Dear Editor,
Human respiratory syncytial virus (HRSV) is the leading viral agent that causes acute lower respiratory infections (ALRIs) in infants, young children, and vulnerable adults. HRSVs have been classified as subgroups A and B by both antigenic and genetic analyses. Both HRSV subgroups A and B have been circulating in China and have caused large outbreaks of severe ALRIs in young children.1,2 Studies on the molecular epidemiology of HRSVs have mainly focused on the second hypervariable region (HVR2), which contains multiple epitopes, in the ectodomain of the major attachment (G) protein.
To date, 11 HRSV-A genotypes (GA1-GA7, SAA1, NA1-NA2, and ON1) and 20 HRSV-B genotypes (GB1-GB4, BA1-BA10, SAB1-SAB4, URU1, and URU2) have been identified based on the sequence analyses of HVR2.3,4 Genotype BA of subgroup B, with a 60-nucleotide duplication in HVR2, was first identified in Buenos Aires and rapidly became the predominant strain of subgroup B.5 Recently, a new genotype, ON1, for subgroup A, with a 72-nucleotide duplication in HVR2, was first reported in Canada.4 However, it is not known whether this new genotype could replace the current genotype and become the predominant one or whether this emerging ON1 could show more pathogenesis, which is information that would be of high interest. This letter reports the first identification of genotype ON1 sequences in clinical specimens from children with ALRI in Beijing in 2012 during the investigation of the subgroup distribution and genetic variability of HRSV in paediatric patients. The rapid global transmission and genetic variation of the ON1 genotype are also discussed.
A total of 3391 nasal pharyngeal aspirates (NPAs) were collected from children with ALRIs hospitalised at the Affiliated Children's Hospital, Capital Institute of Pediatrics in Beijing, China, during the period from July 2012 to December 2012. Of these NPA samples, 285 (8.4%, 285/3391) were positive for HRSV by direct fluorescent assay (Diagnostic Hybrids, Ohio, USA), including 11 samples for which the virus was successfully isolated in Hep-2 cells. A total of 99 of the 285 HRSV-positive NPA samples were randomly selected for subtyping using multiplex reverse transcription polymerase chain reaction; 10 and 79 NPA samples were identified as subgroup A and subgroup B, respectively. The 11 HRSV strains isolated by cell culture consisted of 2 from subgroup A and 9 from subgroup B, suggesting that subgroup B was predominant in the 2012/2013 HRSV season.
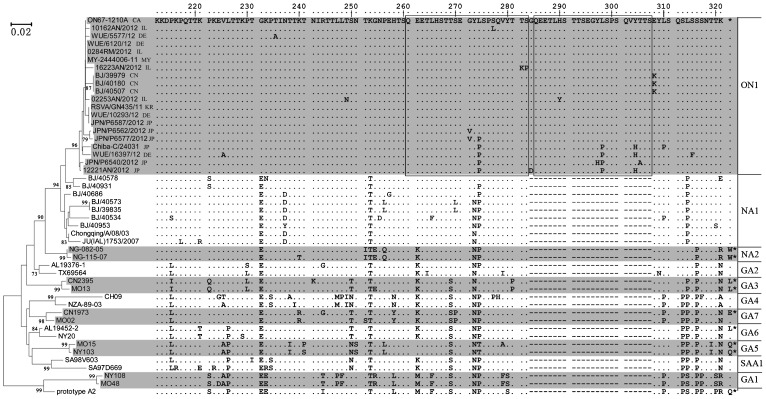
The nucleotide sequences of HVR2 from the Beijing subgroup A-positive specimens were obtained and aligned with the HVR2 sequences from other reference genotypes (Figure 1). An unrooted phylogenetic tree was constructed using the ClustalW program and the neighbour-joining method in the MEGA5 software package. Of the 10 Beijing HRSV subgroup A isolates, 7 belonged to the NA1 genotype, and 3 (BJ/39979, BJ/40507, and BJ/40180) isolated in November 2012 were identified as genotype ON1, containing the 72-nucleotide duplication in HVR2 (Figure 1). NA1 has been reported as the prevailing genotype for subgroup A in Japan,6 Cambodia,3 Malaysia,7 and Canada.4 The 3 Beijing ON1 strains had identical nucleotide sequences for the complete G genes and shared 99.4% nucleotide similarity with prototype ON1 strain ON67-1210A. All 3 of these HRSV ON1-infected patients (2 boys and 1 girl) were younger than 6 months and diagnosed with pneumonia.
Figure 1.
Unrooted phylogenetic tree for HRSV subgroup A nucleotide sequences of HVR2 and their deduced amino acid alignment. The Beijing strains are indicated with “BJ/”, followed by their isolation number (GenBank accession numbers KC461212, KC461213, and KC559441-KC559448). The reference HVR2 sequences of strains with identified genotypes were downloaded from GenBank. The ON1 strain names are followed by their country of collection: Canada (CA), China (CN), Germany (DE), Italy (IL), Japan (JP), South Korea (KR), and Malaysia (MY). The tree was constructed using the neighbour-joining method, with 1000 bootstrap replicates, in Mega5 software. Only bootstrap values of >70% are shown at the branch nodes. The scale bar indicates the number of nucleotide substitutions per site. The amino acid numbering is shown relative to genotype ON1 strain ON67-1210A (GenBank accession no. JN257693). Identical residuals, alignment gaps, and stop codons are indicated with dots, dashes, and asterisks, respectively. Two copies of the duplicated segments in the ON1 strains are framed with rectangles. The genotype assignment is indicated at the end of the alignment.
The HRSV genotype ON1 was first identified in specimens collected at the end of 2010 in Canada.4 To the best of our knowledge, the earliest ON1 sequence from a non-Canadian isolate was detected in Malaysia in November 2011 and had an HVR2 nucleotide sequence that was identical to that of the prototype ON1 strain ON67-1210A.7 In the ensuing years, the ON1 genotype has been found in South Korea,8 Italy (GenBank accession numbers JX988439-JX988452), Japan (GenBank accession numbers AB761609-AB761611 and AB698559), Germany (GenBank accession numbers JX912356-JX912364) and South Africa,9 indicating its rapid global dissemination (Figure 1 and Supplementary Figure S1). Furthermore, surveillance work is needed to investigate whether genotype ON1 is able to cause larger outbreaks and become the predominate genotype for HRSV subgroup A worldwide. Indeed, the BA genotype of subgroup B, with the 60-nucleotide duplication, was found worldwide within a few months since it was first identified in Buenos Aires and has become the predominant genotype of subgroup B.2,5,10
Despite first being identified in Canada, the geographic and temporal origin of the ON1 genotype remains unclear. We did not find any genotype ON1 isolate through the complete G genes sequence analysis in the 189 subgroup A-positive clinical specimens collected between 2007 and 2011 (data not shown). Thus, it is of interest to know whether HRSV with a genotype ON1 sequence emerged in Beijing, China, in 2012.
The alignment of the deduced amino acids of the ON1 HVR2 sequences available in GenBank revealed that amino acid substitutions have occurred in the duplicated segment (Figure 1). For example, three strains from Japan (Chiba-C/24031, JPN/P6540/2012, and 12221/AN/2012) and one German strain (WUE/16397/12) contained 3 to 4 amino acid substitutions in the duplicated region and are, thus, less closely related to the original ON1 strain than the 3 Beijing ON1 strains. This difference reflects the high genetic variability of the ON1 strains to escape herd immunity in different populations. It can be predicted that, as found for the BA genotype,10 several branches of the ON1 genotype will form in the future due to the rapid accumulation of changes in the sequence.
The virulence and immunogenicity of the ON1 strains may change because of the 72 nucleotide G gene duplication. Ongoing surveillance of this genotype around the world is necessary to understand not only the evolution of this important virus but also the relationship between epidemic progress and pathogenesis.
This study was reviewed and approved by the Institutional Review Board of the Capital Institute of Pediatrics.
Acknowledgments
This work was supported by grant no. Z111107056811041 from the Beijing Municipal Science and Technology Commission.
Supplementary Information
References
- Deng J, Qian Y, Liu C, et al. [Etiological study on an outbreak of epidemic asthma-like pneumonia of infants and children in Ruyang county, Henan province, China.] Chin J Pediatr 20013973–75.Chinese. [Google Scholar]
- Deng J, Qian Y, Zhu R, Wang F, Zhao L.[Surveillance for respiratory syncytial virus subtypes A and B in children with acute respiratory infections in Beijing during 2000 to 2006 seasons.] Zhonghua Er Ke Za Zhi 200644924–927.Chinese. [PubMed] [Google Scholar]
- Arnott A, Vong S, Mardy S, et al. A Study of the Genetic Variability of Human Respiratory Syncytial Virus (HRSV) in Cambodia Reveals the Existence of a New HRSV Group B Genotype. J Clin Microbiol. 2011;49:3504–3513. doi: 10.1128/JCM.01131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A, Duvvuri VR, Lai R, et al. Genetic variability of human respiratory syncytial virus a strains circulating in ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One. 2012;7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trento A, Casas I, Calderon A, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84:7500–7512. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobugawa Y, Saito R, Sano Y, et al. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J Clin Microbiol. 2009;47:2475–2482. doi: 10.1128/JCM.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor CS, Sam IC, Hooi PS, Chan YF. Displacement of predominant respiratory syncytial virus genotypes in Malaysia between 1989–2011. Infect Genet Evol. 2013;14:357–360. doi: 10.1016/j.meegid.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Kim YJ, Kim DW, Lee HS, Lee HY, Kim K. Complete genome sequence of human respiratory syncytial virus genotype a with a 72-nucleotide duplication in the attachment protein G gene. J Virol. 2012;86:13810–13811. doi: 10.1128/JVI.02571-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley-Omar Z, Muloiwa R, Hu NC, Eley B, Hsiao NY. Novel Respiratory Syncytial Virus Subtype ON1 among Children, Cape Town, South Africa, 2012. Emerg Infect Dis. 2013;19:668–670. doi: 10.3201/eid1904.121465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trento A, Viegas M, Galiano M, et al. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol. 2006;80:975–984. doi: 10.1128/JVI.80.2.975-984.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.