Abstract
The movement of positively charged S4 segments through the electric field drives the voltage-dependent gating of ion channels. Studies of prokaryotic sodium channels provide a mechanistic view of activation facilitated by electrostatic interactions of negatively charged residues in S1 and S2 segments, with positive counterparts in the S4 segment. In mammalian sodium channels, S4 segments promote domain-specific functions that include activation and several forms of inactivation. We tested the idea that S1–S3 countercharges regulate eukaryotic sodium channel functions, including fast inactivation. Using structural data provided by bacterial channels, we constructed homology models of the S1–S4 voltage sensor module (VSM) for each domain of the mammalian skeletal muscle sodium channel hNaV1.4. These show that side chains of putative countercharges in hNaV1.4 are oriented toward the positive charge complement of S4. We used mutagenesis to define the roles of conserved residues in the extracellular negative charge cluster (ENC), hydrophobic charge region (HCR), and intracellular negative charge cluster (INC). Activation was inhibited with charge-reversing VSM mutations in domains I–III. Charge reversal of ENC residues in domains III (E1051R, D1069K) and IV (E1373K, N1389K) destabilized fast inactivation by decreasing its probability, slowing entry, and accelerating recovery. Several INC mutations increased inactivation from closed states and slowed recovery. Our results extend the functional characterization of VSM countercharges to fast inactivation, and support the premise that these residues play a critical role in domain-specific gating transitions for a mammalian sodium channel.
INTRODUCTION
Voltage-gated sodium channels initiate the action potential in excitable tissues (Armstrong and Hille, 1998; Catterall, 2012). These channels respond to membrane depolarization by activating, followed rapidly by an inactivating transition that limits the duration of the action potential (Hodgkin and Huxley, 1952). The gene sequence for the sodium channel reveals a pattern of regularly spaced arginine and lysine residues in conserved S4 transmembrane segments (Noda et al., 1984). Channel gating is perturbed with mutations of these positively charged residues, suggesting that S4 segments act as voltage sensors to promote activation and initiate fast inactivation (Stühmer et al., 1989). Additional experimental approaches have supported the premise that S4 segments carry the gating charge that translocates outward in response to membrane depolarization in sodium and other voltage-gated ion channels (Shao and Papazian, 1993, Perozo et al., 1994; Yang et al., 1996; Horn et al., 2000). Domain-specific roles for sodium channel S4 segments are suggested by experiments demonstrating that S4 translocation is coupled to fast inactivation (Chahine et al., 1994; Chen et al., 1996; Cha et al., 1999; Sheets et al., 1999), to an intracellular activation gate (Muroi et al., 2010), and to the selectivity filter (Capes et al., 2012).
Electrostatic interactions of S4 positive charges with countercharges in surrounding segments S1, S2, and S3 have been proposed as the stabilizing mechanism facilitating S4 segment translocation. In this structural view, S1–S4 segments comprise a voltage sensor module (VSM) that surrounds a central, S5–S6 pore module (Chanda and Bezanilla, 2008). Experimental evidence for functional interactions within the VSM was first described for Shaker potassium channel (Papazian et al., 1995) and subsequently extended to explain gating transitions in Shaker and other potassium channels (Tiwari-Woodruff et al., 2000; Campos et al., 2007a; Zhang et al., 2007; Wu et al., 2010; Pless et al., 2011). In sodium channels, specific interactions of S4 residues with negative countercharges in S1 and S2 segments have been identified in the prokaryotic channel NachBac (DeCaen et al., 2008, 2009, 2011). There, interactions within the VSM define the resting, closed state of the channel (Paldi and Gurevitz, 2010), with sequential ion pair interactions during depolarization that stabilize a series of activated states (Chakrapani et al., 2010; Yarov-Yarovoy et al., 2012).
In this work, we sought to determine the functional roles for putative countercharges in the VSM of a mammalian sodium channel, hNaV1.4. Eukaryotic voltage-gated sodium channels are comprised of four homologous domains with asymmetric S4 charge content (Trimmer et al., 1989). We wished to identify the contributions of negative countercharges in S1–S3 segments of hNaV1.4 toward domain-specific channel functions. To do this, we built sequence alignments and constructed homology models for each of four VSMs in hNaV1.4, based on the structural data from bacterial sodium channels NaVAb (Payandeh et al., 2011, 2012) and NaVRh (Zhang et al., 2012). Our models predict that for each domain, negative countercharges in S1, S2, and S3 segments are positioned appropriately to interact with positive charges in the S4 segment. We then tested the role of VSM countercharges in hNaV1.4 using site-directed mutagenesis and voltage clamp electrophysiology. We found that charge-reversing mutations in S1–S3 segments alter sodium channel activation and fast inactivation in a manner consistent with our hypothesis that negative countercharges are an important biophysical substrate of domain-specific voltage gating in the mammalian sodium channel. Portions of this work have appeared in abstract form (Groome, J.R., and V. Winston. 2012. Biophysical Society 56th Annual Meeting. Abstr. 425b).
MATERIALS AND METHODS
Alignments and homology modeling
The crystal structure of the bacterial voltage gated sodium channel NaVAb (Payandeh et al., 2011) was downloaded from the Protein Database site (accession no. 3RVY; http://www.rcsb.org). Using DeepView (Swiss pdb viewer http://spdbv.vital-it.ch/), the VSM of the A chain of 3RVY.pdb was saved as a separate file (3RVY_A_sensor.pdb), used as a template for subsequent modeling. The amino acid sequence of the α subunit of human sodium channel protein type 4 (NP_000325) was downloaded from GenBank. Sequences of VSMs for domains I–IV were identified and saved as individual FASTA formatted files.
Domain I–IV sequences were aligned to each other and to 3RVY_A_sensor.pdb, using multiple sequence alignment in MUSCLE (http://www.drive5.com/muscle/). Each of the hNaV1.4 VSM sequences was paired with the aligned amino acid sequence of 3RVY_A_sensor.pdb, using the PIR format required by Modeller. Using the Modeller script model-single.py, a homology model structure was predicted for each hNaV1.4 VSM. “Best” structures were identified using Modeller-derived GA341 scores and aligned with bacterial structures using the VMD multiseq plugin.
Homology models of the VSMs in NaVAb(2) (inactivated state of NaVAb; Payandeh et al., 2012) and NaVRh (NachBac orthologue; Zhang et al., 2012) were constructed using the same approach as for NaVAb. Template structures were derived from 4EKW.pdb (accession no. 4EKW; NaVAb(2)) or 4DXW.pdb (accession no. 4DXW; NaVRh) to construct homology models of the VSM for hNaV1.4 in each domain.
Molecular biology
Mutations of the α subunit of human skeletal muscle sodium channel SCN4A were created with QuikChange XL II Site Directed Mutagenesis kits (Agilent Technologies) by incorporating mutant oligonucleotides into template SCN4A in vector pSP64T. Messenger RNA for α subunit was constructed from template linearized with EcoR1, using SP6 mMESSAGE mMACHINE in vitro transcription kits (Ambion). For all constructs, we coexpressed α subunit with β1 subunit that associates with sodium channels and modulates fast inactivation (Yang et al., 1993). β subunit in vector pgH19 was linearized with HindIII, and transcribed with T3 RNA polymerase. mRNA was injected into Xenopus laevis oocytes at a ratio of 3:1 β1 to α subunit in a volume of 50 nL. Animals were maintained and surgical procedures were performed using protocols approved by the IACUC committee at Idaho State University. Oocytes were maintained in sterile media containing 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 4 mM CaCl2, 5 mM Hepes, 5 mM sodium pyruvate, 3% horse serum, and 100 mg/liter gentamicin, pH 7.4, with gentle agitation at 17.5°C, for 3–7 d before recordings.
Electrophysiology
Experiments were performed at 15°C, in the cut-open oocyte clamp configuration, with a CA-1B amplifier (Dagan Corporation). External solution consisted of 120 mM N-methyl d-glucamine, 2 mM Ca(OH)2, and 5 mM Hepes, pH 7.4. Internal solution consisted of 120 mM N-methyl d-glucamine, 2 mM EGTA, and 5 mM Hepes, pH 7.4. Before voltage clamp, the oocyte was permeabilized with 0.3% saponin to provide electrical access. Membrane potential was held at −120 mV, with capacitance and leak subtraction from a p/8 protocol, using Pulse 8.67 (HEKA). Sodium currents were analyzed with PulseFit 8.67 (HEKA) and Igor Pro 6.02 (WaveMetrics). Statistical analyses were done with Instat 2.0 (GraphPad Software) using Student’s t tests of equilibrium parameters (Table 1), and of kinetic parameters at a given voltage (Table 2).
Table 1.
Equilibrium parameters for wild-type and mutant sodium channels
| Mutation | Location | V0.5 | Slope factor | ΔΔGo | h0.5 | Slope factor |
| mV | kcal/mol | mV | ||||
| hNaV1.4 | 18.3 ± 1.2 | 1.39 ± 0.03 | −65.8 ± 1.1 | 4.24 ± 0.12 | ||
| ENC | ||||||
| S151K | Domain I S1 | 23.2 ± 0.8b | 1.37 ± 0.06 | 0.6 | −66.5 ± 0.7 | 4.67 ± 0.06b |
| D152R | Domain I S1 | 23.2 ± 0.9b | 1.37 ± 0.03 | 0.6 | −64.1 ± 0.6 | 4.11 ± 0.25 |
| E161R | Domain I S2 | 23.8 ± 1.0c | 1.57 ± 0.03d | 1.1 | −60.5 ± 0.6c | 3.82 ± 0.08b |
| E598K | Domain II S1 | 36.3 ± 0.9d | 1.59 ± 0.05b | 3.0 | −59.8 ± 1.4b | 2.90 ± 0.14d |
| N614K | Domain II S2 | 30.3 ± 0.8d | 1.52 ± 0.03b | 1.9 | −61.0 ± 0.6d | 3.84 ± 0.20 |
| E1051R | Domain III S1 | 26.0 ± 0.7d | 1.60 ± 0.02d | 1.5 | −48.4 ± 1.2d | 3.16 ± 0.16d |
| E1051D | Domain III S1 | 25.7 ± 1.3d | 1.39 ± 0.03 | 1.0 | −62.1 ± 1.2a | 3.98 ± 0.15 |
| D1069K | Domain III S2 | 30.0 ± 0.8d | 1.61 ± 0.04c | 2.1 | −49.3 ± 1.9d | 3.56 ± 0.13c |
| D1069E | Domain III S2 | 19.2 ± 1.2 | 1.43 ± 0.02 | 0.2 | −64.4 ± 0.7 | 2.88 ± 0.11d |
| E1373K | Domain IV S1 | 15.2 ± 1.0 | 1.73 ± 0.03d | 0.1 | −50.3 ± 0.7d | 2.47 ± 0.07d |
| E1373N | Domain IV S1 | 18.1 ± 1.2 | 1.60 ± 0.02d | 0.3 | −49.9 ± 0.9d | 2.41 ± 0.06d |
| E1373D | Domain IV S1 | 20.4 ± 0.9a | 1.39 ± 0.02 | 0.3 | −65.8 ± 0.7 | 2.81 ± 0.07d |
| N1389K | Domain IV S2 | 18.6 ± 1.1 | 1.65 ± 0.03d | 0.5 | −48.7 ± 0.9d | 2.58 ± 0.12d |
| N1389E | Domain IV S2 | 21.8 ± 1.0a | 1.31 ± 0.01b | 0.3 | −71.5 ± 1.0c | 4.59 ± 0.11a |
| N1389D | Domain IV S2 | 19.2 ± 0.7 | 1.33 ± 0.02 | 0.0 | −75.4 ± 0.6d | 2.66 ± 0.03d |
| HCR | ||||||
| N144K | Domain I S1 | 35.4 ± 1.2d | 1.58 ± 0.07a | 2.8 | −61.7 ± 0.9b | 3.69 ± 0.70b |
| N144R | Domain I S1 | 34.2 ± 1.2d | 1.40 ± 0.03 | 2.1 | −62.3 ± 0.5b | 4.06 ± 0.09 |
| N144D | Domain I S1 | 20.2 ± 0.7 | 1.19 ± 0.01d | −0.1 | −72.0 ± 0.5d | 5.13 ± 0.09d |
| N144E | Domain I S1 | 20.1 ± 0.9 | 1.29 ± 0.02b | 0.1 | −67.7 ± 0.3 | 5.06 ± 0.09d |
| N591K | Domain II S1 | 34.3 ± 1.7d | 1.30 ± 0.03a | 1.8 | −58.5 ± 1.0d | 2.89 ± 0.13d |
| N591R | Domain II S1 | 34.4 ± 1.1d | 1.38 ± 0.03 | 2.0 | −61.0 ± 0.4c | 5.93 ± 0.15d |
| N591D | Domain II S1 | 32.0 ± 1.1d | 1.50 ± 0.04a | 2.1 | −61.2 ± 0.4c | 4.54 ± 0.07a |
| N591E | Domain II S1 | 18.8 ± 1.3 | 1.42 ± 0.02 | 0.1 | −63.8 ± 0.7 | 4.52 ± 0.11 |
| S1044K | Domain III S1 | 23.6 ± 0.8c | 1.43 ± 0.03 | 0.8 | −57.9 ± 0.6d | 4.00 ± 0.13 |
| S1044R | Domain III S1 | 30.4 ± 0.7d | 1.42 ± 0.02 | 1.6 | −49.8 ± 0.6d | 4.22 ± 0.10 |
| S1044N | Domain III S1 | 22.8 ± 0.6b | 1.39 ± 0.02 | 0.6 | −60.6 ± 0.6c | 4.55 ± 0.07a |
| S1044D | Domain III S1 | 21.4 ± 1.2 | 1.32 ± 0.02a | 0.2 | −70.9 ± 0.5c | 3.99 ± 0.09 |
| N1366K | Domain IV S1 | 19.3 ± 1.6 | 1.41 ± 0.02 | 0.2 | −64.2 ± 1.2 | 3.88 ± 0.08a |
| N1366R | Domain IV S1 | 21.5 ± 1.4 | 1.51 ± 0.11c | 0.7 | −58.3 ± 0.8d | 2.64 ± 0.04d |
| N1366D | Domain IV S1 | 26.3 ± 1.7c | 1.25 ± 0.04b | 0.7 | −85.8 ± 0.6d | 3.37 ± 0.09d |
| N1366E | Domain IV S1 | 21.8 ± 1.0a | 1.41 ± 0.02 | 0.5 | −71.2 ± 1.3b | 3.44 ± 0.08d |
| INC | ||||||
| E171R | Domain I S2 | 36.7 ± 2.1d | 1.24 ± 0.05b | 1.9 | −75.7 ± 1.7d | 3.79 ± 0.25 |
| K175E | Domain I S2 | 22.1 ± 1.2b | 1.37 ± 0.02a | 0.5 | −65.0 ± 0.9 | 4.24 ± 0.14 |
| N194K | Domain I S3 | 19.1 ± 1.0 | 1.33 ± 0.02 | 0.0 | −68.7 ± 0.8a | 2.88 ± 0.04d |
| D197R | Domain I S3 | 35.1 ± 1.5d | 1.28 ± 0.04b | 1.8 | −76.1 ± 1.1d | 4.44 ± 0.24 |
| E624K | Domain II S2 | 26.5 ± 1.2d | 1.44 ± 0.03 | 1.2 | −63.7 ± 0.7 | 5.13 ± 0.14d |
| K628E | Domain II S2 | 18.3 ± 0.9 | 1.34 ± 0.02c | −0.1 | −68.9 ± 0.9a | 3.95 ± 0.11 |
| N643K | Domain II S3 | 17.7 ± 0.4 | 1.41 ± 0.02 | 0.0 | −61.2 ± 0.8b | 4.51 ± 0.08 |
| D646K | Domain II S3 | 27.6 ± 1.2d | 1.41 ± 0.03 | 1.2 | −64.2 ± 0.6 | 3.95 ± 0.13 |
| E1079R | Domain III S2 | 21.6 ± 0.9a | 1.57 ± 0.05b | 0.8 | −63.4 ± 1.1 | 3.86 ± 0.16 |
| K1083E | Domain III S2 | 20.7 ± 1.0a | 1.39 ± 0.02 | 0.3 | −68.9 ± 0.7b | 3.71 ± 0.11a |
| C1098K | Domain III S3 | 23.1 ± 1.0b | 1.27 ± 0.02c | 0.4 | −75.7 ± 0.6d | 3.03 ± 0.11d |
| D1101V | Domain III S3 | 20.2 ± 1.3 | 1.44 ± 0.02 | 0.3 | −63.4 ± 1.0 | 4.54 ± 0.10 |
| E1399K | Domain IV S2 | 20.2 ± 0.8 | 1.47 ± 0.02a | 0.4 | −61.0 ± 0.9b | 3.29 ± 0.08d |
| K1403E | Domain IV S2 | 20.7 ± 1.6 | 1.50 ± 0.04 | 0.5 | −63.9 ± 1.0 | 3.03 ± 0.11d |
| N1417K | Domain IV S3 | 24.2 ± 0.9c | 1.38 ± 0.01 | 0.7 | −69.4 ± 0.6b | 3.77 ± 0.06b |
| D1420K | Domain IV S3 | 22.4 ± 1.2a | 1.57 ± 0.04c | 0.9 | −70.1 ± 0.8b | 2.83 ± 0.09d |
Mean values ± SEM from Boltzmann fits of I/V relations in 10–24 experiments. The free energy change in wild-type and mutant channels is calculated from 4zFV0.5. The change in free energy of activation produced by mutation is given as ΔΔG0.
P ≤ 0.05.
P ≤ 0.01.
P ≤ 0.001.
P ≤ 0.0001.
Table 2.
Kinetic parameters for activation (10–90% time to peak current, 20 mV), time constants of entry (20 mV), and recovery from fast inactivation (−100 mV), and delay in onset to recovery (−100 mV)
| Mutation | Location | Time to peak activation | Fast inactivation tau | Recovery tau | Recovery delay |
| ms | ms | ms | ms | ||
| hNaV1.4 | 1.05 ± 0.03 | 1.26 ± 0.03 | 7.15 ± 0.54 | 1.04 ± 0.05 | |
| ENC | |||||
| S151K | Domain I S1 | 1.33 ± 0.03d | 1.23 ± 0.05 | 11.3 ± 0.61d | 2.18 ± 0.16d |
| D152R | Domain I S1 | 1.04 ± 0.04 | 1.15 ± 0.06 | 8.09 ± 0.48 | 1.42 ± 0.19a |
| E161R | Domain I S2 | 1.18 ± 0.03b | 1.31 ± 0.07 | 5.34 ± 0.25b | 0.95 ± 0.06 |
| E598K | Domain II S1 | 1.09 ± 0.07 | 2.13 ± 0.27b | 5.17 ± 0.48a | 1.09 ± 0.10 |
| N614K | Domain II S2 | 1.15 ± 0.06 | 1.65 ± 0.10b | 5.50 ± 0.38a | 0.86 ± 0.05a |
| E1051R | Domain III S1 | 1.55 ± 0.14b | 3.25 ± 0.55b | 3.61 ± 0.24d | 0.79 ± 0.07b |
| E1051D | Domain III S1 | 1.05 ± 0.04 | 1.23 ± 0.06 | 5.51 ± 0.30a | 1.17 ± 0.10 |
| D1069K | Domain III S2 | 1.18 ± 0.09 | 2.92 ± 0.46b | 3.42 ± 0.16d | 0.84 ± 0.05b |
| D1069E | Domain III S2 | 1.18 ± 0.05a | 1.94 ± 0.16b | 6.31 ± 0.36 | 1.15 ± 0.09 |
| E1373K | Domain IV S1 | 1.65 ± 0.07d | 5.51 ± 0.31d | 3.42 ± 0.39d | 0.44 ± 0.07d |
| E1373N | Domain IV S1 | 1.54 ± 0.07d | 4.94 ± 0.18d | 3.70 ± 0.23d | 0.46 ± 0.02d |
| E1373D | Domain IV S1 | 1.24 ± 0.03d | 2.74 ± 0.22d | 4.85 ± 0.22c | 0.81 ± 0.05b |
| N1389K | Domain IV S2 | 1.85 ± 0.12c | 4.14 ± 0.29d | 4.31 ± 0.42c | 0.42 ± 0.02d |
| N1389E | Domain IV S2 | 1.26 ± 0.04d | 1.79 ± 0.07d | 8.36 ± 0.55 | 1.25 ± 0.07a |
| N1389D | Domain IV S2 | 1.21 ± 0.02d | 1.52 ± 0.04d | 10.66 ± 0.39d | 1.21 ± 0.10 |
| HCR | |||||
| N144K | Domain I S1 | 1.65 ± 0.11d | 2.51 ± 0.30c | 6.19 ± 0.37 | 1.46 ± 0.48c |
| N144R | Domain I S1 | 1.54 ± 0.02d | 1.70 ± 0.08d | 7.63 ± 0.30 | 0.94 ± 0.25 |
| N144D | Domain I S1 | 1.02 ± 0.01 | 1.11 ± 0.04c | 9.60 ± 1.1a | 1.39 ± 0.11c |
| N144E | Domain I S1 | 1.05 ± 0.02 | 1.15 ± 0.04a | 7.19 ± 0.60 | 1.09 ± 0.05 |
| N591K | Domain II S1 | 1.56 ± 0.09d | 2.62 ± 0.21d | 5.68 ± 0.31a | 0.97 ± 0.06 |
| N591R | Domain II S1 | 1.43 ± 0.02d | 1.33 ± 0.04 | 5.51 ± 0.28a | 0.88 ± 0.07 |
| N591D | Domain II S1 | 1.25 ± 0.03d | 1.09 ± 0.03c | 6.90 ± 0.43 | 1.00 ± 0.06 |
| N591E | Domain II S1 | 1.15 ± 0.04a | 1.11 ± 0.03b | 7.90 ± 0.59 | 1.53 ± 0.12c |
| S1044K | Domain III S1 | 1.20 ± 0.03b | 1.63 ± 0.17a | 6.46 ± 0.41 | 1.14 ± 0.04 |
| S1044R | Domain III S1 | 1.30 ± 0.03d | 1.55 ± 0.03d | 4.49 ± 0.25c | 0.67 ± 0.03d |
| S1044D | Domain III S1 | 1.23 ± 0.04c | 1.84 ± 0.1d | 9.94 ± 0.33d | 1.96 ± 0.13d |
| S1044N | Domain III S1 | 1.10 ± 0.03 | 1.27 ± 0.04 | 5.82 ± 0.33a | 0.95 ± 0.07 |
| N1366K | Domain IV S1 | 1.17 ± 0.03b | 1.83 ± 0.10d | 5.89 ± 0.38 | 0.94 ± 0.05 |
| N1366R | Domain IV S1 | 1.91 ± 0.07d | 4.09 ± 0.16d | 14.62 ± 0.58d | 0.40 ± 0.07d |
| N1366D | Domain IV S1 | 1.09 ± 0.03 | 1.60 ± 0.03d | 13.02 ± 0.88d | 2.99 ± 0.13d |
| N1366E | Domain IV S1 | 1.37 ± 0.03d | 2.05 ± 0.08d | 6.45 ± 0.35 | 1.37 ± 0.06c |
| INC | |||||
| E171R | Domain I S2 | 1.09 ± 0.04 | 1.41 ± 0.06a | 9.59 ± 0.48b | 2.20 ± 0.21d |
| K175E | Domain I S2 | 1.13 ± 0.02a | 1.45 ± 0.09 | 6.21 ± 0.36 | 1.05 ± 0.06 |
| N194K | Domain I S3 | 1.52 ± 0.04d | 4.64 ± 0.15d | 6.18 ± 0.30 | 1.13 ± 0.04 |
| D197R | Domain I S3 | 1.02 ± 0.04 | 1.56 ± 0.08b | 16.1 ± 1.43d | 2.80 ± 0.37d |
| E624K | Domain II S2 | 1.26 ± 0.04d | 1.57 ± 0.09b | 6.11 ± 0.28 | 0.91 ± 0.08 |
| K628E | Domain II S2 | 1.12 ± 0.03 | 1.45 ± 0.07a | 6.72 ± 0.15 | 1.09 ± 0.04 |
| N643K | Domain II S3 | 1.24 ± 0.04c | 1.49 ± 0.05c | 6.04 ± 0.37 | 0.96 ± 0.11 |
| D646K | Domain II S3 | 1.50 ± 0.04d | 2.09 ± 0.32a | 7.13 ± 0.30 | 1.05 ± 0.09 |
| E1079R | Domain III S2 | 1.54 ± 0.05d | 2.24 ± 0.18d | 7.13 ± 0.36 | 1.07 ± 0.09 |
| K1083E | Domain III S2 | 1.16 ± 0.04a | 1.31 ± 0.05 | 8.22 ± 0.33 | 2.18 ± 0.25d |
| C1098K | Domain III S3 | 1.11 ± 0.02 | 1.36 ± 0.04a | 10.4 ± 0.60c | 1.61 ± 0.11c |
| D1101V | Domain III S3 | 1.09 ± 0.02 | 1.23 ± 0.07 | 7.63 ± 0.38 | 1.26 ± 0.05b |
| E1399K | Domain IV S2 | 1.57 ± 0.04d | 3.25 ± 0.08d | 7.75 ± 0.91 | 1.21 ± 0.12 |
| K1403E | Domain IV S2 | 1.19 ± 0.06 | 2.47 ± 0.30b | 5.44 ± 0.22 | 1.10 ± 0.06 |
| N1417K | Domain IV S3 | 1.23 ± 0.03d | 1.57 ± 0.06d | 7.50 ± 0.32 | 1.25 ± 0.08a |
| D1420K | Domain IV S3 | 1.45 ± 0.05d | 3.52 ± 0.48c | 7.32 ± 0.31 | 0.80 ± 0.11a |
Mean values ± SEM from 10–24 experiments.
P ≤ 0.05.
P ≤ 0.01.
P ≤ 0.001.
P ≤ 0.0001.
We measured activation of hNaV1.4 or mutant channels depolarized with 20-ms test pulses to voltages ranging from −90 mV to 60 mV. Current amplitude in response to each test pulse was normalized to maximum, and the resulting curve was fit with a Boltzmann distribution according to the equation I/IMAX = 1/(1 + exp[zF(V0.5 – VM)/RT]), where I is the response to a test pulse, IMAX is the maximum response, z is the slope factor, F is Faraday’s constant, V0.5 is the midpoint voltage, VM is the command voltage for a given test pulse, R is the universal gas constant, and T is temperature in degrees Kelvin. From this equation, we derived parameters of midpoint (V0.5) and slope factor of activation, and calculated the free energy change ΔG as 4zFV0.5 according to a four state model as described in Chowdhury and Chanda (2012). We measured activation kinetics as 10–90% rise time to peak current over a voltage range from −20 mV to 60 mV.
We determined steady-state fast inactivation by preconditioning oocyte membrane potential for 300 ms to voltages ranging from −120 mV to 25 mV and then delivering depolarizing test pulses to 30 mV. Peak current amplitude in response to each test pulse was normalized against the maximum, and the resulting h∞ curve was fit with a Boltzmann distribution according to the following equation: I/IMAX = 1 − 1/(1 + exp[zF(h0.5 – VM)/RT]), where VM is the conditioning voltage, to derive parameters of midpoint (h0.5) and slope factor for inactivation. Kinetics of entry into fast inactivation were obtained from exponential fits to decays of sodium current over the voltage range of −90 mV to 60 mV according to the equation It = K0 + K1 exp(−t/tauH), where I is sodium current at time t, K0 is the asymptote, K1 is initial amplitude of sodium current, and tauH is time constant of fast inactivation.
Parameters for recovery of channels from fast inactivation were determined using a double pulse protocol (Fig. S1). Channels were inactivated with a 30-mV depolarization for 30 or 50 ms, followed by command hyperpolarization for durations up to 53 ms, at voltages ranging from −120 mV to −70 mV, and test pulses to 30 mV. The normalized recovery curve was fit with the previous equation to obtain tauH for recovery. Delay in onset to recovery from fast inactivation was taken as time at which normalized recovery current became positive (Kuo and Bean, 1994).
Online supplemental material
Electrophysiology.
An illustration of methodology for calculating recovery time constant and its delay from the double pulse protocol is provided in Fig. S1. Figs. S4–S6 provide the voltage dependencies of recovery delay for mutations tested in this work.
Modeling.
Template and domain specific VSM models are available as text files: (1) NaVAb: 3RVY_A_sensor, 3RVY_NaV_DI, 3RVY_NaV_DII, 3RVY_NaV_DIII, 3RVY_NaV_DIV, (2) NaVAb(2) 4EKW_sensor, 4EKW_NaV_DI, 4EKW_NaV_DII, 4EKW_NaV_DIII, 4EKW_NaV_DIV, and (3) NaVRh: 4DKW_sensor, 4DKW_NaV_DI, 4DKW_NaV_DII, 4DKW_NaV_DIII, 4DKW_NaV_DIV.
We include homology models of the VSM for each hNaV1.4 domain, based on structural data provided by NaVAb(2) and NaVRh as Fig. S2. We also provide homology models based on NaVAb that include residues not investigated in this study but relevant to this work, in Fig. S3.
Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201210935/DC1.
RESULTS
Identification of putative countercharges in hNaV1.4
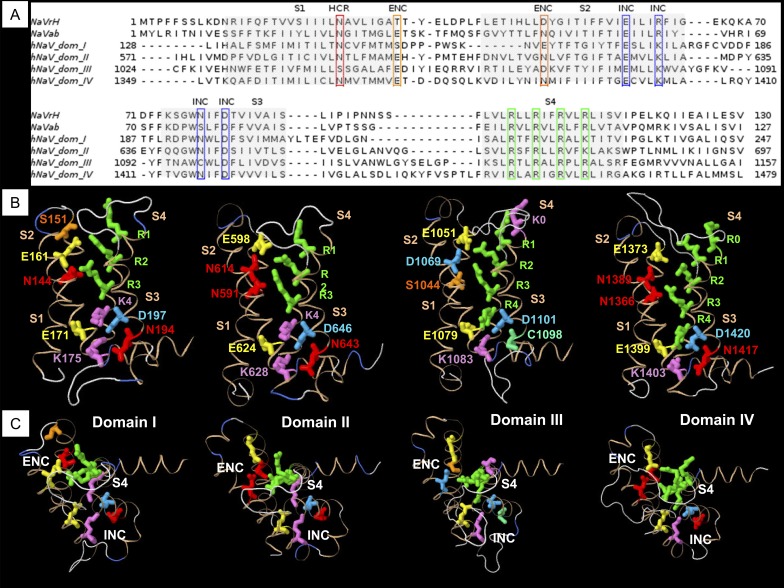
Alignments of the VSM from domains I–IV of hNaV1.4 to bacterial sodium channels NaVAb (Payandeh et al., 2011) and NachBac orthologue NaVRh (Zhang et al., 2012) are shown in Fig. 1 A. Conserved sequences representing putative countercharges in S1, S2, and S3 segments are outlined in boxes. These alignments identified amino acid residues in hNaV1.4 homologous to bacterial sodium channel VSM residues described in crystal structure, characterized for ion-pair interactions, or suggested by structural modeling but not yet functionally characterized.
Figure 1.
Homology modeling of sodium channel VSM. (A) Sequence alignments of VSM from bacterial channels NaVRh and NaVAb, and domains I–IV of mammalian sodium channel hNaV1.4. (B) Homology models of each domain of hNaV1.4 based on the NaVAb template, showing relative positions of putative countercharges in S1, S2, and S3 segments, and positively charged residues through R4 in S4 segments. (C) View of homology models in B with S4 residues R1–R3 aligned in each, to show orientation of countercharges to the S4 segment.
From the bacterial NaVAb template, homology models of the VSM for each domain of hNaV1.4 were created. Fig. 1 B shows the position of each of the conserved residues in the mammalian VSM, with respect to positively charged residues in S4. With the S4 segment aligned along the R1–R3 backbone (Fig. 1 C), side chains for mammalian putative countercharges in domains I–IV are each oriented similarly, facing toward the S4 segment. Models created from the NaVAb(2) and NaVRh templates are shown in Fig. S2. The progressively activated state of the channels in these three models is observed with the position of R4 beneath (NaVAb), traversing (NaVAb(2)), and above (NaVRh) the gating pore. The orientation of putative countercharges in the VSM was similar in each model, with the exception of the outer S1 residue in the extracellular negative charge cluster (ENC), which was observed in a nonstructured region in the NaVRh model.
The sequence alignments and resulting homology models for each domain suggested to us that conserved residues in S1–S3 might influence voltage-dependent functions of hNaV1.4. Therefore, we characterized each of these residues with mutagenesis, using voltage clamp experiments. We compared steady-state and kinetic parameters between wild-type hNaV1.4 channels and mutations in the ENC, intracellular negative charge cluster (INC), and a conserved charge just above the putative gating pore, or hydrophobic charge region (HCR). The position of that HCR S1 residue relative to the gating pore is illustrated in Fig. S3. Also shown are positions of isoleucine, valine, and aromatic residues in each domain of hNaV1.4, homologous to those proposed in NaVAb to comprise the hydrophobic constriction site (HCS; Payandeh et al., 2011; Yarov-Yarovoy et al., 2012).
ENC mutations
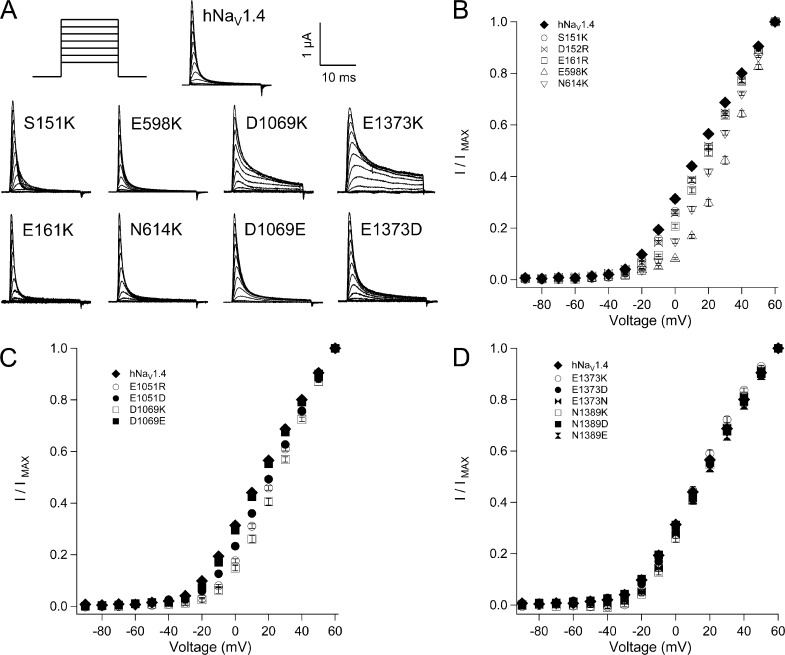
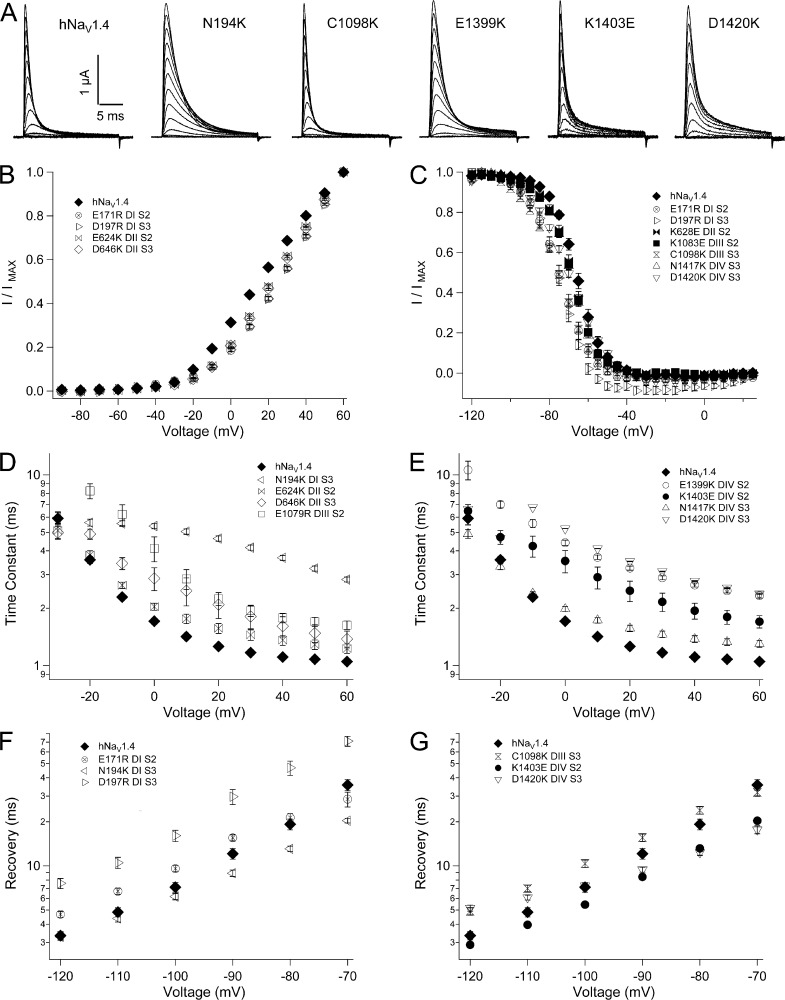
In the bacterial sodium channel NachBac, negatively charged residues in the ENC pair sequentially with positively charged residues in S4 during activation (DeCaen et al., 2008, 2009, 2011). Our models predicted homologous ENC countercharges in each domain of the mammalian channel hNaV1.4, with some question as to the roles of S151, D152, or both as countercharges in domain I. We compared the effects on channel function for charge-reversing mutations of the outer residue at S1 (including S151K and D152R) in domain I, E598K in domain II, E1051R in domain III, and E1373K in domain IV, and at S2 (E161R in domain I, N614K in domain II, D1069K in domain III, and N1389K in domain IV). For ENC residues in domains III and IV, we also tested the effects of charge-conserving mutations.
Mutations of ENC residues in domains I–III inhibited activation (Fig. 2 and Tables 1 and 2). In domain I, activation midpoint (V0.5) was depolarized by 5 mV with mutation at S1 (S151K, D152R) or at S2 (E161R). Activation was slowed by S151K and E161R, and E161R also increased slope factor. Larger depolarizing shifts in I/V relations were observed for domain II ENC mutations E598K (S1; 18 mV) and N614K (S2; 12 mV). In domain III, significant depolarizing shifts of V0.5 were produced by E1051R (S1, 8 mV) and E1051D (7 mV). E1051R increased slope factor and slowed rise time to peak activation. D1069K (S2) significantly depolarized V0.5 by 12 mV and increased slope factor, whereas D1069E produced activation midpoint and slope factor similar to hNaV1.4. For domain IV, charge reversal or substitution did not affect the probability of channel opening, as I/V relations for E1373K/N/D (S1) or N1389K/D/E (S2) were similar to hNaV1.4. However, each of these mutations significantly slowed rise time to peak activation. Slope factor was increased by mutations E1373K, E1373N, and N1389K and decreased by N1389E.
Figure 2.
Conductance in hNaV1.4 and ENC mutations. (A)Traces for wild type and mutant channels in response to depolarizing commands to voltages ranging from −90 mV to +60 mV. I/V relations are shown for ENC mutations in domain I and domain II (B), domain III (C), and domain IV (D). Values represent mean ± SEM (error bars) from 10–22 experiments.
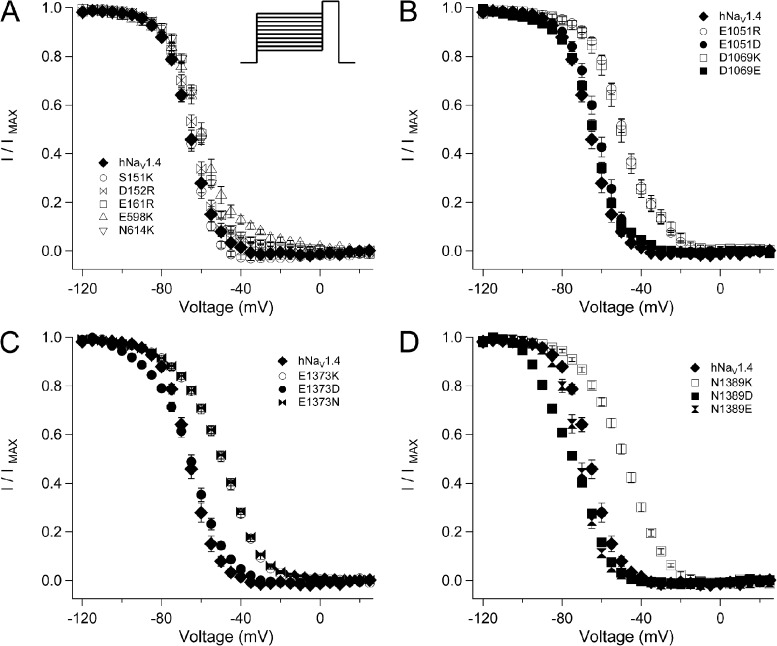
Most ENC charge-reversing mutations produced a depolarizing shift in the midpoint (h0.5) of the steady-state fast inactivation (h∞) curve (Fig. 3 and Table 1). Mutations E161R (domain I S2), E598K (domain II S1), and N614K (domain II S2) depolarized midpoint by 4–6 mV, and E598K also decreased slope factor (Fig. 3 A). Charge-reversing mutations at S1 or S2 in the ENC of domain III (E1051R and D1069K) or domain IV (E1373K and N1389K) produced the largest effects on steady-state inactivation, with 15–17-mV depolarizing shifts of the h∞ curve and significantly decreased slope factor (Fig. 3, B–D). Charge-substituting mutations E1051D and D1069E produced h0.5 values similar to hNaV1.4. E1051D, but not D1069E, produced a slope factor similar to the wild-type channel. At domain IV S1, aspartate substitution at E1373 produced h0.5 similar to hNaV1.4, whereas asparagine substitution produced an effect similar to E1373K, and both mutations significantly decreased slope factor. At domain IV S2, glutamate or aspartate substitution at N1389 produced a significant hyperpolarizing shift of the h∞ curve. Slope factor for N1389E, but not N1389D, was similar to hNaV1.4. These findings indicate that the charge content of domain III and domain IV ENC residues is an important determinant of fast inactivation.
Figure 3.
Steady-state fast inactivation in hNaV1.4 and for ENC mutations. Channels were conditioned to voltages ranging from −120 mV to +25 mV for 300 ms before test pulses to 30 mV. I/V relations (h∞ curves) are shown for ENC mutations in domain I and domain II (A), domain III (B), domain IV S1 (C), and domain IV S2 (D). Values represent mean ± SEM (error bars) from 10–21 experiments.
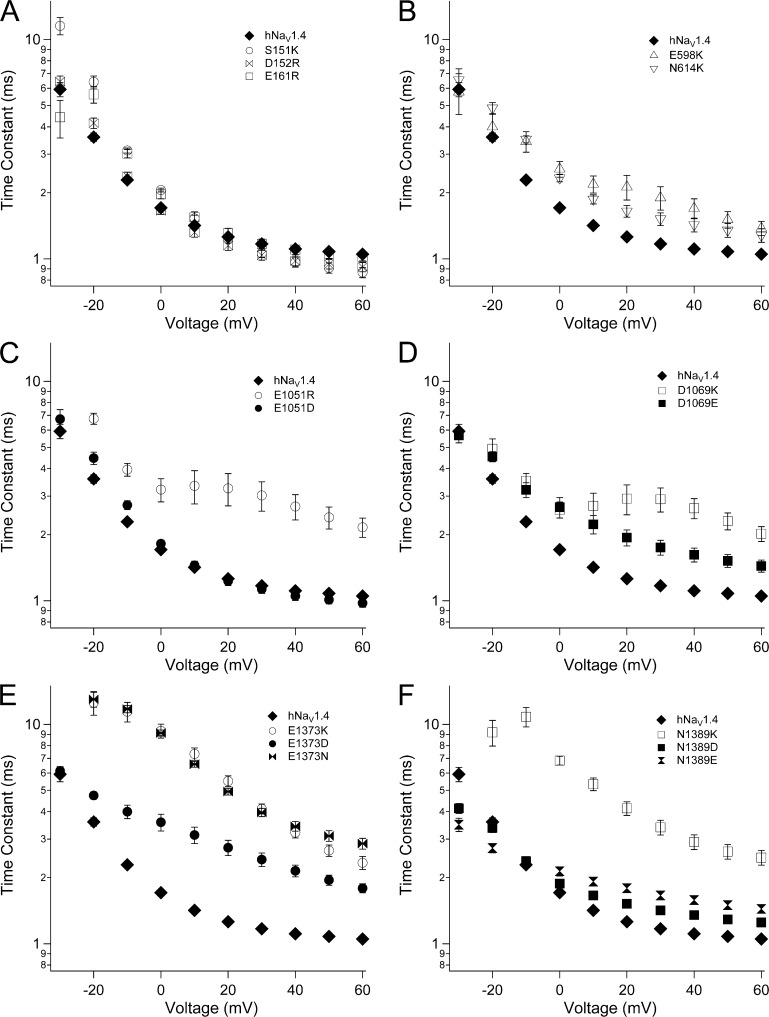
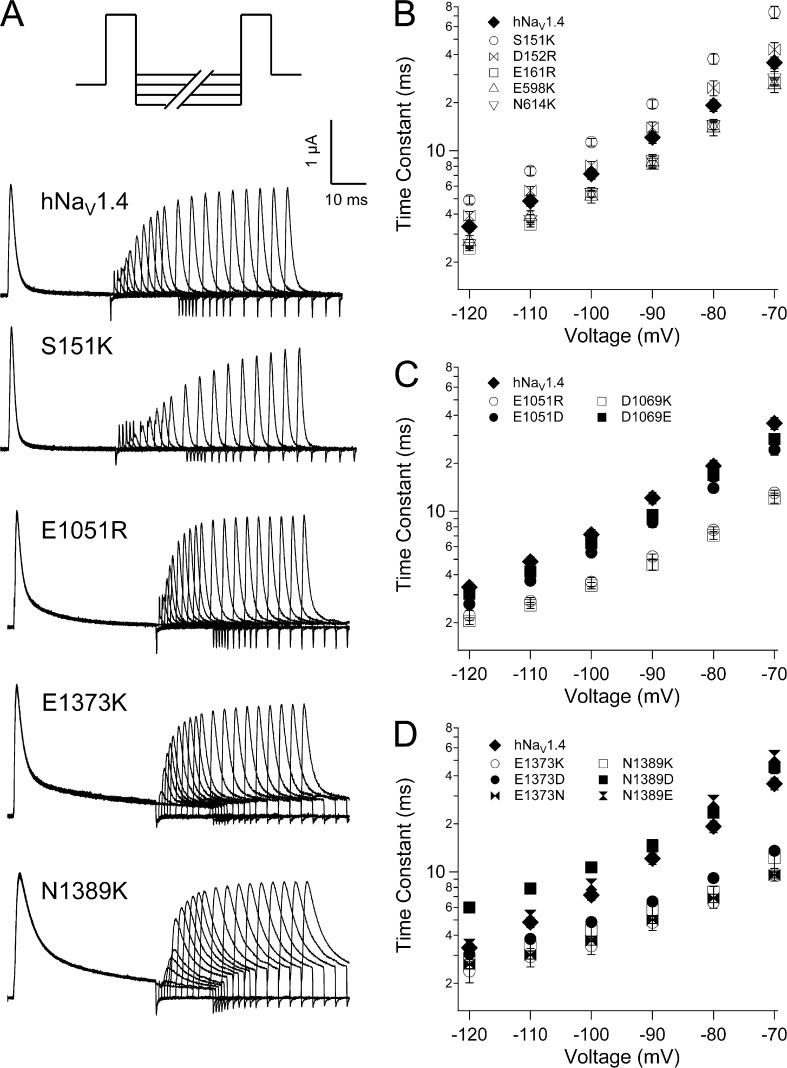
Fast inactivation was slowed by reversal of charge in the ENC for domains II–IV (Fig. 4 and Table 2). In domain II, E598K (S1) and N614K (S2) each slowed the entry of channels into fast inactivation, at voltages more positive than −20 mV (Fig. 4 B). In domain III, E1051R (S1) and D1069K (S2) slowed fast inactivation over this voltage range, and decreased its voltage dependence (Fig. 4, C and D). E1051D fully restored, and D1069E partially restored, wild-type fast inactivation kinetics. In domain IV, charge reversal at S1 (E1373K) or at S2 (N1389K) dramatically slowed fast inactivation at all voltages tested (Fig. 4, E and F). E1373N slowed fast inactivation similar to E1373K, whereas E1373D produced kinetics intermediate between E1373K and hNaV1.4. At domain IV S2, N1389D/E slowed fast inactivation to a lesser extent than N1389K.
Figure 4.
Fast inactivation kinetics in hNaV1.4 and for ENC mutations. Time constants of current decay are shown for ENC mutations in domain I (A), domain II (B), domain III (S1, C; S2, D), and domain IV (S1, E; S2, F). Values represent mean ± SEM (error bars) from 10–22 experiments.
Recovery from fast inactivation was measured at voltages ranging from −120 mV to −70 mV (Fig. 5 A). At domain I S1, S151K slowed recovery and D152R was without effect (Fig. 5 B). Each of the remaining seven ENC charge-reversing mutations significantly accelerated recovery (Table 2). E161R (domain I S2), E598K (domain II S1), and N614K (domain II S2) produced an equivalent and moderate effect to accelerate recovery (Fig. 5 B). E1051R (domain III S1), D1069K (domain III S2), E1373K (domain IV S1), and N1389K (domain IV S2) produced marked acceleration of channel recovery (Fig. 5, C and D). For domain III, these effects were charge dependent, as recovery time constants for E1051D (S1) and D1069E (S2) were similar to hNaV1.4. In domain IV, E1373N and E1373D (S1) each accelerated recovery to a similar extent as for E1373K. N1389E (S2) produced recovery kinetics similar to hNaV1.4, and N1389D significantly slowed recovery at more hyperpolarized potentials, resulting in shallow voltage dependence.
Figure 5.
Recovery kinetics in hNaV1.4 and for ENC mutations. (A) Double pulse protocol used to inactivate and recover channels. Traces are shown for recovery up to 50 ms, with some sweeps removed for clarity. Recovery time constants are shown for ENC mutations in domain I and domain II (B), domain III (C), and domain IV (D). Values represent mean ± SEM (error bars) from 10–18 experiments.
Deactivation kinetics were measured as delay in onset to recovery from fast inactivation (Fig. S1). Effects of ENC mutations on recovery delay paralleled those for recovery (Fig. S4 and Table 2). For example, ENC mutation S151K (domain I S1), which slowed recovery, also prolonged recovery delay. ENC mutations that accelerated recovery shortened the delay to onset. Exceptions were mutations E161R and N1389D, which had no effect on recovery delay. Together, our findings indicate that domain III and domain IV ENC residues, as a consequence of their negative charge content, stabilize the fast-inactivated state.
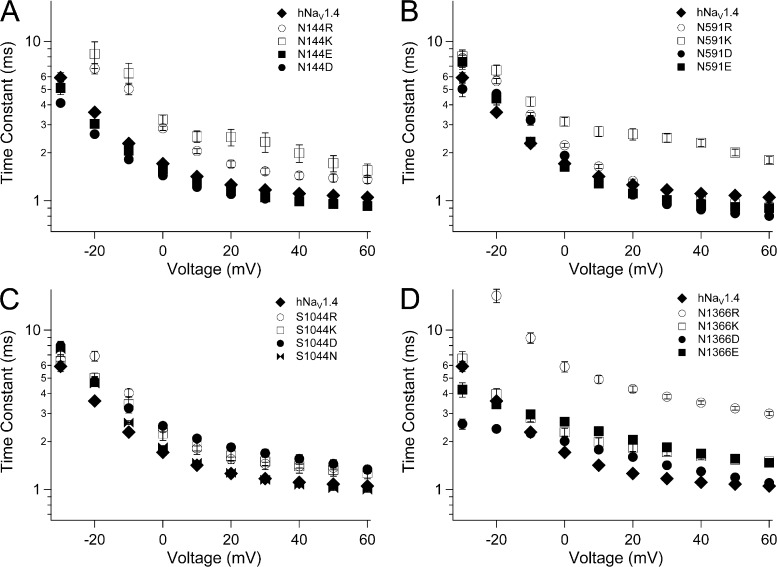
HCR mutations
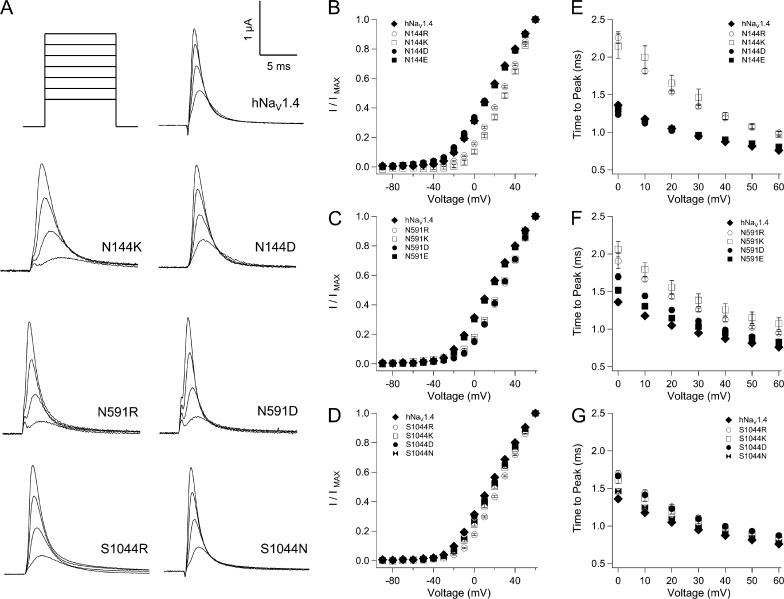
Putative countercharges in the bacterial VSM include S1 residue N36 in NachBac (Yarov-Yarovoy et al., 2012). The homologous residue N25 is observed in the crystal structure of NaVAb as part of a hydrogen bond network with M21 in S1, S83 in S3, and R105 (R3) in S4 (Payandeh et al., 2011). We identified homologous residues in hNaV1.4 (Fig. S3), and characterized the putative countercharge in S1. To do this, we compared the effects of mutations at N144 (domain I), N591 (domain II), S1044 (domain III), and N1366 (domain IV). These loci were considered to be the HCR.
HCR mutations in domains I–III inhibited activation (Fig. 6 and Tables 1 and 2). Charge-reversing mutations N144R/K (domain I; Fig. 6, B and E) or N591R/K (domain II; Fig. 6, C and F) produced 16–17-mV depolarizing shifts of V0.5 and slowed channel activation. For domain I HCR mutations, effects were charge dependent, as N144D/E produced V0.5 values and activation kinetics similar to hNaV1.4. In domain II, glutamate substitution at N591 rescued wild-type V0.5, whereas aspartate substitution produced a depolarizing shift of 14 mV, similar to N591R/K. Both N591E and N591D partially restored wild-type activation kinetics. In domain III, S1044R produced a depolarizing shift of 12 mV, with S1044K/N producing a 5-mV shift (Fig. 6 D). Activation was slowed by S1044R/K, but also by S1044D (Fig. 6 G). For the domain IV HCR locus N1366, aspartate substitution produced a significant, depolarizing shift of the conductance curve (Table 1). N1366D also decreased slope factor, whereas charge reversal (N1366R) increased slope factor.
Figure 6.
Activation parameters for mutations at S1 HCR locus in hNaV1.4. (A) Traces shown are responses to depolarization to 0 mV, 20 mV, 40 mV, and 60 mV for mutations in domains I–III. Plots in B, D, and F show I/V relations for mutations in these domains; plots in C, E, and G show activation kinetics. Values represent mean ± SEM (error bars) from 11–24 experiments.
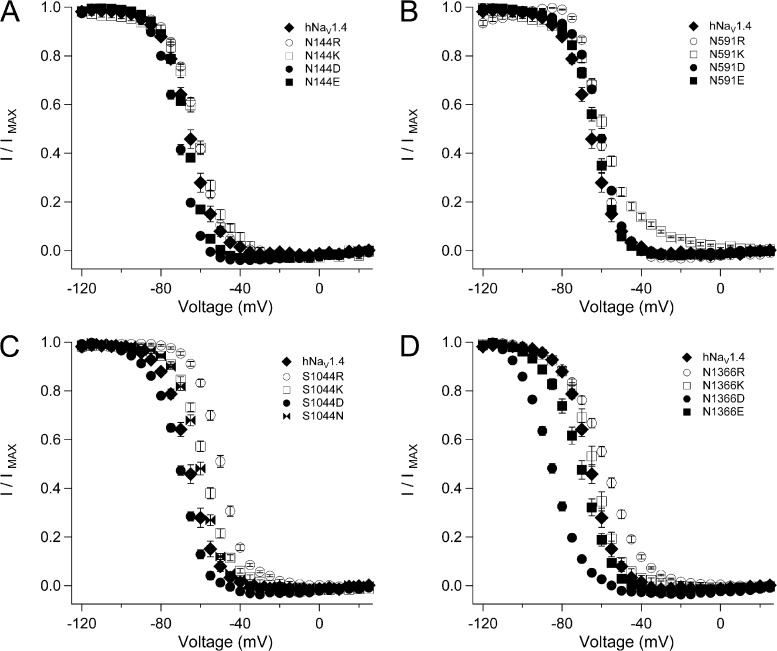
Figs. 7–9 show effects of HCR mutations on steady-state fast inactivation, kinetics of entry into, and recovery from that state. Lysine or arginine replacement at N144 (domain I), N591 (domain II), and S1044 (domain III) produced a significant depolarizing shift of the h∞ curve, as did arginine replacement at N1366 (domain IV; Fig. 7 and Table 1). N144D, S1044D, and N1336D/E each produced a significant hyperpolarizing shift of the h∞ curve. N591D and S1044N produced a depolarizing effect, and N144E and N591E produced h0.5 values similar to hNaV1.4. Slope factor was increased by N144D/E and by N591R, and decreased by N144K, N591K, and each of the mutations at N1366.
Figure 7.
Steady-state fast inactivation for HCR mutations. Plots show h∞ curves for hNaV1.4 and S1 mutations in domain I (N144, A), domain II (N591, B), domain III (S1044, C), and domain IV (N1366, D). Values represent mean ± SEM (error bars) from 11–24 experiments.
Figure 9.
Recovery from fast inactivation for HCR mutations. (A) Traces showing inactivating and recovery sweeps for hNaV1.4 and select aspartate or arginine substitutions in domains I, III, and IV. (B–D) Kinetics of recovery at voltages from −120 mV to −70 mV for mutations in these domains. Values represent mean ± SEM (error bars) from 10–22 experiments.
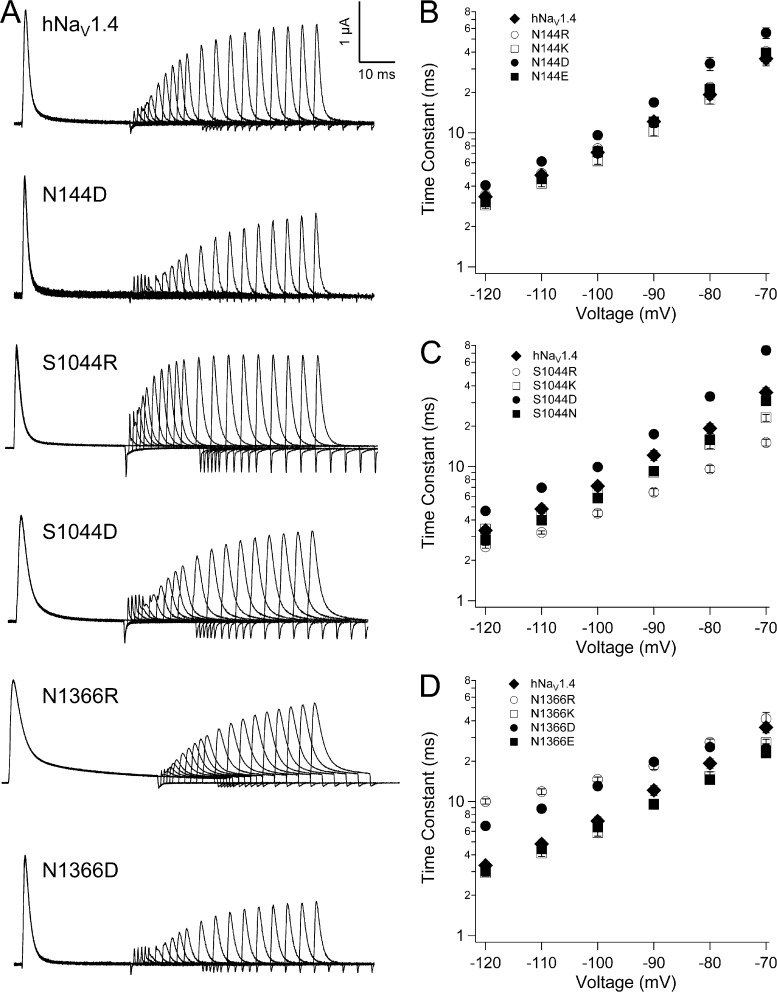
Most charge-reversing mutations at the S1 HCR locus slowed entry of channels into the fast-inactivated state (Fig. 8). N144R/K (domain I) and N591K (domain II) slowed fast inactivation, and both N144K and N591K reduced voltage dependence at voltages more positive than 0 mV. Charge substitutions at these loci produced inactivation kinetics similar to, or accelerated compared with, hNaV1.4. In domains III and IV, charge-reversing mutations (S1044R, domain III; N1366R/K, domain IV) as well as charge-substituting mutations (S1044D, domain III; N1366D/E, domain IV) slowed entry of channels into the fast-inactivated state. N1366R slowed fast inactivation to the greatest extent across the voltage range tested. N1366D decreased the voltage dependence of fast inactivation, with accelerated kinetics at voltages more negative than 0 mV.
Figure 8.
Kinetics of fast inactivation for HCR mutations. Plots show time constants for hNaV1.4 and S1 mutations in domain I (N144, A), domain II (N591, B), domain III (S1044, C), and domain IV (N1366, D). Values represent mean ± SEM (error bars) from 11–24 experiments.
Recovery and its delay were most affected by HCR mutations in domains III and IV (Fig. 9, Fig. S5, and Table 2). Addition of a positive charge at domain III HCR (S1044R) significantly accelerated recovery and its delay, whereas addition of a negative charge (S1044D) prolonged recovery and delay. For the domain IV HCR locus N1366, effects of mutations were less predictable. For example, N1366R slowed recovery, especially at hyperpolarized voltages, resulting in reduced voltage dependence. However, N1366R accelerated recovery delay, with increased voltage dependence. N1366E accelerated recovery at depolarized voltages but prolonged recovery delay.
INC mutations
In NachBac, residue E70, located in the inner portion of the S2 helix, sequentially interacts with positive charges R3 and R4 during activation (DeCaen et al., 2008, 2009). The homologous residue E59 in NaVAb is observed as part of a hydrogen bond network between charged residues in S2, S3, and R108 (R4) in S4, as the INC (Payandeh et al., 2011). Alignments of bacterial sodium channel sequences with domain sequences from mammalian channels NaV1.2 (Yarov-Yarovoy et al., 2012) or NaV1.4 (Fig. 1; Zhang et al., 2012) show another conserved negative charge at the base of S3. Collectively, these INC residues are highly conserved in hNaV1.4 (Fig. 1, A and B). Our alignment shows a glutamate one helical turn under the HCS region, in S2 (E171, E624, E1079, E1399). Each of these negative charges in S2 is followed by a lysine (K175, K628, K1083, K1403) near the base of the helix. In S3, an inner residue, typically asparagine (N194, N643, C1098, N1417) is followed by a conserved aspartate (D197, D646, D1101, D1420). We constructed charge-reversing mutations at each of these loci in S2 and S3, and characterized their effect on channel function. As neither D1101R nor D1101K expressed in oocytes, we tested valine replacement at this locus.
Activation was inhibited by several mutations in the INC (Fig. 10 B and Tables 1 and 2). Overall, effects were notable only for charge reversals in the first two domains, and for residues closest to the hydrophobic region surrounding the gating pore. For example, charge reversal of INC residues at domain I S2 (E171R) or domain I S3 (D197R) resulted in a depolarizing shift of V0.5 of 15 mV or more, but did not affect activation kinetics. Charge reversal at the homologous residues in domain II produced significant depolarizing shifts of V0.5 (E624K, 8 mV; D646K, 10 mV) and slowed activation rise time. Lesser depolarizing shifts in V0.5 were observed with mutation of the inner S3 residue in domain III (C1098K; 5 mV) or in domain IV (N1417K; 6 mV); N1417K also slowed activation. Other mutations in the INC, which did not affect activation probability, did prolong rise time to peak activation of channels, including N194K (D1 S3), N643K (domain II S3), E1079R (domain III S2), E1399K (domain IV S2), and D1420K (domain IV S3).
Figure 10.
Biophysical characterization of INC mutations. (A) Sodium currents in response to command depolarization to voltages from −90 mV to +60 mV. Effects of INC mutations are shown for activation (B), steady-state fast inactivation (C), entry into fast inactivation (D and E), and recovery (F and G). Legends identify residues and locus by domain and segment. Values represent mean ± SEM (error bars) from 10–23 experiments.
Several INC mutations affected one or both parameters of the h∞ curve (Fig. 10 C and Table 1). In domain I, E171R (S2) and D197R (S3) each produced a hyperpolarizing shift of 10 mV, and N194K (S3) significantly reduced the slope factor, compared with wild-type channels. In domain II, N643K (S3) produced a 5-mV hyperpolarizing shift of the h∞ curve, and E624K (S2) increased the slope factor. In domain III, C1098K (S3) produced a 10-mV hyperpolarizing shift of h0.5 and decreased slope factor. Mutations at INC loci in domain IV decreased slope factor, and N1417K and D1420K (S3) produced a 4-mV hyperpolarizing shift.
Charge-reversing INC mutations in each domain slowed entry of channels into the fast-inactivated state (Fig. 10, D and E; and Table 2). For domains I–III, effects were largest for N194K (domain I S3), D646K (domain II S3), and E1079R (domain III S2), and N194K also reduced voltage dependence (Fig. 10 D). E171R (domain I S1), D197R (domain I S3), E624K and K628E (domain II S2), and N643K (domain II S3) slowed fast inactivation to a lesser extent. In domain IV, charge-reversing mutations at each of the four INC loci slowed fast inactivation (Fig. 10 E). E1399K (domain IV S2) and D1420K (domain IV S3), located near the gating pore constriction, slowed fast inactivation to a greater extent than K1403E and N1417K, located near the bases of S2 and S3, respectively.
INC mutations that produced 10 mV or greater hyperpolarizing shifts of the h∞ curve (E171R, domain I S2; D197R, domain I S3; C1098K, domain III S3) slowed recovery (Fig. 10, F and G; and Table 2). Each of these mutations also prolonged the delay in onset to recovery (Fig. S6 and Table 2). Several INC mutations that slowed the entry of channels into fast inactivation (N194K, domain I S3; K1403E, domain IV S2; D1420K, domain IV S3) accelerated recovery, especially at depolarized voltages. D1420K slowed recovery at hyperpolarized voltages, resulting in decreased voltage dependence.
DISCUSSION
The gating particles proposed by Hodgkin and Huxley (1952) as the biophysical mechanism providing voltage sensitivity to excitable membranes have been identified as a repeating motif of positively charged residues in homologous S4 segments of the voltage-gated ion channel superfamily (Yu et al., 2005). Results from experimental approaches using mutagenesis of S4 charge, site-specific toxins that trap S4 segments in resting or activated states, thiosulfonate reagents that discriminate access of S4 residues to the intracellular or extracellular space, or fluorescence measurements of tagged S4 segments all support the premise that S4 segments move outward in response to membrane depolarization (for reviews see Bezanilla, 2008; Catterall, 2012).
A challenge to sliding helix or helical screw models of S4 translocation is how voltage sensor movement is achieved in the interior of a highly hydrophobic membrane environment. Current models describe an S1–S4 VSM that dictates the opening or closing of S5–S6 segments comprising a pore module. This view of ion channel gating is supported with experimental evidence in prokaryotic sodium channels that negative countercharges in S1–S3 directly participate in S4 translocation (DeCaen et al., 2008, 2009, 2011; Paldi and Gurevitz, 2010; Yarov-Yarovoy et al., 2012), but has not yet been rigorously tested for eukaryotic sodium channels.
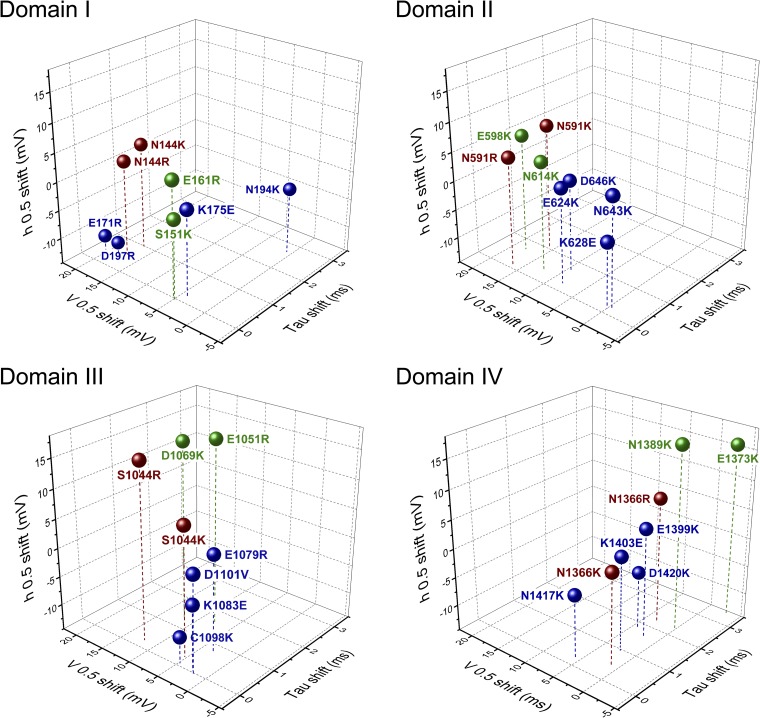
The mechanisms underlying voltage gating in mammalian sodium channels are crucial to our understanding of channelopathies in excitable tissues (George, 2005; Cannon, 2006; Jurkat-Rott et al., 2010). Investigations of these disease-causing mutations have contributed significantly to our understanding of the gating mechanisms underlying activation and fast inactivation. Crystal structures of bacterial channels have provided additional power toward the use of computational models to suggest mechanisms of voltage gating (Vargas et al., 2012). Using the structural data provided by NaVAb and NaVRh, we created homology models of each domain of the mammalian skeletal muscle sodium channel and identified putative countercharges in S1, S2, and S3 segments oriented toward the S4 backbone. Our characterization of mutations at these loci show that activation is promoted by countercharges in domains I–III, and that sodium channel fast inactivation is differentially regulated by countercharges depending on their location with respect to domain and the gating pore constriction. A view of domain-specific sodium channel functions is suggested in Fig. 11, showing the effects of VSM charge-reversing mutations. Activation is most affected by mutations in domains I and II, with fast inactivation most affected in domain IV. Domain III mutations increase (INC) or decrease (HCR, ENC) steady-state fast inactivation. In general, mutations at ENC and HCR loci produce larger effects than those at INC loci.
Figure 11.
Three-dimensional plot showing the effect of charge-reversing mutations of residues in ENC (green), HCR (red), and INC (blue) regions. Shown for each domain are shifts in inactivation kinetics at 20 mV (x axis), activation probability (y axis), and inactivation probability (z axis).
Negative countercharges facilitate activation of mammalian sodium channels
In NachBac, transient pairings of residues E43 (S1) and D60 (S2) with S4 residues promote activation (DeCaen et al., 2008, 2009, 2011; Yarov-Yarovoy et al., 2012). We identified homologues in the VSM of each domain in hNaV1.4. Our results strongly support a role in activation for S1 and S2 ENC residues in domains I (S151, E161), II (E598, N614), and III (E1051, D1069). Charge-reversing mutations at these loci produced a depolarizing shift of V0.5, and S151K, E161R, and E1051R also slowed activation kinetics. These results are consistent with the hypothesis that ENC residues in domains I–III interact with positively charged residues in S4 segments of these domains to stabilize S4 translocation in response to membrane depolarization during activation.
The NachBac S1 residue N36 has not yet been functionally characterized, but structural modeling suggests that this residue also participates as a countercharge in the VSM (Yarov-Yarovoy et al., 2012). We identified homologous S1 residues in hNaV1.4 located one helical turn above the HCS (Fig. 1 and Fig. S3). Our results show that hNaV1.4 HCR S1 residues contribute significantly to domain specificity of activation in hNaV1.4. Mutational perturbation of activation was greatest for HCR residue N144 in domain I, and effects were charge dependent. Decreasing effects on activation were produced by mutations of HCR residues N591, S1044, and N1366 in domains II–IV. The effect of HCR charge-reversing mutations in hNaV1.4, inhibiting activation, are consistent with a model of the activation process in NacBac (Yarov-Yarovoy et al., 2012), showing that the homologous S1 residue is part of a dynamic hydrogen bond network that facilitates S4 translocation during activation.
In the INC, activation probability was reduced by reversal of charge at E171 (domain I S2), D197 (domain I S3), E624 (domain II S2), and D646 (domain II S3). Each of these mutations is located one helical turn under the HCS. Mutations of INC residues located two helical turns under the HCS had no effect on activation. Collectively, our results indicate that ENC and HCR negative countercharges in domains I and II play the most critical role in activation, with contribution from domain III.
Although the precise interactions of VSM countercharges in hNaV1.4 with S4 counterparts are uncertain, a comparison of our results to other studies suggests that the interaction of countercharge with outer positive charges in S4 is crucial to activation. Our free energy change calculations for the effect of point mutations of hNaV1.4 countercharges yielded values of 1–3 kcal/mol, representing the energy of 1–2 hydrogen bonds in an α helix (Sheu et al., 2003). These values are similar to those for point mutations of NachBac S4 positive charges R1, R3, and R4 (Yarov-Yarovoy et al., 2012), for which interaction of countercharge with S4 has been verified with disulfide locking experiments. Molecular dynamics simulations using homology models of rNaV1.4 suggest interaction of R1 with a single ENC countercharge, and R2–R4 with ENC and INC countercharges during the activation process (Gosselin-Badaroudine et al., 2012). Given the relatively small change in activation free energy associated with mutations of countercharge in hNaV1.4, it is likely that interactions of S4 charge with residues nearest the gating pore constriction are the activation determinant.
Each of the domain IV ENC mutations at S1 (E1373) or at S2 (N1389) slowed activation without affecting conductance. This apparent conflict is most likely explained by the dramatic effect of these mutations to prolong fast inactivation. In response to depolarization, sodium channels activate and fast inactivate, with overlapping time courses at the macroscopic level (Aldrich et al., 1983). Activation kinetics measured as time to peak activation do not discriminate between slowed activation per se and prolonged time to peak current amplitude resulting from impaired entry into fast inactivation.
Simulations in rNaV1.4 suggest interaction of countercharges in S1–S3 with each of the first six positive charges in domain IV S4 during transitions between intermediate states leading to activation (Gosselin-Badaroudine et al., 2012). In our homology models, these S4 residues are found in the α helical region of S4, with K7 and R8 located in the S4–S5 helix (unpublished data). From this computational data, and given the dramatic effect of domain IV ENC mutations on fast inactivation, it is likely that domain IV countercharges do interact with S4 residues during membrane depolarization, but that this interaction is not a determinant of pore opening. Our results indicating that negative countercharges in sodium channel domains I–III promote activation are consistent with previous studies showing that activation is dictated by S4 translocation in these domains (Chanda and Bezanilla, 2002; Armstrong, 2006).
Negative countercharges dictate fast inactivation gating
Fast inactivation of the sodium channel and its subsequent recovery determine the upper limit to action potential frequency in axon and muscle fiber membrane signaling. Our results show that negative countercharges are a crucial determinant in sodium channel fast inactivation and provide a greater understanding of its mechanism. ENC residues in domains III (E1051, D1069) and IV (E1373, N1389) appear to play the most significant role in fast inactivation of hNaV1.4. Charge reversal at each of these residues produced a depolarizing shift of the h∞ curve, slowed entry of channels into fast inactivation, and accelerated their recovery. A previous study of domain IV S2 in the rat skeletal muscle sodium channel found that N1382C (Ma et al., 2009) produces the same pattern of effects on fast inactivation as we observed with charge reversal at the homologous locus in hNaV1.4 (N1389K).
Current models of sodium channel fast inactivation posit that voltage sensor translocation in response to membrane depolarization provides access for the inactivation particle to receptor sites in the S4–S5 linker of domain III (Smith and Goldin, 1997) and of domain IV (Lerche et al., 1997). Salt bridge formation between ENC residues in hNaV1.4 and S4 residues during the depolarizing translocation preceding inactivation may explain the charge-dependent effects of domain III and domain IV ENC mutations on fast inactivation. However, the molecular basis of hNaV1.4 countercharge interaction with S4 remains to be shown experimentally. Additionally, our finding that E1373N (domain IV S1) reiterates each of the effects of E1373K on fast inactivation indicates that the structure of the native residue is relevant. Length and orientation of the side chain of ENC countercharges glutamate, aspartate, and asparagine are not equivalent and might confer specific constraints for salt bridge formation between these countercharges and putative S4 pairing partners in the native channel.
Charge-reversing mutations in domain I produced moderate effects on parameters for fast inactivation. One interesting exception was S151K’s effect of slowing recovery of channels from fast inactivation and to delay its onset. Assuming full absorption of S151K into the fast-inactivated state, effects on recovery are independent from slowed activation produced by this mutation. The prolonged delay in onset to recovery in S151K suggests that S151 promotes the deactivating transition of domain I S4, as channels leave the inactivated state and close (Kuo and Bean, 1994). A testable hypothesis is that this residue facilitates return of the nonimmobilized gating charge in that domain (Cha et al., 1999).
In domain II, E598K and N614K produced effects on fast inactivation similar to those produced by mutations of ENC residues in domain III and domain IV, albeit lesser in magnitude. Voltage sensor mutations in all four domains of the sodium channel influence fast inactivation (Kontis and Goldin, 1997), and our results suggest that ENC residues in domain II, as well as in domain III and domain IV, play a role in fast inactivation. However, it is not certain whether destabilization of the fast-inactivated state observed with E598K and N614K is a direct effect on fast inactivation per se. For example, domain II S4 gating transitions are coupled to other voltage sensor movements (Campos et al., 2007b), and domain II ENC mutations might affect that coupling.
HCR residues also appear to play a role in sodium channel fast inactivation. Charge reversal at N144, N591, S1044, and N1366 decreased the probability that channels inactivate, and generally slowed the entry of channels into the fast-inactivated state. However, we found that predictions of charge substitution at these loci to produce wild-type fast inactivation parameters were not met, especially in terms of recovery and its delay. These results suggest that HCR residues may not influence fast inactivation by forming salt bridges with an S4-positive charge. An alternative possibility, hydrogen bonding, is suggested from the crystal structure of NaVAb (Payandeh et al., 2011). In the closed-activated position, S4 residue R105 (R3) makes hydrogen bonds with N25 (S1 HCR), M29 (S1), and S87 (S3). Homologous residues in proximity to R3 are reiterated in the mammalian channel (Fig. S3). Further work is needed to investigate the specific targets of negative countercharges in the mammalian VSM, and structural modeling may help to clarify the roles of putative salt bridges or dynamic hydrogen bonding in hNaV1.4 gating transitions. Nonetheless, our results show that ENC and HCR residues are each important determinants of fast inactivation of the mammalian sodium channel.
Several effects of INC mutations on fast inactivation are noteworthy. First, charge reversal or addition of positive charge at several loci produced a hyperpolarizing effect on the h∞ curve, and none of the 12 mutations tested produced the depolarizing shift that was consistently observed with mutations of ENC or HCR loci. An interpretation of these effects is that at least some of the countercharges in the INC serve to restrict closed-state voltage sensor movement, whereas HCR and ENC charges facilitate S4 translocations, leading directly to pore opening. INC mutations that increased the likelihood of steady-state fast inactivation also slowed channel recovery, suggesting that the INC contributes to stability of the fast-inactivated state through its impact on closed-state transitions.
Each of four charge-reversing mutations in domain IV INC slowed entry of channels into the fast-inactivated state. A previous study showed that domain IV S2 mutation E1392C in rNaV1.4 (Ma et al., 2009) slows fast inactivation, similar our finding with charge reversal at the homologous residue E1399. In a study of domain IV S3 in hNaV1.4, cysteine substitution at D1420 slows fast inactivation to a greater extent than at N1417 (Nguyen and Horn, 2002), similar to our results with charge-reversing mutations D1420K and N1417K. These findings support the hypothesis that intersegmental interactions of countercharges with domain IV S4 are the crucial determinant of fast inactivation. However, our finding that domain I S3 mutation N194K slows fast inactivation to the same extent as observed for domain IV INC mutations suggests that countercharges in other domains also contribute to fast inactivation.
Examination of crystal structures provided by bacterial sodium channels suggests that hydrogen bonding may be an important component of VSM interactions in the INC (Payandeh et al., 2011), in addition to the HCR. We observed a greater diversity of mutational perturbation of fast inactivation in each of these countercharge regions (HCR and INC) compared with the ENC. Future experimental work and structural modeling may provide additional insight into the mechanisms through which countercharges in each of these regions of the mammalian VSM (ENC, HCR, and INC) affect fast inactivation.
Supplementary Material
Acknowledgments
The authors wish to thank J. Abbrusseze (Biology Department, Idaho State University) for reviewing a draft of this manuscript.
This work was supported by National Institutes of Health (NIH) grant 1R15NS064556-01 to J.R. Groome and NIH grant P20 RR016454 to Idaho State University.
Sharona E. Gordon served as editor.
Footnotes
Abbreviations used in this paper:
- ENC
- extracellular negative charge cluster
- HCR
- hydrophobic charge region
- HCS
- hydrophobic constriction site
- INC
- intracellular negative charge cluster
- VSM
- voltage sensor module
References
- Aldrich R.W., Corey D.P., Stevens C.F.. 1983. A reinterpretation of mammalian sodium channel gating based on single channel recording. Nature. 306:436–441. 10.1038/306436a0 [DOI] [PubMed] [Google Scholar]
- Armstrong C.M. 2006. Na channel inactivation from open and closed states. Proc. Natl. Acad. Sci. USA. 103:17991–17996. 10.1073/pnas.0607603103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C.M., Hille B.. 1998. Voltage-gated ion channels and electrical excitability. Neuron. 20:371–380. 10.1016/S0896-6273(00)80981-2 [DOI] [PubMed] [Google Scholar]
- Bezanilla F. 2008. Ion channels: from conductance to structure. Neuron. 60:456–468. 10.1016/j.neuron.2008.10.035 [DOI] [PubMed] [Google Scholar]
- Campos F.V., Chanda B., Roux B., Bezanilla F.. 2007a. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. USA. 104:7904–7909. 10.1073/pnas.0702638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos F.V., Chanda B., Beirão P.S.L., Bezanilla F.. 2007b. β-Scorpion toxin modifies gating transitions in all four voltage sensors of the sodium channel. J. Gen. Physiol. 130:257–268. 10.1085/jgp.200609719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S.C. 2006. Pathomechanisms in channelopathies of skeletal muscle and brain. Annu. Rev. Neurosci. 29:387–415. 10.1146/annurev.neuro.29.051605.112815 [DOI] [PubMed] [Google Scholar]
- Capes D.L., Arcisio-Miranda M., Jarecki B.W., French R.J., Chanda B.. 2012. Gating transitions in the selectivity filter region of a sodium channel are coupled to the domain IV voltage sensor. Proc. Natl. Acad. Sci. USA. 109:2648–2653. 10.1073/pnas.1115575109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A. 2012. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J. Physiol. 590:2577–2589. 10.1113/jphysiol.2011.224204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha A., Ruben P.C., George A.L., Jr, Fujimoto E., Bezanilla F.. 1999. Voltage sensors in domains III and IV, but not I and II, are immobilized by Na+ channel fast inactivation. Neuron. 22:73–87. 10.1016/S0896-6273(00)80680-7 [DOI] [PubMed] [Google Scholar]
- Chahine M., George A.L., Jr, Zhou M., Ji S., Sun W., Barchi R.L., Horn R.. 1994. Sodium channel mutations in paramyotonia congenita uncouple inactivation from activation. Neuron. 12:281–294. 10.1016/0896-6273(94)90271-2 [DOI] [PubMed] [Google Scholar]
- Chakrapani S., Sompornpisut P., Intharathep P., Roux B., Perozo E.. 2010. The activated state of a sodium channel voltage sensor in a membrane environment. Proc. Natl. Acad. Sci. USA. 107:5435–5440. 10.1073/pnas.0914109107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B., Bezanilla F.. 2002. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol. 120:629–645. 10.1085/jgp.20028679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B., Bezanilla F.. 2008. A common pathway for charge transport through voltage-sensing domains. Neuron. 57:345–351. 10.1016/j.neuron.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Chen L.-Q., Santarelli V., Horn R., Kallen R.G.. 1996. A unique role for the S4 segment of domain 4 in the inactivation of sodium channels. J. Gen. Physiol. 108:549–556. 10.1085/jgp.108.6.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Chanda B.. 2012. Estimating the voltage-dependent free energy change of ion channels using the median voltage for activation. J. Gen. Physiol. 139:3–17. 10.1085/jgp.201110722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Zhao Y., Scheuer T., Catterall W.A.. 2008. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc. Natl. Acad. Sci. USA. 105:15142–15147. 10.1073/pnas.0806486105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Sharp E.M., Scheuer T., Catterall W.A.. 2009. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl. Acad. Sci. USA. 106:22498–22503. 10.1073/pnas.0912307106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Scheuer T., Catterall W.A.. 2011. Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc. Natl. Acad. Sci. USA. 108:18825–18830. 10.1073/pnas.1116449108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A.L., Jr 2005. Inherited disorders of voltage-gated sodium channels. J. Clin. Invest. 115:1990–1999. 10.1172/JCI25505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin-Badaroudine P., Delemotte L., Moreau A., Klein M.L., Chahine M.. 2012. Gating pore currents and the resting state of Nav1.4 voltage sensor domains. Proc. Natl. Acad. Sci. USA. 109:19250–19255. 10.1073/pnas.1217990109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin A.L., Huxley A.F.. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117:500–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn R., Ding S., Gruber H.J.. 2000. Immobilizing the moving parts of voltage-gated ion channels. J. Gen. Physiol. 116:461–476. 10.1085/jgp.116.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkat-Rott K., Holzherr B., Fauler M., Lehmann-Horn F.. 2010. Sodium channelopathies of skeletal muscle result from gain or loss of function. Pflugers Arch. 460:239–248. 10.1007/s00424-010-0814-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontis K.J., Goldin A.L.. 1997. Sodium channel inactivation is altered by substitution of voltage sensor positive charges. J. Gen. Physiol. 110:403–413. 10.1085/jgp.110.4.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-C., Bean B.P.. 1994. Na+ channels must deactivate to recover from inactivation. Neuron. 12:819–829. 10.1016/0896-6273(94)90335-2 [DOI] [PubMed] [Google Scholar]
- Lerche H., Peter W., Fleischhauer R., Pika-Hartlaub U., Malina T., Mitrovic N., Lehmann-Horn F.. 1997. Role in fast inactivation of the IV/S4-S5 loop of the human muscle Na+ channel probed by cysteine mutagenesis. J. Physiol. 505:345–352. 10.1111/j.1469-7793.1997.345bb.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Kong J., Kallen R.G.. 2009. Studies of alpha-helicity and intersegmental interactions in voltage-gated Na+ channels: S2D4. PLoS ONE. 4:e7674. 10.1371/journal.pone.0007674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi Y., Arcisio-Miranda M., Chowdhury S., Chanda B.. 2010. Molecular determinants of coupling between the domain III voltage sensor and pore of a sodium channel. Nat. Struct. Mol. Biol. 17:230–237. 10.1038/nsmb.1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.P., Horn R.. 2002. Movement and crevices around a sodium channel S3 segment. J. Gen. Physiol. 120:419–436. 10.1085/jgp.20028636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M.S., Shimizu S., Tanabe T., Takai T., Kayano T., Ikeda T., Takahashi H., Nakayama H., Kanaoka Y., Minamino N., et al. 1984. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature. 312:121–127. 10.1038/312121a0 [DOI] [PubMed] [Google Scholar]
- Paldi T., Gurevitz M.. 2010. Coupling between residues on S4 and S1 defines the voltage-sensor resting conformation in NaChBac. Biophys. J. 99:456–463. 10.1016/j.bpj.2010.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian D.M., Shao X.M., Seoh S.-A., Mock A.F., Huang Y., Wainstock D.H.. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301. 10.1016/0896-6273(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Gamal El-Din T.M., Scheuer T., Zheng N., Catterall W.A.. 2012. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 486:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E., Santacruz-Toloza L.C., Stefani E., Bezanilla F., Papazian D.M.. 1994. S4 mutations alter gating currents of Shaker K channels. Biophys. J. 66:345–354. 10.1016/S0006-3495(94)80783-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless S.A., Galpin J.D., Niciforovic A.P., Ahern C.A.. 2011. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat. Chem. Biol. 7:617–623. 10.1038/nchembio.622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X.M., Papazian D.M.. 1993. S4 mutations alter the single-channel gating kinetics of Shaker K+ channels. Neuron. 11:343–352. 10.1016/0896-6273(93)90189-X [DOI] [PubMed] [Google Scholar]
- Sheets M.F., Kyle J.W., Kallen R.G., Hanck D.A.. 1999. The Na channel voltage sensor associated with inactivation is localized to the external charged residues of domain IV, S4. Biophys. J. 77:747–757. 10.1016/S0006-3495(99)76929-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu S.-Y., Yang D.-Y., Selzle H.L., Schlag E.W.. 2003. Energetics of hydrogen bonds in peptides. Proc. Natl. Acad. Sci. USA. 100:12683–12687. 10.1073/pnas.2133366100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R., Goldin A.L.. 1997. Interaction between the sodium channel inactivation linker and domain III S4-S5. Biophys. J. 73:1885–1895. 10.1016/S0006-3495(97)78219-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stühmer W., Conti F., Suzuki H., Wang X.D., Noda M., Yahagi N., Kubo H., Numa S.. 1989. Structural parts involved in activation and inactivation of the sodium channel. Nature. 339:597–603. 10.1038/339597a0 [DOI] [PubMed] [Google Scholar]
- Tiwari-Woodruff S.K., Lin M.A., Schulteis C.T., Papazian D.M.. 2000. Voltage-dependent structural interactions in the Shaker K(+) channel. J. Gen. Physiol. 115:123–138. 10.1085/jgp.115.2.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer J.S., Cooperman S.S., Tomiko S.A., Zhou J.Y., Crean S.M., Boyle M.B., Kallen R.G., Sheng Z.H., Barchi R.L., Sigworth F.J., et al. 1989. Primary structure and functional expression of a mammalian skeletal muscle sodium channel. Neuron. 3:33–49. 10.1016/0896-6273(89)90113-X [DOI] [PubMed] [Google Scholar]
- Vargas E., Yarov-Yarovoy V., Khalili-Araghi F., Catterall W.A., Klein M.L., Tarek M., Lindahl E., Schulten K., Perozo E., Bezanilla F., Roux B.. 2012. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 140:587–594. 10.1085/jgp.201210873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Delaloye K., Zaydman M.A., Nekouzadeh A., Rudy Y., Cui J.. 2010. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 135:595–606. 10.1085/jgp.201010408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Bennett P.B., Makita N., George A.L., Jr, Barchi R.L.. 1993. Expression of the sodium channel β 1 subunit in rat skeletal muscle is selectively associated with the tetrodotoxin-sensitive α subunit isoform. Neuron. 11:915–922. 10.1016/0896-6273(93)90121-7 [DOI] [PubMed] [Google Scholar]
- Yang N., George A.L., Jr, Horn R.. 1996. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 16:113–122. 10.1016/S0896-6273(00)80028-8 [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V., DeCaen P.G., Westenbroek R.E., Pan C.-Y., Scheuer T., Baker D., Catterall W.A.. 2012. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. USA. 109:E93–E102. 10.1073/pnas.1118434109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.H., Yarov-Yarovoy V., Gutman G.A., Catterall W.A.. 2005. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol. Rev. 57:387–395. 10.1124/pr.57.4.13 [DOI] [PubMed] [Google Scholar]
- Zhang L., Sato Y., Hessa T., von Heijne G., Lee J.K., Kodama I., Sakaguchi M., Uozumi N.. 2007. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc. Natl. Acad. Sci. USA. 104:8263–8268. 10.1073/pnas.0611007104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., et al. 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134. 10.1038/nature11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.