Abstract
The presence of close to 100% large-headed multi-tailed spermatozoa in the ejaculate has been described as a rare phenotype of male infertility with a very poor prognosis. We demonstrated previously that most cases were caused by a homozygous mutation (c.144delC) in the Aurora Kinase C gene (AURKC) leading to the absence or the production of a nonfunctional protein. AURKC deficiency in these patients blocked meiosis and resulted in the production of tetraploid spermatozoa unsuitable for fertilization. We describe here the study of two brothers presenting with large-headed spermatozoa. Molecular analysis of the AURKC gene was carried out in two brothers presenting with a typical large headed spermatozoa phenotype. Both affected brothers were heterozygous for the c.144delC mutation. After complete sequencing of the gene a new heterozygous variant, c.436-2A>G, was identified in both patients. This mutation is located in the acceptor consensus splice site of exon 5. AURKC transcripts were extracted from one of the patient’s leukocytes and Reverse Transcription Polymerase Chain Reaction (RT-PCR) could be realised showing the presence of a truncated transcript indicating that c.436-2A>G leads to the skipping of exon 5.
These results indicate that AURKC molecular analysis of patients with large-headed spermatozoa should not be stopped in the absence of a homozygous recurrent mutation on exon 3 but complete sequence analysis should be performed. This diagnosis is important as the identification of AURKC mutations in patients indicates that all spermatozoa will be chromosomally abnormal and that ICSI should not be attempted.
Keywords: Base Sequence; DNA Mutational Analysis; Exons; Heterozygote; Humans; Infertility, Male; diagnosis; genetics; Male; Meiosis; genetics; Molecular Sequence Data; Mutation; Pedigree; Polymerase Chain Reaction; Protein-Serine-Threonine Kinases; genetics; Siblings; Spermatogenesis; genetics; Spermatozoa; metabolism; pathology; Tunisia
INTRODUCTION
Infertility concerns a minimum of 70 million couples worldwide. An important proportion of cases is believed to have a genetic component, yet few causal genes have been identified so far. Patients with large-headed multiflagellar spermatozoa or “macrozoospermia” present with a majority of large-headed, multiflagellar polyploid spermatozoa in the ejaculate. This syndrome was first described in 1977 (Nistal et al., 1977) and cases have been described regularly ever since (Benzacken et al., 2001; Devillard et al., 2002; Escalier, 1983; In’t Veld et al., 1997; Mateu et al., 2006; Pieters et al., 1998), these studies highlighting a highly abnormal chromosomal contempt in these patient’s spermatozoa. In 2007 we demonstrated that a homozygous mutation (c.144delC) in the AURORA KINASE C (AURKC) gene was found in a large majority of macrozoocephalic patients (Dieterich et al., 2007). A carrier frequency of 1/50 was established from individuals from the Maghrebian general population, comparable to that of Y-microdeletions, thus far the only known recurrent genetic event altering spermatogenesis. We then could demonstrate that large-headed spermatozoa from AURKC deficient patients were tetraploid indicating that without a functional AURKC protein, meiosis could not be completed (Dieterich et al., 2009).
A few other genes have been linked with other infertility phenotypes and in particular globozoospermia, a phenotype characterized by the production of small round spermatozoa devoid of acrosome. We recently showed that three quarters of patients tested presenting with type I (pure) globozoospermia were homozygously deleted for the DPY19L2 gene (Harbuz et al., 2011). DPY19L2 is almost exclusively expressed in the testes and is involved in sperm elongation and acrosome formation. Overall, these findings strengthen the importance of gene defects in the etiology of male non-obstructive infertility and let foresee that many more yet undiscovered genes are likely liked to other dysfunctions of spermatogenesis.
Here we studied two brothers who presented with typical macrozoospermia with a few non-megaloheaded spermatozoa. In total eleven ICSI had been unsuccessful attempted. Molecular analysis revealed the presence of an unknown AURKC mutation. Transcript analysis confirmed the pathogenicity of the newly identified mutation, showing that the encoded protein would lack one of its seven exons. Furthermore, this strengthens the prognostic value of AURKC genotyping for patients with large headed spermatozoa, reinforcing the fact that ICSI should not be attempted for AURKC mutated patients.
MATERIALS AND METHODS
Patients and control subjects
Both patients (II.1, II.2) are brothers (Figure 1) from Tunisian descent and were treated for infertility at the CPSR les Jasmins in Tunis. They were diagnosed with macrozoospermia following routine sperm analysis. Both had close to 100% large headed spermatozoa with a sperm count <1 M/ml (Table 1). After centrifugation and careful examination, a few normal looking spermatozoa which could fit into an injection pipette could be identified for each patient. Six and 5 ICSI were carried out for patient 1 and 2 respectively between 1999 and 2005 (Table 2).
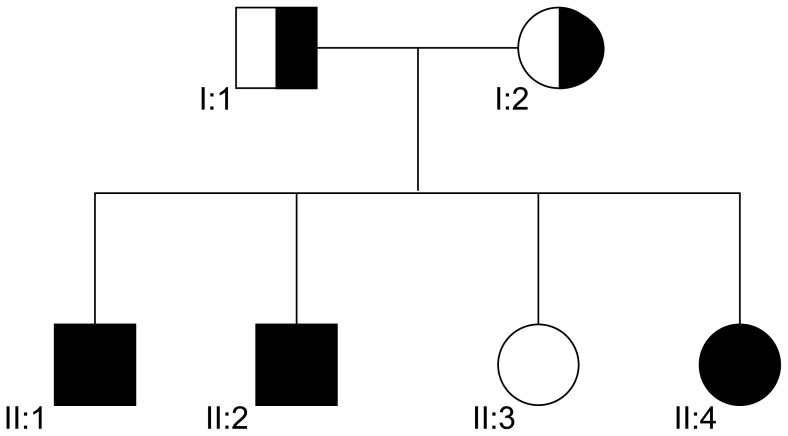
Figure 1. Family tree.
II:1 and II:2 and II:4 are compound heterozygous carrying c.144delC and c.436-2A>G
Table 1.
Sperm parameters (average of 4 separate analyses for each patient).
| Patients: | II:1 | II:2 |
|---|---|---|
| Sperm volume (ml) | 3 | 3 |
| Nb spz (× 106 per ml) | 0.9 | 0.8 |
| Round cells (×106 cells) | 12 | 1 |
| Motility A+B, 1 h (%) | 8 | 7 |
| Vitality (%) | 35 | 48 |
| Large-headed (%) | 100 | 100 |
| Multiflagellar (%) | 28 | 52 |
| Multiple Anomalies Index | 3.52 | 3.6 |
Table 2.
Details of the 11 ICSI realised (Cumulus-oocyte complexes (COCs)).
| P1.1 | P1. 2 | P1.3 | P1. 4 | P1.5 | P1.6 | P2.1 | P2. 2 | P2.3 | P2. 4 | P2.5 | Av. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COCs collected | 14 | 8 | 10 | 8 | 2 | 6 | 34 | 28 | 18 | 9 | 21 | 14,4 |
| M2 oocytes | 8 | 6 | 6 | 3 | 0 | 6 | 13 | 20 | 13 | 5 | 8 | 8 |
| 2pn | 3 | 4 | 6 | 1 | / | 5 | 5 | 7 | 7 | 3 | 6 | 4,7 |
| grade 1/2 D2 | 2 | 2 | 3 | 0 | / | 3 | 4 | 5 | 5 | 2 | 4 | 3 |
| grade 3 D2 | 1 | 2 | 3 | 1 | / | 1 | 1 | 2 | 2 | 1 | 2 | 1,6 |
| Transf. D2/3 | 3 | 4 | 3 | 1 | / | 3 | 4 | 3* | 3 | 2 | 0** | 2,9 |
Two additional embryos were transferred after thawing.
A blastocyst transfer was planned but none of the 6 embryos reached that stage.
Both patient’s wives had normal reproductive parameters. A conventional GnRH-agonist (Lupron) long-protocol, was carried out for ovarian hyperstimulation. Final oocyte maturation and the induction of ovulation were performed with 10,000 IU hCG (Ferring Arzneimittel GmbH) upon observation of at least three follicles >17 mm. Thirty-six hours after hCG injection, oocyte retrieval took place using transvaginal ultrasound guided follicle aspiration.
The patients sisters II:3 and II:4 are single and have not yet tried to have children.
DNA was extracted from saliva samples from the two brothers and their first degree relatives. Control DNAs were extracted from blood from French anonymous fertile donors originating from North Africa (Algeria, Morocco or Tunisia). All patients, family members and anonymous donors gave their written informed consent, all national laws and regulations were respected.
Molecular analyses
DNA Genomic DNA was extracted from peripheral blood leucocytes using a guanidium chloride extraction procedure. Patients and family member’s DNA was extracted from saliva using Oragene DNA Self-Collection Kit (DNAgenotech, Canada) using the manufacturer’s recommendations.
The seven AURKC exons and intronic boundaries were amplified as described previously (Dieterich et al., 2007). All analyses were carried out using the BigDye Terminator v3.1 sequencing kit and an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Primers and protocols are as published previously (Dieterich et al., 2007).
RNA was extracted from nucleated cells isolated from the whole blood using Ficoll 400 by Sigma-Aldrich Corporation (St. Louis, MO, USA) following the manufacturer’s protocol. RNA extraction was carried out on isolated white blood cells using Macherey Nagel NucleoSpin microRNA II columns (Macherey Nagel, Hoerdt, France) using the manufacturer’s protocol. Reverse transcription (RT) was carried out with 5 μL of extracted RNA (approximately 500 ng). Hybridization of the oligo (dT) was realized by incubating for 5 min at 65°C the following mix: 5 μL of RNA, 3 μL of poly T oligo primers (dT)12–18 (10 mM, Pharmacia), 3 μL of the four dNTPs (0.5 mM, Rochediagnostics) and 2.2 μL of H2O followed by ice quenching. RT was then carried out during 30 min at 55 °C after the addition of 4 μL of 5X buffer, 0.5 μL RNase inhibitor and 0.5 μL of Transcriptor Reverse Transcriptase (Roche Diagnostics). Five μL of the obtained cDNA mix was used for the subsequent polymerase chain reaction (PCR).
Primers were designed to amplify exons 4-6 from cDNA to characterize the consequences of the c.436-2A>G mutation. The 5′ primer was located on exon 4 (CAATATCCTGCGCCTGTATAACT) and the 3′ primer on exon 6 (TCATTTCTGGCGGCAAGT). Two microliters of the reversed transcribed RNA was amplified with these primers (40 cycles) at an elongation temperature of 58°C.
High resolution melting (HRM) analysis was performed with the LightCycler 480 (Roche), using the LightCycler 480 HRM master kit. Results were analyzed with the Gene scanning software (Roche) as described in (Harbuz et al., 2010).
RESULTS
Both patients (II.1, II.2) were treated for infertility at the Clinique des Jasmin in Tunis between 1999 and 2005. They were diagnosed with macrozoospermia following routine sperm analysis. Both had 100% large headed spermatozoa with a sperm count <1 M/ml (Table 1). However, after centrifugation and careful examination, a few smaller, “normal looking” spermatozoa which could fit into an injection pipette were identified in all semen samples analysed from each patients. Six and five ICSI cycles were attempted for patients 1 and 2 respectively (Table 2) before the characterization of the role of AURKC in macrozoospermia (Dieterich et al., 2007) and the identification of the two AURKC mutations in the patients. There was no major difference observed in the ICSI results obtained from both brothers (Table 2), we therefore comment on the figures obtained from data averaged from the 11 ICSI cycles. Transfers were realised on D2 unless D2 was a Sunday in which case transfer was carried the next day on D3. There was one attempt of blastocyst transfer on D5 but none of the 6 embryos developed to the blastocyst stage, so there were no embryos transferred (Table 2 attempt P2.5). The average number of cumulus–oocyte complexes (COCs) retrieved and of M2 oocytes obtained was good, with an average of 14.4 and 8 respectively. The overall fertilization rate (59%) was lower than what was achieved with testicular sperm for azoospermic patients (around 70%, (De Croo et al., 2000)). After fertilisation most zygotes cleaved and reached the four cell-stage (96%) with 65% of grade 1 or 2 embryos (table 2). One to four embryos were transferred, and in cycle P2.2 (Table 2) an additional transfer of thawed embryos was carried out. There were no pregnancies initiated from any of these cycles.
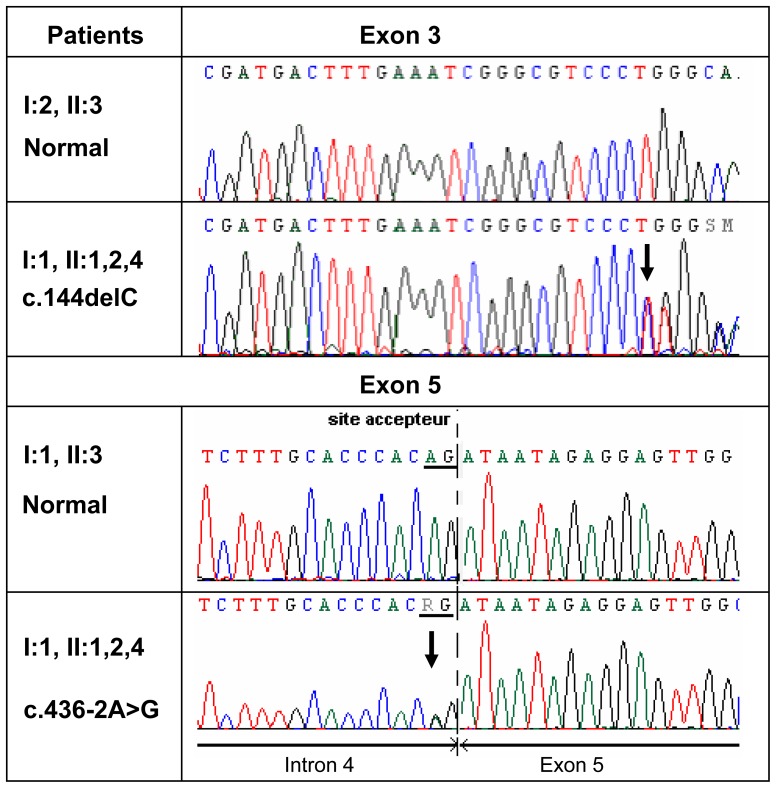
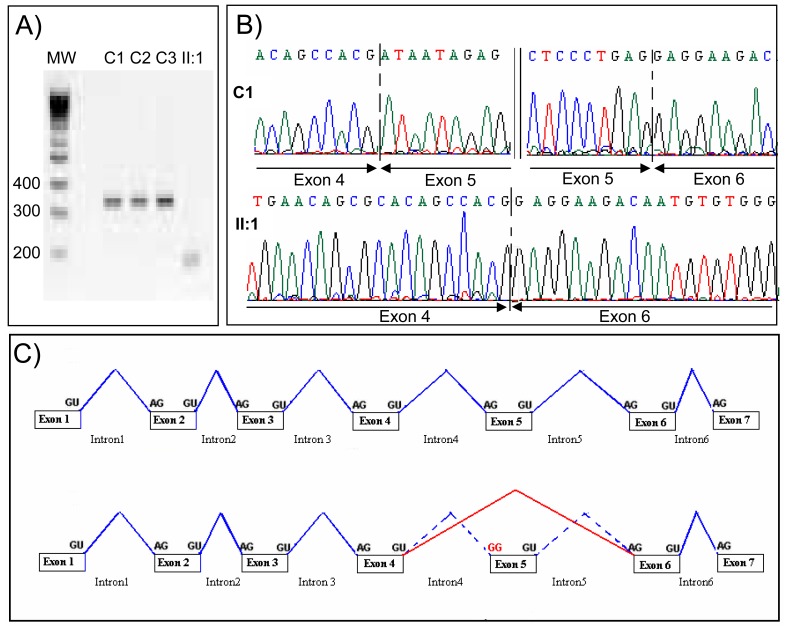
Saliva samples were collected from both patients II:1 and II:2 and family members for AURKC analysis. Sequencing of the AURKC exon 3 containing the c.144delC mutation revealed a single copy of the recurrent deletion in both brothers (Figure 2). The other exons were sequenced and the c.436-2A>G variant was identified in both brothers (Figure 2). This substitution takes place two nucleotides prior to exon 5 and alters a AG consensus acceptor splice signal likely to be crucial for adequate splicing. Both splice-prediction programs http://www.fruitfly.org/seqtools/splice.html and http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi indicated that the substitution would obliterate the acceptor site and that the splicing machinery would skip exon 5. To verify this hypothesis we performed RT-PCR on cDNA obtained from controls and patient II:1. Amplification of a sequence ranging from cDNA exon 4-6 yielded a normal band of 329 bp in controls whereas a single smaller band of 179bp was obtained from II:1 cDNA (Figure 3A). Sequence analysis of the amplified products confirmed that the mRNA from patient II:1 was indeed devoid of exon 5 (Figure 3B) as shown in diagram 3C.
Figure 2. Electropherogram of AURKC exon 3 and 6.
Electropherogram showing the presence of heterozygous mutations c.144delC and c.436-2A>G in individuals I:1, II:1,2,4 and in I:1, II:1,2,4 respectively.
Figure 3. Transcript analysis from control subjects from the general population (C1-C3) and patient II:1, [c.436-2A>G, c.144delC].
A) Electrophoresis showing the RT-PCR amplification of AURKC exon 4-6. Controls C1-C3 yield a normal fragment of 329bp and patient II:1 a shortened fragment of 180bp devoid of exon 5. B) Electropherogram showing the exons boundaries of the bands showed in pannel A. Sequence analysis indicates that exons 4-6 are present in control C1 whereas exon 5 is removed from II:1 transcript. C) Illustrates the exon 5 skipping as observed in patient II:1.
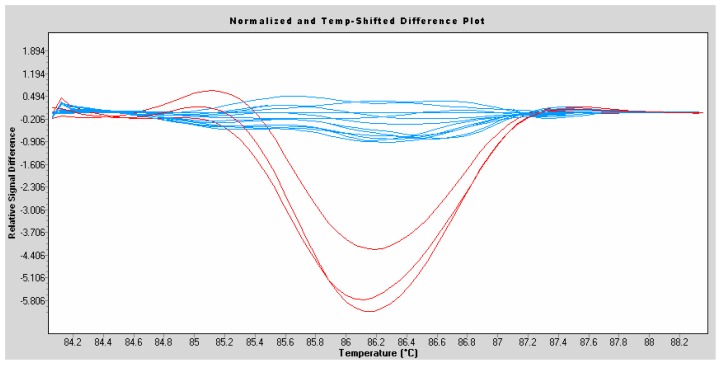
Furthermore, to exclude the possibility that c.436-2A>G may be a common variant in the population studied we analysed AURKC exon 5 from 100 individuals of North African descent. The analysis was carried out by High Resolution Melting (HRM) which highlights the presence of variants in amplified fragments. Three c.436-2A>G heterozygous individuals (II:1,2,4) were tested by HRM and showed a characteristic red profile clearly different from that of all the tested control individuals giving a flat profile in blue (Figure 4). This indicated that none of the 100 North African controls tested had any nucleotide variant in exon 5 or in its bordering intronic sequences.
Figure 4.
High Resolution Melting (HRM) analysis of AURKC exon 5 on subjects from the North African general population (blue) and c.436-2A>G heterozygotes (II:1,2,4) (red).
DISCUSSION
We have demonstrated previously that all patients with a typical macrozoospermia phenotype carried an AURKC mutation. In a series of 32 typical macrozoocephalic patients 31 were homozygous for the c.144delC mutation and one was a compound heterozygote carrying the recurrent mutation and a missense mutation in exon 6, p.Cys229Tyr (Dieterich et al., 2009). Here we characterized two brothers carrying the recurrent mutation and the new c.436-2A>G variant. This mutation is located on a consensus acceptor splice site signal predicted to be necessary for adequate splicing of AURKC mRNA. We demonstrate that a normal mRNA could be consistently amplified from control subjects whereas only a truncated transcript devoid of exon 5 could be amplified by RT-PCR from patient II:1. This confirms the results predicted by the splice site prediction softwares tested which predicted the abrogation of the acceptor site leading the prolongation of intron 5 until the following acceptor site preceding exon 6 (Figure 3C). Sequence analysis of exon 5 truncated product indicates that there is no disruption of the reading frame caused by this splicing mutation. The mutant protein will lack the 50 amino acid coded by exon 5 but there will be no premature stop codon introduced in the coding frame. Amino acid 146-195 will be missing from the protein. They are localised in the middle of the catalytic domain and their absence is therefore very likely to severely hamper the functionality of the protein.
We could also observe that there was no band of normal size corresponding to the c.144del allele. This indicates that the c.144del mutation induced non-sense mRNA decay, at least in leukocytes. Nonsense-mediated mRNA decay (NMD) is a cellular surveillance mechanism that results in the degradation of transcripts containing premature translation termination codons. It also influences the expression of certain wild-type transcripts (for review see (Maquat, 2004)). This mechanism is important to limit the presence of truncated protein which could have a dominant negative effect and are likely to be much more damageable to the cell and the organism than a half amount of gene product expected in heterozygotes. In our previous work we had demonstrated that the surrexpression of a c.144delC mutant protein in Hela cells lead to the production of a 71aa truncated protein stopped by the first stop codon read after the frame-shift deletion. In the Hela cells the mRNA concentration was however approximately 50 times greater in cells transfected with the normal AURKC sequence than with the c.114del mutated clone, indicating the presence of a strong mRNA decay of the c.144delC allele in Hela cells (Dieterich et al., 2007). Here, this mRNA decay is confirmed in vivo on blood leukocytes. This suggests that c.144delC homozygous patients (the vast majority of macrozoocephalic patients yet described) do not have any AURKC protein. As stated previously the new mutation described here does not create any non-sense signal/stop codon in the mRNA sequence and logically we did not observe any non-sense mRNA decay of the mutant allele.
Aurora kinases (AURK A, B and C) are cell cycle regulatory serine/threonine kinases essential to the successful execution of mitotic cell division by ensuring the formation of a bipolar spindle and accurate chromosome segregation (Bischoff et al., 2002). AURKC, shares a high amino-acid sequence identity with AURKB but its expression is almost testis specific (Bernard et al., 1998; Tang et al., 2001) where it is involved in chromatin condensation and proper attachment of homologous chromosomes during the first meiotic division (Tang et al., 2006). Abnormal cell division was observed in vitro upon depletion of AURKB as well as upon overexpression of AURKB and AURKC mutant proteins (Honda et al., 2003; Tatsuka et al., 1998). In each case large multinucleated cells accumulated, reminiscent of the large-headed spermatozoa observed in macrozoospermia. AURKC could rescue the AURKB-silenced multinucleation phenotype, suggesting that its function can overlap with and complement AURKB during mitosis (Sasai et al., 2004). Aurora C knockout mice are viable and males have normal testis weights, but reduced litter size, with some males being sterile. Homozygous male have a higher rate of morphologically abnormal spermatozoa (21%) compared to normal controls (5%) (Kimmins et al., 2007). Abnormalities include heterogeneous chromatin condensation, loose acrosomes and blunted heads. Original work suggested that there might be several AURKC copies in the mouse genome (Hu et al., 2000) and this could explain the mild phenotype observed in AURKC −/− mice (Avo Santos et al.; Yang et al.). Careful search of the updated mouse genome however does not support this information. We therefore believe that the milder phenotype observed in mouse compared to human is likely due to a greater overlap of AURB and C functions in mice spermatogenesis compared to human, allowing for AURKB to compensate the absence of AURKC only in mice. AURKC is also described to be present in both human and mice oocytes. The fecundity of Aurkc−/− female mice was not discussed (Kimmins et al., 2007), presumably because it was not affected, which is what was observed in human (Dieterich et al., 2009). Recent work however indicates that microinjection of a kinase-deficient Aurora-C (AurC-KD) mRNA into mouse oocytes led to the production of cytokinesis failure in meiosis I, resulting in producing large polyploid oocytes, a pattern similar to Aurora-C deficiency human spermatozoa (Yang et al., 2010). The authors conclude that Aurkc but not Aurkb plays an essential role in mouse oogenesis. One can however wonder whether the injected deficient mRNA and the protein it subsequently produces would not interfere with both Aurkc and b, potentially by saturating the physiological localization of both native proteins. The observed phenotype would therefore be equivalent of oocyte specific Aurk b and c double mutant, thus explaining the effect on oogenesis. Whether the action AURKC is necessary for oogenesis therefore remains to be clarified. Interestingly AURKC has also been described to be highly expressed in early human preimplantation embryos (Avo Santos et al., 2011). The authors logically suggest that AURKC is likely involved in chromosome segregation in the first few embryonic divisions and speculate that it could be linked with the high aneuploidy rate observed in preimplantation embryos (Avo Santos et al., 2011). Overall these data confirms the implication of AURKC in gametogenesis and early human reproduction. The question that now need to be addressed is what is the specificity of AURKC compared to AURKB in male meiosis ?
The brothers studied here have a typical macrozoocephalic phenotype but with a low sperm concentration (<1M/ml) whereas the average sperm concentration measured in a series of 32 c.144delC homozygous men was 7.2M/ml. This could indicate that the presence of the abnormal protein – with a truncation of exon 5 – has an effect on cell concentration. On the other hand a few normal looking spermatozoa could be observed and selected for each ICSI attempts. Such spermatozoa were seldom observed in c.144delC homozygous patients indicating that the truncated protein might preserve a small functionality permitting a few spermatozoa to pass meiosis. Careful selection by Motile Sperm Organelle Morphology Examination (MSOME) has previously been applied to select the more normal looking spermatozoa in AURKC c.144delC deleted patients (Chelli et al., 2010). In that study only 6 normal looking spermatozoa were selected and FISH analysis was carried out on these spermatozoa. All six were aneuploid confirming that ICSI should not be attempted for AURKC mutated patients even after a very thorough morphological selection (Chelli et al., 2010). Here, in the course of 11 ICSI attempts a total of 88 normal looking spermatozoa could be used for ICSI. Sperm morphology was assessed by standard light microscopy. As almost all spermatozoa were abnormal, the main criteria for selection and injection was whether the spermatozoa could fit into the ICSI pipette. Some spermatozoa with a slight defect in head or flagella morphology were therefore sometimes selected. Fertilization could be achieved by 47 of these gametes but no pregnancy was obtained after 8 embryo transfer. We can speculate that these embryos were most likely carrying some gross chromosomal abnormalities which prevented sustained embryo development. Our results therefore reinforce our view that ICSI should not be attempted for AURKC mutated men.
We identified here a third mutation in the AURKC gene causing macrozoospermia opening the door for yet more allelic variants of this gene. We demonstrate again that there can be no hope of autologous fertilization for AURKC deficient men who can only be directed towards gamete donation or adoption. Other groups propose a rapid screening of exon 3 only to detect the presence of the recurrent c.144delC mutation in patients with macrozoospermia (El Kerch et al., 2011). Our findings indicate that the molecular analysis should not be stopped after a negative screening of exon 3 and that it is important to sequence the whole of AURKC coding sequence for men with macrozoospemia to assure that no rarer genetic variants will be missed and to avoid unnecessary ICSI cycles for these patients and their spouses. The prognosis for non-mutated men - normally with milder forms of the pathology - is more open. To better assess the reproductive potential of these patients a FISH analysis should be performed on spermatozoa. Depending on the results, ICSI, potentially accompanied by Preimplantation Genetic Diagnosis (PGD) could be proposed as mentioned and realised previously (Kahraman et al., 2004).
Acknowledgments
We thank our patients and family members for their participation. We thank Nabila Ben Khoud for her precious help in sample collection.
FUNDING
This work is part of the project “Identification and Characterization of Genes Involved in Infertility (ICG2I)” funded by the program GENOPAT 2009 from the French Research Agency (ANR). This work was also funded in part by program CIBLE 2009 from the Rhône-Alpes Région.
AUTHOR’S ROLES
MBK and RH undertook all the molecular work. RZ supervised all the sperm analyses and embryo work. LH and RZ oversaw sample collection and recruitment and supervised all clinical aspects of the work, PR, CA, JL, RZ contributed to data analysis, PFR supervised all molecular laboratory work and interpreted the results. PFR designed the overall study, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to the report.
References
- Avo Santos M, van de Werken C, de Vries M, Jahr H, Vromans MJ, Laven JS, Fauser BC, Kops GJ, Lens SM, Baart EB. A role for Aurora C in the chromosomal passenger complex during human preimplantation embryo development. Hum Reprod. 2011 doi: 10.1093/humrep/der111. [DOI] [PubMed] [Google Scholar]
- Benzacken B, Gavelle FM, Martin-Pont B, Dupuy O, Lievre N, Hugues JN, Wolf JP. Familial sperm polyploidy induced by genetic spermatogenesis failure: case report. Hum Reprod. 2001;16:2646–2651. doi: 10.1093/humrep/16.12.2646. [DOI] [PubMed] [Google Scholar]
- Bernard M, Sanseau P, Henry C, Couturier A, Prigent C. Cloning of STK13, a third human protein kinase related to Drosophila aurora and budding yeast Ipl1 that maps on chromosome 19q13.3-ter. Genomics. 1998;53:406–409. doi: 10.1006/geno.1998.5522. [DOI] [PubMed] [Google Scholar]
- Bischoff FZ, Sinacori MK, Dang DD, Marquez-Do D, Horne C, Lewis DE, Simpson JL. Cell-free fetal DNA and intact fetal cells in maternal blood circulation: implications for first and second trimester non-invasive prenatal diagnosis. Hum Reprod Update. 2002;8:493–500. doi: 10.1093/humupd/8.6.493. [DOI] [PubMed] [Google Scholar]
- Chelli MH, Albert M, Ray PF, Guthauser B, Izard V, Hammoud I, Selva J, Vialard F. Can intracytoplasmic morphologically selected sperm injection be used to select normal-sized sperm heads in infertile patients with macrocephalic sperm head syndrome? Fertil Steril. 2010;93:1347 e1341–1345. doi: 10.1016/j.fertnstert.2008.10.059. [DOI] [PubMed] [Google Scholar]
- De Croo I, Van der Elst J, Everaert K, De Sutter P, Dhont M. Fertilization, pregnancy and embryo implantation rates after ICSI in cases of obstructive and non-obstructive azoospermia. Hum Reprod. 2000;15:1383–1388. doi: 10.1093/humrep/15.6.1383. [DOI] [PubMed] [Google Scholar]
- Devillard F, Metzler-Guillemain C, Pelletier R, DeRobertis C, Bergues U, Hennebicq S, Guichaoua M, Sele B, Rousseaux S. Polyploidy in large-headed sperm: FISH study of three cases. Hum Reprod. 2002;17:1292–1298. doi: 10.1093/humrep/17.5.1292. [DOI] [PubMed] [Google Scholar]
- Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sele B, Jouk PS, et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet. 2007;39:661–665. doi: 10.1038/ng2027. [DOI] [PubMed] [Google Scholar]
- Dieterich K, Zouari R, Harbuz R, Vialard F, Martinez D, Bellayou H, Prisant N, Zoghmar A, Guichaoua MR, Koscinski I, et al. The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet. 2009;18:1301–1309. doi: 10.1093/hmg/ddp029. [DOI] [PubMed] [Google Scholar]
- El Kerch F, Lamzouri A, Laarabi FZ, Zahi M, Ben Amar B, Sefiani A. Confirmation of the high prevalence in Morocco of the homozygous mutation c.144delC in the aurora kinase C gene (AURKC) in the teratozoospermia with large-headed spermatozoa. J Gynecol Obstet Biol Reprod (Paris) 2011 doi: 10.1016/j.jgyn.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Escalier D. Human spermatozoa with large heads and multiple flagella: a quantitative ultrastructural study of 6 cases. Biol Cell. 1983;48:65–74. doi: 10.1111/j.1768-322x.1984.tb00203.x. [DOI] [PubMed] [Google Scholar]
- Harbuz R, Lespinasse J, Boulet S, Francannet C, Creveaux I, Benkhelifa M, Jouk PS, Lunardi J, Ray PF. Identification of new FOXP3 mutations and prenatal diagnosis of IPEX syndrome. Prenat Diagn. 2010;30:1072–1078. doi: 10.1002/pd.2613. [DOI] [PubMed] [Google Scholar]
- Harbuz R, Zouari R, Pierre V, Ben Khelifa M, Kharouf M, Coutton C, Merdassi G, Abada F, Escoffier J, Nikas Y, et al. A Recurrent Deletion of DPY19L2 Causes Infertility in Man by Blocking Sperm Head Elongation and Acrosome Formation. Am J Hum Genet. 2011;88:351–361. doi: 10.1016/j.ajhg.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HM, Chuang CK, Lee MJ, Tseng TC, Tang TK. Genomic organization, expression, and chromosome localization of a third aurora-related kinase gene, Aie1. DNA Cell Biol. 2000;19:679–688. doi: 10.1089/10445490050199063. [DOI] [PubMed] [Google Scholar]
- In’t Veld PA, Broekmans FJ, de France HF, Pearson PL, Pieters MH, van Kooij RJ. Intracytoplasmic sperm injection (ICSI) and chromosomally abnormal spermatozoa. Hum Reprod. 1997;12:752–754. doi: 10.1093/humrep/12.4.752. [DOI] [PubMed] [Google Scholar]
- Kahraman S, Sertyel S, Findikli N, Kumtepe Y, Oncu N, Melil S, Unal S, Yelke H, Vanderzwalmen P. Effect of PGD on implantation and ongoing pregnancy rates in cases with predominantly macrocephalic spermatozoa. Reprod Biomed Online. 2004;9:79–85. doi: 10.1016/s1472-6483(10)62114-1. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Crosio C, Kotaja N, Hirayama J, Monaco L, Hoog C, van Duin M, Gossen JA, Sassone-Corsi P. Differential functions of the aurora-B and aurora-C kinases in Mammalian spermatogenesis. Mol Endocrinol. 2007;21:726–739. doi: 10.1210/me.2006-0332. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Mateu E, Rodrigo L, Prados N, Gil-Salom M, Remohi J, Pellicer A, Rubio C. High incidence of chromosomal abnormalities in large-headed and multiple-tailed spermatozoa. J Androl. 2006;27:6–10. doi: 10.2164/jandrol.05033. [DOI] [PubMed] [Google Scholar]
- Nistal M, Paniagua R, Herruzo A. Multi-tailed spermatozoa in a case with asthenospermia and teratospermia. Virchows Arch B Cell Pathol. 1977;26:111–118. doi: 10.1007/BF02889540. [DOI] [PubMed] [Google Scholar]
- Pieters MH, Speed RM, de Boer P, Vreeburg JT, Dohle G, In’t Veld PA. Evidence of disturbed meiosis in a man referred for intracytoplasmic sperm injection. Lancet. 1998;351:957. [PubMed] [Google Scholar]
- Sasai K, Katayama H, Stenoien DL, Fujii S, Honda R, Kimura M, Okano Y, Tatsuka M, Suzuki F, Nigg EA, et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil Cytoskeleton. 2004;59:249–263. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- Tang CJ, Chuang CK, Hu HM, Tang TK. The zinc finger domain of Tzfp binds to the tbs motif located at the upstream flanking region of the Aie1 (aurora-C) kinase gene. J Biol Chem. 2001;276:19631–19639. doi: 10.1074/jbc.M100170200. [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev Biol. 2006;290:398–410. doi: 10.1016/j.ydbio.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Tatsuka M, Katayama H, Ota T, Tanaka T, Odashima S, Suzuki F, Terada Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58:4811–4816. [PubMed] [Google Scholar]
- Yang KT, Li SK, Chang CC, Tang CJ, Lin YN, Lee SC, Tang TK. Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell. 2010;21:2371–2383. doi: 10.1091/mbc.E10-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]