A6 and 1A7 at relatively high concentrations (data not shown) was consistent with our previously published observations (Laub et al., 2010; Lavine et al., 2005). We thus turned to FICZ, an endogenous AHR agonist that displays much greater potency than TCDD with both X. laevis (Laub et al., 2010) and mammalian AHRs (Wincent et al., 2009). Because FICZ is readily metabolized by XLK-WG and other cell lines, exposure time was reduced to 3 h to preserve the sensitivity of the response (Laub et al., 2010; Wincent et al., 2009). A representative experiment is shown in Fig. 4. FICZ induced CYP1A6 mRNA more than 700-fold, and the maximal induction of CYP1A7 was even greater (Fig. 4A and B). CYP1B mRNA was also clearly induced, although the degree of induction was dramatically lower (~10-fold, vs. 700- to 1500-fold for the CYP1As; Fig. 4C). Surprisingly, CYP1C mRNA induction was barely detectable, even at the high concentrations of FICZ used in these experiments.
Fig. 4.
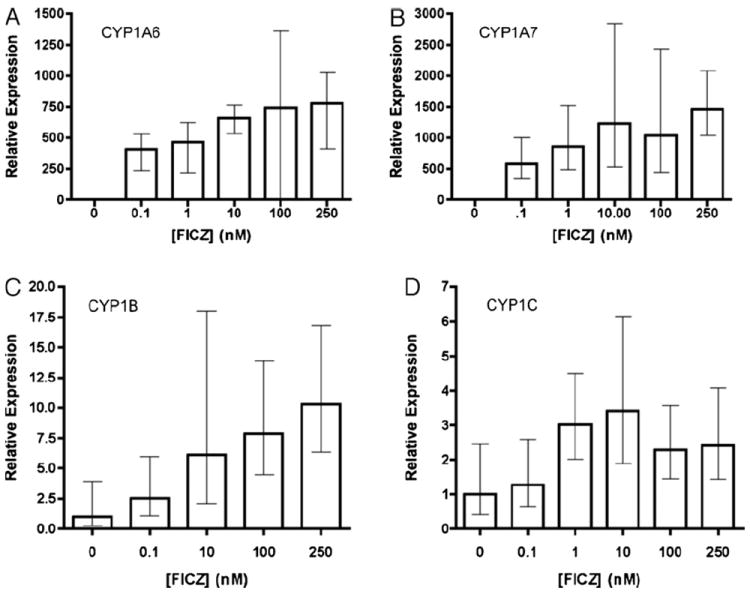
FICZ Induction of CYP1 mRNAs in XLK-WG cells. Near confluent cells were treated for 3 h with DMSO or the indicated concentrations of FICZ. Levels of the indicated transcript were determined by qPCR and calculated using the 2−ΔΔCt method. Error bars define the range of fold induction defined by one standard error of the mean ΔΔCt. Ct values were normalized to the endogenous control β-actin. Experiments were repeated at least three times; a representative result is presented. (A) CYP1A6, (B) CYP1A7, (C) CYP1B, (D) CYP1C.
Fig. 4 depicts induction of each X. laevis CYP1 mRNA relative to its own constitutive expression. To compare mRNA levels between paralogs, we repeated the experiments using a single 96-well plate and normalized expression to CYP1C, the least abundant transcript. A representative experiment is show in Fig. 5. Presuming equal efficiency of each primer set, these experiments reveal that CYP1B mRNA was 5- to 75-fold more abundant than the other transcripts (Fig. 5A). In contrast, CYP1A7 mRNA was most highly expressed following FICZ treatment (Fig. 5B). The degree to which mRNA expression relates to actual protein expression will require the development of specific antibodies for each protein. Meanwhile, it seems reasonable to hypothesize that CYP1C makes a minimal contribution to induced EROD and MROD activities, while CYP1A6 and 1A7 likely contribute the bulk of these activities in XLK-WG cells. Although CYP1B mRNA is expressed at relatively high levels in untreated cells, these cells exhibit very little constitutive AROD activity (Figs. 1 and 2), suggesting a minimal role for the encoded enzyme.
Fig. 5.
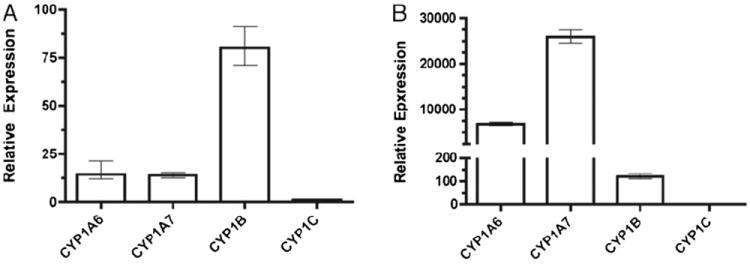
Relative expression of CYP1s in XLK-WG cells. Near confluent cells were treated for 3 h with (A) DMSO or (B) 250 nM FICZ. Levels of the indicated transcript were determined by qPCR and calculated using the 2−ΔΔCt method. Error bars define the range of fold induction defined by one standard error of the mean ΔΔCt. Ct values were normalized to the endogenous control β-actin. Relative expression was normalized to CYP1C, the least abundant transcript. Experiment was repeated three times; a representative result is presented.
4. Discussion
CYP1 induction represents an extensively used biomarker of exposure of vertebrates to contaminant ligands of the AHR (Hahn, 2002). In this study we sought to identify inducible CYP1 genes from X. laevis, the most widely employed amphibian model in aquatic toxicology, and to discern the relationship between the induction of X. laevis CYP1s at the mRNA and enzyme activity levels.
4.1. CYP1 gene multiplicity and evolution
We identified two new members of the CYP1 family: X. laevis CYP1B and CYP1C. These complement the previously described CYP1A6 and 1A7 (Fujita et al., 1999). Sequencing of multiple clones strongly indicated that only one form each of CYP1B and CYP1C were present. X. laevis is a pseudotetraploid species (Hughes and Hughes, 1993), and it is perhaps surprising that we did not isolate duplicate forms of these genes. However, we have no reason to believe that our degenerate primers would favor one closely related paralog over another, and a similar approach taken by our lab successfully amplified multiple paralogs of both AHRs and both ARNTs in this species (Lavine et al., 2005; Rowatt et al., 2003). Furthermore, teleosts were also subject to lineage specific genome duplications (Hoegg et al., 2004; Meyer and Van de Peer, 2005), yet zebrafish (and other teleosts) have only one copy each of CYP1A, 1B, and 1D (Godard et al., 2005; Goldstone et al., 2009). Thus, the existence of multiple CYP1Bs and CYP1Cs in X. laevis is not a foregone conclusion, and the paralogs resulting from the historic genome duplication in this species (Hughes and Hughes, 1993) are either not expressed in this cell line or lost altogether as functional genes. Until the search for CYP1B and 1C paralogs is exhaustively concluded, we have refrained from assigning a number to the names of these genes, as has been done for their X. tropicalis orthologs CYP1B1 and 1C1.
CYP1D genes comprise the remaining CYP1 subfamily. A CYP1D ortholog exists in X. tropicalis (Jönsson et al., 2011a) and likely in X. laevis as well. Because CYP1D is not induced by AHR agonists in other species (Goldstone et al., 2009; Jönsson et al., 2011a, 2007; Zanette et al., 2009), we did not search for X. laevis CYP1D sequences in this study.
4.2. Induction of CYP1B and CYP1C mRNAs: X. laevis vs. X. tropicalis
Although CYP1A6 and 1A7 mRNAs are strongly induced by TCDD in XLK-WG cells, we were unable to detect induction of CYP1B and CYP1C, except by the more potent AHR agonist, FICZ. In contrast, Jönsson et al. (2011a) demonstrated that PCB126 (>100 nM) induced both CYP1B1 and CYP1C1 in X. tropicalis prometamorphic tadpoles. The WHO TEF value for PCB126 is only 0.1 (van den Berg et al., 1998). It is thus surprising that expression of these genes was not responsive to TCDD in the X. laevis cell line. This discrepancy could reflect differences in the PCB126 affinity of AHRs from each species, species-specific differences in promoter structure and/or chromatin conformation, or tissue-specific effects. Previous studies in our lab using transactivation assays in COS-7 cells (M.E. Kalnoske and W.H. Powell, unpublished data) demonstrated the capacity of X. laevis AHR1α and AHR1β to mediate PCB126-induced transcriptional activation of luciferase under control of the mouse CYP1A1 enhancer region (Garrison et al., 1996). In zebrafish kidney, neither CYP1C1 nor 1C2 is strongly induced by PCB 126 (Zanette et al., 2009), suggesting that low inducibility of CYP1C in XLK-WG cells may reflect similar kidney-specific regulation in X. laevis.
4.3. CYP1 mRNA vs. AROD expression
Based on parallel expression patterns of mRNA and enzyme activities, we suggest that CYP1A6 and/or CYP1A7 perform the bulk of EROD and MROD induced by AHR agonists in XLK-WG cells. Heterologous expression will be necessary to determine the catalytic preferences of the individual enzymes. Meanwhile, examination of the amino acid sequences at positions homologous to the active site residues of orthologous enzymes allows development of some hypotheses. Valine at position 382 of human CYP1A1 [within putative sequence recognition site 5 (SRS-5; Gotoh, 1992)] is associated with alkyl resorufin substrate specificity. CYP1A2 contains leucine at this position, and the reciprocal mutation of each enzyme switches the catalytic preference for ethoxy- or methoxyresorufin (Liu et al., 2004). X. laevis CYP1A6 is identical to CYP1A1 with valine at this position (V390), while CYP1A7 contains methionine (Fig. 6 and Figure S1). CYP1A7 thus resembles chicken CYP1A5, which reportedly exhibits low AROD activity (Rifkind et al., 1994; Sinclair et al., 1997; Verbrugge et al., 2001), suggesting that CYP1A7 may be similarly inactive with these substrates. Kubota et al. (2009) made a similar argument to explain the disparity in AROD activities exhibited by CYP1A5 in cormorants and chickens. Compared with CYP1A4, chicken CYP1A5 preferentially metabolizes arachadonic acid epoxygenation and uroporphyrinogen oxidation (Rifkind et al., 1994; Sinclair et al., 1997). If methionine at this position contributes to catalytic preference for physiologically relevant substrates in X. laevis CYP1A7, it could represent an interesting case of convergent evolution, since CYP1A7 and CYP1A5 arose from different gene duplication events during vertebrate evolution (Goldstone and Stegeman, 2006 and Fig. 3).
Fig. 6.
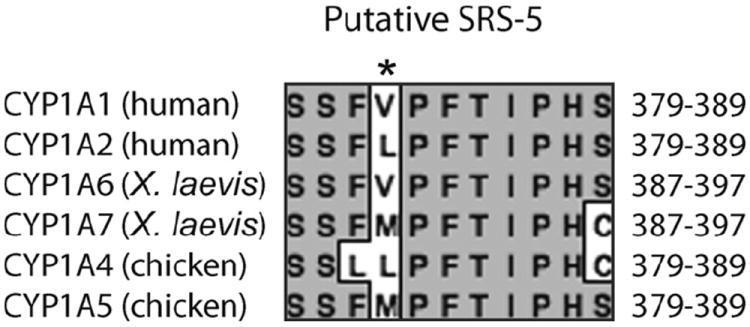
Amino acid alignment of putative SRS-5 from human, frog, and chicken CYP1As. Positions of included residues are indicated at right. *Position of variability that impacts AROD substrate preference in human CYP1A1 and 1A2. See Figure S1 (on-line supplemental material) for a complete amino acid alignment of these proteins.
5. Conclusions
In this study, we characterized the inducibility of EROD, MROD, BROD, and PROD activities in XLK-WG, a X. laevis cell line. These activities are biomarkers of CYP1 expression induced in vertebrates by exposure to a wide range of contaminants, including PHAH and PAH. We also cloned cDNAs encoding two CYP1 family members, CYP1B and CYP1C, comparing their induction by AHR agonists to the previously identified family members, CYP1A6 and CYP1A7. Our observations demonstrate that CYP1A6 and 1A7 mRNAs are much more strongly induced than CYP1B and CYP1C and thus suggest the hypothesis that the corresponding enzymes are likely responsible for the bulk of induced EROD and MROD activity in this cell line. Characterizing induced CYP1 transcripts and AROD activities represents a significant step toward clarifying the relationship of these mRNA and enzyme biomarkers of contaminant exposure in X. laevis, the most commonly used amphibian model system in aquatic toxicology. The tissue culture approach represents a valuable complement to the long standing use of frog embryos and tadpoles in toxicological studies, providing a well suited model system to determining the detailed molecular mechanisms underlying the regulation of these toxicologically important genes.
Supplementary Material
Acknowledgments
We thank Leo Laub for assistance and expert technical support and Kenyon colleagues Chris Gillen and Harry Itagaki for advice in preparing the manuscript.
Role of the funding source
This work was funded by the National Institutes of Health (R15 ES011130) and the Kenyon College Summer Science Scholars Program. The real time PCR instrument was purchased with an education grant from the Howard Hughes Medical Institute. Content of this manuscript is solely the responsibility of the authors and does not necessarily represent the views of the funding agencies.
Abbreviations
- CYP1
cytochrome P450 family I
- PHAH
planar halogenated aromatic hydrocarbons
- PAH
polynuclear aromatic hydrocarbons
- PCB
polychlorinated biphenyl
- AHR
aryl hydrocarbon receptor
- EROD
7-ethoxyresorufin-O-deethylase
- MROD
7-methoxyresorufin-O-deethylase
- BROD
7-benzyloxyresorufin-O-deethylase
- PROD
7-pentyloxyresorufin-O-deethylase
- AROD
7-alkyloxyresorufin-O-deethylase
- FETAX
frog embryo teratogenesis assay – Xenopus
- FICZ
6-formylindolo[3,2b]carbazole
- TCDD
2,3,7,8 tetrachlorodibenzo-p-dioxin
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.aquatox.2012.02.028.
Conflict of interest
None.
References
- American Society of Testing and Materials. Annual Book of ASTM Standards. American Society of Testing and Materials; Philadelphia, PA: 1998. Standard Guide for conducting the frog embryo teratogenesis assay—Xenopus (FETAX) pp. 826–836. [Google Scholar]
- Burke MD, Mayer RT. Differential effects of phenobarbitone and 3-methylcholanthrene induction on the hepatic mirosomal metabolism and cytochrome P450-binding of phenoxazone and a homologous series of its n-alkyl ethers (alkoxyresorufins) Chem Biol Interact. 1983;45:243–258. doi: 10.1016/0009-2797(83)90072-8. [DOI] [PubMed] [Google Scholar]
- Burke MD, Thompson S, Weaver RJ, Wolf CR, Mayer RT. Cytochrome-P450 specificities of alkoxyresorufin O-dealkylation in human and rat-liver. Biochem Pharmacol. 1994;48:923–936. doi: 10.1016/0006-2952(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Crespi CL, Penman BW, Steimel DT, Smith T, Yang CS, Sutter TR. Development of a human lymphoblastoid cell line constitutively expressing human CYP1B1 cDNA: substrate specificity with model substrates and promutagens. Mutagenesis. 1997;12:83–89. doi: 10.1093/mutage/12.2.83. [DOI] [PubMed] [Google Scholar]
- Degitz SJ, Holcombe GW, Flynn KM, Kosian PA, Korte JJ, Tietge JE. Progress towards development of an amphibian-based thyroid screening assay using Xenopus laevis. Organismal and thyroidal responses to the model compounds 6-propylthiouracil methimazole, and thyroxine. Toxicol Sci. 2005;87:353–364. doi: 10.1093/toxsci/kfi246. [DOI] [PubMed] [Google Scholar]
- Diani-Moore S, Labitzke E, Brown R, Garvin A, Wong L, Rifkind AB. Sunlight generates multiple tryptophan photoproducts eliciting high efficacy CYP1A induction in chick hepatocytes and in vivo. Toxicol Sci. 2006;90:96–110. doi: 10.1093/toxsci/kfj065. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Ohi H, Murayama N, Saguchi K, Higuchi S. Molecular cloning and sequence analysis of cDNAs coding for 3-methylcholanthrene-inducible cytochromes P450 in Xenopus laevis liver. Arch Biochem Biophys. 1999;371:24–28. doi: 10.1006/abbi.1999.1425. [DOI] [PubMed] [Google Scholar]
- Garrison PM, Tullis K, Aarts JMMJG, Brouwer A, Giesy JP, Denison MS. Species-specific recombinant cell lines as bioassay systems for the detection of 2,3,7,8-tetrachlorodibenzo-p-dioxin-like chemicals. Fund Appl Toxicol. 1996;30:194–203. doi: 10.1006/faat.1996.0056. [DOI] [PubMed] [Google Scholar]
- Godard CA, Goldstone JV, Said MR, Dickerson RL, Woodin BR, Stegeman JJ. The new vertebrate CYP1C family: cloning of new subfamily members and phylogenetic analysis. Biochem Biophys Res Commun. 2005;331:1016–1024. doi: 10.1016/j.bbrc.2005.03.231. [DOI] [PubMed] [Google Scholar]
- Goldstone HM, Stegeman JJ. A revised evolutionary history of the CYP1A subfamily: gene duplication, gene conversion, and positive selection. J Mol Evol. 2006;62:708–717. doi: 10.1007/s00239-005-0134-z. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, Goldstone HM, Morrison AM, Tarrant A, Kern SE, Woodin BR, Stegeman JJ. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol Biol Evol. 2007;24:2619–2631. doi: 10.1093/molbev/msm200. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, Jonsson ME, Behrendt L, Woodin BR, Jenny MJ, Nelson DR, Stegeman JJ. Cytochrome P450 1D1: a novel CYP1A-related gene that is not transcriptionally activated by PCB126 or TCDD. Arch Biochem Biophys. 2009;482:7–16. doi: 10.1016/j.abb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Gutleb AC, Appelman J, Bronkhorst MC, van den Berg JH, Spenkelink A, Brouwer A, Murk AJ. Delayed effects of pre- and early-life time exposure to polychlorinated biphenyls on tadpoles of two amphibian species (Xenopus laevis and Rana temporaria) Environ Toxicol Pharmacol. 1999;8:1–14. doi: 10.1016/s1382-6689(99)00023-x. [DOI] [PubMed] [Google Scholar]
- Hahn ME. Biomarkers and bioassays for detecting dioxin-like compounds in the marine environment. Sci Total Environ. 2002;289:49–69. doi: 10.1016/s0048-9697(01)01016-6. [DOI] [PubMed] [Google Scholar]
- Hestermann EV, Stegeman JJ, Hahn ME. Relative contributions of affinity and intrinsic efficacy to aryl hydrocarbon receptor ligand potency. Toxicol Appl Pharmacol. 2000;168:160–172. doi: 10.1006/taap.2000.9026. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hughes MK, Hughes AL. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol Biol Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- Jelaso AM, Lehigh-Shirey E, Means J, Ide CF. Gene expression patterns predict exposure to PCBs in developing Xenopus laevis tadpoles. Environ Mol Mutagen. 2003;42:1–10. doi: 10.1002/em.10173. [DOI] [PubMed] [Google Scholar]
- Jönsson ME, Berg C, Goldstone JV, Stegeman JJ. New CYP1 genes in the frog Xenopus (Silurana) tropicalis: induction patterns and effects of AHR agonists during development. Toxicol Appl Pharmacol. 2011a;250:170–183. doi: 10.1016/j.taap.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3,3′,4,4′,5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicol Appl Pharmacol. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson ME, Woodin BR, Stegeman JJ, Brunstrom B. Cytochrome p450 1 genes in birds: evolutionary relationships and transcription profiles in chicken and Japanese quail embryos. PLoS ONE. 2011b;6:e28257. doi: 10.1371/journal.pone.0028257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung RE, Walker MK. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on development of anuran amphibians. Environ Toxicol Chem. 1997;16:230–240. [Google Scholar]
- Kennedy SW, Jones SP. Simultaneous measurement of cytochrome P4501A catalytic activity and total protein concentration with a fluorescence plate reader. Anal Biochem. 1994;222:217–223. doi: 10.1006/abio.1994.1476. [DOI] [PubMed] [Google Scholar]
- Kubota A, Kim EY, Iwata H. Alkoxyresorufin (methoxy-, ethoxy-, pentoxy- and benzyloxyresorufin O-dealkylase activities by in vitro-expressed cytochrome P450 1A4 and 1A5 from common cormorant (Phalacrocorax carbo) Comp Biochem Physiol C: Toxicol Pharmacol. 2009;149:544–551. doi: 10.1016/j.cbpc.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Laub LB, Jones BD, Powell WH. Responsiveness of a Xenopus laevis cell line to the aryl hydrocarbon receptor ligands 6-formylindolo[3,2-b]carbazole (FICZ) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Chem Biol Interact. 2010;183:202–211. doi: 10.1016/j.cbi.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine JA, Rowatt AJ, Klimova T, Whitington AJ, Dengler E, Beck C, Powell WH. Aryl hydrocarbon receptors in the frog Xenopus laevis: two AhR1 paralogs exhibit low affinity for 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Toxicol Sci. 2005;88:60–72. doi: 10.1093/toxsci/kfi228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ericksen SS, Sivaneri M, Besspiata D, Fisher CW, Szklarz GD. The effect of reciprocal active site mutations in human cytochromes P450 1A1 and 1A2 on alkoxyresorufin metabolism. Arch Biochem Biophys. 2004;424:33–43. doi: 10.1016/j.abb.2003.12.040. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mahata SC, Mitsuo R, Aoki JY, Kato H, Itakura T. Two forms of cytochrome P450 cDNA from 3-methylcholanthrene-treated European eel Anguilla anguilla. Fish Sci. 2003;69:615–624. [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gammaTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Morrison HG, Oleksiak MF, Cornell NW, Sogin ML, Stegeman JJ. Identification of cytochrome P-450 1A (CYP1A) genes from two teleost fish, toadfish (Opsanus tau) and scup (Stenotomus chrysops), and phylogenetic analysis of CYP1A genes. Biochem J. 1995;308:97–104. doi: 10.1042/bj3080097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, Weil EJ, Karchner SI, Sogin ML, Stegeman JJ. Molecular cloning of CYP1A from the estuarine fish Fundulus heteroclitus and phylogenetic analysis of CYP1A genes: update with new sequences. Comp Biochem Physiol. 1998;121C:231–240. doi: 10.1016/s0742-8413(98)10044-0. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) Garland Publishing, Inc.; New York, London: 1994. [Google Scholar]
- Oberg M, Bergander L, Hakansson H, Rannug U, Rannug A. Identification of the tryptophan photoproduct 6-formylindolo[3,2-b]carbazole, in cell culture medium, as a factor that controls the background aryl hydrocarbon receptor activity. Toxicol Sci. 2005;85:935–943. doi: 10.1093/toxsci/kfi154. [DOI] [PubMed] [Google Scholar]
- Okey AB. An aryl hydrocarbon receptor odyssey to the shores of toxicology: the Deichmann Lecture, International Congress of Toxicology-XI. Toxicol Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- Poland A, Glover E. Genetic expression of aryl hydrocarbon hydroxylase by 2,3,7,8-tetrachlorodibenzo-p-dioxin: evidence for a receptor mutation in genetically non-responsive mice. Mol Pharmacol. 1975;11:389–398. [Google Scholar]
- Rabergh CM, Vrolijk NH, Lipsky MM, Chen TT. Differential expression of two CYP1A genes in rainbow trout (Oncorhynchys mykiss) Toxicol Appl Pharmacol. 2000;165:195–205. doi: 10.1006/taap.2000.8941. [DOI] [PubMed] [Google Scholar]
- Rifkind AB, Kanetoshi A, Orlinick J, Capdevila JH, Lee C. Purification and biochemical characterization of two major cytochrome-P-450 isoforms induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in chick embryo liver. J Biol Chem. 1994;269:3387–3396. [PubMed] [Google Scholar]
- Rowatt AJ, DePowell JJ, Powell WH. ARNT gene multiplicity in amphibians: characterization of ARNT2 from the frog Xenopus laevis. J Exp Zool. 2003;300B:48–57. doi: 10.1002/jez.b.45. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scornaienchi ML, Thornton C, Willett KL, Wilson JY. Functional differences in the cytochrome P450 1 family enzymes from Zebrafish (Danio rerio) using heterologously expressed proteins. Arch Biochem Biophys. 2010;502:17–22. doi: 10.1016/j.abb.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair PR, Gorman N, Walton HS, Sinclair JF, Lee CA, Rifkind AB. Identification of CYP1A5 as the CYP1A enzyme mainly responsible for uroporphyrinogen oxidation induced by AH receptor ligands in chicken liver and kidney. Drug Metab Dispos. 1997;25:779–783. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Uehara S, Murayama N, Yamazaki H. CYP1D1, pseudogenized in human, is expressed and encodes a functional drug-metabolizing enzyme in cynomolgus monkey. Biochem Pharmacol. 2011;81:442–450. doi: 10.1016/j.bcp.2010.11.003. [DOI] [PubMed] [Google Scholar]
- van den Berg M, Birnbaum L, Bosveld BTC, Brunström B, Cook P, Feeley M, Giesy JP, Hanberg A, Hasegawa R, Kennedy SW, Kubiak T, Larsen JC, Leeuwen FXR, Liem v, Nolt AKD, Peterson C, Poellinger RE, Safe L, Schrenk S, Tillitt D, Tysklind D, Younes M, Waern M, Zacharewski FT. Toxic equivalency factors (TEFs) for PCBs PCDDs, and PCDFs for humans and wildlife. Environ Health Perspect. 1998;106:775–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugge LA, Giesy JP, Verbrugge DA, Woodin BR, Stegeman JJ. Catalytic and immunochemical properties of hepatic cytochrome P450 1A in three avian species treated with beta-naphthoflavone or isosafrole. Comp Biochem Physiol C: Toxicol Pharmacol. 2001;130:67–83. doi: 10.1016/s1532-0456(01)00221-6. [DOI] [PubMed] [Google Scholar]
- Wilkie TM, Simon MI. Cloning multigene families with degenerate PCR primers. Methods: Companion Methods Enzymol. 1991;2:32–41. [Google Scholar]
- Wincent E, Amini N, Luecke S, Glatt H, Bergman J, Crescenzi C, Rannug A, Rannug U. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem. 2009;284:2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- Zanette J, Jenny MJ, Goldstone JV, Woodin BR, Watka LA, Bainy AC, Stegeman JJ. New cytochrome P450 1B1, 1C2 and 1D1 genes in the killifish Fundulus heteroclitus: basal expression and response of five killifish CYP1s to the AHR agonist PCB126. Aquat Toxicol. 2009;93:234–243. doi: 10.1016/j.aquatox.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


