Abstract
The origin and the effects of homochirality in the biological world continuously stimulate numerous hypotheses and much debate. This work attempts to look at the biohomochirality issue from a different angle—the mechanical properties of the bulk biomaterial and their relation to nanoscale structures. Using a pair of oppositely charged peptides that co-assemble into hydrogels, we systematically investigated the effect of chirality on the mechanical properties of these hydrogels through different combinations of syndiotactic and isotactic peptides. It was found that homochirality confers mechanical advantage, resulting in higher elastic modulus and strain yield value. Yet, heterochirality confer kinetic advantage, resulting in faster gelation. Structurally, both homochiral and heterochiral hydrogels are made of fibers interconnected by lappet-like webs, but the homochiral peptide fibers are thicker and denser. The result highlights the possible role of biohomochirality in the evolution and/or natural selection of biomaterials.
Keywords: hydrogels, mechanical properties, homochirality, heterochirality, dynamic rheometry, NMR spectroscopy, small-angle X-ray scattering, small-angle neutron scattering
Introduction
The phenomenon of homochirality, its origin and implications, is one of the most enigmatic questions in the life sciences.1 From a fundamental viewpoint, of greatest interest is the core controversy of the preservation of mirror symmetry in the non-biological world—i.e., the existence of racemic mixtures of enantiomers (or heterochirality)—and the break of mirror symmetry in the biological world—i.e., the existence of enantiopure systems (or homochirality).2 Natural biomacromolecules are homochiral, with proteins comprised exclusively of L-amino acids, while nucleic acids and most polysaccharides contain only D-sugars. The origin of the homochirality of life is a matter of much debate, and numerous hypotheses and speculation exist in the literature.3 As to the advantages conferred by homochirality, the discussion has focused on the molecular aspects of homochirality, such as protein folding,4 chiroselective replication,5 enzyme catalysis6 and DNA recognition.7 However, de novo design efforts have shown that introducing D-amino acids into proteins greatly expands structural motifs and even lead to enhanced stability.8 Such studies put the molecular advantage of homochirality into question.
In comparison, little is known about the material aspects of homochirality, i.e., whether homochirality confers advantages to material properties such as mechanical strength. One might expect that such effects could be profound and lead to significant morphological differences of the resulting bulk material.9 In this work, we investigate whether homochirality confers mechanical advantages to soft biomaterials. Mechanical properties are important functional parameters for biomaterials.10 Specifically, we will compare the viscoelastic properties of homochiral vs. heterochiral hydrogels. Hydrogels are viscoelastic soft biomaterials that have many natural (e.g., collagen) and manmade (e.g., contact lenses) examples. It has been proposed that life may have originated in a hydrogel environment.11 Hence, the effect of homochirality on the mechanical properties of hydrogels might have implications for the origin of life.
In organogels, three scenarios have been observed: homochirality leads to better gelling ability,9,12 homochirality and heterochirality make no difference in gelling ability,13 and heterochirality leads to better gelling ability.14 However, the dominant situation is that homochirality leads to better gelling ability.15 In these studies of organo-gelators, the gelling ability was measured by either the minimum gelation concentration or the gelation temperature. There was no report on the mechanical properties of homochiral vs. heterochiral organogels.
In contrast to organogels, where homochirality in most cases leads to better gelling ability, hydrogels appear to show a preference for heterochirality in many cases, a phenomenon called stereo-complexation.16 For example, stereocomplexes are formed between poly(D-lactic) and poly-(L-lactic) acids through stereospecific interlocking.16 It has been shown that the formation of the heterochiral stereocomplexes from D- and L-enantiomers is more rapid and more complete as opposed to homochiral enantiomers; and the polyethylene glycol gels formed in the presence of heterochiral stereocomplexes of the poly(lactic) acids are capable to hold more water and are more stable with higher transition temperatures than gels formed in the presence of homochiral poly(lactic) acids.16 It has been shown that hydrogels resulting from the stereocomplexation of 50/50 mixtures of D- and L-enantiomers of poly(lactic) acid with polyethylene glycol demonstrate much higher elastic moduli G' as compared to the hydrogels where the ratio of D- and L-enantiomers was shifted towards greater homochirality (e.g., 84/16).17 Similarly, photoinduced hydrogelation of stereocomplexes constructed from poly(lactic) acid, polyethylene glycol and methacrylate results in mechanically stronger hydrogels for the D+L mixture of poly(lactic) acid as compared to the pure L-poly(lactic) acid material (the elastic modulus G' is ca. two orders of magnitude higher).18 Hydrogelation induced by poly(lactic) acid stereocomplexation persists after poly(lactic) acids are grafted to dextran.19
Such preference of heterochiral stereocomplexation is also found in other aqueous systems. For example, it was found that L-peptides stereocomplex with poly(D-lactic) acid but not with poly(L-lactic) acid.20 Also, pectate, a D-polysaccharide, stereocomplexes more efficiently with poly-(L-lysine) than with poly(D-lysine).21 In all these cases, heterochirality (D−L complexation) was preferred over homochirality (L-L or D-D complexation). In addition, heterochiral 1,3,5-cyclohexyl-tricaroboxamide-phenyl-alanines (LLD or DDL) are excellent hydrogelators while their homochiral counterparts (LLL or DDD) do not gelate.22 However, in these studies, mechanical properties were not reported.
In peptide hydrogels, it has been recently reported that heterochiral hydrogels assembled from a pair of D-, L-peptides have higher elastic modulus than the parent homochiral hydrogels.23 Previously, it was reported that poly(D-lysine) and poly(L-lysine) form amyloid-like fibrils while each individual enantiomer remains a clear solution,24 although no rheological data were reported for these poly(D-lysine) + poly(L-lysine) stereocomplexes. In the case of actual amyloid fibers, somewhat contradictory observations have been reported. On the one hand, it was reported that amyloid fibers demonstrate homochiral stereospecificity in that L-peptides or proteins deposit onto preexisting L-fibers but not onto preexisting D-fibers.25 On the other hand, it was reported that inhibitors of amyloid fibers demonstrate heterochiral stereospecificity in that D-oligopeptides better inhibit the fribrillization of L-β-amyloid peptide than L-oligopeptides of the same sequence.26
This observed preference for heterochriality over homochirality in an aqueous environment raises the following question: does homochirality confer advantages to biomaterials? After all, natural protein biomaterials are homochiral. In this work, we address this question using a pair of self-repulsive but mutually attractive peptides that can co-assemble into hydrogels. Different chiral combinations are explored in a systematic fashion. Using dynamic rheometery, we found that heterochirality indeed leads to quicker gelation and higher elastic modulus in the first few hours of gelation. Afterwards, homochiral gels outpace heterochiral gels and homochirality eventually leads to a much higher elastic modulus and slightly higher strain yield than heterochirality. Thus, for these peptide hydrogels, homochirality confers mechanical advantage while heterochirality confers kinetic advantage. Using a combination of NMR spectroscopy, small-angle X-ray and neutron scattering techniques (SAXS and SANS), we explored the mechanism underlying these observations.
Experimental Section
Peptide design and synthesis
Oppositely charged undecapeptide modules (11 amino acids long) have been designed in accordance with our earlier approach27 whereby a positively charged module and a negatively charged module co-assemble into a hydrogel in phosphate-buffered saline (PBS) when mixed. The positive module contains alternating positively charged (lysine, K) and neutral (tryptophan, W; and alanine, A) amino acids, while the negative module contains alternating negatively charged (glutamate, E) and neutral (tryptophan, W; and alanine, A) amino acids. Such general design separates positively and negatively charged amino acids into different peptide chains. As a result of the electrostatic repulsions inherent within each peptide module, spontaneous hydrogelation and/or self-assembly due to slight pH, temperature and ionic strength changes are avoided. In order to study the chirality effect of the hydrogels, both syndiotactic and isotactic peptides were made: two syndiotactic peptides comprised of alternating D- and L-amino acids, denoted D,L-K and D,L-E, respectively; two isotactic peptides comprised of all L-amino acids, denoted L-K and L-E, respectively; and two isotactic peptides comprised of all D-amino acids, denoted D-K and D-E, respectively (Table 1). The N-, C-termini of each peptide were acetylated (Acetyl-) and amidated (-amide), respectively, to block terminal charges. In this work, all peptide sequences are palindromic.
Table 1.
Tacticity, sequence and molecular weight of undecapeptides modules.a
| Peptide | Tacticity | Peptide Sequence | M.W. (Da) |
|---|---|---|---|
| D,L-K | syndiotactic | acetyl-LK-DW-LK-DA-LK-DA-LK-DA-LK-DW-LK-amide | 1,413 |
| D,L-E | syndiotactic | acetyl-LE-DW-LE-DA-LE-DA-LE-DA-LE-DW-LE-amide | 1,419 |
| D-K | isotactic | acetyl-DK-DW-DK-DA-DK-DA-DK-DA-DK-DW-DK-amide | 1,413 |
| D-E | isotactic | acetyl-DE-DW-DE-DA-DE-DA-DE-DA-DE-DW-DE-amide | 1,419 |
| L-K | isotactic | acetyl-LK-LW-LK-LA-LK-LA-LK-LA-LK-LW-LK-amide | 1,413 |
| L-E | isotactic | acetyl-LE-LW-LE-LA-LE-LA-LE-LA-LE-LW-LE-amide | 1,419 |
A, alanine; E, glutamic acid; K, lysine; W, tryptophan. The N-, C-termini of each peptide were acetylated (acetyl-) and amidated (-amide), respectively. Modular material assembly is achieved by pairing a positive module with a negative module. Five pairs were made: pair 1 D,L-K + D,L-E; pair 2 L-K + D-E; pair 3 D-K + L-E; pair 4 D-K + D-E; pair 5 L-K + L-E.
All peptides were synthesized on Rink-amide MBHA resin by means of a CEM microwave synthesizer using standard solid-phase Fmoc-chemistry.28 The crude peptides were cleaved by a cocktail containing 95 % trifluoroacetic acid (TFA), 2.5 % triisopropylsilane and 2.5 % water volume fraction. The side chains were also deprotected during cleavage. TFA was removed by rotary evaporation under reduced pressure. The crude peptides were precipitated and washed twice by cold diethyl ether. The crude peptides were dissolved in water and lyophilized before purification.
Preparative reverse-phase HPLC (RPLC) method was used to purify the crude peptides. In the purification of positive charged peptides (L-K, D-K and D,L-K), solvent A was 0.1 % mass fraction HCl in water and solvent B was 0.1 % mass fraction HCl in MeOH. In the purification of negatively charged peptides (L-E, D-E and D,L-E), solvent A was 20 mM NH4HCO3 in water (pH 7.0), solvent B was 20 mM NH4HCO3 (pH 7.0) in MeOH/water (8:2, volume ratio). The chromatographic method of peptide purification was as follows: 0–40 % B in 0–60 min, 40–100 % B in 60–90 min with a linear gradient for each segment. The purity of each peptide was verified by analytical RPLC methods with the same solvents used for preparative RPLC. Molecular weights of all peptides were verified by ESI-MS in positive and negative modes, respectively.
Purified peptides were dissolved in PBS composed of 50 mM phosphate buffer and 100 mM sodium chloride, pH 7.4. The final concentrations of the individual peptide stock solutions were 16.0 mM, determined on the basis of the molar absorptivity of tryptophan at 280 nm (ε280 = 5690 M−1·cm−1).29 From the six peptide modules presented in Table 1, five mixtures were made: pair 1: D,L-K + D,L-E; pair 2: L-K + D-E; pair 3: D-K + L-E; pair 4: D-K + D-E; pair 5: L-K + L-E. The five pairs were characterized by a combination of NMR spectroscopy, dynamic rheometry, small-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS) techniques, all at 25°C.
NMR spectroscopy measurements
To induce hydrogelation, 300 µL solution of 16 mM positively charged peptide module (L-K, or D-K, or D,L-K) was mixed with 300 µL solution of 16 mM negatively charged peptide module (L-E, or D-E, or D,L-E, respectively), so the final concentration of each peptide in the mixture was 8 mM. Individual peptide solutions were pre-equilibrated at room temperature, then mixed in a 1.5 mL plastic centrifuge tube and transferred into a 5 mm NMR tube by a long glass disposable pipette. All samples contained 10 % mass fraction D2O to provide the deuterium lock signal.
All NMR experiments were carried out on a Varian INOVA 500 spectrometer equipped with a triple resonance indirect detection probe with Z-gradient. The temperature of the NMR spectrometer probe was pre-set to 25°C. NMR data acquisition started about 5 min after mixing of the two oppositely charged peptides (the 5-min delay was due to instrument tuning, shimming and 90° pulse calibration).
The 1H peak at 3.0 ppm (ε-H from side chain of lysine residues) was used to monitor the gelation process. Due to the very short transverse relaxation time T2 of gelled peptides, only free peptides can be detected by NMR spectroscopy.30 With more and more peptides incorporated into the gel matrix, the 1H signals from peptides decrease. To monitor gelation, 1D 1H spectra were acquired every 5 minutes during the first 2 hours for pairs 4 and 5; and during the first hour for pairs 2 and 3. Afterwards, 1D 1H spectra were recorded every hour until the monitored signal intensity reached a plateau (~ 14 hr). For pair 1, 1D 1H spectra were acquired every 30 minutes in the first 3 hours, and then recorded every hour until 8 hours after mixing. 6 months later, the 1D 1H spectrum of pair 1 was recorded again. In order to compare the signal intensities from different 1D 1H spectra, the same calibrated 90° pulse and the same receiver gain were used in all cases and the relaxation delay was set to a value larger than 5 times that of the T1 relaxation time of the 1H signal.
Pair 1 shows no sign of gelation. To investigate whether peptides in pair 1 form clusters, diffusion coefficients of peptides (based on the 1H signals from amino acid side chains) and TFA (based on the 19F signal from the -CF3 group) in the mixture were measured by the BPP-LED method,31 with its two parent solutions serving as references. Experiments were carried out on a Varian Inova 400 with Z-gradient, equipped with a broadband probe. Diffusion time was 400 ms for peptide measurements and 200 ms for TFA measurements. The pulsed field gradient strength increased linearly to gain sufficient signal decay in 16 steps. Since diffusion is a behavior of the whole molecule, diffusion coefficients of peptides were calculated based on the average of 5 1H signals attributed to different side chains of the peptides.
Dynamic Rheometry
All sample preparation procedures and measurements were performed at 25 °C and the final pH for all samples dissolved in the PBS buffer was 7.4. 16 mM solutions of two oppositely charged peptide modules were centrifuged separately for 10 min at 8,000 rpm. 200 µL of each K- and E-peptides were mixed through a Y-shaped connector in the sealed cell of a rheometer, immediately followed by monitoring of the gelation process.
Dynamic rheological measurements were performed using a NOVA Rheometer (REOLOGICA Instruments, Inc., Sweden) featuring a null balance system which allows for nano-torque and nano-strain measurement control and analysis. The instrument is also equipped with a sealed-cell geometry which prevents dehydration of the water-based samples during prolonged measurements. In addition, to exclude possible dehydration of the samples at 25 °C, a simple in-house designed system was used to humidify the incoming air used for the sealed-cell bearing (see Figure S9, Supporting Information). Rheological characterizations of the samples were performed using a 25 mm diameter cone-and-plate steel geometry (4° cone angle). The detailed description of the sequential rheological experiments could be found elsewhere.32 Briefly, time-sweep measurements were conducted at 0.2 % strain amplitude and 1 rad/s angular frequency followed by frequency-sweep measurements at 0.2 % strain amplitude, while the frequency was varied from 0.01 to 100 rad/s in a log mode with 18 data points per frequency decade. After the frequency-sweep measurements and before the strain-sweep measurements, a time-sweep of 3 hr was performed on the gel at 0.2 % strain amplitude, 1 rad/s frequency to confirm that the gel remains undisturbed by the frequency-sweep (see Figure S12, Supporting Information). Strain-sweep measurements were then performed with a single integration cycle at 1 rad/s frequency, within the range of strain amplitudes from 0.1 % to 100 % in a log mode with 23 data points per decade.
Small-Angle X-Ray Scattering (SAXS) and Small-Angle Neutron Scattering (SANS)
Initial peptide solutions were prepared as for rheological experiments, except that the PBS buffer was prepared in D2O both for SAXS and SANS measurements. To prepare samples for SAXS experiments, 10–15 µL equal volumes of each peptide (10–15 µL) were centrifuged into (20 sec at 500 RPM) a cylindrical glass capillary (Charles Supper Co.) with a diameter of 1.0 mm and a wall thickness of 0.01 mm. Scattering data were collected at 0.3, 1.5, 2.5, 4, 6, 24, 48, and 72 hrs after mixing for pairs 2 and 3 and 0.5, 1.5, 3, 5, 7, 24, 48, and 72 hrs after mixing for pairs 4 and 5, and 72 h after mixing for pair 1. For SANS experiments, the respective oppositely charged peptide modules were mixed inside a titanium cell with 1-mm path length and quartz windows 30 mm in diameter, which is routinely used for SANS measurements at the National Institute of Standards and Technology (NIST) Center for Neutron Research (NCNR).
SAXS data were collected using the beamline 12ID-B of the Advanced Photon Sources (APS) at the Argonne National Laboratory. For every measurement, the monochromic X-ray beam (λ = 0.689 Å) with a size of 0.07 mm × 0.20 mm was adjusted to pass through the centers of the capillaries. The exposure time for all samples was set to 0.2 sec to avoid detector saturation and radiation damage to the samples. X-ray scattering intensities were collected using the 2D detector Pilatus 2M (DECTRIS Ltd). The 2D scattering images were converted into 1D scattering profiles of I(Q) vs. Q in the Q-range from 0.007 Å−1 to 0.6 Å−1 by means of azimuthal averaging after solid angle correction. The resulting 1D profiles were normalized over the intensity of the transmitted X-ray beam, using the software package at the beamline 12ID-B. I(Q) is the scattering intensity of X-rays, and Q is the scattering vector amplitude which is related to the X-ray wavelength λ and the scattering angle θ by
| (1) |
Subtraction of the solvent scattering (PBS in D2O) involved normalization based on the ratio of incident and transmitted X-ray photon counts to account for the slight differences in the thickness of different capillaries. Also additional background scattering correction was performed in accordance with the generally accepted published procedure.33
SANS data were collected using the 30 m SANS instrument (NG-7) at NIST.34 Monochromatic neutrons at λ = 6 Å with a wavelength spread (Δλ/λ) of 0.14 were detected on a 64 cm × 64 cm two-dimensional detector. Data on SANS intensity were collected with a Q-range from 0.001 Å−1 to 0.4 Å−1. The low-Q configuration used neutron focusing lenses and an 8 Å neutron wavelength. Scattering intensities were normalized using direct beam transmission measurements and were reduced according to published protocols.33,35 Both SAXS and SANS instruments have pinhole geometry.
The combined usage of SAXS and SANS techniques to characterize hydrogel structures has evident advantages. The high flux of the X-ray beam from the synchrotron allows one to reduce the data collection time down to 0.2 sec, thus facilitating the use of SAXS to follow the gelation process in real time. In comparison, the data collection time in SANS is about 1–2 hours, making it unsuited to monitor the gelation process. On the other hand, due to the different wavelength parameters (0.689 Å for SAXS vs. 6 Å and 8 Å for SANS) and detector setup, the SANS instrument allows one to get down to much lower Qmin values as compared to the SAXS (0.001 Å−1 for SANS vs. 0.007 Å−1 for SAXS). Lower Q values provide the possibility to reliably observe molecular assemblies of much greater size (up to ~2000 Å for SANS vs. up to ~500 Å for SAXS).
Pair 1 of the syndiotactic peptides D,L-K and D,L-E does not gel. Instead, clusters of finite size are formed. The solution structure of pair 1 at 72 h was measured by SAXS and the data were processed using the ATSAS software.36,37 The analysis of pair-wise distance distribution functions for globular particles P(r) (Eq. 2) was performed using the linear regularization method of indirect Fourier-transformation using the program GNOM.36
| (2) |
P(r) reflects the probability that two randomly chosen points in a scattering particle are at r distance apart from each other, and P(r) = 0 happens at the maximum linear dimension of the scattering particle, dmax. P(r) also provides the information about the distances between the electrons in the scattering particle, and could be calculated from the electron density distribution:
| (3) |
where γ(r) is the spherically (whole volume) averaged correlation function of the electron density reflecting the difference in the scattering intensity between the positions r and r+x. In the case of homogeneous particles when ρ(r) = ρ(r+x) = Δρ, which is the difference in electron densities between the scattering particle and the solvent and is a constant
| (4) |
where γ0(r) is the normalized correlation function or so-salled characteristic function38 defined only by the geometry of the scattering particles, γ0(r) = 1, for r =0; and γ0(r) = 0, for r > dmax. The radius of gyration of the scattering globular particle, Rg, is derived from the second moment of P(r) as:
| (5) |
Rg is the root mean square distance of all unit-volume elements from the center of gravity of the scattering particle, and in the case of X-rays, the distribution of the mass is defined by the electron density distribution within the scattering particle. A simulated annealing algorithm was used to restore low resolution 3D structures of D,L-K+D,L-E clusters in solution built from densely packed dummy atoms implemented in the DAMMIN program.39 To build the most probable and reliable 3D model, multiple DAMMIN shape solutions (at least 20 runs) were aligned with respect to their principal axes of inertia followed by the structural discrepancy minimization using SUPCOMB program40 and averaging by means of the DAMAVER routine.41
ATSAS software36,37 also has been used to estimate the zero-angle scattering intensity I(0) from the Guinier analysis of lnI(Q) vs. Q2 plots. Since I(0) characterizes the mass of the scattering particles, it was used to monitor the gelation process of all hydrogels over time. At the same time, the fibers formed by pairs 2–5 have one dimension (length) much greater than the other two (cross-section). The length of the fibers exceeded the upper detection limits of our techniques (~500 Å for SAXS and ~2000 Å for SANS). Hence we analyzed the scattering data of pairs 2–5 in terms of the cross-sectional dimensions of the scattering particles using the standard approach of multiplying I(Q) by Q. This approach essentially removes information about the length of the scattering particles.42 A simulated annealing algorithm, analogous to that used for restoring the 3D shape for pair 1,39 was used to restore the 2D cross-sections for pairs 2–5. Here, we model the 2D cross-sections of the fibers formed by pairs 2–5 using the algorithm in a purpose-written program that is described elsewhere.43 In SAXS data processing, the dummy atoms were arranged on a flat grid of 20 × 20 close-packed dummy atoms, each 3 Å in diameter. In SANS data processing, where bigger 2D cross-sectional patterns were modeled, the grid has 20 × 50 close-packed dummy atoms, each 3 Å in diameter to model dimensions of about 200 Å, and 30 × 150 close-packed dummy atoms, each 5 Å in diameter to model dimensions of about 800 Å. This allows one to model pictorial cross-sectional slices of the hydrogels showing the fibers and how they are connected into fibrous network. The program calculated the pair distance distribution function, Pc(r), for the model cross-section composed of the dummy atoms. Pc(r) is the distribution of distances between area elements in the cross-section, weighted by the scattering density at each radial distance, r. The optimization procedure is in general described elsewhere.32,43 After optimization, the radius of gyration of the cross-section Rc in Å, the maximum cross-sectional dimension dmax in Å, the cross-section area Sc in Å2, and the zero-angle scattering intensity Ic(0), which is proportional to the mass per unit length of the fiber (in arbitrary unit per Å), were determined from Pc(r). dmax is the r value at which Pc(r) goes to 0. The zeroth and the second moments of Pc(r) yield Ic(0) and Rc values, respectively. Rc is the contrast-weighted mean distance of all area elements from the center of scattering density. The program also outputs the model cross-sections as atomic coordinate files in the Protein Data Bank format which allowed their pictorial presentation. Based on these coordinates and the grid dimensions, the area of the cross-section, Sc, can be calculated. Corrections for scaling and incoherent background were applied to the model scattering profile so it could be compared directly with experimental scattering data.43
Results and Discussion
Of the five peptide pairs, pair 1 is made of syndiotactic peptides; pairs 2 and 3 are made of homochiral peptides of opposite chiralities, and pairs 4 and 5 are made of homochiral peptides of the same chirality (Table 1). Pairs 2 and 3 are mirror images of each other while pairs 4 and 5 are mirror images of each other. Figure 1 shows the photographs of the five pairs in NMR tubes: pair 1 does not gel and remains a clear solution; pairs 2 and 3 form opaque gels; pairs 4 and 5 form translucent gels. From visual observation, gelation is instantaneous for pairs 2 and 3 but much more gradual for pairs 4 and 5. As it has been mentioned in the Experimental Section, the electrostatic repulsion prevents the hydrogelation and/or self-assmbly of the individual peptide modules. This is also confirmed by the narrow 1H NMR signals in the spectra of all individual peptide modules as well as by the absence of their scattering in the control SAXS experiments (see Supporting Information).
Figure 1.
Hydrogels co-assembled from a pair of oppositely charged peptide modules. Left to right: syndiotactic pair 1: D,L-K+D,L-E; heterochiral pair 2: D-K+L-E; heterochiral pair 3: L-K+D-E; homochiral pair 4: D-K+D-E; homochiral pair 5: L-K+L-E. Pair 1 does not form a hydrogel. Pairs 2–5 form hydrogels.
NMR Monitoring of the Assembly Process
The above visual observations were confirmed by NMR spectroscopy. Figure 2 shows the 1H NMR spectra for pair 1 (6 months after mixing) and pairs 2–5 (17 hours after mixing). For pair 1, the peptide 1H signals are still very sharp after 6 months, consistent with no gelation. For pairs 2–5, hardly any peptide 1H signals are left, consistent with extensive gelation because gelled peptides are no longer detectable by solution NMR due to their extremely short T2 values.30 Figure 3 shows the gelation process monitored by NMR through the 1H signal from the ε-H of the lysine side chains. The 1H signal intensity of pair 1 did not change for 6 months, again consistent with no gelation. Of the four gelling pairs, the two heterochiral pairs 2 and 3 gelled much faster than the two homochiral pairs 4 and 5. However, the eventual extent of gelation, measured by the 1H signal intensity from non-incorporated peptides, is almost the same for all four gelling pairs after 18 hrs of gelation (ca. 98 %). Unfortunately, the repetitiveness of the amino acid sequences in pair 1 makes it impossible to study the structure of its assemblies using multidimensional NMR. Therefore, to confirm that pair 1 forms clusters of finite size, the diffusion coefficient, D, of the peptides was measured, with a small molecule TFA as the reference point. Figure 4 shows the diffusion coefficient data in the mixture and in each parent peptide solution. The total peptide concentration in all three solutions is 16 mM. Keep in mind that, at pH 7.4, D,L-K carries six positive charges, D,L-E carries six negative charges, and TFA carries one negative charge. Hence, in the D,L-K solution, it is highly likely that the positively charged peptide associates with several copies of the negatively charged TFA. But such peptide-TFA association is unlikely in the D,L-E solution as both the peptide and TFA are negatively charged. This difference in peptide-TFA association explains why both peptide and TFA have smaller diffusion coefficients in the D,L-K solution than that in the D,L-E solution. In the mixture, the (D,L-K + TFA) complex is replaced by the (D,L-K + D,L-E) complex. The replacement of TFA by D,L-E can be attributed to three factors: (i) D,L-E might interact with D,L-K more strongly because it has multiple carboxylic groups, hydrophobic groups and H-bond donors/acceptors while TFA has only one of each; (ii) D,L-E (8 mM) is in great excess of TFA (trace amount); (iii) gelation is kinetically much less reversible than TFA binding. Consistent with such replacement, the diffusion of the (D,L-E + D,L-K) complex is slower than both D,L-E and (D,L-K + TFA) as shown in Figure 4. In contrast, diffusion of TFA lies between that of the two parent solutions, indicating that there might still be a portion of TFA bound to the (D,L-E + D,L-K) complex.
Figure 2.
Manifestations of hydrogelation in 1H NMR spectra. (A) 1H spectrum of the syndiotactic pair 1, 6 months after mixing. The sharp 1H signals indicate there is no gelation in this pair. (B) 1H spectra of the heterochiral pairs 2 and 3 and homochiral pairs 4 and 5. In the gelled state, the 1H signals from the peptides are too broad to be observed due to extremely short transverse relaxation time T2. The almost complete disappearance of peptide 1H signals indicate that gelation is near completion 17 hrs after mixing.
Figure 3.
Gelation process monitored by 1H NMR spectroscopy. The 1H signal intensity was converted to the concentration of unincorporated peptide, Cpeptide, based on the boundary condition that the initial concentration of unincorporated peptide was 8 mM. For the syndiotactic pair, the 1H signal intensity does not decrease with time, even after 6 months (right panel), which suggests no gelation for this pair. Cyan: pair 1; red: pair 2; green: pair 3; orange: 4; blue: pair 5.
Figure 4.
Diffusion coefficients (D) of syndiotactic peptides (cyan square) and TFA (black square) in different solutions. Statistical error bars correspond to one standard deviation and represent error in the diffusion coefficients estimation.
Rheological Characterization of Heterochiral and Homochiral Materials
Time-sweep rheological monitoring of the gelation process was entirely consistent with visual observations and NMR measurements. No gelation was observed for the syndiotactic pair 1, where the detected values of elastic (G') and viscous (G") moduli were very low (see Supporting Information). In contrast, the two heterochiral pairs 2 and 3 showed fast gelation with G' reaching plateau within 5–10 h (~ 5 kPa) with very slight growth afterwards. Gelation of the two homochiral pairs 4 and 5 was significantly slower with G' of both gels reaching plateau values around 48 hrs (~ 90 kPa, Figure 5(A)). From Figure 5(B), it can be seen clearly that the heterochiral pairs gelled faster initially but were outpaced by the homochiral pairs around 4 – 4.5 hrs. All heterochiral and homochiral hydrogels appear to be fairly stiff materials with the elastic modulus G' significantly higher than the viscous modulus G" (phase angle δ = arctan (G"/G') ~ 4–7°, see Supporting Information). The frequency spectra for all four pairs point to the formation of very stable materials (Figure 5(C)). Log G'(ω) profiles show very small dependence on the angular frequency within the range from 0.01 to 100 rad/s. This is consistent with the build-up of solid-like hydrogel networks for all gelling pairs, which are characterized by very short relaxation times and little or no mobility at the longest measurement duration (t = 2π/ω, and for ω = 0.01 rad/s, t ~ 600 s). Strain-sweep experiments (Figure 5(D)) show that the homochiral pairs have slightly higher strain yield value than the heterochiral pairs (γyield ~ 4 % vs. 3 %).
Figure 5.
Dynamic rheological characterization of heterochiral and homochiral peptide pairs. (A) Homochiral pairs lead to higher G′ values after 48 hs of gelation; (B) heterochiral pairs gelate faster and lead to higher G′ values within the first 4.5 hrs; (C) frequency sweep; (D) strain sweep. Red: pair 2; green: pair 3; orange: pair 4; blue: pair 5.
In summary, for this class of self-repulsive but mutually attractive oligopeptides, chirality has a great impact on their gelation behavior with homochirality leading to stronger gels and heterochirality leading to faster gelation. This is in sharp contrast to poly-(lactic) acids, where heterochirality leads to both stronger gels and faster gelation than homochirality.17,44 The difference in mechanical properties at 72 hr is not caused by the extent of gelation, which is ca. 98% for all 4 gelling pairs as shown by NMR spectroscopy (Figure 3). To understand the underlying mechanism for stronger gel but slower gelation caused by homochirality, the nano-scale structures of both homo- and hetero-chiral hydrogels are explored by SAXS and SANS. SAXS was used to monitor the gelation process over time and SANS was used to investigate the structures of the hydrogel fibers at larger scale.
Structural Analysis of Heterochiral and Homochiral Aggregates Using SAXS and SANS
As it has been already mentioned above, the structure of pair 1 assemblies cannot be obtained from NMR spectroscopy due to the sequence repetitiveness of the peptides, therefore, we resort to SAXS to investigate its solution structure. The SAXS scattering profile for pair 1 at 72 hrs after mixing demonstrates a scattering intensity significantly higher than the individual peptides, consistent with the formation of larger molecular aggregates (Figure 6(A)). To gain further structural insight into the oligomers formed by pair 1, we resort to Guinier analysis. Often, the analysis of Guinier region is capable to corroborate the formation of the aggregates of finite size, while the nonlinearity of Guinier plots typically suggests the presence of very different large assemblies, sometimes even of the size unresolved by SAXS (do you mean beyond the detection limit of SAXS?). In the case of pair 1, Guinier plots for rod-like particles (ln QI(Q) vs. Q2, bottom inset in Figure 6(A)) is linear (Q ~ 0.13–0.22 Å−1) and shows the characteristic upturn pointing to the formation of elongated aggregates of finite length.45 Here, the scattering data are analyzed only from the viewpoint of the cross-sectional dimensions of the scattering particles since the multiplication of I(Q) by Q essentially removes the length data of the scattering particle.42 Guinier plot lnI(Q) vs Q2 (characterizing the particles of the arbitrary shape, or sometimes called globular particles) shown in the top inset in Figure 6(A) is also linear (Q ~ 0.02–0.05 Å−1), and this suggests that pair 1 assembles into finite aggregates with fairly similar dimensional characteristics. The indirect Fourier transform of the experimental scattering data I(Q) vs. Q using GNOM36 results in pair-wise distance distribution function P(r) characteristic for elongated assemblies with Rg = 13.4 ± 0.4 Å and dmax = 55 Å (Figure 6(B)). Of note, we are fully aware that despite such dimensional similarities of the assemblies, aggregation of two oppositely charged peptides could result in differently shaped particles. Still, in order to get a pictorial understanding of possible 3D shapes of the aggregates, we used the ab initio low resolution shape reconstruction program DAMMIN.39 This program which is based on the simulated annealing algorithm uses the dummy atom model to reconstruct the shape of the particle in solution by minimization of the differences between the model scattering and the experimental X-ray scattering data I(Q) vs. Q. A series of more than 20 separate runs of DAMMIN were performed resulting in a set of different 3D shapes which, of course, are not unique and are not necessarily absolutely identical with each other. These resulting different 3D models were superimposed using the best-matching alignment program SUPCOMB.40 This program starts from the inertia-axis alignment of our modeled 3D objects; such alignment is then refined by minimization of the normalized structural discrepancy (NSD) parameter. NSD value is a quantitative indicator of the structural similarity of the aligning models. NSD = 0, for identical structures, and NSD > 1, for systemically different structures. All our more than 20 shapes have NSD ~ 0.4, which speaks in favor of their structural similarity. Their average low resolution 3D model (inset in Figure 6(B)), is not unique, but gives general illustration of the possible average shape of the pair 1 aggregate in solution of finite size. The formation of oligomers of finite size from pair 1 is also in a good agreement with NMR results which show pair 1 has sharp 1H spectrum (Figure 2A) but reduced diffusion coefficients (Figure 4).
Figure 6.
SAXS analysis of the heterodimers formed by the syndiotactic pair 1. (A) Scattering profiles I(Q) vs. Q of pair 1 (D,L-K+D,L-E), cyan, 72 h after mixing; and of the individual peptide D,L-K, light green (another peptide, D,L-E, gives almost identical profile). Insets: Guinier plot for globular particles, ln I(Q) vs. Q2, for pair 1 (top); Guinier plot for rod-like particles, ln QI(Q) vs. Q2, for pair 1 (bottom); black solid lines show linear regions (R2 ~ 0.95). Statistical error bars correspond to one standard deviation and represent error in the scattering intensity estimation. (B) Pair-wise distance distribution function P(r) of pair 1 (quality fit parameter ~0.7–0.8, for ideal fit it is 1.0); the inset shows the ab initio restored low resolution 3D shape of peptide clusters.
Contrary to pair 1, the mixing of all other four pairs resulted in the formation of fibrous hydrogel networks. The growth of this fibrous network is evidenced by the growth in SAXS scattering intensity I(Q) vs. Q within 72 hrs of the gelation monitoring as well as from the changes in the zero-angle scattering intensity I(0) from the Guinier analysis of lnI(Q) vs. Q2 plot (Figure 7).
Figure 7.
SAXS scattering profiles I(Q) vs. Q of the heterochiral and homochiral pairs showing the consistent growth in the scattering intensity over time. (A) Heterochiral pair 2 from 0.3 h to 72 h. (B) Heterochiral pair 3 from 0.3 h to 72 h. (C) Homochiral pair 4 from 0.5 h to 72 h. (D) Homochiral pair 5 from 0.5 h to 72 h. Insets in all plots show the changes in the corresponding Guinier plots for rod-like particles, lnQ×I(Q) vs. Q2, at the start and at the end of monitoring period. (Center) Time dependence of the zero-angle scattering intensities I(0) from Guinier analysis of lnI(Q) vs. Q2 plots for the four peptide pairs. The lines represent a basic B-spline fit of the data. Red: pair 2; green: pair 3; orange: pair 4; blue: pair 5.
Fully consistent with NMR and rheological data, the homochiral pairs initially have lower I(Q) and I(0) values than the heterochiral pairs, which are indicative of slower initial fiber growth for the homochiral pairs. However, the growth of homochiral fibers soon outpaces that of the heterochiral fibers and the homochiral pairs have larger I(Q) and I(0) values than the heterochiral pairs after a few hrs. At 72 hrs, the zero-angle scattering intensity I(0) from pairs 4 and 5 is about 2 times that of pairs 2 and 3, and about 70 times that of pair 1 (Figure 6(A) and Figure 7(Center)). From these data, it is clear that stronger gel but slower gelation of the homochiral pairs is related to the morphology of the peptide fibers.
As to the shape and dimension of the peptide fibers, the linearity of the Guinier plots for rod-like particles (see the insets in all panels in Figure 7) points to the formation of elongated asymmetrical assemblies in all gelling pairs, indicative of fiber formation from the very beginning of SAXS monitoring. The length of these fibers is beyond the maximum resolved size of our SAXS setup, which is ~ 500 Å. However, the cross-section of the fibers is within the detection limit of SAXS, therefore we modeled the cross-sections of the peptide fibers that best fit the scattering data and tracked the changes in these cross-sections over time. The cross-section pair-wise distance distribution functions, Pc(r), as well as the cross-section shape are shown in Figure 8 for all four pairs. From Pc(r) and from the modeled cross-sectional shapes, Rc, dmax, Sc, Ic(0) as well as fiber density ρ = Ic(0)/Sc, can be obtained for various time points and are plotted in Figure 9. All the cross-sectional parameters for the hetero-chiral fibers have already reached stable values within 30 min of gelation and only grew slightly between 30 min and 72 hrs. In contrast, the cross-sectional parameters of the homochiral fibers grew steadily between 30 min and 72 hrs. All four cross-sectional parameters, dmax, Rc, Sc and ρ, behave similarly to the elastic modulus G′: within the first few hours, the homochiral values are lower than the heterochiral values; but eventually, the homochiral values are much higher than heterochiral values. This feature provides a structural explanation to the observed stronger gel but slower gelation associated with homochirality: the heterochiral fibers are formed quicker than the homochiral fibers, but eventually the homochiral fibers are thicker than the heterochiral fibers.
Figure 8.
SAXS monitoring of the gelation process. Left and right columns show the time evolution of the 2D average cross-section of the peptide fibers. Four central panels show the consistent growth in the corresponding pair-wise distance distribution function of the cross-section, Pc(r) over time (in all model calculations χ2 ~ 0.5–0.8). Red: pair 2; green: pair 3; orange: pair 4; blue: pair 5.
Figure 9.
Time evolution of the cross-sectional parameters of peptide fibers. (A) Radius of gyration of the cross-section, Rc; (B) maximum cross-section dimension, dmax; (C) cross-section surface area, Sc; (D) fiber density, ρ, calculated as Ic(0)/Sc, where Ic(0) is the zero-angle scattering intensity of the cross-section proportional to mass per unit length of the fiber. The lines represent a basic B-spline fit of the data. Red: pair 2; green: pair 3; orange: pair 4; blue: pair 5.
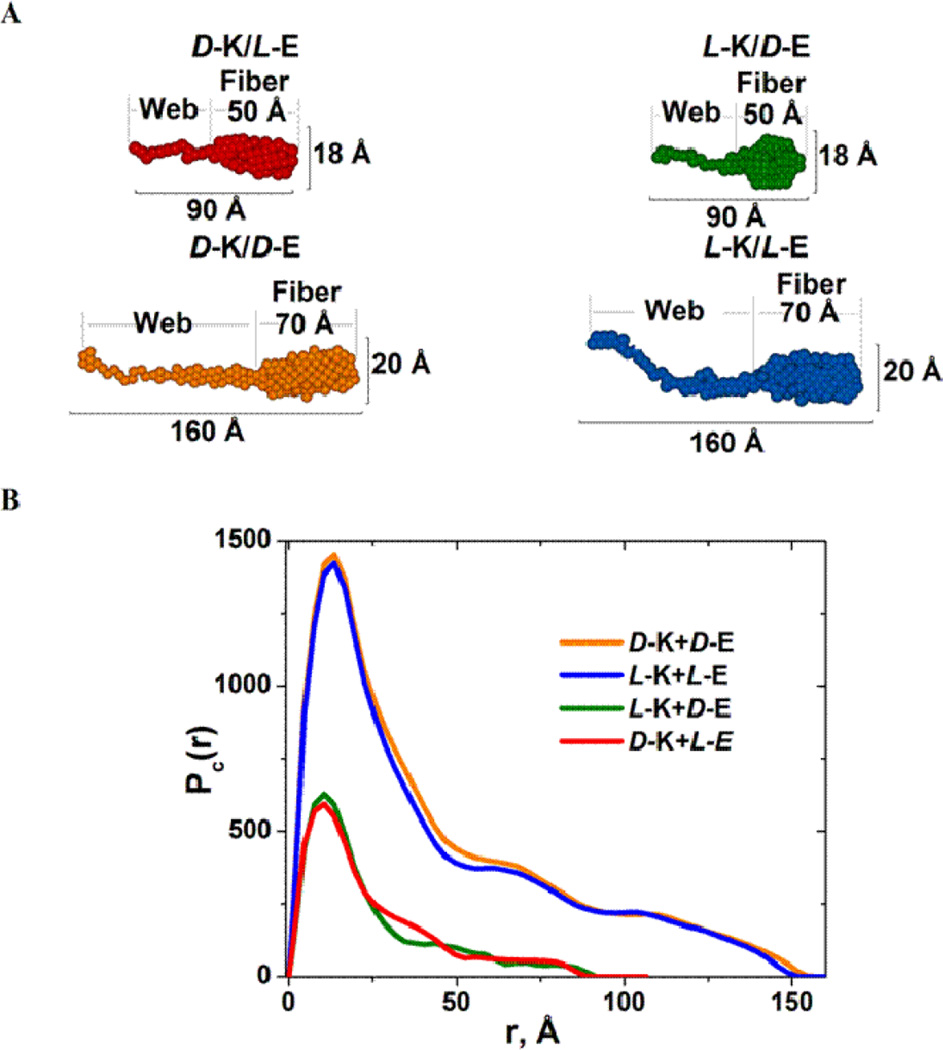
However, the exact homochiral-heterochiral crossing point of Rc, dmax, Sc and ρ (Figure 9) are 2–3 hrs earlier than that of G′ (Figure 5(B) vs. Figure 9). This suggests that structural features at a scale larger than the upper detection limit of SAXS (~500 Å) also contribute the mechanical properties of these hydrogels. To explore structural features at a larger scale, we resort to SANS, which has an upper detection limit of ~2000Å. SANS data were collected after 96 h of gelation. In full agreement with SAXS results, SANS scattering profiles I(Q) vs. Q (Figure 10(A)) demonstrate evident distinction between heterochiral (2 and 3) and homochiral (4 and 5) hydrogels. As expected, the two heterochiral pairs 2 and 3 are very similar to each other, and the same is true for the two homochiral pairs 4 and 5 (Figure 10(A)). As seen from the linearity of Guinier plots for rod-like particles lnQI(Q) vs. Q2 (inset in Figure 10(A)), both heterochiral and homochiral hydrogels are made of elongated asymmetrical fibers. Interesting structural feature of both homochiral and heterochiral hydrogels follows from the Guinier plots for flat particles lnQ2I(Q) vs. Q2 (Figure 10(B)). Here, the distinct upturn in the region Q > 0.06 Å−1 (see, the region at Q2 > 0.0036 Å−2) suggests the presence of flat structural elements in all hydrogels in addition to the network formed by elongated fibers. Indeed, based on the SANS data in the Q-range starting from 0.01 Å−1 and the grid of 20 × 50 dummy atoms (3 Å each), the modeling of average 2D cross-section of the hydrogel fibers reveals such flat webs attached to the fibers. As seen from the 2D shapes (Figure 11(A)) and their corresponding pair distance distribution functions (Figure 11(B)), the cross-sections of both heterochiral and homochiral hydrogel networks includes the fibers per se with the attached lappet-like webs growing from the side surface of the fibers. The fiber cross-section dimensions obtained from SANS are identical to those obtained from SAXS (compare Figure 11 vs. Figure 8 at 72 hr). One could reasonably suggest that the hydrogel networks are formed by the individual fibers interconnected with each other by flat, lappet-like webs. In an attempt to model the greater cross-sectional slice of such interconnected hydrogel networks, we used the SANS data from the lowest Q-range starting from Qmin ~ 0.005 Å−1 (inaccessible to our SAXS setup) and the grid of 30 × 150 dummy atoms (5 Å each). The resulting pictorial structures (Figure 12) vividly demonstrate that homochiral networks are formed by thicker fibers interconnected by thicker webs than the heterochiral networks and this translates into higher mechanical strength for the homochiral networks. At present, it is not clear, however, why some of the peptide modules do not incorporate into the fibers, and form the web-like connections instead. One possible explanation could be that the residual surface charges of the fibers serve as nuclei for lateral growth resulting in flat, interconnecting structures. To the same extent, since the length of the fiber, as it has been mentioned above, could not be reliably determined from our SAX(N)S data, it is hard to estimate the relationship between the fiber length and the longitudinal extension of the webs illustrated by the schematic cartoon in Figure 12(C).
Figure 10.
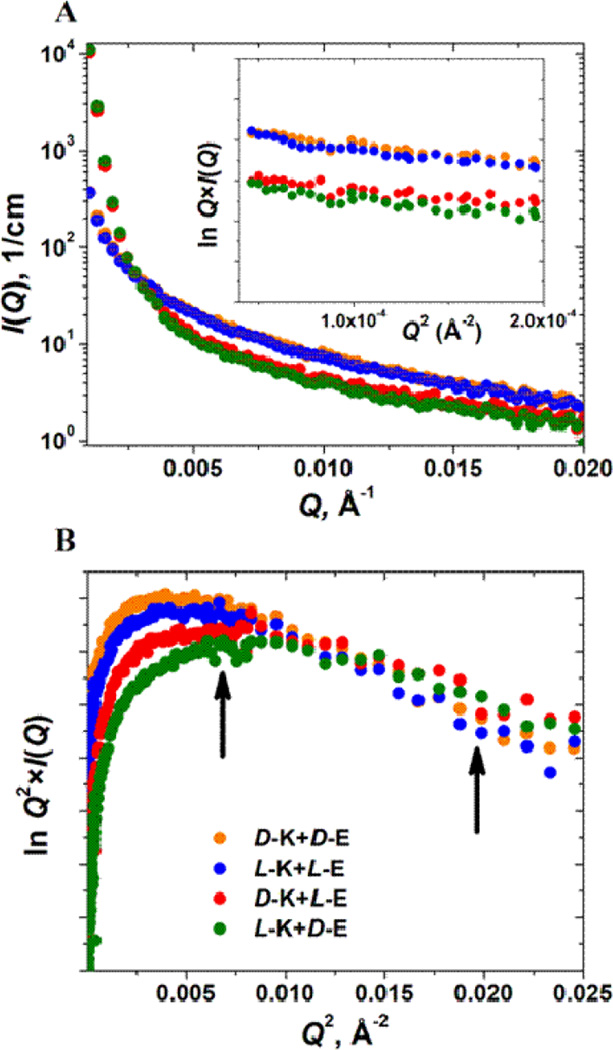
SANS data collected after 96 hrs of gelation. (A) Scattering profiles I(Q) vs. Q of all four gelling pairs. Guinier plots for rod-like particles, lnQ×I(Q) vs. Q2, are shown in the inset, and the linearity in this region (for Q, from 0.007 to 0.014 Å−1) indicates the formation of elongated fibers in all hydrogels. (B) Guinier plots for flat particles, lnQ2×I(Q) vs. Q2, and the linearity in this region (for Q, from 0.08 to 0.14 Å−1 shown by two arrows in the figure) indicates the formation of flat, lappet-like webs interconnecting the peptide fibers in all hydrogels. Statistical error bars correspond to one standard deviation and represent error in the scattering intensity estimation. Red: pair 2; green: pair 3; orange: pair 4; blue: pair 5.
Figure 11.
Analysis of the SANS data. (A) 2D average cross-section of individual peptide fibers, including the flat, lappet-like webs interconnecting the peptide fibers. (B) Corresponding cross-section pair-wise distance distribution functions, Pc(r), for respective homochiral and heterochiral fibers shown in (A) (in all model calculations χ2 ~ 1.0–1.3). Red: pair 2; green: pair 3; orange: pair 4; blue: pair 5.
Figure 12.
Pictorial presentation of the 3D slice of the hydrogels under study showing the cross-sections of the individual fibers interconnected with flat, “lappet-like” webs shown in gray. Reconstruction from the 2D cross-shape restored from SANS data with the low Qmin values (~ 0.003–0.005 Å−1): (A) for the heterochiral hydrogel pair 2 (D-K + L-E). (B) for the homochiral hydrogel pair 5 (L-K + L-E). Cross-sectional total size of the slice in both cases is 170 Å × 500 Å, in all model calculations χ2 ~ 1.0–1.3. (C) shows pictorial cartoon illustrating the schematical fibrous network organization.
Of note, the combined application of NMR, dynamic rheometry, and small-angle scattering techniques to monitor the gelation process show a discrepancy with regard to the time point of the completion of gelation (cf. Figure 3, Figure 5(A), and Figure 9). For NMR, which traces the decay of the free peptide concentration as a result of its incorporation into the hydrogel fiber, this amounts to about 14 hours (Figure 3). Dynamic rheometry, on the other hand, shows that the mechanical strength (G′) of the hydrogels do not change significantly after about 30 hours and reaches plateau at about 48 hours (Figure 5(A)). As for SAX(N)S, the dimensional parameters of the fiber cross-section appear to grow up to 72 hours and remain unchanged between within the interval 72–96 hours (Figure 9 and Figure 11). Based on these observations, one might suggest that after the incorporation of the peptides into the fiber, accompanied by the growth of the fibrous network, further restructuring of the fibers per se do not contribute significantly to the mechanical properties of bulk material. Yet, it is quite possible that in addition to the individual fiber parameters, other factors also contribute to the mechanical properties of the hydrogels. One of such factors, for example, could be the persistence length of the fibers, Lp, see, e.g., MacKintosh theory,46 and the issue of the determination of Lp in hydrogel fibers from the SAX(N)S data is worthy of future careful study.
In summary, the time course and the morphology of the peptide aggregates in the five pairs provide a structural explanation to the differences in their material properties. The syndiotactic pair 1 forms finite size aggregates (heterodimer) with no fibrous network. Macroscopically, this pair remains a clear solution. Both heterochiral pairs and homochiral pairs form fibrous networks and the fibers are interconnected by flat lappet-like webs. Macroscopically these pairs exist as solid-like hydrogels. As shown by SAXS analysis, the heterochiral pairs quickly form fibers, but these fibers show limited growth after a few hrs. In contrast, the homochiral pairs have slow initial fibrillization but the fibers grow steadily for up to 72 hrs. Eventually, the homochiral fibers outgrow the heterochiral fibers, resulting in a network made of thicker and denser fibers, which leads to higher G′ and γyield.
As to why the syndiotactic pair 1 forms oligomers of finite size, and, among the isotactic pairs, why the homochiral pairs 4 and 5 form thicker fibers than the heterochiral pairs 2 and 4, the NMR, SAXS and SANS analyses conducted in this work cannot provide a definitive answer. One possible explanation could be that, similar to amyloid-β-proteins Aβ-40 and Aβ-42,47 some peptide pairs form “open” oligomers that are prone to grow while some peptide pairs form “close” oligomers that are difficult to grow. Such “openness” or “closeness” could be defined by the degree of electrostatic charge compensation in the aggregates as well as by the availability of probable β-sheet links and/or hydrophobic contacts—all stemming from the structural differences between L- and D-enantiomers. Our more detailed research in this area is underway.
Conclusions
For hydrogels co-assembled from a pair of self-repulsive but mutually attractive oppositely charged oligopeptides, chirality is shown to be an influential factor in determining the rate of gelation as well as the mechanical properties of the resulting biomaterial. Homochirality is associated with stronger gels but slower gelation. In other words, homochirality confers mechanical advantage but heterochirality confers kinetic advantage to this class of biomaterials. Structurally, homochiral peptide pairs form networks made of thicker and denser fibers while heterochiral peptides pairs form networks more quickly. The observed mechanical advantage posed by homochirality provides another angle to assess its role in biology. The generality of this observation and its relevance to the origin of life awaits further investigation.
Supplementary Material
Acknowledgments
Financial support provided by the NIH (EB004416) is gratefully acknowledged. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Beamtime was awarded through the program of General User Proposals to GUP-24524. We also thank Drs. J. Ilavsky and Xiaobing Zuo (ANL) for assistance. The identification of commercial products does not imply endorsement by the National Institute of Standards and Technology nor does it imply that these are the best for the purpose. This work is based upon activities supported in part by the National Science Foundation under Agreement No. DMR-0944772.
Footnotes
Supporting Information Available. Analytical HPLC and ESI-MS spectra of the peptides; the photo of the in-house designed simple humidifier for sealed-cell rheometer; detailed rheological data on all peptide systems, NMR spectra and SAXS data on the individual peptides and model fits to the expertimental SANS and SAXS data.
References
- 1.(a) Blackmond DG. Cold Spring Harbor Perspec. Biol. 2010;2:a002147. doi: 10.1101/cshperspect.a002147. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fitz D, Reiner H, Plankensteiner K, Rode BM. Curr. Chem. Biol. 2007;1:41–52. [Google Scholar]; (c) Kuhn H. Curr. Opin. Coll. Interface Sci. 2008;13:3–11. [Google Scholar]; (d) Weissbuch I, Illos RA, Bolbach G, Lahav M. Acct. Chem. Res. 2009;42:1128–1140. doi: 10.1021/ar900033k. [DOI] [PubMed] [Google Scholar]
- 2.Avetisov V, Goldanskii V. Proc. Natl. Acad. Sci. USA. 1996;93:11435–11442. doi: 10.1073/pnas.93.21.11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Fitz D, Reiner H, Plankensteiner K, Rode BM. Curr. Chem. Biol. 2007;1:41–52. [Google Scholar]; (b) Green MM, Jain V. Orig. Life Evol. Biosph. 2010;40:111–118. doi: 10.1007/s11084-009-9180-7. [DOI] [PubMed] [Google Scholar]; (c) Joyce GF, Visser GM, Van Boeckel CAA, Van Boom JH, Orgel LE, Van Westrenen J. Nature. 1984;310:602–604. doi: 10.1038/310602a0. [DOI] [PubMed] [Google Scholar]
- 4.(a) Nanda V, Andrianarijaona A, Narayanan C. Protein Sci. 2007;16:1667–1675. doi: 10.1110/ps.072867007. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kumar A, Ramakrishnan V, Ranbhor R, Patel K, Durani S. J. Phys. Chem. B. 2009;113:16435–16442. doi: 10.1021/jp906811k. [DOI] [PubMed] [Google Scholar]
- 5.Saghatelian A, Yokobayashi Y, Soltani K, Ghadiri MR. Nature. 2001;409:797–801. doi: 10.1038/35057238. [DOI] [PubMed] [Google Scholar]
- 6.(a) Ladik JJ, Szerekes Z. J. Mol. Model. 2006;12:462–467. doi: 10.1007/s00894-005-0073-z. [DOI] [PubMed] [Google Scholar]; (b) Milton RCdeL, Milton SCF, Kent SBH. Science. 1992;256:1445–1448. doi: 10.1126/science.1604320. [DOI] [PubMed] [Google Scholar]; (c) Urata H, Shinohara K, Ogura E, Ueda Y, Akagi M. J. Am. Chem. Soc. 1991;113:8174–8175. [Google Scholar]
- 7.Reich Z, Schramm O, Brumfeld V, Minsky A. J. Am. Chem. Soc. 1996;118:6345–6349. [Google Scholar]
- 8.(a) Clark TD, Buriak JM, Kobayashi K, Isler M, Ghadiri MR. J. Am. Chem. Soc. 1998;120:8949–8962. [Google Scholar]; (b) Haack T, Gonzalez MJ, Sanchez Y, Giralt E. Letters in Peptide Science. 1997;4:377–386. doi: 10.1002/(SICI)1099-1387(199707)3:4%3C299::AID-PSC121%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]; (c) Durani S. Acc. Chem. Res. 2008;41:1301–1308. doi: 10.1021/ar700265t. [DOI] [PubMed] [Google Scholar]; (d) Anil B, Song B, Tang Y, Raleigh DP. J. Am. Chem. Soc. 2004;126:13194–13195. doi: 10.1021/ja047119i. [DOI] [PubMed] [Google Scholar]
- 9.Hirst AR, Smith DK, Feiters MG, Geurts HPM. Chem. Eur. J. 2004;10:5901–5910. doi: 10.1002/chem.200400502. [DOI] [PubMed] [Google Scholar]
- 10.Bao G, Suresh S. Nature Materials. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 11.(a) Trevors JT, Pollack GH. Progr. Biophys. Mol. Biol. 2005;89:1–8. doi: 10.1016/j.pbiomolbio.2004.07.003. [DOI] [PubMed] [Google Scholar]; (b) Trevors JT. C. R. Biol. 2011;334:269–272. doi: 10.1016/j.crvi.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 12.(a) Luo X, Liu B, Liang Y. Chem. Commun. 2001:1556–1557. doi: 10.1039/b104428c. [DOI] [PubMed] [Google Scholar]; (b) Beceril J, Escuder B, Miravet JF, Gavara R, Luis SV. Eur. J. Org. Chem. 2005:481–485. [Google Scholar]; (c) Džolić Z, Wolsperger K, Žinić M. New J. Chem. 2006;30:1411–1419. [Google Scholar]
- 13.Hirst AR, Huang B, Castelletto V, Hamley IW, Smith DK. Chem. Eur. J. 2007;13:2180–2188. doi: 10.1002/chem.200601665. [DOI] [PubMed] [Google Scholar]
- 14.(a) Frkanec L, Žinić M. Chem. Commun. 2010;46:522–537. doi: 10.1039/b920353m. [DOI] [PubMed] [Google Scholar]; (b) Makarević J, Jokić M, Frkanec L, Vujičić NS, Žinić M. Beilstein J. Org. Chem. 2010;6:945–959. doi: 10.3762/bjoc.6.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brizard A, Oda R, Huc I. Topics Curr. Chem. 2005;256:814–826. doi: 10.1007/b107174. [DOI] [PubMed] [Google Scholar]
- 16.Slager J, Domb AJ. Adv. Drug Delivery Rev. 2003;55:549–583. doi: 10.1016/s0169-409x(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 17.Hiemstra C, Zhong Z, Li L, Dijkstra PJ, Feijen J. Biomacromolecules. 2006;7:2790– 2795. doi: 10.1021/bm060630e. [DOI] [PubMed] [Google Scholar]
- 18.Hiemstra C, Zhou W, Zhong Z, Wouters M, Feijen J. J. Am. Chem. Soc. 2007;129:9918–9926. doi: 10.1021/ja072113p. [DOI] [PubMed] [Google Scholar]
- 19.(a) de Jong SJ, de Smedt SC, Demeester J, van Nostrum CF, Kettenes-van den Bosch JJ, Hennink WE. J. Control Release. 2001;72:47–56. doi: 10.1016/s0168-3659(01)00261-9. [DOI] [PubMed] [Google Scholar]; (b) de Jong SJ, de Smedt SC, Wahls MWC, Demeester J, Kettenes-van den Bosch JJ, Hennink WE. Macromolecules. 2000;33:3680–3686. [Google Scholar]
- 20.(a) Slager J, Cohen Y, Khalfin R, Talmon Y, Domb AJ. Macromolecules. 2003;36:2999– 3000. [Google Scholar]; (b) Slager J, Domb AJ. Biomaterials. 2002;23:4389–4396. doi: 10.1016/s0142-9612(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 21.(a) Paradossi G, Chiessi E, Malovikova A. Biopolymers. 1999;50:201–209. doi: 10.1002/(SICI)1097-0282(199908)50:2<201::AID-BIP9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]; (b) Paradossi G, Chiessi E, Malovikova A. Macromolecules. 2001;34:8179–8186. [Google Scholar]
- 22.Friggeri A, van der Pol C, van Bommel KJC, Heers A, Stuart MCA, Feringa BL, van Esch J. Chem. Eur. J. 2005;11:5353–5361. doi: 10.1002/chem.200500007. [DOI] [PubMed] [Google Scholar]
- 23.Nagy KJ, Giano MC, Jin A, Pochan DJ, Schneider JP. J. Am. Chem.Soc. 2011;133:14975–14977. doi: 10.1021/ja206742m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Fuhrhop J-H, Krull M, Bueldt G. Angew. Chem. Int. Ed. Engl. 1987;26:699–700. [Google Scholar]; (b) Dzwolak W, Ravindra R, Nicolini C, Jansen R, Winter R. J. Am. Chem. Soc. 2004;126:3762–3768. doi: 10.1021/ja039138i. [DOI] [PubMed] [Google Scholar]
- 25.(a) Esler WP, Stimson ER, Fishman JB, Ghilardi JR, Vinters HV, Mantyh PW, Maggio JE. Biopolymers. 1999;49:505–514. doi: 10.1002/(SICI)1097-0282(199905)49:6<505::AID-BIP8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; (b) Wadai H, Yamaguchi KI, Takahashi S, Kanno T, Kawai T, Naiki H, Goto Y. Biochemistry. 2005;44:157–164. doi: 10.1021/bi0485880. [DOI] [PubMed] [Google Scholar]
- 26.(a) Chalifour RJ, McLaughlin RW, Lavoie L, Morisette C, Trebmlay N, Boule M, Sarrazin P, Stea D, Lacombe D, Tremblay P, Gervais F. J. Biol. Chem. 2003;278:34874–34881. doi: 10.1074/jbc.M212694200. [DOI] [PubMed] [Google Scholar]; (b) Grillo-Bosch D, Carulla N, Cruz M, Sánchez L, Pujol-Pina R, Madurga S, Rabanal F, Giralt E. ChemMedChem. 2009;4:1488–1494. doi: 10.1002/cmdc.200900191. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran S, Tseng Y, Yu YB. Biomacromolecules. 2005;6:1316–1321. doi: 10.1021/bm049284w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. New York: Oxford University Press; 2000. pp. 1–75. [Google Scholar]
- 29.Gill SC, von Hippel PH. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y, Taraban M, Yu YB. Soft Matter. 2011;7:9890–9893. doi: 10.1039/C1SM06389H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu DH, Chen AD, Johnson CS. J. Magn. Reson. A. 1995;115:260–264. [Google Scholar]
- 32.Hyland LL, Taraban MB, Feng Y, Hammouda B, Yu YB. Biopolymers. 2012;97:177–188. doi: 10.1002/bip.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitten A, Trewhella J. Micro and Nano Technologies in Bioanalysis. New York: Humana; 2009. Small-Angle Scattering and Neutron Contrast Variation; pp. 307–322. [DOI] [PubMed] [Google Scholar]
- 34.Glinka CJ, Barker JG, Hammouda B, Krueger S, Moyer JJ, Orts WJ. J. Appl. Crystallogr. 1998;31:430–445. [Google Scholar]
- 35.Kline SR. J. Appl. Chrystallogr. 2006;39:895–900. [Google Scholar]
- 36.Svergun DI. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 37.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 38.Porod G. Kolloid-Z. 1951;124:83–114. [Google Scholar]
- 39.Svergun DI. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozin MB, Svergun DI. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 41.Volkov VV, Svergun DI. J. Appl. Cryst. 2003;36:860–864. [Google Scholar]
- 42.Glatter O, Kratky O, editors. Small Angle X-Ray Scattering. London: Academic Press; 1982. [Google Scholar]
- 43.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Proc. Natl. Acad. Sci. USA. 2008;105:18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiemstra C, Zhong ZY, Dijkstra P, Feijen J. Macromol. Symp. 2005;224:119–131. [Google Scholar]
- 45.Guinier A. Ann Phys (Paris) 1939;12:161–236. [Google Scholar]
- 46.MacKintosh FC, Käs J, Janmey PA. Phys. Rev. Lett. 1995;75:4425–4428. doi: 10.1103/PhysRevLett.75.4425. [DOI] [PubMed] [Google Scholar]
- 47.Bernstein SL, Dupuis NF, Lazo NL, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT. Nature Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.